Abstract
Theory suggests that the risk of extinction by mutation accumulation can be comparable to that by environmental stochasticity for an isolated population smaller than a few thousand individuals. Here we show that metapopulation structure, habitat loss or fragmentation, and environmental stochasticity can be expected to greatly accelerate the accumulation of mildly deleterious mutations, lowering the genetic effective size to such a degree that even large metapopulations may be at risk of extinction. Because of mutation accumulation, viable metapopulations may need to be far larger and better connected than would be required under just stochastic demography.
Kimura, Maruyama, and Crow (1) first noted that mildly deleterious mutations may create a considerably larger mutational load in small populations than more deleterious mutations. Deleterious mutations impose a load on populations through a reduction in the mean survivorship and/or reproductive rates of individuals (2–5). In very large, effectively infinite populations, an equilibrium mutation load exists that is independent of the mutational effect (2, 3, 5, 6). However, in sufficiently small isolated populations, mildly deleterious mutations may be a potent extinction force, because individually they are nearly invisible to natural selection, although causing an appreciable cumulative reduction in population viability (1, 5, 7–11). Theory suggests that the accumulation of mildly deleterious mutations can be comparable to environmental stochasticity in causing extinction of populations smaller than a few thousand individuals (8, 12, 13).
All previous theoretical work on extinction caused by mutation accumulation has focused on a single panmictic population, but most populations have some degree of subdivision, which may greatly magnify the stochastic development of mutation load (14, 15). Moreover, most theoretical work on extinction has focused on only one or two extinction mechanisms at a time, for reasons of mathematical tractability. But demographic (16, 17), environmental (16, 17), and genetic stochasticity (8, 10, 11) operate simultaneously in natural populations, and their synergy may have a strong impact on the probability of extinction (12, 13, 18–22).
Genetic and Demographic Model
We simulate the dynamics of metapopulation extinction using a biologically realistic model that includes stochastic demographic (17, 23), environmental (17, 23), and genetic (24) mechanisms. Our individual-based model (25) includes population dynamics within each patch, explicit spatial structure, and an explicit representation of each diploid genome (10, 11, 17, 23, 24).
We parameterize the genetic mechanisms in our model using values from a broad array of empirical studies. Independent data on a diversity of organisms suggest that the genomic deleterious mutation rate, U, is frequently on the order of 0.1 to 1 per individual per generation in multicellular eukaryotes, and that the average homozygous and heterozygous effects of such mutations are typically less than 5% (26). In flies, data suggest U ≈ 1, with the average mutation decreasing fitness by about 2% in the heterozygous state (26), although some suggest that U is much smaller, and that the average mutational effect is much larger (27). U may commonly be on the order of 1 in vertebrates and flowering plants (26, 28).
Within-Patch Life Cycle.
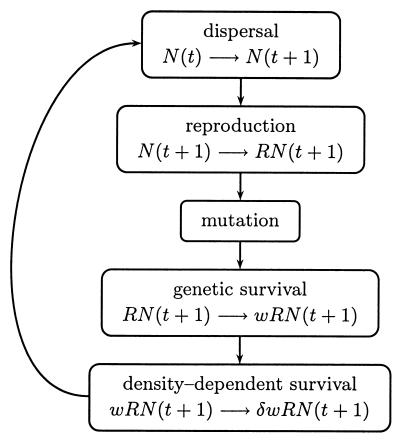
Generations are nonoverlapping, and the within-patch life cycle is seen in Fig. 1. For randomly mating monoecious populations,† each generation RN zygotes are created, where R is individual fecundity, and N is the number of adults. A zygote is formed by randomly drawing, with replacement, two gametes from N parents. For dioecious populations with separate sexes, each generation, polygynous mating between a randomly drawn male (with replacement) and a randomly drawn female (without replacement) produces a Poisson-distributed number of zygotes with expectation 2R. Thus, mating continues until the pool of unmated females (approximately N/2 individuals) is exhausted. We note that the average overall population fecundity is the same for monoecious and dioecious mating. The genome consists of 10,000 loci placed on 8 chromosomes. The number of crossover events per chromosome per generation is Poisson distributed with expectation 1, and the number of new mutations per diploid genome per generation is Poisson distributed with expectation U = 1. Selection culls zygotes, with the probability of genetic survival of each individual determined by the viability function w(i, j) = (1 − hs)i(1 − s)j, where s is the fractional reduction in viability caused by a homozygous mutation, i and j are the numbers of loci in the individual that are heterozygous and homozygous for deleterious mutations, and h is the dominance coefficient. We let h(s) = 1/(2 + 20s), so that mutations of large effect are almost recessive (as s → 1, h → 0.045), whereas those of small effect are almost additive (as s → 0, h → 1/2), consistent with observed data (26, 29, 30). The number of zygotes surviving selection varies stochastically both because the genetic survival of each individual is probabilistic and because the probability, w(i, j), itself is a random variable that differs among individuals because of mutation load. For simplicity, we denote the number of zygotes surviving selection by wRN, but it should be kept in mind that each of the RN zygotes has its own genetic viability, wk, (1 ≤ k ≤ RN).
Figure 1.
The metapopulation life cycle begins with dispersal between patches by Eqs. 2, 3, or 4. Reproduction follows with the creation of RN zygotes that receive U new mutations per zygote. Genetic survival is determined by the absolute viability w, and density-dependent survival is determined by δ (Eq. 1). Although we present a deterministic population-level life cycle for ease of exposition, our probabilistic model actually tracks each individual through the entire life cycle (17, 25).
After selection, the density-dependent survival probability, δ, is determined by the number of zygotes, wRN, that survived selection and the carrying capacity, K, of a local population, δ = min(K/wRN,1) (Eq. 1). We note that our choice for population regulation is conservative, because the deterministic version does not generate fluctuations. By contrast, some commonly used forms for density dependence have the potential for highly variable cyclic and chaotic dynamics (cf. ref. 31), which, because of population bottlenecks, would decrease the genetic effective population size (32, 33).
Metapopulation Structure.
Metapopulation structure is modeled as a linear array of patches connected by either nearest-neighbor (stepping stone), global (island), or intermediate dispersal. For ease of exposition, we present the deterministic equations for dispersal but, because individuals are discrete, their movement between patches is, in fact, probabilistic.‡ For nearest-neighbor dispersal at generation t + 1, in patch i, the total number of individuals is
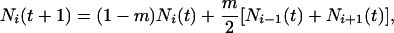 |
2 |
where m is the fraction of a local population that emigrates every generation. Because we assume uniform dispersal over the source patch and its two nearest neighbors, m = 2/3. Dispersal is nonuniform for other values of m; for m < 2/3, individuals tend to remain in the source patch; for m > 2/3, individuals tend to avoid returning to the source patch. Because there is a balance between emigration and immigration, m is also the fraction of a local population that is replaced by immigrants every generation.
For global dispersal,
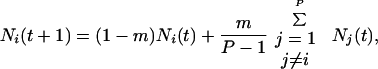 |
3 |
where m = (P − 1)/P, and P is the total number of patches in the metapopulation. The value for m reflects our assumption that dispersal is uniform over all of the patches in the metapopulation.
For intermediate dispersal (i.e., the dispersal neighborhood is larger than nearest-neighbor yet smaller than global),
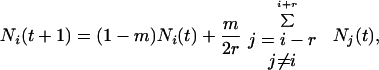 |
4 |
where r is the maximum dispersal distance (measured in patch numbers along the array) from a source patch, and m = 2r/(2r + 1). The value for m reflects uniform dispersal over all 2r + 1 patches in a dispersal neighborhood. We assume reflecting boundary conditions for both nearest-neighbor and intermediate dispersal. We emphasize that for all three dispersal formulations, mixing among the local populations is unrestricted within the dispersal neighborhood.
Results
Extinction Genetic Effective Size—the Benchmark Population.
We begin by examining the extinction behavior of a simple monoecious population with random mating and demographic stochasticity.§ This model provides a benchmark for explaining the extinction time behavior of the more realistic models we examine later. Our approach is inspired by the genetic effective population size concept, where the magnitude of random genetic drift in more realistic models is compared to a theoretically ideal population (32–34).
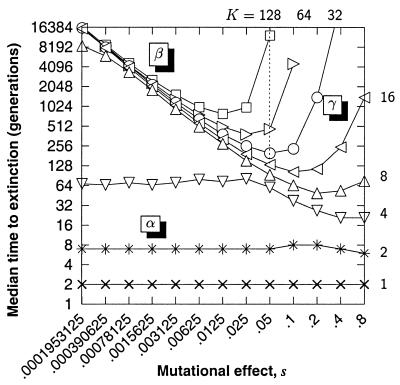
We find that extinction time-scaling¶ behavior falls roughly into three categories that are determined by the population carrying capacity and the magnitude of the mutational effect (see α, β, and γ in Fig. 2). Extinction of small populations that are subject to very mild mutational effects (α-region) is because of demographic stochasticity and is extremely rapid. Within the α-region, time to extinction is independent of the mutational effect, suggesting that the probability of extinction is not changed by accumulating mutations on the short time scale on which extinction occurs. Within the β-region, extinction is because of the accumulation of very mild deleterious mutations and is extremely slow. The time to extinction does depend on the mutational effect, with milder effects taking longer to cause extinction. Population carrying capacity has very little impact on extinction time, indicating that the accumulation dynamics of mutations are almost entirely governed by random genetic drift, with the rate of mutation accumulation being ≃ U for any K. Within the γ-region, extinction is also because of the accumulation of deleterious mutations; there, however, the time scale of extinction is extremely sensitive to the population carrying capacity and the magnitude of the mutational effect. As the carrying capacity drops, even mutations of large effect rapidly accumulate by genetic drift and hasten the extinction of the population. If the mutational effect is very large, and the carrying capacity is not too low, then natural selection has a strong influence on the time to extinction.
Figure 2.
The extinction time behavior of a simple population with monoecious random mating and demographic stochasticity. K is the carrying capacity, the fecundity, R = 5, and the homozygous mutational effect is s. Simulations start without a mutational load—starting with the mutational load of an infinite population at mutation-selection balance would shorten the time to extinction but would not change the overall form of the figure. The α, β, and γ regions are explained in the text.
The larger populations in Fig. 2 clearly demonstrate that the minimum time to extinction, tmin, occurs for some intermediate magnitude of the mutational effect s. Table 1 displays the values of s and K that minimize the time to extinction. The value of the mutational effect that minimizes the median extinction time is s = 1.6/K. Our result agrees very well with Crow's theory that, for a specified population size, there is a mutational effect that minimizes fitness, and that Ks is of order 1 (15). Theoretical work by Lande also predicts such an inverse relationship for s (8). Furthermore, Table 1 also shows there is an approximate doubling of tmin for successive pairs of s and K.
Table 1.
Mutational effect that minimizes the median extinction time at various carrying capacities
| s | K | tmin |
|---|---|---|
| 0.2 | 8 | 49 |
| 0.1 | 16 | 103 |
| 0.05 | 32 | 200 |
| 0.025 | 64 | 382 |
| 0.0125 | 128 | 785 |
Data are from Fig. 2.
Single Population with Polygynous Mating.
Most population genetic models assume a random union of gametes, but this mating system is in reality rather rare (36). In most animal mating systems, individuals (rather than gametes) come together to mate (36). A more realistic mating system for many species is lottery polygyny, where all males attempt to mate many times, but females mate only once or a few times (36). Mating behavior approximating lottery polygyny is found in such diverse organisms as Drosophila (36), wood frogs (36), banner-tailed kangaroo rats (37), and grizzly bears (37).
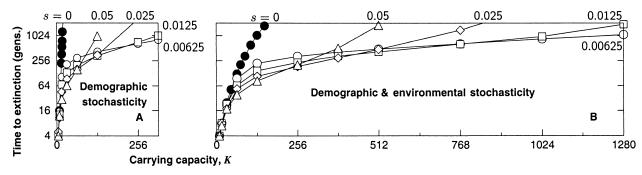
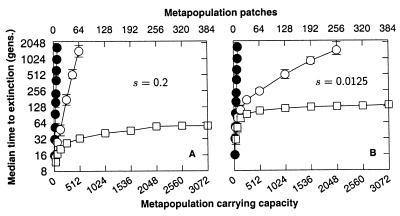
Theory predicts that lottery polygyny should produce an effective population size that is 2/3 of the actual size (36, 38). Under lottery polygyny, we find extinction times equivalent to those of the benchmark population with roughly 2/3 as many individuals, in agreement with theory. For example, with K = 128 and s = 0.05, lottery polygyny gives a median extinction time of 997 generations (Fig. 3A), whereas the benchmark population (Fig. 2), with 2/3 as many individuals (K = 85), produces a median time to extinction of 1,096 generations.
Figure 3.
The median time to extinction for a single population with polygynous mating and (A) demographic stochasticity and (B) demographic and environmental stochasticity [K is lognormally distributed over time with CV(K) = 1]. The fecundity, R = 10. Simulations of populations with mutation accumulation (open symbols) start with the mutational load of an infinite population at mutation-selection balance. Because of the initial mutational load, the initial average genetic viability is 0.37 (= e−U), in accordance with Haldane's theory (2). The homozygous mutational effect is s. Populations without mutation accumulation are denoted (●, s = 0). To equalize the initial growth rates of populations with and without mutation accumulation, we fix the genetic viability (w; see Fig. 1) of individuals in populations without mutation accumulation at 0.37. Because such individuals are genetically inert, individual genetic viability remains at 0.37 for all succeeding generations.
Single Population with Polygynous Mating and Environmental Stochasticity.
Fluctuations in the population carrying capacity can cause a drastic reduction in the genetic effective population size (32, 33). We find that fluctuations in the carrying capacity do indeed cause a large reduction in the extinction genetic effective population size with a consequent decline in the time to extinction (compare Fig. 3 A–B). For example, returning to our reference set of parameters K = 128 and s = 0.05, we find that the fluctuating carrying capacity drops the time to extinction to just 70 generations (Fig. 3B). Comparing this extinction time to our ideal benchmark population (Fig. 2), we find that the effective size of the fluctuating population in this particular example is close to an ideal population of size Ke = 8.
Accelerated Extinction Caused by Mutation Accumulation.
The relative risk of extinction because of demographic and genetic factors has received much attention in conservation biology (cf. ref. 19). With this risk in mind, we investigate the extinction–time scaling for a population without mutation and for an identical population that is also subject to the accumulation of new mild mutations. For the population without mutation, increased carrying capacity causes the time to extinction to grow very rapidly (●, Fig. 3A). Extinction is unlikely for all but the smallest populations. Although environmental stochasticity retards the very rapid growth of extinction time somewhat (●, Fig. 3B), increased carrying capacity still rapidly reduces the risk of extinction.
We find that the accumulation of new mildly deleterious mutations fundamentally alters the scaling of extinction time (open symbols, Fig. 3), causing the extinction of populations that would be deemed safe on the basis of demography alone (●, Fig. 3). Increased carrying capacity still provides additional insurance against extinction by increasing the efficiency of selection against deleterious mutations. In fact, at very low K, the rise in the extinction time for populations with and without mutation accumulation is the same. However, the rise in extinction time with carrying capacity is rather slow over an intermediate range of K. Ultimately, as the carrying capacity increases further, natural selection finally becomes efficient, and the time to extinction rises rapidly (because our focus is on shorter time scales, this phase is not prominent in Fig. 3). Moreover, the range of carrying capacities vulnerable to accelerated mutational extinction depends on the mutational effect, s, becoming larger as the mutational effect becomes smaller.
Metapopulation Structure.
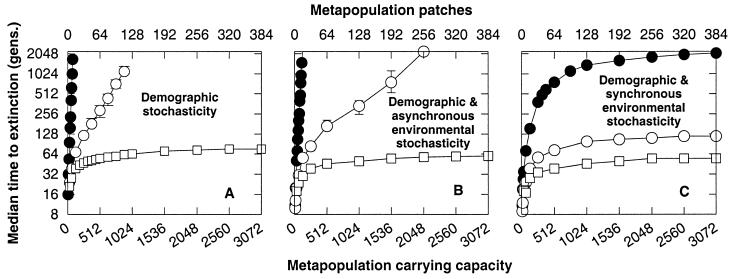
Metapopulation extinction time (Fig. 4A) is highly sensitive to changes in the number of metapopulation patches and their connectivity. Increasing the number of patches or enlarging the dispersal neighborhood increases the median time to extinction. This result parallels what we found for a single population, where increasing the carrying capacity increased the median time to extinction (Fig. 3). In both situations, the median time to extinction increases because natural selection becomes more efficient.
Figure 4.
The time to extinction for a metapopulation with (A) demographic stochasticity, (B) demographic and spatially asynchronous environmental stochasticity, and (C) demographic and spatially synchronous environmental stochasticity. Globally dispersing metapopulations without mutation are denoted (●). Metapopulations with mutation accumulation are denoted (○, global dispersal; □, nearest-neighbor dispersal). The carrying capacity of a patch, K = 8. For B, K is lognormally distributed with CV(K) = 0.2. Spatially asynchronous environmental stochasticity is modeled by independently sampling K for each patch, each generation. For C, K is lognormally distributed over time, with CV(K) = 0.2. Spatially synchronous environmental stochasticity is modeled by sampling a single value of K for all patches, once per generation. Polygynous mating, fecundity R = 10. The homozygous (heterozygous) mutational effect, s = 0.05 (0.017). See Fig. 3 for initial conditions.
The dispersal neighborhood size can have a strong effect on mutation accumulation. Global dispersal markedly enhances the efficiency of natural selection and produces the longest median times to extinction (○, Fig. 4A). Nearest-neighbor dispersal hampers natural selection and produces the shortest median times to extinction (□, Fig. 4A). However, in both cases, for metapopulations with more than a few patches, mutation accumulation accelerates extinction time by many orders of magnitude, compared to a globally dispersing metapopulation without mutation accumulation (●, Fig. 4A). Furthermore, extinction because of mutation accumulation can be quite rapid, on the order of tens of generations.
The extinction time scaling in Fig. 4A implies that the extinction genetic effective population size is highly sensitive to the dispersal range of individuals within the metapopulation. Under global dispersal (○, Fig. 4A), the extinction genetic effective size grows rapidly with the number of metapopulation patches, causing extinction time to increase. Adding patches increases the overall number of individuals in the metapopulation, which directly increases the effective size because global dispersal thoroughly mixes the metapopulation. By contrast, under nearest- neighbor dispersal (□, Fig. 4A) the extinction genetic effective size asymptotes as the number of metapopulation patches increases, causing the extinction time to approach a constant. Comparing the asymptotic extinction time of the nearest-neighbor metapopulation (75 generations; □, Far Right, Fig. 4A) to our ideal benchmark population (s = 0.05, Fig. 2), we find that the effective size is close to an ideal population of size Ke = 6. Remarkably, a nearest-neighbor metapopulation of census size K = 3,072 is able to resist mutation accumulation only like an ideal population of size Ke = 6 in this particular example.
The extinction genetic effective size, under nearest-neighbor dispersal, is not made larger by adding patches to the metapopulation, because mixing between distant patches is poor. Assortative mating increases the likelihood of inbreeding depression and accelerates the rate of mutation accumulation. Moreover, the local mutation accumulation process is much faster than the diffusive process that transports alleles across the metapopulation. To increase the extinction genetic effective size of the metapopulation, the dispersal range must be increased. Our results are consistent with Whitlock and Barton's finding that spatial subdivision usually decreases the genetic effective size of a population (39).
Metapopulation Structure and Environmental Stochasticity.
Environmental stochasticity influences the scaling of extinction time with a potency that depends on synchrony among the metapopulation patches. Asynchronous environmental fluctuations (Fig. 4B) simply stretch the scaling that was found in a constant environment (Fig. 4A). The range of carrying capacities vulnerable to mutation accumulation under global dispersal expands (○, Fig. 4A), but otherwise the overall form of the scaling is quite similar.
Synchronous environmental fluctuations are devastating to metapopulations with and without mutation accumulation (Fig. 4C). In both cases, the effective size of the metapopulation approaches a constant as the number of patches grows. Beyond a certain size, neither type of metapopulation benefits from adding patches, as is indicated by the asymptotic behavior of extinction time.
Under synchronous environmental fluctuations, the acceleration of extinction caused by mutation accumulation is striking (Fig. 4C). For a large metapopulation without mutation accumulation, the extinction time is about 2,000 generations (●), whereas for a large globally dispersing metapopulation with mutation accumulation, the extinction time is just slightly longer than 100 generations (○). Moreover, the nearest-neighbor dispersing metapopulation with mutation accumulation lasts only 55 generations (□).
Habitat Fragmentation and the Collapse of Metapopulation Viability.
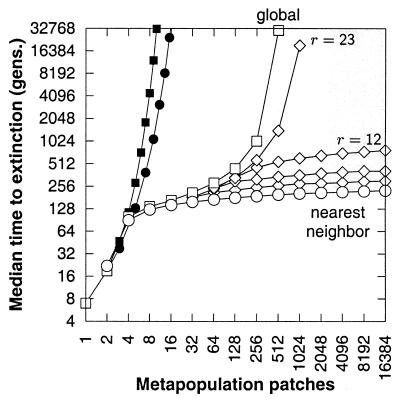
For metapopulations, we find that mutation accumulation and extinction time are highly sensitive to habitat fragmentation. We model habitat fragmentation by altering the connectivity among patches by changing the size of the dispersal neighborhood. Our assumption is that barriers reduce dispersal efficiency in fragmented natural systems. Extinction time falls precipitously when the size of the dispersal neighborhood drops below a critical threshold (compare successive diamond plots, Fig. 5). This threshold indicates that there is a critical level of habitat connectivity that must be maintained for efficient selection against deleterious mutations. Previous work has shown that purely demographic models of a territorial species also exhibit abrupt changes in persistence/extinction behavior, associated with the ratio of unsuitable to suitable habitat (40).
Figure 5.
The collapse of metapopulation viability in fragmented habitat. Habitat fragmentation is modeled by systematically altering the dispersal range of individuals from global (□) to nearest neighbor (○). The time to extinction can collapse suddenly as the dispersal range (see Eq. 3) decreases below a threshold value (compare r = 23 and r = 12). For the diamond plots, the dispersal radii are (Upper to Lower) r = 23, 12, 6, and 3. For comparison, the extinction times for metapopulations without mutation accumulation are also plotted (■, global dispersal; ●, nearest-neighbor dispersal). Each patch corresponds to the territory of a mating pair (40). Monoecious random mating, fecundity R = 5. The homozygous (heterozygous) mutational effect is 0.025 (0.01). Initially, individuals are mutation free, which inflates the extinction time scaling.
Interaction of the Mutational Effect with Habitat Fragmentation.
For a metapopulation in unfragmented habitat, mildly deleterious mutations are more damaging than highly deleterious mutations (compare the extinction time scaling of the globally dispersing metapopulations (○, Fig. 6) for the effects s = 0.2, 0.0125). Because unfragmented habitat provides conditions that are closest to system-wide random mating, the effective size of the metapopulation is relatively large, enhancing selection against highly deleterious mutations. Yet mildly deleterious mutations are nearly neutral, accumulate relatively rapidly, and produce a more rapid extinction time scaling. This situation is similar to what we found in the benchmark single population case for larger carrying capacities (Fig. 2). Just as in the single population case with large carrying capacity, the mild mutational effects are the most damaging, causing the minimum time to extinction.
Figure 6.
Interaction of the mutational effect with habitat fragmentation. The homozygous (heterozygous) mutational effect is (A) s = 0.2 (0.03), (B) s = 0.05 (0.017), and (B) s = 0.0125 (0.0056). See Fig. 4A for details.
For a metapopulation in fragmented habitat, highly deleterious mutations are the most damaging, leading to the most rapid extinction time scaling (compare the extinction time scaling of the nearest-neighbor dispersing metapopulations (□, Fig. 6) for the effects s = 0.2, 0.0125). Because the effective population size of fragmented habitat is quite low, even a mutational effect s = 0.2 is nearly neutral and causes the most rapid erosion of metapopulation viability. This situation is similar to what we found in the single population case for smaller carrying capacities (Fig. 2). Just as in the single population case with small carrying capacity, the larger mutational effects are the most damaging, causing the minimum time to extinction.
Variable Time to Extinction in Homogeneous Habitat.
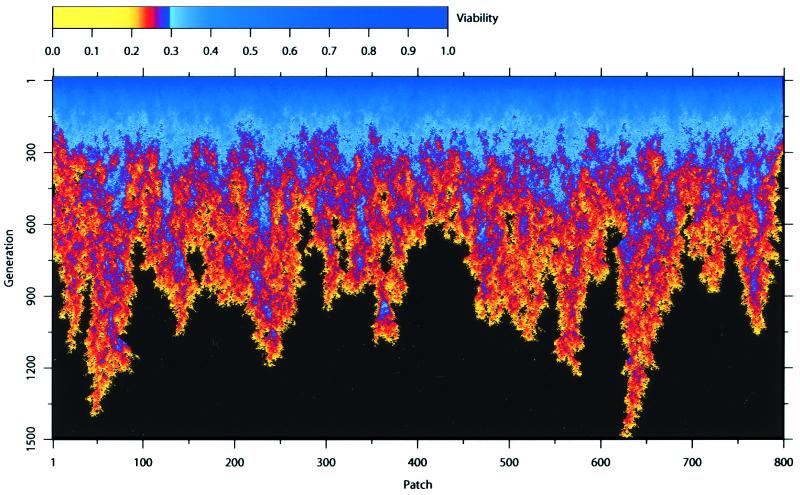
We find that the decline and extinction of a metapopulation because of mutation accumulation can be a strikingly complex spatiotemporal process (Fig. 7). Fluctuations in viability because of stochastic feedback between selection, mutation, segregation, linkage, crossing-over, inbreeding depression, dispersal, density-dependence, and finite population size generate an extremely heterogeneous pattern of extinction. All of the heterogeneity in Fig. 7 is due to endogenous biological mechanisms—the environment is entirely homogeneous.
Figure 7.
The decline and extinction of a metapopulation caused by mutation accumulation. Initially, each of the 800 patches is filled to carrying capacity (K = 10) with mutation-free individuals. In the initial phase (<150 generations), mutations accumulate relatively rapidly because the variance in viability, and consequently the response to selection, is low. This causes the average viability within patches to plummet throughout the metapopulation. Around generation 300, the rate of mutation accumulation becomes highly variable among regions, resulting in the relatively rapid extinction of some regions. However, the inexorable onslaught of new mutations and their accumulation eventually cause the extinction of the entire metapopulation. By contrast, in a single population with the same carrying capacity (8,000), we would expect viability to reach a quasisteady state and the time to extinction to be extremely long, if not indefinite. Nearest-neighbor dispersal with emigration fraction, m = 1/3. Monoecious random mating with fecundity, R = 5. The homozygous (heterozygous) mutational effect is 0.025 (0.01).
Discussion
Early work suggested that demography is usually of more immediate importance to biological conservation than population genetics in determining the minimum viable sizes of wild populations (19). More recently, theory has shown that the accumulation of deleterious mutations may threaten small isolated populations on time scales relevant to conservation (8, 10–13). Here we have shown that accumulation of deleterious mutations may be a significant threat to large metapopulations and would be expected to exacerbate the effect of habitat loss or fragmentation on metapopulation viability. From a genetic perspective, a single large fragmented metapopulation is much more vulnerable to extinction than a panmictic population of the same overall number of individuals. Because the interaction between mutation accumulation and metapopulation demography is synergistic, an assessment of metapopulation viability based only on demographic forces is especially likely to underestimate the risk of extinction.
The decline in the scaling of extinction time because of habitat fragmentation is especially troublesome from a conservation perspective. Because the decline is sudden but extinction itself still takes a while to occur, the metapopulation may be completely inviable on intermediate or long time scales, although appearing healthy on short time scales. In other words, a sharp reduction in time to extinction can occur for a gradual increase in the degree of habitat fragmentation, and the looming fate of the fragmented metapopulation will not be immediately obvious. Fortunately, because extinction is delayed, there might be sufficient time for habitat remediation that would presumably restore efficient selection against deleterious mutations.
Acknowledgments
We thank C. Baer, J. Colbourne, J. Conery, J. F. Crow, R. Lande, M. O'Hely, I. Saccheri, and M. Sherrill for their valuable help with this research. Research funding was provided by a National Science Foundation Postdoctoral Research Fellowship in Biosciences Related to the Environment (DBI-9804267), with additional funding provided by the Oregon Department of Fish and Wildlife, the National Institutes of Health, and the Academy of Finland.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Because individuals are both male and female, mating can occur between any pair of individuals, including self-mating. The amount of self fertilization is the random amount—two uniting gametes are as likely to come from the same individual as from any other two.
In the simulation, we dispersed each of the individuals from a patch by drawing, with equal probability, a random destination patch from within the originating patch's dispersal neighborhood. For a dispersal neighborhood of size n, on average, the proportion of dispersed individuals in each of the n patches is 1/n.
Demographic stochasticity is the fluctuation of population size because of the random survival and birth of discrete individuals. Demographic stochasticity is an inherent feature of our model because it is individual based (17).
¶ We define “extinction time-scaling” as the overall graphical trend displayed by the median time to extinction over some range of a key parameter. Median extinction times were estimated by repeatedly simulating the model dynamics for each specified set of parameters. Ninety-five percent confidence intervals were obtained by the bootstrap (35). In most cases, the confidence intervals are smaller than the symbols used to plot the median.
References
- 1.Kimura M, Maruyama T, Crow J F. Genetics. 1963;48:1303–1312. doi: 10.1093/genetics/48.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haldane J B S. Am Nat. 1937;71:337–349. [Google Scholar]
- 3.Muller H J. Am J Hum Genet. 1950;2:111–176. [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace B. J Hered. 1987;78:134–142. doi: 10.1093/oxfordjournals.jhered.a110345. [DOI] [PubMed] [Google Scholar]
- 5.Lynch M, Gabriel W. Evolution (Lawrence, KS) 1990;44:1725–1737. doi: 10.1111/j.1558-5646.1990.tb05244.x. [DOI] [PubMed] [Google Scholar]
- 6.Hopf F A, Michod R E, Sanderson M J. Theor Popul Biol. 1988;33:243–265. doi: 10.1016/0040-5809(88)90015-9. [DOI] [PubMed] [Google Scholar]
- 7.Gabriel W, Lynch M, Bürger R. Evolution (Lawrence, KS) 1993;47:1744–1757. doi: 10.1111/j.1558-5646.1993.tb01266.x. [DOI] [PubMed] [Google Scholar]
- 8.Lande R. Evolution (Lawrence, KS) 1994;48:1460–1469. doi: 10.1111/j.1558-5646.1994.tb02188.x. [DOI] [PubMed] [Google Scholar]
- 9.Gabriel W, Bürger R. In: Conservation Genetics. Loeschcke V, Tomiuk J, Jain S K, editors. Basel: Birkhäuser; 1994. pp. 69–84. [Google Scholar]
- 10.Lynch M, Conery J, Bürger R. Am Nat. 1995;146:489–518. [Google Scholar]
- 11.Lynch M, Conery J, Bürger R. Evolution (Lawrence, KS) 1995;49:1067–1080. doi: 10.1111/j.1558-5646.1995.tb04434.x. [DOI] [PubMed] [Google Scholar]
- 12.Lande R. Conserv Biol. 1995;9:782–791. [Google Scholar]
- 13.Lande R. Res Popul Ecol. 1998;40:259–269. [Google Scholar]
- 14.Wright S. Evolution and the Genetics of Populations; a Treatise. Vol. 3. Chicago: Univ. of Chicago Press; 1977. p. 485. [Google Scholar]
- 15.Crow J F. In: Oxford Surveys in Evolutionary Biology. Futuyma D, Antonovics J, editors. Vol. 9. Oxford, U.K.: Oxford Univ. Press; 1993. pp. 3–42. [Google Scholar]
- 16.Lande R. Am Nat. 1993;142:911–927. doi: 10.1086/285580. [DOI] [PubMed] [Google Scholar]
- 17.Higgins K. Nat Resour Mod. 1999;12:65–108. [Google Scholar]
- 18.Gilpin M E, Soulé M E. In: Conservation Biology. Soulé M E, editor. Sunderland, MA: Sinauer; 1986. pp. 19–34. [Google Scholar]
- 19.Lande R. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- 20.Boyce M S. Annu Rev Ecol Syst. 1992;23:481–506. [Google Scholar]
- 21.Caughley G. J Anim Ecol. 1994;63:215–244. [Google Scholar]
- 22.Frankham R. Annu Rev Genet. 1995;29:305–327. doi: 10.1146/annurev.ge.29.120195.001513. [DOI] [PubMed] [Google Scholar]
- 23.Higgins K, Hastings A, Sarvela J N, Botsford L W. Science. 1997;276:1431–1435. [Google Scholar]
- 24.Conery J S, Lynch M. Bioinformatics. 1999;15:85–86. doi: 10.1093/bioinformatics/15.1.85. [DOI] [PubMed] [Google Scholar]
- 25.Caswell H, Nisbet R M, de Roos A M, Tuljapurkar S. In: Structured-Population Models in Marine, Terrestrial and Freshwater Systems. Tuljapurkar S, Caswell H, editors. New York: Chapman & Hall; 1997. pp. 3–17. [Google Scholar]
- 26.Lynch M, Blanchard J, Houle D, Kibota T, Schultz S, Vassilieva L, Willis J. Evolution (Lawrence, KS) 1999;53:645–663. doi: 10.1111/j.1558-5646.1999.tb05361.x. [DOI] [PubMed] [Google Scholar]
- 27.Fry J D, Keightley P D, Heinsohn S L, Nuzhdin S V. Proc Natl Acad Sci USA. 1999;96:574–579. doi: 10.1073/pnas.96.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondrashov A S. Genetica (The Hague) 1998;102/103:183–197. [PubMed] [Google Scholar]
- 29.Greenberg R, Crow J F. Genetics. 1960;45:1153–1168. doi: 10.1093/genetics/45.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillespie J H. Population Genetics: A Concise Guide. Baltimore: Johns Hopkins Univ. Press; 1998. [Google Scholar]
- 31.Higgins K, Hastings A, Botsford L W. Am Nat. 1997;149:247–269. [Google Scholar]
- 32.Hartl D L, Clark A G. Principles of Population Genetics. 3rd Ed. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 33.Crow J F, Kimura M. An Introduction to Population Genetics Theory. New York: Harper & Row; 1970. [Google Scholar]
- 34.Wright S. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efron B, Tibshirani R J. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 36.Nunney L. Evolution (Lawrence, KS) 1993;47:1329–1341. doi: 10.1111/j.1558-5646.1993.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 37.Nunney L, Elam D R. Conserv Biol. 1994;8:175–184. [Google Scholar]
- 38.Caballero A. Heredity. 1994;73:657–679. doi: 10.1038/hdy.1994.174. [DOI] [PubMed] [Google Scholar]
- 39.Whitlock M C, Barton N H. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lande R. Am Nat. 1987;130:624–635. [Google Scholar]