The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens
Schofield and colleagues demonstrate that a functional HIF system is present in the simplest animal, Trichoplax adhaerens. Their results imply that the HIF system is conserved in all animals, and reveal conservation of biochemical properties in the oxygen-sensing machinery
Keywords: evolution, HIF, hypoxia, oxygen sensing, Trichoplax
Abstract
The hypoxic response in humans is mediated by the hypoxia-inducible transcription factor (HIF), for which prolyl hydroxylases (PHDs) act as oxygen-sensing components. The evolutionary origins of the HIF system have been previously unclear. We demonstrate a functional HIF system in the simplest animal, Trichoplax adhaerens: HIF targets in T. adhaerens include glycolytic and metabolic enzymes, suggesting a role for HIF in the adaptation of basal multicellular animals to fluctuating oxygen levels. Characterization of the T. adhaerens PHDs and cross-species complementation assays reveal a conserved oxygen-sensing mechanism. Cross-genomic analyses rationalize the relative importance of HIF system components, and imply that the HIF system is likely to be present in all animals, but is unique to this kingdom.
Introduction
Maintaining oxygen homoeostasis is a challenge for all aerobic life and particularly for animals, due to their greater mass and energy demands. In humans, the hypoxic response involves a gene array that is regulated by the heterodimeric hypoxia-inducible transcription factor (HIF), which contains basic helix–loop–helix (bHLH) and Per–Arnt–Sim (PAS) domains. HIF regulates fundamental processes including glycolysis, the tricarboxylic acid cycle and specialized oxygen delivery systems in higher animals (for a review, see Kaelin & Ratcliffe, 2008).
The stability and transcriptional activity of HIFα are regulated by post-translational hydroxylations that are catalysed by prolyl-hydroxylase domain enzymes (PHDs/EGLNs) and the asparaginyl hydroxylase factor inhibiting HIF (FIH). Hydroxylation of two HIFα prolyl residues in amino- and carboxy-terminal oxygen-dependent degradation domains (NODD and CODD, respectively) promotes binding to the von Hippel Lindau protein (VHL) elongin B/C ubiquitin ligase complex which signals proteasomal degradation (Kaelin & Ratcliffe, 2008). HIF activity is reduced by FIH-catalysed asparaginyl hydroxylation in the HIFα C-terminal transcriptional activation domain (CAD). The HIF hydroxylases are 2-oxoglutarate (2OG)-dependent oxygenases (Hausinger, 2004; Loenarz & Schofield, 2010); their oxygen dependence as well as other properties enable them to act as oxygen sensors.
Functional HIF systems have been characterized in Caenorhabditis elegans and Drosophila, but the evolutionary development of HIF signalling has been unclear. The presence of PHD-related oxygenases in many life forms, including protists and prokaryotes (van der Wel et al, 2005; McDonough et al, 2006; Lee et al, 2009), has led to a proposal that they mediate an oxygen-sensing mechanism, which is “conserved throughout evolution in all organisms since bacteria” (Leite et al, 2008).
We report that the main components of the human HIF system—HIF, PHD and VHL—function in the simplest known animal, Trichoplax adhaerens, but probably not in non-metazoans. The targets of HIF in T. adhaerens suggest a role for it in the adaptation of basal animals to changing oxygen levels in the early Cambrian period (Holland, 2006). Characterization of the components of the T. adhaerens HIF system shows the ‘core' conserved features of oxygen sensing and—together with results from bioinformatic analyses—their relative importance. Our results—which provide the first detailed analysis of a biochemical pathway in T. adhaerens—rationalize the evolution of the HIF system in all animals and highlight the central role of prolyl hydroxylation in hypoxic signalling.
Results and Discussion
Evidence for a HIF system in T. adhaerens
Analyses of the human genome have shown an increased frequency of hypoxia-response elements (HREs) in promoter regions (Mole et al, 2009; Xia et al, 2009), suggesting HREs have been selected for in HIF-containing organisms. On performing cross-genomic analyses of 50 eukaryotes (supplementary Fig S1A online), we found no HRE enrichment in promoter regions of protists (eukaryotic microorganisms), but HRE enrichment was observed in promoter regions of vertebrates and invertebrates. The short nature of the HRE sequence (5′-RCGTG-3′, in which if R=A, it is not preceded by C), and the possibility that it binds to factors other than HIF, make the significance of this enrichment unclear. We therefore focused on investigating the presence of HIF at the unicellular protist–multicellular animal boundary.
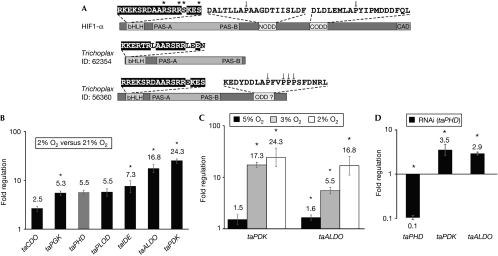
No proteins with both bHLH and PAS domains were found in the choanoflagellate Monosiga brevicollis (the unicellular organisms closest to animals; Abedin & King, 2008) or other protists, implying a lack of HIF but we identified candidate HIFα and HIFβ genes in the simplest known animal, T. adhaerens (Fig 1A; supplementary Fig S1B online). However, the T. adhaerens HIFα (taHIFα) oxygen-dependent degradation domain (ODD) differs substantially from human HIF1α NODD and CODD (15% and 35% identity over 20 residues, respectively), contains four prolyl residues, but lacks the consensus LXXLAP prolyl-hydroxylation site of HIFα proteins that have been studied (Kaelin & Ratcliffe, 2008).
Figure 1.
Hypoxic regulation of Trichoplax adhaerens genes. (A) Domain structures of T. adhaerens bHLH-PAS proteins; sequence 56360 (Srivastava et al, 2008) corresponds to the likely taHIFα homologue. Asterisks indicate (predicted) DNA-interacting residues on the basis of homology modelling (PDB 1AN4). Known and putative HIFα ODD sequences are shown; arrows indicate known and putative prolyl-hydroxylation sites. (B) RT–qPCR analysis of T. adhaerens showing fold regulation in hypoxia (2% O2) relative to normoxia (normalized to β-actin; n=3; ±s.e.m.; *P<0.05). (C) Increasing degrees of hypoxia increased expression of taALDO and taPDK (normalized to β-actin; n=3; ±s.e.m.; *P<0.05). (D) RT–qPCR analysis of RNAi against taPHD in T. adhaerens. Whereas taPHD levels are reduced, expression of hypoxia-inducible genes (taPDK and taALDO) is increased (normalized to β-actin; n=3; ±s.e.m.; *P<0.05). ALDO, fructose-biphosphate aldolase; bHLH, basic helix–loop–helix; CDO, cysteine dioxygenase; HIF1α, hypoxia-inducible transcription factor 1α; IDE, insulin-degrading enzyme; ODD, oxygen-dependent degradation domain; PDK, pyruvate dehydrogenase kinase; PGK, phosphoglycerate kinase; PLOD, procollagen lysine hydroxylase; RNAi, RNA interference; RT–qPCR, reverse transcription–quantitative PCR; taHIFα, Trichoplax adhaerens hypoxia-inducible transcription factor-α; taPHD, Trichoplax adhaerens prolyl-hydroxylase.
The response of T. adhaerens to hypoxia has not been previously investigated. Observation of sustained movement showed that incubation of T. adhaerens in hypoxic seawater (5% atmospheric O2) did not affect viability for >2 days; at 3, 2 and 1% O2, T. adhaerens survived for ∼1.5 days, >16 and ∼5 h, respectively. To determine whether T. adhaerens contains an HIF system, we identified candidate genes for reverse transcription–quantitative PCR (RT–qPCR) analysis. Selection was based on the presence of ⩾1 putative HRE −300 to +200 bp from the predicted translational start site (supplementary Fig S1A online) and on the similarity with a human homologue of defined role. Exposure of T. adhaerens to hypoxia (2% O2, 12 h) upregulated putative HIF target genes (Fig 1B); the degree of upregulation ranged from modest to very significant (∼20-fold; P<0.01) for fructose-biphosphate aldolase (taALDO) and pyruvate dehydrogenase kinase (taPDK). Upregulation of both taALDO and taPDK expression increased significantly when oxygen levels were progressively decreased from 5% to 2% (Fig 1C).
Identification of features of oxygen sensing through HIF
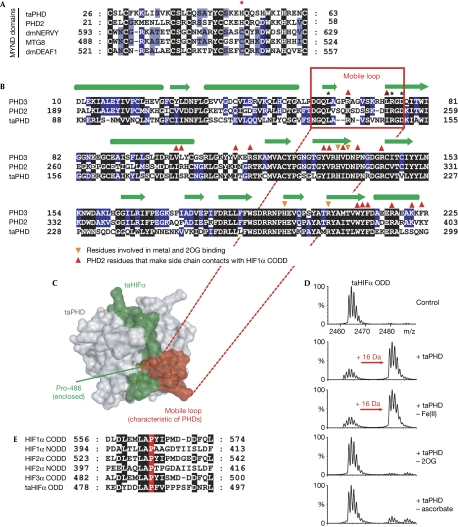
To test for oxygenase involvement in the T. adhaerens hypoxic response, we used the hydroxylase inhibitor dimethyloxalylglycine (DMOG). Dose–response measurements (n=8 T. adhaerens; 0.1, 1, 5 and 10 mM DMOG) resulted in a reduction in size (after 5, 2, 0.5 and 0.5 h, respectively); at 5 and 10 mM DMOG, non-adherence to the surface (after 3 and 1 h, respectively) was followed by lysis (after 6 and 4 h, respectively). As observed in hypoxia, DMOG treatment (1 mM, 48 h) in normoxia significantly upregulated taALDO (4.8-fold; P<0.01). By using bioinformatics we identified a single putative PHD homologue (taPHD; Fig 2A,B), which, similarly to PHD2 but not PHD1/3, has a non-canonical MYND-type zinc finger. A ‘mobile loop' that, in PHD2, is involved in substrate binding (Chowdhury et al, 2009) is conserved in taPHD (Fig 2C). The complete loop sequence is not conserved in non-metazoan PHD-related enzymes (van der Wel et al, 2005; Leite et al, 2008), suggesting that they do not act on HIF. RNA interference with taPHD significantly reduced taPHD transcript levels to ∼10% relative to control, and significantly increased taPDK and taALDO mRNA levels (∼3.5- and 2.9-fold, respectively), supporting the idea that it is involved in hypoxic signalling (Fig 1D).
Figure 2.
Trichoplax adhaerens PHD has conserved substrate-binding features and is active as a taHIFα prolyl hydroxylase. (A) Comparison of the MYND finger for PHD2/taPHD with stereotypical MYND finger sequences. Note that one of the cysteines is replaced by a histidine in the PHDs (indicated by an asterisk; GenBank entries: dmNERVY, 45445680; MTG8, 4757916; dmDEAF1, 7293736). (B) Sequence comparison of human PHD2/3 with taPHD; sequences corresponding to the mobile region are boxed. Secondary structures: α-helices, green cylinders; β-strands, green arrows. (C) Homology model for the binding of taHIFα (green) by taPHD (grey; using PDB 3HQR). Note that a mobile loop (red) is conserved in taPHD and appears ‘anchored' to the active site by electrostatic (Arg 148/Asp 150) and hydrophobic interactions (Leu 138/Ile 147). (D) Mass spectrometric analyses showing taPHD-catalysed hydroxylation of taHIFα ODD (16-Da mass shift). taPHD co-purifies with Fe(II) and its activity is stimulated by ascorbate. (E) Alignment of T. adhaerens and human HIFα ODD domains. CODD, C-terminal ODD; NODD, N-terminal ODD; ODD, oxygen-dependent degradation domain; 2OG, 2-oxoglutarate; PHD, prolyl hydroxylase; taHIFα, T. adhaerens hypoxia-inducible transcription factor-α; taPHD, T. adhaerens prolyl-hydroxylase.
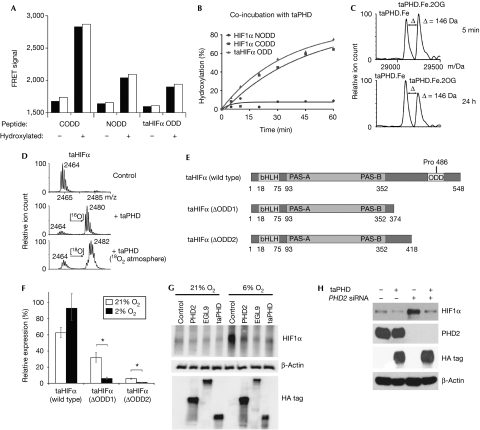
We then prepared recombinant taPHD in Escherichia coli; taPHD co-purified with iron and stimulated uncoupled 2OG turnover in the presence of ascorbate. Analysis by mass spectrometry after incubation with the predicted taHIFα ODD revealed that taPHD catalysis (with or without its MYND finger) produces a 16-Da mass shift (Fig 2D); fragmentation mass spectrometry identified the hydroxylation site as Pro 486 (Fig 2E; supplementary Fig S2 online). Bioinformatic analyses indicated the presence of a homologous VHL complex (34% identity over 84 residues with human VHL; 53% and 85% identity with human elongins B and C, respectively). Homology modelling studies predict that binding of taHIFα to taVHL is similar, with the role of Ser 111 and His 115 in human VHL taken over by taVHL Thr 60 and His 64 (supplementary Fig S3B online). Indeed, taPHD-catalysed hydroxylation at taHIFα Pro 486 promoted binding to the human VHL complex (Fig 3A; supplementary Fig S3A online).
Figure 3.
T. adhaerens prolyl-hydroxylase and prolyl hydroxylase 2 have conserved functions in the hypoxic response. (A) FRET assay showing that binding of human and T. adhaerens HIFα peptides to the VHL complex depends on prolyl trans-4-hydroxylation (two replicates). (B) taPHD-catalysed hydroxylation of an equimolar mixture of HIF1α NODD, CODD and taHIFα ODD (NODD hydroxylation was observed after incubation overnight). (C) Incubation of taPHD with equimolar Fe(II) and 2OG (mass: 146 Da) leads to a stable complex in the absence of substrate (half-life >24 h; note the lack of succinate formation), as shown by non-denaturing electrospray ionization mass spectrometry. (D) Hydroxylation of taHIFα in 18O2 proceeds with 18O incorporation. (E) Domain analysis of taHIFα splice variants. (F) Reverse transcription–quantitative PCR analysis of the effect of hypoxia on relative taHIFα splice variant levels (n=3; ±s.e.m.; *P<0.05). See supplementary Fig S4 online for splice sites. (G) Immunoblot of human 293T cells transfected with haemagglutinin-tagged human PHD22−426, C. elegans EGL92−723 and T. adhaerens taPHD, showing that all enzymes cause reduction in endogenous HIF1α levels in hypoxia. (H) Immunoblot of human 293T cells showing that taPHD is sufficient to suppress endogenous HIF1α levels in the absence of PHD2. Cells were transfected with or without haemagglutinin-tagged T. adhaerens taPHD plus control or PHD2 siRNA. CODD, C-terminal ODD; FRET, fluorescence resonance energy transfer; HA, haemagglutinin; HIFα, hypoxia-inducible transcription factor-α; NODD, N-terminal oxygen-dependent degradation domain; 2OG, 2-oxoglutarate; PHD, prolyl hydroxylase; siRNA, small interfering RNA; taPHD, T. adhaerens prolyl-hydroxylase; VHL, von Hippel Lindau protein.
Further analyses of taPHD revealed conservation of biochemical properties with PHD2. taPHD hydroxylated HIF1α CODD and NODD, but the former was a more efficient substrate; efficiency was similar to taHIFα ODD (Fig 3B). Both taHIFα and CODD peptides bound more tightly to taPHD than to NODD in non-denaturing electrospray ionization mass spectrometry analyses (the ratio of bound:unbound ODD to the taPHD was 6:4 for taHIFα; 1:1 for CODD and <1:20 for NODD). Consistent with normoxic upregulation of hypoxia-inducible genes in T. adhaerens by DMOG, taPHD was inhibited by N-oxalylglycine, fumarate and other known 2OG oxygenase inhibitors (supplementary Fig S3C online). PHD2 is atypical in that it reacts unusually slowly with oxygen and forms a stable complex with Fe(II) and 2OG (Flashman et al, 2010). Similarly to PHD2, taPHD bound to Fe(II) tightly, formed a stable enzyme.Fe(II).2OG complex under aerobic conditions in the absence of substrate (half-life>24 h; Fig 3C), was stereoselective for the production of trans-4-hydroxyproline and catalysed hydroxylation with the incorporation of oxygen from O2 (Fig 3D). Together, these results suggest that the nature of the PHD reactions with oxygen and 2OG might be intrinsic to their oxygen-sensing role.
In mammalian cells, a feedback loop involving hypoxia/HIF-mediated upregulation of PHD2/3 regulates HIF activity (Cioffi et al, 2003; Olga et al, 2004; Minamishima et al, 2009). Similarly, the taPHD mRNA level was upregulated ∼5.5-fold on exposure of T. adhaerens to hypoxia (2% O2; Fig 1B), suggesting the presence of a conserved feedback loop. In addition to full-length taHIFα, we also observed alternatively spliced taHIFα forms that could act as negative HIF regulators because they lack the ODD (ΔODD1–2; Fig 3E). RT–qPCR analyses of full-length and ΔODD1–2 taHIFα abundance demonstrated that the latter decrease significantly on hypoxic exposure (Fig 3F; supplementary Fig S4 online), suggesting that oxygen-regulated splicing produces functionally distinct taHIFα isoforms, as is found for human HIF3α and Drosophila HIFα (Makino et al, 2002; Gorr et al, 2004).
To test whether taPHD regulates full-length endogenous human HIF1α, genes encoding for haemagglutinin-tagged human PHD2, C. elegans EGL-9 (egg-laying defective 9; its PHD homologue) and taPHD were expressed ectopically in human cells; all the constructs reduced HIF1α levels to a similar extent in moderate hypoxia (6% O2), in which there is reduced endogenous PHD activity (Fig 3G). In the presence of small interfering RNA against PHD2, taPHD strongly suppressed endogenous HIF1α levels (Fig 3H).
Evolution of the HIF system in animals
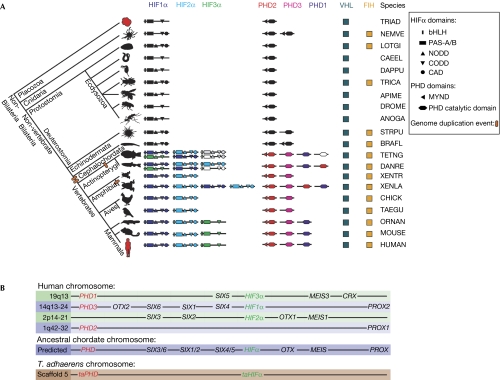
We then used cross-genomic bioinformatic analyses to test whether the HIF system is conserved in animals, and to investigate how multiple sites of hydroxylation have accrued during evolution (supplementary Table S2 online). Our results (Fig 4A) imply that all animals contain ⩾1 HIFα, with bHLH-PAS domains and ⩾1 ODD (more closely related to human CODD than to NODD), ⩾1 PHD, with an N-terminal non-canonical MYND finger, and VHL. These features define a ‘minimal' HIFα-ODD/MYND-PHD/VHL system in all animals with annotated genomes. Most of the analysed animals (20 of 24) contain two putative HIFα prolyl-hydroxylation sites (supplementary Table S2 online), and the sequence that is more similar to HIFα NODD always precedes the ‘CODD'. HIFα asparaginyl-hydroxylation sites (CAD) were observed in 16 of 24 species, including all deuterostomes, and the presence of such a site correlates with predicted FIH homologues. To test FIH/CAD assignments, we analysed the most basal animal in which both FIH and the CAD are conserved: Nematostella vectensis. We found that its HIFα CAD (VKALFPYVTQSDAEVNAPV) was hydroxylated by human FIH (a 16-Da mass shift from 2,045 to 2,061 Da was observed).
Figure 4.
Evolutionary analysis of the hypoxia-inducible transcription factor system. (A) Phylogenetic domain analysis of HIF system gene products across metazoans (not to scale). Ovals represent genome duplications; likely pseudogenes are not shown; uncoloured domains indicate no isoform assignment; grey domains reflect predictions necessitated by incompletely sequenced genomes. Profile HMMs were used to search for more distant homologues. In some deposited genomes, protein domains of interest were annotated as introns (possibly due, in part, to their low sequence conservation), requiring both gene re-annotation and profile HMM searches considering all likely genome translations. See supplementary Tables S2 and S3 online for sequences of HIFα subdomains and abbreviations. (B) Comparison of the relative locations of human HIFα and PHD genes (not to scale), with prediction of their relative position in an ancestral chordate genome, and T. adhaerens gene location (note that basal animals contain few homeobox genes). In the human genome, HIFα and PHD genes map to four related chromosome regions, close to homeobox genes of the SIX1/2, SIX3/6, SIX4/5, PROX, MEIS and OTX gene families. bHLH, basic helix–loop–helix; CAD, C-terminal transcriptional activation domain; CODD, C-terminal oxygen-dependent degradation domain; FIH, factor inhibiting HIF; HIF, hypoxia-inducible transcription factor; HMM, hidden Markov model; NODD, N-terminal ODD; PAS, Per–Arnt–Sim; PHD, prolyl hydroxylase; VHL, von Hippel Lindau protein.
As the selectivity of human PHDs for CODD over NODD (Hirsila et al, 2003; Chan et al, 2005; Flashman et al, 2008) is conserved in taPHD (Fig 3B), we investigated whether this is true for other animals. Among ODDs from eight further species, most (9 of 14) were found to undergo hydroxylation by taPHD and PHD2 (supplementary Fig S3D online); in cases in which there were both NODD and CODD, the latter was hydroxylated more efficiently.
Invertebrates contain single HIFα and (with few exceptions) PHD genes, whereas vertebrates contain multiple HIFα and PHD genes (Fig 4A). Comparison between the relative genomic positions of human HIFα/PHD genes with those of homeobox genes (Fig 4B)—whose multiplication history is linked to the two genome duplication events in vertebrate evolution (Holland et al, 2007)—implies that the HIFα/PHD genes duplicated twice at the base of the vertebrate subphylum to give four genes—HIF1–4α and PHD1–4—one of which was subsequently lost. Additional genome duplications in the teleost fish lineage and Xenopus laevis rationalize the presence of more than three HIFα and four PHD genes in these organisms. The positions of the human HIFα/PHD genes suggest that they were close (probably within ∼1 Mb) on the ancestral chordate chromosome. Intriguingly, the T. adhaerens taPHD and taHIFα genes are located within <20 kb of each other (Fig 4B), indicating that their proximity might have evolutionary significance.
Conclusions
Taken together, our results suggest that the HIF–PHD–VHL triad is conserved in all animals, including HIFα with a CODD-like domain and PHD with a non-stereotypical MYND finger domain. This triad is not present in choanoflagellates such as M. brevicollis or in other protists, suggesting that there is a boundary between these unicellular organisms and metazoans. Analysis of sponge and ctenophore genomes will soon be possible, which might further refine our knowledge about this transition. Some animals have additional regulatory interfaces, including FIH/HIFα CAD, and/or a HIFα NODD. The PHD–ODD dyad is thus more important than the FIH–CAD dyad, in terms of both its distribution and, probably, its role in individual animals. The progenitor ODD in non-bilateral animals more closely resembles CODD than NODD; thus, NODD probably evolved after CODD. After NODD evolution, some invertebrate (and vertebrate) HIFα genes seem to have lost this ODD. Studies of taPHD and PHD2 with fragments of HIFα ODDs from animals at different evolutionary stages imply that the preference of human PHDs for HIFα CODD over NODD substrates is conserved, supporting our proposal that an ODD more closely related to CODD evolved before NODD.
In humans, in addition to HIF1/2α CAD asparaginyl hydroxylation, FIH catalyses asparaginyl hydroxylation of ankyrin repeats, a ubiquitous eukaryotic protein–protein interaction motif (Loenarz & Schofield, 2010). Whereas T. adhaerens does not contain the FIH–CAD dyad, it occurs in the non-bilateral animal Nematostella (Fig 4A). It is therefore unclear whether the HIFα CAD evolved after the PHD–ODD dyad. Nevertheless, the apparent absence of FIH in animals lacking an HIFα CAD (including Drosophila and C. elegans) implies either a stronger selection pressure for CAD than for ankyrin hydroxylation, or that these two types of hydroxylation are linked.
In higher animals, HIF targets remodel metabolism such that it is optimized for hypoxia and regulate highly differentiated systemic functions (Kaelin & Ratcliffe, 2008). The role of HIF in basal animals, in which many of these roles are not relevant, has not been addressed. In T. adhaerens, genes encoding the glycolytic enzymes taALDO and phosphoglycerate kinase (taPGK) were found to be hypoxically induced; their human homologues were among the first discovered HIF targets and helped to define HIF as a mediator of oxygen homeostasis in humans (Kaelin & Ratcliffe, 2008). Interestingly, taPDK was among the strongest hypoxically induced genes, and human PDK1 is also a HIF target; by deactivating the pyruvate dehydrogenase-catalysed conversion of pyruvate to acetyl-CoA, PDK1 directs pyruvate away from the tricarboxylic acid cycle. The hypoxic upregulation of taPDK and glycolysis enzymes suggests that HIF-regulated direction of metabolism towards glycolysis is conserved in T. adhaerens. As in humans, in which 2OG oxygenases other than the PHDs themselves have been shown to be HIF targets (Xia et al, 2009), we found that a collagen lysyl hydroxylase and cysteine dioxygenase are hypoxically induced. This is interesting from an evolutionary perspective, because the rise of multicellular animals has been linked to that of collagen (Towe, 1970).
Overall, these results suggest that there might be a core set of genes that are hypoxically regulated through the HIF system in all animals. We propose that the HIF system helped to enable animal life to respond to metabolic challenges, including the increase in atmospheric oxygen levels on the Earth over the course of animal evolution.
Methods
T. adhaerens cultures. T. adhaerens were fed on Pyrenomonas helgolandii and maintained in petri dishes with Ultramarine synthetic seawater (Waterlife Ltd, UK) of 3.5% salinity (19°C) at a light:dark rhythm of 16:8 h. For RNA extraction, 20–100 individuals at any vegetative developmental stage were washed in seawater and starved for 18 h. For hypoxia studies, T. adhaerens were grown under normoxia (21% O2) or hypoxia in an Invivo2 hypoxic workstation (Ruskinn Technologies, UK).
T. adhaerens RNA isolation and RT–qPCR analysis. T. adhaerens RNA was extracted using the RNeasy Micro Kit (Qiagen) and treated with DNase I; complementary DNA was obtained by using the AffinityScript QPCR cDNA Synthesis Kit (Stratagene). RT–qPCR was performed on a MiniOpticon real-time–PCR system (Bio-Rad) using Brilliant II SYBR Green qPCR Master Mix (Stratagene). Fold changes were determined by the ΔCt method, normalized to β-actin (taACTB) and analysed using Miner 2.2 (http://www.miner.ewindup.info). For details, see supplementary information online.
Human cell culture, transfections and immunoblots. HEK 293T cells were grown in normoxia (21% O2/5% CO2) or hypoxia (6% O2/5% CO2 for 4 h) in an Invivo2 hypoxic workstation. Plasmid transfections used FuGENE 6 (Roche). For RNA interference, cells were transfected with haemagglutinin-taPHD pEF6 plasmid plus PHD2 small interfering RNA oligonucleotide or 50 nM control (Drosophila HIFα) using DharmaFECT Duo (Fisher Scientific). For details, see supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council, the Wellcome Trust and the German Science Foundation (B.S.) for support, and acknowledge a Rhodes Scholarship and a William R. Miller Junior Research Fellowship (both to C.L.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abedin M, King N (2008) The premetazoan ancestry of cadherins. Science 319: 946–948 [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen SE, Giaccia AJ (2005) Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1α. Mol Cell Biol 25: 6415–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, McDonough MA, Mecinovi J, Loenarz C, Flashman E, Hewitson KS, Domene C, Schofield CJ (2009) Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases. Structure 17: 981–989 [DOI] [PubMed] [Google Scholar]
- Cioffi CL, Qin Liu X, Kosinski PA, Garay M, Bowen BR (2003) Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem Biophys Res Commun 303: 947–953 [DOI] [PubMed] [Google Scholar]
- Flashman E, Bagg EAL, Chowdhury R, Mecinovic J, Loenarz C, McDonough MA, Hewitson KS, Schofield CJ (2008) Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J Biol Chem 283: 3808–3815 [DOI] [PubMed] [Google Scholar]
- Flashman E, Hoffart LM, Hamed RB, Bollinger JM Jr, Krebs C, Schofield CJ (2010) Evidence for the slow reaction of hypoxia-inducible factor prolyl hydroxylase 2 with oxygen. FEBS J 277: 4089–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorr TA, Tomita T, Wappner P, Bunn HF (2004) Regulation of Drosophila hypoxia-inducible factor (HIF) activity in SL2 cells. J Biol Chem 279: 36048–36058 [DOI] [PubMed] [Google Scholar]
- Hausinger RP (2004) Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39: 21–68 [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem 278: 30772–30780 [DOI] [PubMed] [Google Scholar]
- Holland HD (2006) The oxygenation of the atmosphere and oceans. Phil Trans R Soc Lond B Biol Sci 361: 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PWH, Booth HAF, Bruford EA (2007) Classification and nomenclature of all human homeobox genes. BMC Biol 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr, Ratcliffe PJ (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30: 393–402 [DOI] [PubMed] [Google Scholar]
- Lee C-YS, Stewart EV, Hughes BT, Espenshade PJ (2009) Oxygen-dependent binding of Nro1 to the prolyl hydroxylase Ofd1 regulates SREBP degradation in yeast. EMBO J 28: 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite RB, Brito AB, Cancela ML (2008) An oxygen molecular sensor, the HIF prolyl 4-hydroxylase, in the marine protist Perkinsus olseni. Protist 159: 355–368 [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ (2010) Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci doi:10.1016/j.tibs.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L (2002) Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J Biol Chem 277: 32405–32408 [DOI] [PubMed] [Google Scholar]
- McDonough MA et al. (2006) Cellular oxygen sensing: crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc Natl Acad Sci USA 103: 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG Jr (2009) A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol Cell Biol 29: 5729–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ (2009) Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 284: 16767–16775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olga A, Gadisetti VRC, Matthew W, James RV, Joseph R, Jodi KM, Linehan WM, Barrett JC (2004) Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J Cell Biochem 92: 491–501 [DOI] [PubMed] [Google Scholar]
- Srivastava M et al. (2008) The Trichoplax genome and the nature of placozoans. Nature 454: 955–960 [DOI] [PubMed] [Google Scholar]
- Towe KM (1970) Oxygen-collagen priority and the early metazoan fossil record. Proc Natl Acad Sci USA 65: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel H, Ercan A, West CM (2005) The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-α class of animal prolyl 4-hydroxylases. J Biol Chem 280: 14645–14655 [DOI] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL (2009) Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA 106: 4260–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.