Abstract
Molecular chaperones are central to cellular protein homeostasis. In mammals, protein misfolding diseases and aging cause inflammation and progressive tissue loss, in correlation with the accumulation of toxic protein aggregates and the defective expression of chaperone genes. Bacteria and non-diseased, non-aged eukaryotic cells effectively respond to heat shock by inducing the accumulation of heat-shock proteins (HSPs), many of which molecular chaperones involved in protein homeostasis, in reducing stress damages and promoting cellular recovery and thermotolerance. We performed a meta-analysis of published microarray data and compared expression profiles of HSP genes from mammalian and plant cells in response to heat or isothermal treatments with drugs. The differences and overlaps between HSP and chaperone genes were analyzed, and expression patterns were clustered and organized in a network. HSPs and chaperones only partly overlapped. Heat-shock induced a subset of chaperones primarily targeted to the cytoplasm and organelles but not to the endoplasmic reticulum, which organized into a network with a central core of Hsp90s, Hsp70s, and sHSPs. Heat was best mimicked by isothermal treatments with Hsp90 inhibitors, whereas less toxic drugs, some of which non-steroidal anti-inflammatory drugs, weakly expressed different subsets of Hsp chaperones. This type of analysis may uncover new HSP-inducing drugs to improve protein homeostasis in misfolding and aging diseases.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-010-0216-8) contains supplementary material, which is available to authorized users.
Keywords: Chaperone network, Heat shock proteins, Foldase, NSAID, Cellular stress response, Unfolded protein response
Introduction
The term “heat-shock proteins” (HSPs) was first used to describe Drosophila melanogaster proteins that massively accumulate during heat stress (Tissieres et al. 1974). When subject to a sharp increase in temperature, prokaryotes and eukaryotes alike transiently reallocate their general house-keeping protein synthesis machinery to the specific accumulation of a small subset of highly conserved Hsps, initially named according to their molecular weight on sodium dodecyl sulfate polyacrylamide gels: Hsp100 (ClpB/A/C), Hsp90 (HtpG), Hsp70 (DnaK), Hsp60 (GroEL), Hsp40 (DnaJ), the small Hsps (IbpA/B), and Hsp10 (GroES) (E. coli proteins in brackets; Daniels et al. 1984; Tissieres et al. 1974; Kimpel and Key 1985). A general mechanism was then proposed for Hsp70 by Pelham (1986) and for GroEL by Ellis, whereby these two major classes of Hsps may prevent the aggregation of stress-denatured or nascent proteins in the cell and thus “chaperone” the correct native folding and/or assembly of other proteins, without being part of the final native protein structures (Ellis et al. 1989).
Under physiological and stress conditions, the various chaperone families act in a tightly interconnected network (Csermely et al. 2008). Genetic and biochemical studies show that in bacteria, the chaperone network has a key role in housekeeping (Deuerling et al. 1999) and in the cellular response to various stresses (Liberek and Georgopoulos 1993). Eukaryotes may use different subsets of redundant, partially overlapping chaperones to fold and translocate proteins under physiological conditions, to prevent protein misfolding and aggregation during stress (Albanese et al. 2006), and to recover misfolded proteins after stress (Mogk et al. 1999; Tomoyasu et al. 2001). During a noxious heat shock, an overload of the cellular chaperones may occur (Csermely 2001; Nardai et al. 2002), overwhelming chaperone- and protease-based cellular proteostasis (Morimoto 2008). When the stress is over, the cellular protein network is restructured, and the so-called hubs, which, under stress were transiently replaced by molecular chaperones, regain control of cellular functions (Soti et al. 2005; Szabadkai et al. 2006; Szalay et al. 2007). The general purpose of this study was to gain knowledge on HSP- and chaperone-inducing treatments and drugs that best recapitulate natural patterns of HSP chaperone gene expression in tissues challenged by heat or cellular stresses, to improve proteostasis, particularly in deficient tissues, in aging or degenerative diseases associated to protein misfolding (for a review, see Hinault et al. 2006).
Hsp70s and co-chaperones With the exception of some archaea (Large et al. 2009), members of the evolutionary conserved Hsp70 chaperone family are present in all the ATP-containing compartments of living organisms (Macario and de Macario 1999). Thus, in human, the major isoform Hsp72 (HSPA1A) and the heat shock cognate 70 (Hsc70/HSPA8) are located in the cytosol and nucleus, whereas BiP (Grp78/HSPA5) is in the endoplasmic reticulum, mtHsp70 (Grp75/mortalin/HSPA9) is in mitochondria (Hageman and Kampinga 2009), and there are possibly also Hsp70s in peroxisomes (Hageman et al. 2007). In addition, plant chloroplasts and protozoan apicoplasts contain Hsp70s most similar to cyanobacteria (Soll 2002; Tarun et al. 2008). The functional Hsp70 chaperone network entails ATP-driven interactions between many diverse substrate-specific and less specific J-domain co-chaperones (49 in human) that target the fewer Hsp70 isoforms (Kampinga et al. 2009) onto hundreds of protein substrates in the cell and are regulated by various nucleotide exchange factors (NEF) such as GrpE (Harrison 2003), BAG (Kabbage and Dickman 2008), HspBP1 (Kabani et al. 2002), and Hsp110 proteins (Shaner and Morano 2007). These networks are crucial to the co-translational folding of nascent polypeptides, the remodeling of native protein complexes, the transduction of cellular signals, the regulation of the cell cycle, proliferation and apoptosis (Jolly and Morimoto 2000), the regulation of the heat shock response, the unfolding and refolding of stress-denatured proteins, and the import of proteins into the mitochondria (De los Rios et al. 2006), chloroplasts (Shi and Theg 2010), and the endoplasmic reticulum (reviewed in Zimmermann et al. 2010). Moreover, the Hsp70/Hsp40 networks control the stability and activity of native proteins such as σ32 and the oligomeric state of native protein complexes, such as repE (Rodriguez et al. 2008), clathrin cages (Schuermann et al. 2008), and IkB (Weiss et al. 2007) and yeast prions (Wickner 1994; Shorter and Lindquist 2008).
The Hsp100 The Hsp100 chaperones are ATPase members of the AAA+ superfamily, including bacterial ClpB, mitochondrial Hsp78, chloroplast ClpC/D, and eukaryotic orthologues in the cytoplasm of fungi, yeast (Hsp104), and plants (Hsp101), (Mogk et al. 2008). The Hsp100 chaperones share sequence, structural, and functional similarities with the AAA+-gated proteases, such as the lid of the eukaryotic proteasome, the ATPase moiety of the bacterial proteases HslU/V, ClpA/P, ClpX/P, and Lon (for a review, see Sharma et al. 2009). Whereas the sole bacterial Hsp70/Hsp40/NEF chaperone network can effectively disaggregate and unfold small soluble protein aggregates (Diamant et al 2000; Ben-Zvi et al. 2004), it best acts in concert with Hsp100 (ClpB) in bacteria and in the cytoplasm of plants and fungi (but not in animals), to disaggregate large insoluble aggregates into natively refoldable polypeptides (Glover and Lindquist 1998; Goloubinoff et al. 1999; Motohashi et al. 1999). As with the other major classes of molecular chaperone, Hsp100 plays a vital role in the survival of bacteria, yeast, and plant cells during and following exposures to high temperatures or chemical stresses (Sanchez and Lindquist 1990).
Hsp60/10 Whereas the proper folding of nascent proteins in bacteria mostly depends on the activity of the Hsp70/40/NEF network, 10–15% of newly synthesized polypeptides are better substrates for bacterial Hsp60/Hsp10 network (for a review, see Liberek et al. 2008). Class I chaperonins represented by Hsp60 homologues are found in bacteria (GroEL), mitochondria, and chloroplasts. A functional Hsp60/Hsp10 complex comprises 14 identical subunits arranged in two stacked heptameric rings, requiring two heptameric co-chaperones, Hsp10/GroES (Azem et al. 1995). Seminal observations showed that artificially denatured proteins become prevented from aggregating upon binding to purified GroEL and, moreover, become subsequently refolded to the native state in a strict GroES- and ATP-dependent manner (Goloubinoff et al. 1989).The class II of chaperonins, which are present in archaea and in the cytoplasm of eukaryotes (Large et al. 2009), forms TCP-1 ring complex (TRiC, also named CCT for chaperonin-containing TCP1) consisting of two stacked rings with eight different paralogous subunits per ring (Booth et al. 2008). The TRiCs act as ATP-dependent central mediators of cytosolic protein folding and assembly (Hartl and Hayer-Hartl 2009), which are also important to prevent protein aggregation and toxicity (Kitamura et al. 2006).
Hsp90 family and co-chaperones Whereas the physiological and stress-related functions of HtpG, the bacterial Hsp90, and of the mitochondrial and chloroplast Hsp90s remain unclear (Sato et al. 2010; Hasan and Shimizu 2008), in vitro, Hsp90 can prevent protein aggregation in an ATP-independent manner (Wiech et al. 1992). Owing to the early discovery of specific Hsp90 inhibitors (Whitesell et al. 1994), the cellular functions of endoplasmic reticulum (ER) (Grp94), cytosol, and nuclear located Hsp90s in eukaryotes are better known than a role of prokaryotic HtpG. The EEVD motif at the carboxy-terminus of cytoplasmic Hsp90 and some cytoplasmic Hsp70s is a docking site for connecting proteins with tetratricopeptide repeats (TPR) (Blatch and Lassle 1999; van der Spuy et al. 2000). Bridged by TPR-containing co-chaperones, both chaperones can form functional super-complexes that modify in a yet ill-defined ATP-dependent mechanism, the structure, and consequently the function, of hundreds of the so-called native “client” proteins in the cell (Whitesell and Lindquist 2005; Zhao et al. 2005). Thus, the Hop co-chaperone containing three TPR repeats bridges Hsp70 and Hsp90, which, together with Hsp40 and p23 co-chaperone, drive structural and functional changes in native protein complexes in the cell, such as the progesterone receptor (Cintron and Toft 2006; Onuoha et al. 2008). CHIP is another TPR co-chaperone of Hsp90 with a U-box domain, whose activity promotes “protein triage” of Hsp70- or Hsp90-bound proteins fated to proteasomal degradation (Connell et al. 2001). Taken together, in eukaryotic cells, heterocomplexes of Hsp90 with about a dozen co-chaperones (Picard 2006; for an updated list of Hsp90 co-chaperones, see http://www.picard.ch/downloads/Hsp90interactors.pdf) with Hsp70 and protein clients are key to various physiological processes, in particular signal transduction.
Small Hsps Unlike the ATPase chaperones Hsp100, Hsp90, Hsp70, and Hsp60, the small Hsps (sHSPs) have a conserved α-crystalline domain that passively binds misfolded intermediates, independently from ATP hydrolysis (Jakob et al. 1993). Without stress, sHsps are mostly assembled into large oligomeric complexes (Garrido et al. 2006), which, under stress conditions, may dissociate into amphiphilic dimers that prevent misfolding polypeptides from aggregating (Jakob et al. 1993) and protect membranes from heat disruption (Horvath et al. 2008; Haslbeck et al. 2005). sHsps cooperate with Hsp70/Hsp40 and Hsp100 or the GroEL/GroES chaperone networks in refolding of misfolded proteins (for a review, see Nakamoto and Vigh 2007). Human Hsp27 and Hsp70 are often, although not obligatorily, co-expressed in response to a variety of physiological and environmental stimuli (Garrido et al. 2006) (Vigh et al. 2007). As sHsps have strong cytoprotective properties (Garrido et al. 2006), their inhibition is an important target in pharmacological therapies to cancer (Didelot et al. 2007), whereas the upregulation sHsp may prevents liver damage (Kanemura et al. 2009) or pathologies caused by protein misfolding, such as Alzheimer’s (Fonte et al. 2008; Wu et al. 2010), Parkinson’s (Zourlidou et al. 2004), and Huntington’s disease (Perrin et al. 2007).Here, we used published microarray data from Homo sapiens and the land plant Arabidopsis thaliana to perform a meta-analysis of the expression profiles of bioinformatically predicted chaperones, co-chaperones, and foldase genes (together called the chaperome), following various abiotic and chemical stresses. Clustering of induction profiles revealed that heat shock primarily induces cytoplasmic and mitochondrial but not ER chaperone networks, a profile that was best mimicked by isothermal treatments with Hsp90 inhibitors or less faithfully by other compounds, many of which are known as anti-inflammatory drugs. Sequence analysis of HSP promoters showed that canonical heat hock elements (HSEs) were unexpectedly rare in HSP genes. This type of analysis may uncover new HSP-inducing drugs that best recapitulate natural patterns of HSP chaperone gene expression in undamaged tissues, to improve protein homeostasis in defective aging tissues and in protein misfolding pathologies.
Methods
List of human and plant chaperones, co-chaperones, and foldases
Two lists of bio-informatically identified “chaperomes” were compiled for the human and the A. thaliana genomes (Supplemental Tables 1a and 2, respectively), which included all the predicted protein sequences sharing at least 40% homology with one of the conserved canonical chaperone families in eukaryotes and their corresponding, identifiable prokaryotic homologues (prokaryote genes named in brackets): Hsp70 (DnaK), Hsp90 (HtpG), Hsp100/Hsp78 (ClpB/C), Hsp40 (DnaJ), Hsp60 (GroEL), and the α-crystalline domain containing small HSPs (IbpA/B), trigger factors, co-chaperones and nucleotides exchange factors (such as Hsp110, Grp170, Bag2, GrpE, GroES, Cpn10, and Cpn20), all J-domain containing proteins with a conserved HPD motive, over a dozen of Hsp90 co-chaperones (interactors) as listed by Picard (http://www.picard.ch/downloads/Hsp90interactors.pdf). Because peptidyl prolyl isomerases (PPIs) and protein disulfide isomerases (PDIs) are clearly involved in cellular protein homeostasis in general and although they do not belong to the canonical chaperone families, we chose to add them to this analysis (for a review, see Sharma et al. 2009).
Bioinformatic analysis
All data are MIAME compliant. The raw data were extracted from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO). For the A. thaliana chaperome metadata analysis, the microarray data for dithiothreitol (DTT), tunicamycin, and the five heat treatments were extracted from the GEO (ATH1-121501 Affymetrix Arabidopsis ATH1 Genome Array), under the following series accession numbers: GSE4021 (leaf disks), GSE11758 (mature leaves), GSE4760 (seedlings), GSE16222 (seedlings), GSE12619 (seedlings), GSE4062 (shoots), and GSE11758 (mature leaves), respectively. Microarray datasets for salicylic acid (seedlings), ibuprofen (seedlings), 2,3,5-triiodobenzoicacid (TIBA, seedlings), and 2,4,6-trihydroxybenzamide (2,4,6-T, seedlings) were obtained from Genevestigator (Zimmermann et al. 2004). Array metadata for DTT, tunicamycin, and one heat treatment denoted in GEO as GSE16222 were joined to the rest of metadata obtained from Genevestigator as a tab delimited file.
For the human and Arabidopsis chaperome metadata analysis, respectively, 167 and 281 probes (Supplemental Tables 1a and b and 2) corresponding to unique genes were chosen as described (Hageman and Kampinga 2009). The microarray data for the predicted human chaperome was searched in NCBI GEO (HG_U133 Plus 2.0 Affymetrix Human Genome Array): sapphyrin PCI-5002 GSE6962 (A549 tumor), echinomycin GSE7835 (U251 cells), etoposide GSE11954 (hepatic stellate cells), simvastatin GSE4883 (human peripheral blood monocytes macrophages), 2-deoxyglucose GSE13548 (HeLa cells), tunicamycin GSE13548 (HeLa cells), phorbol 12-myristate 13-acetate GSE12736 (K562 cells), cadmium (early) GSE9951 (immortalized human normal prostate epithelial cell line), paclitaxel GSE11552 (294T cells), heat shock study GSE9916 (THP-1 cells), elesclomol study GSE11552 (294T cells), smoking study six brands of cigarettes (early) GSE10718 (normal human bronchial epithelial cells), propiconazole GSE10410 (human primary hepatocytes), N-acetylcystein GSE11552 (294T cells), rifampicin GSE10410 (human primary hepatocytes), myclobutanil GSE10410 (human primary hepatocytes), estrogen (late) GSE11324 (MCF7 cells), dihydrotestosterone GSE7708 (LNCaP cells), doxycycline GSE7678 (SW480 cells), VAF347 GSE10463 (immature monocyte-derived dendritic cells), and apple procyanidin GSE9647 (human vascular endothelial cells). The array metadata for 2-deoxyglucose and tunicamycin were from GEO and joined to the rest of the metadata obtained from Genevestigator as a tab delimited file. Solar ultraviolet, Hsp90 inhibitors and non-steroidal anti-inflammatory drug (NSAID) microarray data for available human Hsp70 and Hsp40 genes were searched directly in Genevestigator (HG_U133A Affymetrix Human Genome Array) and joined with data from the heat shock study (GSE9916) as a tab delimited file.
Transcripts were considered responsive only when showing at least a 2-fold change in response to an investigated treatment. The distance matrix was evaluated using the Pearson correlation coefficient, and clusters were created using the complete linkage method by Cluster 3.0 (http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/cluster/manual/index.html) and visualized by the JavaTreeView (Saldanha 2004) algorithm, exported as a postscript file, processed by Adobe Illustrator (Adobe Systems, Mountain View, CA, USA). Annotation and presumed subcellular localization of chaperones were performed according to the Uniprot database.
In an attempt to generate a significant network best describing the degree of connectivity between the various chaperones, co-chaperones, and foldases in the human chaperome, we used the STRING database and web resource. STRING weights both physical and functional protein–protein interactions and integrates various informations from different metadatabase sources, to produce a network map showing all possible protein–protein interactions in the chaperome (http://string-db.org) (Jensen et al. 2009). Because under non-stressed conditions, the members of the chaperone network are expected to have more but weaker interactions among themselves than under stressful conditions (Csermely et al. 2008), we chose the low confidence factor of 0.15 for this analysis. The interactions between chaperones was visualized using Medusa (Hooper and Bork 2005)
Using the search PromForm program from the Promoter Database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm), a search for HSEs containing direct or inverted repeats of 5′-nGAAn-3′ was performed from 3 kbp upstream to 300 bp downstream from the predicted transcription start site of each bio-informatically identified chaperone gene from mammals (as listed in Supplemental Table 1b).
Results
Most HSPs are not chaperones, and most chaperones are not HSPs
As exemplified by the two seminal reviews, “Molecular chaperone functions of heat-shock proteins” (Hendrick and Hartl 1993) and “Heat-shock proteins as molecular chaperones” (Becker and Craig 1994), the terms “chaperones” and “HSPs” are often indiscriminately used in the literature. In 2009, a survey of 300 articles in PubMed citing both “chaperones” and “Hsp70” in their title, abstract, or introduction, a third stated that molecular chaperones are Hsps and Hsps are molecular chaperones without distinction (data not shown). To estimate the validity of the generally assumed strong linkage between HSPs and molecular chaperones, we examined the microarray responses to mild heat treatments in two very different eukaryotes, a higher plant and a mammal, and compared the messenger RNA (mRNA) expression patterns of bio-informatically identified chaperome genes (listed in Supplemental Tables 1a and 2), to the rest of the corresponding genomes. Because of the high degree of evolutionary conservation, members of the α-crystalline-containing small HSPs and of the four canonical families of ATP-hydrolyzing chaperones Hsp100, Hsp90, Hsp70 and Hsp60 and their respective co-chaperones, as well as the PPIs and the PDIs, were identified by simple bioinformatic analysis (see “Methods”). Hence, of the 23,438 predicted protein-encoding genes in H. sapiens (The Genome Reference Consortium, version GRCh37), 168 genes were identified as belonging to “the human chaperome” (Supplemental Table 1b). Similarly, of the 27,379 predicted protein-encoding genes in the terrestrial plant A. thaliana (The Arabidopsis Information Resource, version TAIR9), 305 genes were identified as “the Arabidopsis chaperome” (Supplemental Table 2).
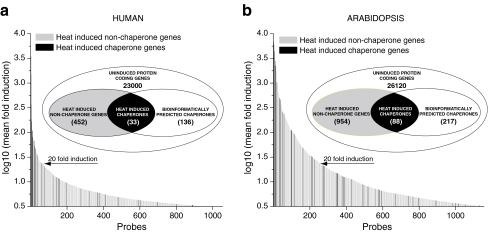
A meta-analysis of microarray data from different organisms and laboratories is always limited by the smallest number of available probes for HSP and chaperome genes, printed on the chips. Thus, we could effectively follow the changes in the expression profiles of up to 167 human chaperome genes and up to 281 Arabidopsis chaperome genes. Microarray data showed that a short sub-lethal heat treatment (37°C–>43°C, 60 min for human, 23°C–>38°C, 90 min for the plant) upregulated more than 3.16-fold (log10 value = 0.5) about 2% of the human genes and 4% of the plant genes, respectively (Fig. 1). Noticeably, in human and plant, chaperome genes were, respectively, 17 and seven times, more likely to be induced by heat than non-chaperone genes. Moreover, while being only 0.7% of the human genome, 20% of the chaperome was massively (>20-fold) induced by heat (Fig. 1a), a remarkable 28-fold enrichment of expressed mRNA levels. Similarly, while being only 1.1% of the plant genome, 16% of the chaperome was most massively (>20-fold) induced by heat, a 14-fold enrichment of mRNA levels (Fig. 1b). Despite this general high propensity of the chaperomes to be induced by heat, a majority of chaperome genes (66% for human and 72% for plant) yet remained uninduced by heat, confirming the importance not to confuse HSPs for chaperones and vice versa. Hence, molecular chaperones and foldases should be specifically referred as such and not as HSPs, especially in a context of their physiological functions in protein homeostasis, cellular trafficking, signaling, or of their induction by other means than heat.
Fig. 1.
Distribution and expression levels of heat-induced chaperome genes in human and plant. Distribution and fold-expression levels of heat-induced genes in (a) human monocyte leukemia THP-1 cells (37°C–>43°C 1 h and (b) plant Arabidopsis thaliana (23°C–>38°C, 90 min). Microarray probes corresponding to bio-informatically predicted chaperome genes are in black and non-chaperome genes are grey. The microarray data for human and plant sets were extracted from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under the series accession nos. GSE9916 and GSE16222, respectively
The heat-inducible chaperones can be upregulated by isothermal chemical treatments
Although heat has a demonstrated strong effect on the induction of a specific subset of chaperome genes, other isothermal chemical or physical stresses may also induce similar or different subsets of chaperome genes (Saidi et al. 2005, 2007).
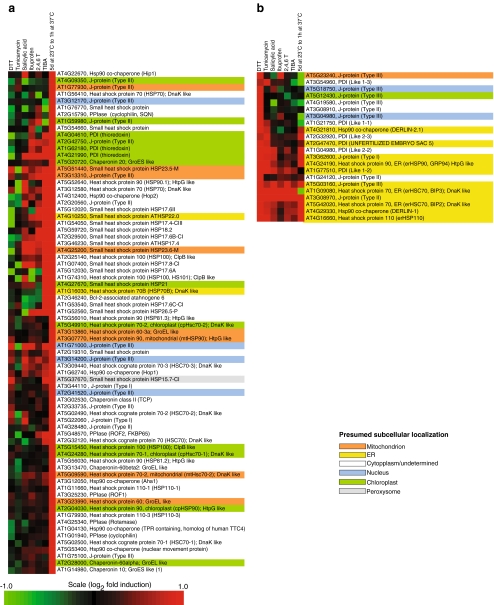
In Arabidopsis, five independent heat treatments (HS) by different laboratories and different temperature conditions showed comparable induction patterns of chaperome genes associated to the cellular stress response (CSR), demonstrating the robustness of our approach (see Supplemental Fig. 1). Isothermal treatments with inhibitors of polar auxin transport, such as TIBA and 2,4,6-T and also with the plant hormone salicylic acid and with ibuprofen, which are both NSAIDs in mammals, showed mildly increased levels of particular chaperones, which, however, only loosely clustered with the heat treatments (Fig. 2, Supplemental Fig. 2). Two main treatment clusters were, however, clearly observed, one following heat or specific chemical treatments, corresponding to a general response associated to the CSR (for a review, see Calabrese et al. 2008; Fig. 2a), in which chaperones mostly targeted to the cytosol, the plasma membrane and the mitochondria were upregulated (Aparicio et al. 2005), and the other following chemical treatments with stressors specific to the ER, corresponding to a response known as the “unfolded protein response” (UPR), in which chaperones mostly targeted to the ER lumen and membranes were upregulated (Schroder and Kaufman 2005, Fig. 2b). Unexpectedly, several plant heat shock cognates, in particular Hsc70s, were also upregulated by heat in plants (Supplemental Fig. 1, arrows) as well as in animal cells. Thus, Hsc70 (HspA8) was induced by heat (Fig. 3, treatment I) or isothermal treatments with N-acetylcycteine or estrogen (Fig. 3, treatment E, T). Hsc70s are thus either wrongly named, or their definition as being non-heat inducible chaperone cognates must be corrected.
Fig. 2.
Clustering of upregulated RNA expression levels in Arabidopsis chaperome under seven abiotic and chemical treatments: dithiothreitol (DTT), tunicamycin, salicylic acid, ibuprofen, 2,3,5- triiodobenzoicacid (TIBA), 2,4,6-trihydroxybenzamide (2,4,6-T), and heat treatment as indicated. Gene clusters typical of (a) the cellular stress response (CSR) or of the (b) unfolded protein response (UPR). The presumed subcellular localizations are indicated with different background colors
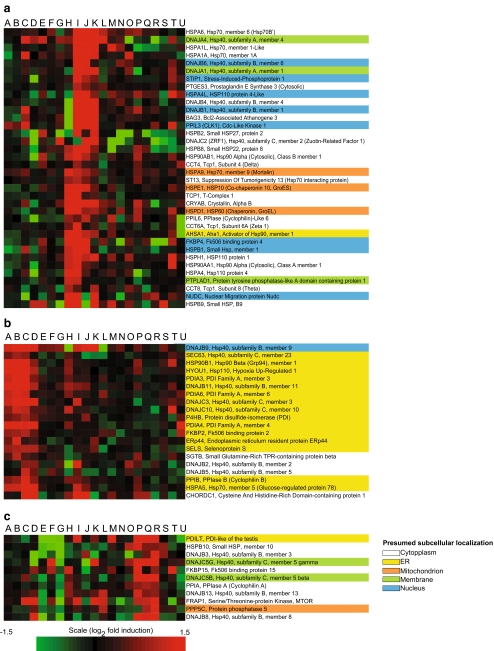
Fig. 3.
Clustering of upregulated RNA expression levels in the human chaperome under 21 treatments: A 2-deoxyglucose, B tunicamycin, C phorbol 12-myristate 13-acetate, D cadmium, E N-acetylcysteine, F paclitaxel, G doxycycline, H echinomycin, I heat shock study, J elesclomol, K smoking, L simvastatin, M etoposide, N VAF347, O sapphyrin PCI-5002, P propiconazole, Q myclobutanil, R rifampicin, S dihydrotestosterone, T estrogen, and U apple procyanidin. Gene clusters typical a of the cellular stress response (CSR), b of the unfolded protein response (UPR), and c of a main less specific cell response are shown. The presumed subcellular localizations are indicated with different background colors of the gene names
In cultured animal cells, several seminal studies using isothermal treatments with amino acid analogs (Hightower 1980) or low concentrations of zinc (Whelan and Hightower 1985) caused expression of molecular chaperones. In a similar manner, isothermal treatments of human cells with Hsp90 inhibitors, UV light, elesclomol, or cigarette smoke best recapitulated the expression pattern of heat shock by inducing a similar subset of about 45 out of 167 chaperome genes (Supplemental Fig. 3), most of which targeted to the cytoplasm, membranes, and mitochondria, but not to the ER (Fig. 3a). In contrast, treatments with tunicamycin, 2-deoxyglucose, or dithiothreitol induced a very different set of human chaperome genes, whose products were targeted to the ER lumen and membranes (Fig. 3b). Interestingly, the antibiotic rifampicin and two antifungal agents, myclobutanil and propiconazole, induced an additional chaperone cluster (Fig. 3c), which was not apparently related to a given subcellular localization. In conclusion, members of UPR gene cluster should be specifically called “UPR chaperones” and of the CSR gene cluster “CSR chaperones”, not HSPs, even when addressed in a context of heat shock.
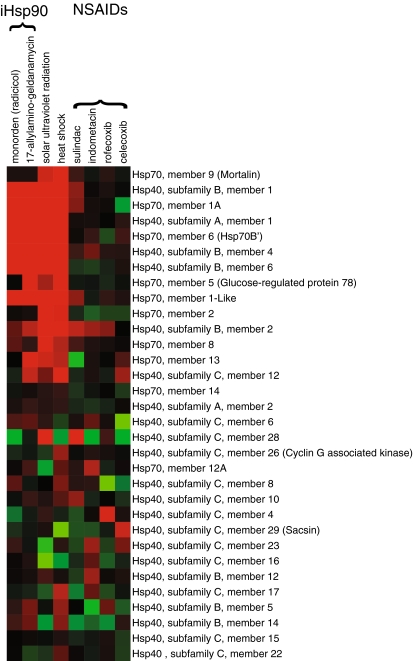
As with plants, treatments of human cells with several NSAIDs induced various chaperones, in particular members of the Hsp70/40 family. However, expression patterns were generally heterogeneous and at lower intensities than with CSR-inducing treatments (Fig. 4). Together with the fact that NSAIDs also induce CSR chaperones in plants (Saidi et al. 2005), this suggests that different drugs commonly classified as NSAIDs may have unconventional activation mechanisms that may not necessarily involve the inhibition of cyclooxygenases of which plants are devoid.
Fig. 4.
Expression profile of human Hsp70 and Hsp40 orthologs under different treatments. Heat and UV induced up- and downregulated human Hsp70 and Hsp40 genes, as compared to isothermal treatments with two Hsp90 inhibitors and four different NSAIDs, as indicated
CSR but not UPR activates the core elements of the chaperone network
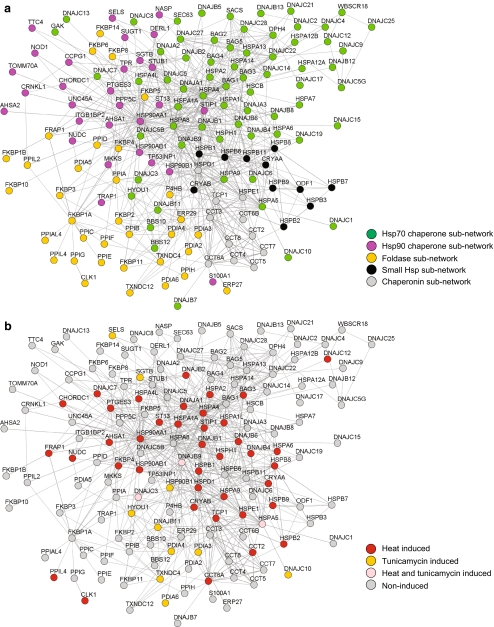
Our clustering analysis of the mammalian chaperome was thus far based only on mRNA expression metadata under various physical and chemical treatments (Figs. 3 and 4). We next estimated the robustness of this result using the STRING database that includes additional criteria, such as physical and known functional interactions (co-expression and experimental view) or computational predictions of homology and text mining co-occurrence (Jensen et al. 2009). The STRING analysis revealed a network with a strong core that cumulated most connections with other members of the chaperome that was mainly composed of Hsp90s (HSP90AA1, HSP90AB1, and HSP90B1) and Hsp70s (HSPA1A HSPA1L, HSPA2, and HSPA8), tightly interconnected to each other through common co-chaperones, such as Hip (ST13) and Hop (STIP1) (Fig. 5a). This chaperone network could be simply subdivided into five chaperone subnetworks comprising most of: (1) the Hsp70s and their co-chaperones (green, Fig. 5a, upper right), (2) the Hsp90s and their co-chaperones (magenta, Fig. 5a, upper left), (3) the PPIase and PDIs (yellow, Fig. 5a, lower left), the chaperonins (grey, Fig. 5a, lower right), and the small HSPs (black, Fig. 5a, lower right). Justifying our choice to include PPIases and PDIs in the chaperome, these foldases were nearly all found to be well-connected to the Hsp90s and the chaperonins subnetworks but, unexpectedly, much less connected to the Hsp70s and the small Hsps subnetworks (Fig. 5a).
Fig. 5.
Chaperone network rearrangements in CSR and UPR. a Interaction network of proteins in different cellular functions from STRING analysis of the chaperome (Supplemental Table 1) showing Hsp70 (green), Hsp90 (magenta), foldases (orange), small Hsp (black) and chaperonin subnetwork (gray). b Stress-induced centralization of chaperone hubs in CSR (red), UPR (orange), or both (pink)
We next examined the positioning of proteins that were at least 2-fold upregulated by heat, as a paradigm of CSR (Fig. 5b, red), or by tunicamycin, as a paradigm of UPR (Fig. 5b, yellow). Most sHSPs, together with the Hsp70s and Hsp90s core of the whole chaperone network, were strongly enriched with CSR-induced but not with UPR-induced proteins. This confirms that CSR chaperones are not evenly scattered over the whole network but rather form a compact, specific core subnetwork (Fig. 5b, red circles). Confirming our transcriptomic analysis (Fig. 3), UPR-induced chaperones were fewer (Fig. 5b, yellow circles) with only three proteins (HSPA5, DNAJC3, and DNAJB9) induced by both treatments. Remarkably, many J-domain, co-chaperones, and Hsp90 co-chaperones and most of the PPIases and PDIs remained uninduced by either heat or ER stressors.
HSEs are frequent but not obligatorily present in promoters of heat-induced chaperone genes
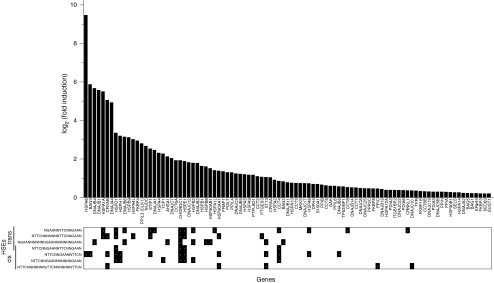
The presence of HSEs is essential to the specific binding of activated heat-shock factors, in particular HSF-1, and to the recruitment of RNA polymerase II at the transcription start sites of HSP genes (Xing et al. 2005; Yao et al. 2006). In animals, yeast, and plants, HSEs are thought to be optimal when composed of three direct repeats of 5′-nGAAn-3′ (Amin et al. 1988; Kroeger and Morimoto 1994; Yamamoto et al. 2009), or less optimal, with inverted repeats of 5′-nTTCn-3′. We next searched for complete or partial, direct or inverted HSE patterns, between positions −3000 and 300 bp from the translation start site of each predicted human chaperome gene (Supplemental Table 1b). Expectedly, HSEs were found to be more frequent in the regulatory sequences of 45 chaperone genes that are induced at least 2-fold by heat (Fig. 6), compared to less- or non-heat induced chaperone genes. Whereas in the CSR chaperone cluster (Fig. 3, cluster a), 17 out of 24 genes (71%) contained an average of two HSEs, in the UPR chaperone cluster (Fig. 3a, cluster b), only two out of 12 genes (17%) contained a single HSE motive. However, a majority of mammalian heat-induced chaperone genes remained devoid of identifiable HSEs, suggesting that HSEs are contributing only partially to the specificity of the heat-shock response and that other specific HSF-binding sites remain to be experimentally identified.
Fig. 6.
Heat shock elements found in heat-induced human chaperome genes. Distribution of fold expression levels of heat-induced chaperone genes in human monocyte leukemia THP-1cells (graph) and the presence of specific HSEs between −3000 and 300 bp of the translation start site of each genes (lower panel). The microarray data for the human chaperome subset (Supplemental Table 1a) is as in Fig. 1a
Our combined microarray, networking and sequence promoter analysis, may contribute to the identification of potential chaperone and co-chaperone matches. Thus, the HSPD1 and HSPE1 genes, respectively encoding for mitochondrial Hsp60 and Hsp10, have similar patterns of mRNA expression (Figs. 3, 5) and were also similar in terms of HSEs patterns in their promoters (Fig. 6). Likewise, the Hsp70 chaperone genes, HSPA1A and HSPA6, had expression profiles similar to the Hsp40 co-chaperones DNAJA4, DNAJB6, DNAJB1, DNAJB4, and DNAJA1 and to the nucleotide exchange factors BAG3 or HSPH1. All but two (DNAJA4 and DNAJB1) had one or two HSEs in their promoters, suggesting that they may preferably collaborate with each other under particular stressful or physiological conditions.
Discussion
The term molecular chaperone is adequate but incomplete
The term “molecular chaperone” was first used by Laskey to describe the properties of nucleoplasmin (Laskey et al. 1978). In 1989, Ellis adorned protein biochemistry with this social term to describe the new molecular function of a class of proteins that mediate the native folding and/or assembly of other polypeptides (Ellis et al. 1989). Whereas a social chaperone has a rather passive function—it merely prevents improper associations between youngsters—molecular chaperones, in particular the ATP-fuelled GroEL (Shtilerman et al. 1999), ClpB (Goloubinoff et al. 1999), and Hsp70 (Ben-Zvi et al. 2004), have since been found to carry an active function of repairing structural damages by unfolding and promoting native protein refolding or degradation (reviewed in Sharma et al. 2009). Hence, the social term “molecular police” might have better reflected the energy-consuming mechanism by which ATPase chaperones first apprehend, then “rehabilitate” strayed-off misfolded proteins, converting them into non-toxic functional proteins of the cell (Hinault and Goloubinoff 2007). Noticeably, alongside maintaining a “state of law” among cellular proteins, called protein homeostasis (or proteostasis, Morimoto 2008) in the crowded protein population of the cell, the network of chaperones, like a highly coordinated police force, is also involved in non-stress housekeeping functions, such as regulating protein trafficking, signaling, and import into organelles. Thus, although incomplete, the term “molecular chaperone” is not entirely wrong and should be maintained, primarily because it is now well-anchored into the scientific community.
The cellular stress response is likely under the control of the plasma membrane
Sensing temperature changes in eukaryotes has been initially attributed to unspecified thermolabile proteins in the cytoplasm, whose unfolding supposedly recruits inhibitory Hsp90 and Hsc70 chaperones, thereby activating formerly chaperone-repressed Hsfs (Voellmy and Boellmann 2007). However, it has been recently shown in plants that ambient temperature upshifts, for example, from 12 to 27°C, can trigger the upregulation of 2,764 HSP genes, many of which are chaperone genes (Kumar and Wigge 2010). Likewise, isothermal treatments with non-toxic chemicals, such as hormones and NSAIDs unlikely to denature proteins in bacteria, yeast, plants, and animals can upregulate specific HSP and chaperone genes (Saidi et al. 2005, 2007; Vigh et al. 2007). Similarly, the generally observed induction of the CSR in various organisms by treatments with membrane fluidizers strongly suggests that subtle changes in the plasma membrane state (fluidity, the raft/non-raft organization) are the most upstream events of the temperature-sensing and signaling pathway, both in prokaryotes and eukaryotes (de Marco et al. 2005; Vigh et al. 1998, 2007). Because abnormally exposed hydrophobic parts of misfolded proteins often wrongly interact with membranes and because, without a Ca2+ entry-dependent clearance signal from the plasma membrane, Hsp90 inhibitors cannot directly induce an isothermal HSR in plants, the plasma membrane also stands as a sensor of intra- and extracellular protein aggregates, activating the CSR chaperones to reduce aggregate toxicity (Saidi et al. 2009).
Analysis of the chaperone network under stress
Molecular chaperones with their co-chaperones form a dynamic network with hundreds of so-called client proteins (Picard 2006). During and after stress, a massive reallocation of the cellular resources occurs, which in turn causes dramatic rearrangements in the chaperone network (Szalay et al. 2007). Pallotai et al. (2008) showed that in the stressed interactome in yeast, novel centralities or important interactions in the network appeared, which in parallel increased the number of unlinked or isolated network elements or modules. As resources diminish during stress, the function of most protein networks becomes transiently arrested, whereas the few that remains active, such as the CSR chaperone network, become more active and abundant and take control on many of the cellular functions in a centralized and highly hierarchical manner (Szalay et al. 2007). Thus, in Escherichia coli, heat shock places the transcription factor σ32 as a central hub regulating other protein networks (El-Samad et al. 2005), while in yeast, refolding of damaged proteins by stress-induced chaperones is prioritized, compared to the folding of newly synthesized proteins indicating a massive rearrangement in the cellular protein networks (Albanese et al. 2006). Our comparative analysis of mRNA co-expression under various physical and chemical treatments revealed a modular organization of the chaperone networks, whereby the UPR, involving only a small set of ER chaperones, can be clearly distinguished from CSR chaperones, mainly located to the cytoplasm and organelles. This was confirmed using additional comparative criteria in the STRING-generated network map that showed that CSR chaperones contribute mostly to a core of Hsp70s, Hsp90s, and sHSPs, and to a much lesser extent to chaperone regulators such as the co-chaperones and to the foldases. Mechanistically, a massive increase in the cellular concentrations of Hsp90s and Hsp70s without an equivalent accumulation of co-chaperones is expected to create an imbalance resulting, at least in the case of Hsp70, in a higher binding/holdase capacity than processive unfolding/folding activity (Sharma et al. 2009). Such a pro-holdase tendency under heat shock is corroborated by our data (Fig. 5b), showing strong over-representation of heat-induced sHSPs, which are holdases par excellence of the whole chaperone network (Veinger et al. 1998). This suggests that the primary concern of heat-stressed cells is to prevent protein misfolding and aggregation during stress, while ATP-consuming unfoldase/refoldase activities depending on co-chaperones are postponed to the recovery phase after the stress, a condition under which the proteins refolded by the chaperones can become and stably remain native.
Treatments that increase the cellular chaperone load
Aging and protein misfolding diseases apparently result from a general failure of the cellular protein homeostasis machinery, which is most relevantly composed of molecular chaperones, foldases, and proteases. Whereas environmental stresses, pollutants, and mutations may increase the rate at which toxic misfolded protein conformers accumulate in cells, aging cells show decreasing abilities to respond to proteotoxic species by upregulating their cellular chaperone load (Ben-Zvi et al. 2009; Hinault et al. 2006). To combat aging and protein misfolding diseases, it is therefore central to identify the reasons for the failure of the CSR in aging and diseased cells. One possible reason could be an increased rigidity of membranes commonly observed in aging cells, as lipid saturation and loss of fluidity may strongly decrease the cell’s ability to sense a heat stress and induce an appropriate HSR (Horváth et al. 1998), implying that in old cells, the CSR too could become deficient when challenged by intra- or extracellular proteotoxic species (Vigh et al. 2007). An additional possibility is that particular toxic protein conformers, such as oligomers or protofibrils of α-synuclein, tau or polyglutamine proteins, respectively found in diseased neurons in Parkinson, Alzheimer, and Huntington diseases, act as non-competitive inhibitors of the various layers of cellular defenses, such as the chaperones themselves, the proteasome, the aggresome, and lysosomal autophagy (reviewed in Kopito 2000; Ross and Poirier 2004). Moreover, toxic protein conformers can directly damage membranes by spontaneously forming pores that cause ion leakage and cell death (Lashuel et al. 2002).
One therapeutic approach would be to identify treatments that can restore the expression levels and the activity of the CSR chaperones in particular. Mild repetitive heat-shocks, as with daily saunas, would probably be the most natural way to induce the accumulation of protective and repairing CSR chaperones in tissues suffering from stress damages or chronic protein misfolding diseases. However, arguing against long-term beneficial effects of thermotherapy (Pouppirt 1929), increased temperatures also increase the propensity of proteins to unfold and form toxic misfolded species. Rather, isothermal exposures to non-toxic HSP-inducing drugs might better treat protein misfolding diseases and aging, especially drugs that cross the blood–brain barrier (Hinault et al. 2006).
Alongside Hsp90 inhibitors that are very toxic and cause apoptosis through oxidative stress (Kirshner et al. 2008) and are therefore used in cancer therapy (Solit and Chiosis 2008), we found that the thiobenzoylhydrazide, elesclomol, best recapitulated in human cells the heat-induced chaperome pattern (Fig. 3). Thus, despite their ability to best mimic the induction of anti-inflammatory, anti-aggregation CSR chaperones load, Hsp90 inhibitors and elesclomol may counter-productively accelerate, rather than delay, neuronal death in protein misfolding diseases.
In contrast, other hydroxylamines, such as arimoclomol and bimoclomol, are apparently much less toxic than elesclomol and are also inducers of CSR chaperones, although not nearly as effective (Vigh et al. 1997; Kieran et al. 2004; Lanka et al. 2009). Likewise, but less pronounced, several NSAIDs, such as aspirin, diclofenac, ibuprofen, and indomethacin, also upregulated the expression of some chaperone genes, albeit not in clearly recognizable clusters, as with elesclomol or tunicamycin. NSAIDs are known to decrease inflammation and fever via inhibitory mechanisms of prostaglandin synthesis, and, in parallel, they induce the accumulation of some CSR chaperones (Hinault et al. 2006; Smalley et al. 1995). It is thus possible that in mammals, specific NSAIDs may decrease the pro-apoptotic consequences of inflammation and oxidative stress while at the same time maintain an artificially elevated load of protective anti-apoptotic chaperones.
Possibly, the use of various NSAID combinations, together with other of CSR-chaperone-inducing drugs, such as hydroxylamine derivatives (Vigh et al. 1997), vanilloids, such as curcumin (Kanitkar and Bhonde 2008), flavonoids, such as quercetin (Aalinkeel et al. 2008), or omega-3-fatty acids (Narayanan et al. 2006), with anti-aging and anti-apoptotic effects on developing brain (Sinha et al. 2009), may improve protein misfolding diseases and aging in general (Gidalevitz et al. 2010). Combinations of non-toxic HSP drugs could directly decrease the cellular concentration of cytotoxic protein conformers and indirectly block neuroinflammation and apoptosis signals, leading to an arrest of tissue loss. This type of mRNA expression meta-analysis, combined with network analysis, thus provides important tools to test known and new drugs for their potential specific chaperone-inducing ability, to combat protein-misfolding diseases and aging.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Correlation of Arabidopsis chaperome transcription in five independent heat shock treatments. Heat shock cognates are indicated by arrows. The microarray data were extracted from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under the series accession numbers GSE4760 (Hsf2 mutant, 4 days at 23°C to 2 h at 38°C), GSE16222 (5 days at 23°C to 1 h at 37°C), GSE12619 (7 days at 22°C to 1 h at 37°C), GSE4062 (15 days at 22°C to 2 h at 37°C), and GSE11758 (mature leaves at 20°C to 1 h at 37°C) (PDF 956 kb)
Clustering of RNA expression levels of the Arabidopsis chaperome under seven abiotic and chemical treatments: dithiothreitol (DTT), tunicamycin, salicylic acid, ibuprofen, 2,3,5- triiodobenzoicacid (TIBA), 2,4,6-trihydroxybenzamide (2,4,6-T), and heat treatment as indicated. Gene clusters typical of the of the cellular stress response (CSR) (a) or of the unfolded protein response (UPR) (b) are indicated with brackets. The presumed subcellular localizations are indicated with different background colors (PDF 1032 kb)
Clustering of RNA expression levels of 168 genes from the human chaperome under 21 treatments: A 2-deoxyglucose, B tunicamycin, C phorbol 12-myristate 13-acetate, D cadmium, E N-acetylcysteine, F paclitaxel, G doxycycline, H echinomycin, I heat shock study, J elesclomol, K smoking, L simvastatin, M etoposide, N VAF347, O sapphyrin PCI-5002, P propiconazole, Q myclobutanil, R rifampicin, S dihydrotestosterone, T estrogen, and U apple procyanidin. Gene clusters typical a of the cellular stress response (CSR), b of the unfolded protein response (UPR), and c of a main less specific cell response are indicated with brackets. The presumed subcellular localizations are indicated with different background colors of the gene names (PDF 442 kb)
List of identified human chaperone genes. (1) Small HSP, GroEL and CCT-like chaperonins, Hsp70, DNAJ, Hsp100 and Hsp90 families have been previously annotated (Kampinga et al. 2009). (2) The Bag family has been previously annotated (Takayama et al. 1999). (3) FKBP and CYP like peptide-prolyl isomerase families have been previously annotated (Barik 2006). (4) Protein disulfide isomerase family has been previously annotated (Ellgaard and Ruddock 2005). (5)Human Hsp90 co-chaperones have been identified according to www.picard.ch (XLS 45 kb)
List of identified heat shock elements (HSEs) in human chaperome. Nucleotide sequences of canonical HSEs in cis and trans orientations were identified between position −3000 and 300 bp from the translation start site of each chaperone gene (XLS 32 kb)
List of identified Arabidopsis thaliana chaperome genes. (1) Small HSPs were obtained by BLAST using α-crystalline domain as queries against the Arabidopsis protein subset of the NCBI database. (2) Chaperonins have been previously annotated (Hill and Hemmingsen 2001). (3) DnaJ proteins were re-annotated as compared to the previous J-protein nomenclature (Rajan and D’Silva 2009, as shown in brackets). All isoforms, splice variants, and paralogous protein according to previous annotations have been removed. The gene loci At2g02200, At5g34895, At2g07010 (MWJ3.6), At1g31210, At2g14930, At2g13940, and At2g24660 (corresponding to AtDjC6, AtDjC7, AtDjC50, and AtDjC58–AtDjC61, respectively) contained transposable elements and were removed as such. (4) The Hsp70 and Hsp110 were obtained by BLAST using DnaK from E. coli as query against the Arabidopsis protein subset of the NCBI database. (5) The Hsp100 were obtained by BLAST using ClpB from E. coli as the query against Arabidopsis protein subset of the NCBI database. Only the most ClpB-like proteins were retained, while ClpA/C proteins that are associated to the ClpP protease were excluded. (6) The Bag family has been previously annotated (Yan 2003). (7) The Hsp90 family has been previously annotated (Krishna and Gloor 2001). (8) PPIase families have been previously annotated (Romano et al. 2005). (9) PDIs families have been previously annotated (Houston et al. 2005). (10) Human Hsp90 co-chaperones were identified in yeast and animals according to http://www.picard.ch and used as queries to identify Arabidopsis orthologs in the protein subset of the NCBI database (XLS 162 kb)
Supplemental references (DOC 39.5 kb)
Acknowledgments
This research was financed in part by grant no. 3100A0-109290 from the Swiss National Science Foundation, the Alzheimer’s Drug Discovery Foundation New York, and the Zwahlen Grant from the Faculty of Biology and Medicine from Lausanne University.
References
- Aalinkeel R, Bindukumar B, Reynolds JL, Sykes DE, Mahajan SD, Chadha KC, Schwartz SA. The dietary bioflavonoid, quercetin, selectively induces apoptosis of prostate cancer cells by down-regulating the expression of heat shock protein 90. Prostate. 2008;68(16):1773–1789. doi: 10.1002/pros.20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Yam AYW, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124(1):75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. Key features of heat-shock regulatory elements. Mol Cell Biol. 1988;8(9):3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio F, Thomas CL, Lederer C, Niu Y, Wang DW, Maule AJ. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005;138(1):529–536. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azem A, Diamant S, Kessel M, Weiss C, Goloubinoff P. The protein-folding activity of chaperonins correlates with the symmetric GroEL(14)(GroES(7))(2) heterooligomer. Proc Natl Acad Sci USA. 1995;92(26):12021–12025. doi: 10.1073/pnas.92.26.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Craig EA. Heat-shock proteins as molecular chaperones. Eur J Biochem. 1994;219(1–2):11–23. doi: 10.1111/j.1432-1033.1994.tb19910.x. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, De los Rios P, Dietler G, Goloubinoff P. Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual Hsp70 chaperones. J Biol Chem. 2004;279(36):37298–37303. doi: 10.1074/jbc.M405627200. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106(35):14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21(11):932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Booth CR, Meyer AS, Cong Y, Topf M, Sali A, Ludtke SJ, Chiu W, Frydman J. Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat Struct Mol biol. 2008;15(7):746–753. doi: 10.1038/nsmb.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Mancuso C, Pennisi G, Calafato S, Bellia F, Bates TE, Stella AMG, Schapira T, Kostova ATD, Rizzarelli E. Cellular stress response: a novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem Res. 2008;33(12):2444–2471. doi: 10.1007/s11064-008-9775-9. [DOI] [PubMed] [Google Scholar]
- Cintron NS, Toft D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J Biol Chem. 2006;281(36):26235–26244. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang JH, Wu YX, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Csermely P. Chaperone overload is a possible contributor to ‘civilization diseases’. Trends Genet. 2001;17(12):701–704. doi: 10.1016/S0168-9525(01)02495-7. [DOI] [PubMed] [Google Scholar]
- Csermely P, Korcsmáros T, Kovács IA, Szalay MS, Soti C (2008) Systems biology of molecular chaperone networks. In: Derek J, Chadwick JG (eds) The biology of extracellular molecular chaperones., pp 45–58 [DOI] [PubMed]
- Daniels CJ, Mckee AHZ, Doolittle WF. Archaebacterial heat-shock proteins. EMBO J. 1984;3(4):745–749. doi: 10.1002/j.1460-2075.1984.tb01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P. Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA. 2006;103(16):6166–6171. doi: 10.1073/pnas.0510496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Vigh L, Diamant S, Goloubinoff P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress Chaperones. 2005;10(4):329–339. doi: 10.1379/CSC-139R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400(6745):693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275(28):21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- Didelot C, Lanneau D, Brunet M, Joly AL, Thonel A, Chiosis G, Garrido C. Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr Med Chem. 2007;14(27):2839–2847. doi: 10.2174/092986707782360079. [DOI] [PubMed] [Google Scholar]
- El-Samad H, Kurata H, Doyle JC, Gross CA, Khammash M. Surviving heat shock: control strategies for robustness and performance. Proc Natl Acad Sci USA. 2005;102(8):2736–2741. doi: 10.1073/pnas.0403510102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, Vandervies SM, Hemmingsen SM. The molecular chaperone concept. Biochem Soc Symp. 1989;55:145–153. [PubMed] [Google Scholar]
- Fonte V, Kipp DR, Yerg J, Merin D, Forrestal M, Wagner E, Roberts CM, Link CD. Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J Biol Chem. 2008;283(2):784–791. doi: 10.1074/jbc.M703339200. [DOI] [PubMed] [Google Scholar]
- Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70. Cell Cycle. 2006;5(22):2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr Opin Struck Biol. 2010;20(1):23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/S0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on 2 chaperonin proteins and Mg-ATP. Nature. 1989;342(6252):884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Ben Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA. 1999;96(24):13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J, Kampinga HH. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones. 2009;14(1):1–21. doi: 10.1007/s12192-008-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman J, Vos MJ, Waarde MAWH, Kampinga HH. Comparison of intra-organellar chaperone capacity for dealing with stress-induced protein unfolding. J Biol Chem. 2007;282(47):34334–34345. doi: 10.1074/jbc.M703876200. [DOI] [PubMed] [Google Scholar]
- Harrison C. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones. 2003;8(3):218–224. doi: 10.1379/1466-1268(2003)008<0218:GANEFF>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol biol. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Hasan CMM, Shimizu K (2008) Effect of temperature up-shift on fermentation and metabolic characteristics in view of gene expressions in Escherichia coli. Microb Cell Fact 7:35. doi:10.1186/1475-2859-7-35 [DOI] [PMC free article] [PubMed]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol biol. 2005;12(10):842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hightower LE. Cultured animal-cells exposed to amino-acid-analogs or puromycin rapidly synthesize several polypeptides. J Cell Physiol. 1980;102(3):407–427. doi: 10.1002/jcp.1041020315. [DOI] [PubMed] [Google Scholar]
- Hinault MP, Goloubinoff P. Molecular crime and cellular punishment: active detoxification of misfolded and aggregated proteins in the cell by the chaperone and protease networks. Adv Exp Mol Biol. 2007;594:47–54. doi: 10.1007/978-0-387-39975-1_5. [DOI] [PubMed] [Google Scholar]
- Hinault MP, Ben-Zvi A, Goloubinoff P. Chaperones and proteases—cellular fold-controlling factors of proteins in neurodegenerative diseases and aging. J Mol Neurosci. 2006;30(3):249–265. doi: 10.1385/JMN:30:3:249. [DOI] [PubMed] [Google Scholar]
- Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21(24):4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- Horváth I, Glatz A, Varvasovszki V, Török Z, Pali T, Balogh G, Kovacs E, Nadasdi L, Benko S, Joo F, Vígh L. Membrane physical state controls the signaling mechanism of the heat shock response in Synechocystis PCC 6803: identification of hsp17 as a “fluidity gene”. Proc Natl Acad Sci USA. 1998;95(7):3513–3518. doi: 10.1073/pnas.95.7.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath I, Multhoff G, Sonnleitner A, Vigh L. Membrane-associated stress proteins: more than simply chaperones. Biochim Biophys Act. 2008;1778(7–8):1653–1664. doi: 10.1016/j.bbamem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Jakob U, Gaestel M, Engel K, Buchner J. Small heat-shock proteins are molecular chaperones. J Biol Chem. 1993;268(3):1517–1520. [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92(19):1564–1572. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and S1s1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531(2):339–342. doi: 10.1016/S0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- Kabbage M, Dickman MB. The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65(9):1390–1402. doi: 10.1007/s00018-008-7535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14(1):105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemura H, Kusumoto K, Miyake H, Tashiro S, Rokutan K, Shimada M. Geranylgeranylacetone prevents acute liver damage after massive hepatectomy in rats through suppression of a CXC chemokine GRO1 and induction of heat shock proteins. J Gastrointest Surg. 2009;13(1):66–73. doi: 10.1007/s11605-008-0604-x. [DOI] [PubMed] [Google Scholar]
- Kanitkar M, Bhonde RR. Curcumin treatment enhances islet recovery by induction of heat shock response proteins, Hsp70 and heme oxygenase-1, during cryopreservation. Life Sci. 2008;82(3–4):182–189. doi: 10.1016/j.lfs.2007.10.026. [DOI] [PubMed] [Google Scholar]
- Kieran D, Kalmar B, Dick JRT, Riddoch-Contreras J, Burnstock G, Greensmith L. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med. 2004;10(4):402–405. doi: 10.1038/nm1021. [DOI] [PubMed] [Google Scholar]
- Kimpel JA, Key JL. Presence of heat-shock mRNAs in field grown soybeans. Plant Physiol. 1985;79(3):672–678. doi: 10.1104/pp.79.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner JR, He SQ, Balasubramanyam V, Kepros J, Yang CY, Zhang M, Du ZJ, Barsoum J, Bertin J. Elesclomol induces cancer cell apoptosis through oxidative stress. Mol Cancer Ther. 2008;7(8):2319–2327. doi: 10.1158/1535-7163.MCT-08-0298. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8(10):1163–1224. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/S0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Kroeger PE, Morimoto RI. Selection of new Hsf1 and Hsf2 DNA-binding sites reveals differences in trimer cooperativity. Mol Cell Biol. 1994;14(11):7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-Containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140(1):136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Lanka V, Wieland S, Barber J, Cudkowicz M. Arimoclomol: a potential therapy under development for ALS. Expert Opin Investig Drugs. 2009;18(12):1907–1918. doi: 10.1517/13543780903357486. [DOI] [PubMed] [Google Scholar]
- Large AT, Goldberg MD, Lund PA. Chaperones and protein folding in the archaea. Biochem Soc Trans. 2009;37:46–51. doi: 10.1042/BST0370046. [DOI] [PubMed] [Google Scholar]
- Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT. Neurodegenerative disease—amyloid pores from pathogenic mutations. Nature. 2002;418(6895):291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Liberek K, Georgopoulos C. Autoregulation of the Escherichia coli heat-shock response by the Dnak and Dnaj heat-shock proteins. Proc Natl Acad Sci USA. 1993;90(23):11019–11023. doi: 10.1073/pnas.90.23.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27(2):328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macario AJL, Macario EC. The archaeal molecular chaperone machine: peculiarities and paradoxes. Genetics. 1999;152(4):1277–1283. doi: 10.1093/genetics/152.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rudiger S, Roder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18(24):6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Hasiberger T, Tessarz P, Bukau B. Common and specific mechanisms of AAA plus proteins involved in protein quality control. Biochem Soc Trans. 2008;36:120–125. doi: 10.1042/BST0360120. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Gen Dev. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA. 1999;96(13):7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Vigh L. The small heat shock proteins and their clients. Cell Mol Life Sci. 2007;64(3):294–306. doi: 10.1007/s00018-006-6321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan NK, Narayanan BA, Bosland M, Condon MS, Nargi D. Docosahexaenoic acid in combination with celecoxib modulates HSP70 and p53 proteins in prostate cancer cells. Int J Cancer. 2006;119(7):1586–1598. doi: 10.1002/ijc.22031. [DOI] [PubMed] [Google Scholar]
- Nardai G, Csermely P, Soti C. Chaperone function and chaperone overload in the aged. A preliminary analysis. Exp Gerontol. 2002;37(10–11):1257–1262. doi: 10.1016/S0531-5565(02)00134-1. [DOI] [PubMed] [Google Scholar]
- Onuoha SC, Couistock ET, Grossmann JG, Jackson SE. Structural studies on the co-chaperone hop and its complexes with Hsp90. J Mol Biol. 2008;379(4):732–744. doi: 10.1016/j.jmb.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Palotai R, Szalay MS, Csermely P. Chaperones as integrators of cellular networks: changes of cellular integrity in stress and diseases. IUBMB Life. 2008;60(1):10–18. doi: 10.1002/iub.8. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Speculations on the functions of the major heat-shock and glucose-regulated proteins. Cell. 1986;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, Deglon N. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington’s disease. Mol Ther. 2007;15(5):903–911. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17(6):229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Pouppirt PS. Treatment of Parkinson’s syndrome with fever produced by baths: report of case. Cal West Med. 1929;31(3):192–195. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor sigma(32) by the DnaK and DnaJ chaperones. Mol Cell. 2008;32(3):347–358. doi: 10.1016/j.molcel.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA (2004) Protein aggregation and neurodegenerative disease. Nat Rev Neurosci 10(Suppl):S10–S17. doi:10.1038/Nm1066 [DOI] [PubMed]
- Saidi Y, Finka A, Chakhporanian M, Zryd JP, Schaefer DG, Goloubinoff P. Controlled expression of recombinant proteins in Physcomitrella patens by a conditional heat-shock promoter: a tool for plant research and biotechnology. Plant Mol Biol. 2005;59(5):697–711. doi: 10.1007/s11103-005-0889-z. [DOI] [PubMed] [Google Scholar]
- Saidi Y, Domini M, Choy F, Zryd JP, Schwitzguebel JP, Goloubinoff P. Activation of the heat shock response in plants by chlorophenols: transgenic Physcomitrella patens as a sensitive biosensor for organic pollutants. Plant Cell Environ. 2007;30(6):753–763. doi: 10.1111/j.1365-3040.2007.01664.x. [DOI] [PubMed] [Google Scholar]
- Saidi Y, Finka A, Muriset M, Bromberg Z, Weiss YG, Maathuis FJM, Goloubinoff P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell. 2009;21(9):2829–2843. doi: 10.1105/tpc.108.065318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Lindquist SL. Hsp104 required for induced thermotolerance. Science. 1990;248(4959):1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H. HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol Microbiol. 2010;76(3):576–589. doi: 10.1111/j.1365-2958.2010.07139.x. [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schuermann JP, Jiang JW, Cuellar J, Llorca O, Wang LP, Gimenez LE, Jin SP, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R. Structure of the Hsp110: Hsc70 nucleotide exchange machine. Mol Cell. 2008;31(2):232–243. doi: 10.1016/j.molcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Morano KA. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones. 2007;12(1):1–8. doi: 10.1379/CSC-245R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SK, Christen P, Goloubinoff P. Disaggregating chaperones: an unfolding story. Curr Prot Pept Sci. 2009;10:432–446. doi: 10.2174/138920309789351930. [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM. A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell. 2010;22(1):205–220. doi: 10.1105/tpc.109.071464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27(20):2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtilerman M, Lorimer GH, Englander SW. Chaperonin function: folding by forced unfolding. Science. 1999;284(5415):822–825. doi: 10.1126/science.284.5415.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha RA, Khare P, Rai A, Maurya SK, Pathak A, Mohan V, Nagar GK, Mudiam MKR, Godbole MM, Bandyopadhyay S. Anti-apoptotic role of omega-3-fatty acids in developing brain: perinatal hypothyroid rat cerebellum as apoptotic model. Int J Dev Neurosci. 2009;27(4):377–383. doi: 10.1016/j.ijdevneu.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Smalley WE, Ray WA, Daugherty JR, Griffin MR. Nonsteroidal antiinflammatory drugs and the incidence of hospitalizations for peptic-ulcer disease in elderly persons. Am J Epidemiol. 1995;141(6):539–545. doi: 10.1093/oxfordjournals.aje.a117469. [DOI] [PubMed] [Google Scholar]
- Solit DB, Chiosis G. Development and application of Hsp90 inhibitors. Drug Discov Today. 2008;13(1–2):38–43. doi: 10.1016/j.drudis.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Soll J. Protein import into chloroplasts. Curr Opin Plant Biol. 2002;5(6):529–535. doi: 10.1016/S1369-5266(02)00296-0. [DOI] [PubMed] [Google Scholar]
- Soti C, Pal C, Papp B, Csermely P. Molecular chaperones as regulatory elements of cellular networks. Curr Opin Cell Biol. 2005;17(2):210–215. doi: 10.1016/j.ceb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzulo R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175(6):901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay MS, Kovacs IA, Korcsmaros T, Bode C, Csermely P. Stress-induced rearrangements of cellular networks: consequences for protection and drug design. FEBS Lett. 2007;581(19):3675–3680. doi: 10.1016/j.febslet.2007.03.083. [DOI] [PubMed] [Google Scholar]
- Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, Daly TM, Bergman LW, Kappe SHI. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci USA. 2008;105(1):305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissieres A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster—relation to chromosome puffs. J Mol Biol. 1974;84(3):389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40(2):397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Spuy J, Kana BD, Dirr HW, Blatch GL. Heat shock cognate protein 70 chaperone-binding site in the co-chaperone murine stress-inducible protein 1 maps to within three consecutive tetratricopeptide repeat motifs. Biochem J. 2000;345:645–651. doi: 10.1042/0264-6021:3450645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273(18):11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- Vigh L, Literati PN, Horvath I, Torok Z, Balogh G, Glatz A, Kovacs E, Boros I, Ferdinandy P, Farkas B, Jaszlits L, Jednakovits A, Koranyi L, Maresca B. Bimoclomol: a nontoxic, hydroxylamine derivative with stress protein-inducing activity and cytoprotective effects. Nat Med. 1997;3(10):1150–1154. doi: 10.1038/nm1097-1150. [DOI] [PubMed] [Google Scholar]
- Vigh L, Maresca B, Harwood JL. Does the membrane’s physical state control the expression of heat shock and other genes? Trends Biochem Sci. 1998;23(10):369–374. doi: 10.1016/S0968-0004(98)01279-1. [DOI] [PubMed] [Google Scholar]
- Vigh L, Horvath I, Maresca B, Harwood JL. Can the stress protein response be controlled by ‘membrane-lipid therapy’? Trends Biochem Sci. 2007;32(8):357–363. doi: 10.1016/j.tibs.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Mol Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- Weiss YG, Bromberg Z, Raj N, Raphael J, Goloubinoff P, Ben-Neriah Y, Deutschman CS. Enhanced heat shock protein 70 expression alters proteasomal degradation of I kappa B kinase in experimental acute respiratory distress syndrome. Crit Care Med. 2007;35(9):2128–2138. doi: 10.1097/01.CCM.0000278915.78030.74. [DOI] [PubMed] [Google Scholar]
- Whelan SA, Hightower LE. Induction of stress proteins in chicken-embryo cells by low-level zinc contamination in amino acid-free media. J Cell Physiol. 1985;122(2):205–209. doi: 10.1002/jcp.1041220207. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, Decosta B, Myers CE, Neckers LM. Inhibition of heat-shock protein Hsp90-Pp60(V-Src) heteroprotein complex-formation by benzoquinone ansamycins—essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci USA. 1994;91(18):8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [Ure3] as an altered Ure2 protein—evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264(5158):566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wiech H, Buchner J, Zimmermann R, Jakob U. Hsp90 chaperones protein folding in vitro. Nature. 1992;358(6382):169–170. doi: 10.1038/358169a0. [DOI] [PubMed] [Google Scholar]
- Wu YJ, Cao ZM, Klein WL, Luo Y. Heat shock treatment reduces beta amyloid toxicity in vivo by diminishing oligomers. Neurobiol Aging. 2010;31(6):1055–1058. doi: 10.1016/j.neurobiolaging.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HY, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong YL, Park-Sarge OK, Sarge KD. Mechanism of Hsp70i gene bookmarking. Science. 2005;307(5708):421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Takemori Y, Sakurai M, Sugiyama K, Sakurai H. Differential recognition of heat shock elements by members of the heat shock transcription factor family. FEBS J. 2009;276(7):1962–1974. doi: 10.1111/j.1742-4658.2009.06923.x. [DOI] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442(7106):1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Zhao RM, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the Hsp90 chaperone. Cell. 2005;120(5):715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136(1):2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Zourlidou A, Smith MDP, Latchman DS. HSP27 but not HSP70 has a potent protective effect against alpha-synuclein-induced cell death in mammalian neuronal cells. J Neurochem. 2004;88(6):1439–1448. doi: 10.1046/j.1471-4159.2003.02273.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Correlation of Arabidopsis chaperome transcription in five independent heat shock treatments. Heat shock cognates are indicated by arrows. The microarray data were extracted from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under the series accession numbers GSE4760 (Hsf2 mutant, 4 days at 23°C to 2 h at 38°C), GSE16222 (5 days at 23°C to 1 h at 37°C), GSE12619 (7 days at 22°C to 1 h at 37°C), GSE4062 (15 days at 22°C to 2 h at 37°C), and GSE11758 (mature leaves at 20°C to 1 h at 37°C) (PDF 956 kb)
Clustering of RNA expression levels of the Arabidopsis chaperome under seven abiotic and chemical treatments: dithiothreitol (DTT), tunicamycin, salicylic acid, ibuprofen, 2,3,5- triiodobenzoicacid (TIBA), 2,4,6-trihydroxybenzamide (2,4,6-T), and heat treatment as indicated. Gene clusters typical of the of the cellular stress response (CSR) (a) or of the unfolded protein response (UPR) (b) are indicated with brackets. The presumed subcellular localizations are indicated with different background colors (PDF 1032 kb)
Clustering of RNA expression levels of 168 genes from the human chaperome under 21 treatments: A 2-deoxyglucose, B tunicamycin, C phorbol 12-myristate 13-acetate, D cadmium, E N-acetylcysteine, F paclitaxel, G doxycycline, H echinomycin, I heat shock study, J elesclomol, K smoking, L simvastatin, M etoposide, N VAF347, O sapphyrin PCI-5002, P propiconazole, Q myclobutanil, R rifampicin, S dihydrotestosterone, T estrogen, and U apple procyanidin. Gene clusters typical a of the cellular stress response (CSR), b of the unfolded protein response (UPR), and c of a main less specific cell response are indicated with brackets. The presumed subcellular localizations are indicated with different background colors of the gene names (PDF 442 kb)
List of identified human chaperone genes. (1) Small HSP, GroEL and CCT-like chaperonins, Hsp70, DNAJ, Hsp100 and Hsp90 families have been previously annotated (Kampinga et al. 2009). (2) The Bag family has been previously annotated (Takayama et al. 1999). (3) FKBP and CYP like peptide-prolyl isomerase families have been previously annotated (Barik 2006). (4) Protein disulfide isomerase family has been previously annotated (Ellgaard and Ruddock 2005). (5)Human Hsp90 co-chaperones have been identified according to www.picard.ch (XLS 45 kb)
List of identified heat shock elements (HSEs) in human chaperome. Nucleotide sequences of canonical HSEs in cis and trans orientations were identified between position −3000 and 300 bp from the translation start site of each chaperone gene (XLS 32 kb)
List of identified Arabidopsis thaliana chaperome genes. (1) Small HSPs were obtained by BLAST using α-crystalline domain as queries against the Arabidopsis protein subset of the NCBI database. (2) Chaperonins have been previously annotated (Hill and Hemmingsen 2001). (3) DnaJ proteins were re-annotated as compared to the previous J-protein nomenclature (Rajan and D’Silva 2009, as shown in brackets). All isoforms, splice variants, and paralogous protein according to previous annotations have been removed. The gene loci At2g02200, At5g34895, At2g07010 (MWJ3.6), At1g31210, At2g14930, At2g13940, and At2g24660 (corresponding to AtDjC6, AtDjC7, AtDjC50, and AtDjC58–AtDjC61, respectively) contained transposable elements and were removed as such. (4) The Hsp70 and Hsp110 were obtained by BLAST using DnaK from E. coli as query against the Arabidopsis protein subset of the NCBI database. (5) The Hsp100 were obtained by BLAST using ClpB from E. coli as the query against Arabidopsis protein subset of the NCBI database. Only the most ClpB-like proteins were retained, while ClpA/C proteins that are associated to the ClpP protease were excluded. (6) The Bag family has been previously annotated (Yan 2003). (7) The Hsp90 family has been previously annotated (Krishna and Gloor 2001). (8) PPIase families have been previously annotated (Romano et al. 2005). (9) PDIs families have been previously annotated (Houston et al. 2005). (10) Human Hsp90 co-chaperones were identified in yeast and animals according to http://www.picard.ch and used as queries to identify Arabidopsis orthologs in the protein subset of the NCBI database (XLS 162 kb)
Supplemental references (DOC 39.5 kb)