Abstract
Seven pharmaceutical heparins were investigated by oligosaccharide mapping by digestion with heparin lyase 1, 2 or 3, followed by high performance liquid chromatography analysis. The structure of one of the prepared mapping standards, ΔUA-Gal-Gal-Xyl-O-CH2CONHCH2COOH, (where ΔUA is 4-deoxy-α-L-threo-hex-4-eno-pyranosyluronic acid, Gal is β-D-galactpyranose, and Xyl is β-D-xylopyranose) released from the linkage region using either heparin lyase 2 or heparin lyase 3 digestion, is reported for the first time. A size-dependent susceptibility of site cleaved by heparin lyase 3 was also observed. Heparin lyase 3 acts on the under sulfated domains of the heparin chain and does not cleave the linkages within heparin’s antithrombin III binding site. Thus, a novel low molecular weight heparin (LMWH) was afforded on heparin lyase 3 digestion of heparin due to this unique substrate specificity, which has anticoagulant activity comparable to that of currently available LMWH.
Introduction
Heparin (Figure 1) is a highly sulfated, linear polysaccharide consisting of repeating uronic acid (1→4)-D-glucosamine disaccharide subunits. Variable substitution of its disaccharide subunits with N-sulfo, O-sulfo and N-acetyl groups give rise to the extremely complex sequences of heparin. The uronic acid is most frequently an α-L-idopyranosyl uronic acid (IdoAa) but can be a β-D-glucopyranosyl uronic acid (GlcA), with or without a 2-O-sulfo group (IdoA2S and GlcA2S). The 2-deoxy-2-amino β-D-glucopyranose (GlcN) residue may be unsubstituted, modified with N-acetyl group (GlcNAc) or most commonly with an N-sulfo group (GlcNS). The GlcN residue is most commonly substituted with a 6-O-sulfo group (GlcNS6S and GlcNAc6S) but only rarely with a 3-O-sulfo group (GlcNS3S and GlcNS3S6S). Of all of these possible disaccharide subunits, IdoA2S-(1→4)-GlcNS6S is the most abundant in heparin. This disaccharide accounts for up to 90% of the total disaccharide units in heparins from bovine lung and up to 70% of the total disaccharide units in heparins from porcine intestine.1
Figure 1.
Schematic representation of heparin and heparin derived oligosaccharides. Heparin is polydisperse with chains of molecular weight 5,000–40,000 Dalton with a degree of polymerization from dp20 to dp160 (n + m = 4–74). Two under sulfated domains (<2 sulfo groups per disaccharide unit), indicated at the reducing end and in the internal portion (when R = H) of the chain, are heparin lyase 3 cleavable. The linkages labeled with A in 4c, C in 6c, D and E in 6d, and F in 6a, can be digested by heparin lyase 3. The linkages labeled B in 6b and G in 8a, each connected at their non-reducing end to a fully sulfated (6 sulfo groups) tetrasaccharide residue, are resistant to heparin lyase 3.
Heparin is found primarily in the granules of connective-tissue type mast cells, where it is biosynthesized as an intracellular proteoglycan, serglycin, to which multiple heparin polysaccharide chains are covalently attached. Following their biosynthesis, the heparin proteoglycan polysaccharide chains and core protein are cleaved by an endoglucuronidase and proteases to give polydisperse mixtures of smaller heparin glycosaminoglycan and peptidoglycan chains that are stored in the cytoplasmic secretory granules of mast cells as non-covalent complexes.1,2
Heparin has a wide range of important biological activities due to its ability to interact with a large number of proteins.3,4 As a clinical anticoagulant, heparin has been one of the most effective and most widely used drugs since its introduction in 1926. Heparin is unique for being one of the first biopolymeric drugs, one of a few polydisperse microheterogenous drugs, and one of the only carbohydrate-based drugs.1,2 Low molecular weight heparins (LMWHs), are a group of heparin-derived anticoagulant / antithrombotic agents that were developed at the end of the 20th century. The introduction of LMWHs primarily resulted from our improved understanding of the molecular basis for coagulation. It was believed that a short heparin chain, containing an antithrombin III (AT) binding site but unable to accommodate thrombin binding, would prevent the formation of a ternary heparin-AT-thrombin, affording a more selective anticoagulant/antithrombotic agent. LMWHs are prepared through controlled chemical and enzymatic depolymerization of heparin. Most of the resulting chains are too small to accommodate thrombin in a ternary complex, and thus, inhibit the coagulation cascade primarily through coagulation factor Xa, which interacts directly with AT bound to a specific pentasaccharide sequence. This AT-binding sequence, -GlcNAc6S-GlcA-GlcNS3S6S-IdoA2S-GlcNS6S- has a central 3-O-sulfo group containing glucosamine residue.5 Despite the factor Xa selectivity of LMWHs (anti-Xa/anti-IIa activity of 4–8), their clinical value comes primarily from their enhanced subcutaneous bioavailability and improved pharmacodynamics.2
The current production of pharmaceutical heparin involves isolation of the raw heparin from porcine intestine and its multi-step purification.6 As a result of a major heparin contamination crisis, which has led to the death of over 100 patients, there has recently been an increased interest in the analysis and structural evaluation of pharmaceutical heparin.6 Improved analytical techniques and methods are required for such purposes.
Heparin lyases isolated from Flavobacterium heparinum also known as Pedobacter heparinus are important members of a class of enzymes called polysaccharide lyases (Enzyme Commission (EC) # 4.2.2) that depolymerize specific acidic polysaccharides.7–9 Previous studies demonstrated that the site in heparin at which heparin lyase 1 acts is →4)-α-D-GlcNS6S-(1→4)-α-L-IdoA2S-(1→.10,11 The site at which heparin lyase 3 acts in heparan sulfate, a glycosaminoglycan structurally related to heparin but with lower sulfation, is →4)-α-D-GlcNAc / GlcNS / GlcNS6S-(1→4)-α-L-IdoA / β-D-GlcA-(1→.10–13 Heparin lyase 2 has a wider range of specificities acting on both heparin and heparan sulfate. It acts at linkages containing either (1→4)-α-L-iduronic acid or (1→4)-β-D-glucuronic acid residues, and accommodates many additional modifications of these polysaccharides as well.10–14 It is generally accepted that tetrasaccharides containing 3-O-sulfo glucosamine reducing end moiety are resistant toward the heparin lyase 1, 2 and 3.15,16 This has posed some problems for completely converting heparin and heparan sulfate to disaccharide products. In addition to the specificity studies, the action patterns of these heparin lyases have been extensively studied.17 But how these action patterns are impacted by substrate size and sequence microheterogeneity is not well understood,18 limiting the development of these enzymes as powerful analytical tools for heparin structure determination.19,20
The saccharide composition of heparin is usually established by disaccharide analysis and nuclear magnetic resonance (NMR) spectra.21–22 A number of chemical and enzymatic methods are available for the cleavage of heparin at specific linkages. Heparin lyase cleavage of heparin affords more restricted cleavage than chemical methods due to enzymatic specificity.10,11 The presence of rare 3-O-sulfo glucosamine residues and the lyase sensitivity of adjacent sites pose additional difficulty in structural analysis. While the linkage to the non-reducing side of a 3-O-sulfo glucosamine residue is resistant to all of the heparin lyases,15,16 the linkage to the reducing side of this residue is extremely sensitive to heparin lyases 1 and 2.18 As a result of this specificity, exhaustive depolymerization of heparin chains with heparin lyase 1 and 2 generates oligosaccharides that contain 3-O-sulfo glucosamine residues at their reducing end but lack intact AT pentasaccharide binding sites.18 The different specificities of heparin lyase 1, 2 and 3 suggest their application as reagents for oligosaccharide mapping.
The goal of oligosaccharide mapping is to separate and analyze oligosaccharide products to assess the distribution of susceptible linkages within the intact heparin chain. Heparin oligosaccharide mapping heparin lyase 1 has been previously used to provide a molecular fingerprint of the polymeric structure and reveals detailed structural information for its sequence determination, and provide oligosaccharide products for further structural analysis.23–25 In the current study, seven pharmaceutical heparins have been examined by oligosaccharide mapping with heparin lyase 1, 2 and 3. While it is generally accepted that heparin lyase 3 does not cut heparin we have discovered that it does indeed act on heparin, offering a new means for the preparation of a novel LMWH.
Materials and Methods
Materials
Seven pharmaceutical sodium porcine intestinal heparins originating from Europe, U.S. and China were obtained for the study with anticoagulant activities ranging from 195 to 209 IU/mg. Sodium heparin used for standard preparation was obtained from porcine intestinal mucosa (176 units/mg, Celsus Laboratories, Cincinnati, OH). Low molecular weight heparin was from Pfizer (New York, NY). Cloning, expression and purification of the recombinant heparin lyase 1 (Enzyme Commission (EC) # 4.2.2.7), heparin lyase 2 (no EC # assigned) and heparin lyase 3 (EC # 4.2.2.8) from F. heparinum were performed essentially as described in previous studies.22–24 Electrophoresis-grade acrylamide, NN', -methylene-bis-acrylamide, sucrose, glycine, ammonium persulfate (APS), NN,N',N'-tetramethylenediamine (TEMED), bromophenol blue and polyacrylamide gels (Bio-Gel P2 and P10) were from Bio-Rad (Hercules, CA, USA). Sodium chloride, boric acid, disodium salt of ethylenediaminetetraacetic acid (disodium EDTA), phenol red, azure A, and alcian blue were from Fisher (Pittsburgh, PA, USA). Deuterium oxide (99.99%) was from Sigma Aldrich (Saint Louis, Missouri, USA).
Heparin mapping
Each individual heparin sample (4.0 mg) was dissolved in 4.0 mL of 50 mM, pH 7.5, sodium phosphate buffer and was incubated in a 30ºC water bath with heparin lyase 1 (1.5 IU, activity against heparin), heparin lyase 2 (350 mIU, activity against heparin) or heparin lyase 3 (5.0 IU activity against heparan sulfate). The reaction completion was monitored by taking out small amount of aliquots of the reaction mixture for polyacrylamide gel electrophoresis (PAGE) analysis (see below). The same amount of each heparin lyase was added to the reaction at each 12 h interval for another two times for an exhaustive digestion on the substrates. Reactions were finally quenched by heating in a 100ºC water bath for 10 min.
Heparin lyase digested heparin samples were analyzed by PAGE using a mini-gel apparatus (Bio-Rad, Hercules, CA). Each sample (5 μL) was mixed with 5 μL of 50 % (w/v) sucrose and loaded into a stacking gel of 5% (w/v total acrylamide) and fractionated on a 15% or 22% resolving gel. Electrophoresis was performed at 200 V for 30 min for 15% gel or 80 min for 22% gel. Gels were fixed and stained with 0.5% (w/v) alcian blue in 2% (v/v) acetic acid and de-stained with methanol water 50% (v/v). Undigested heparin samples were used as a negative control on the 15% PAGE gel.
SAX-HPLC analysis of heparin lyase digestion products were performed on a Shimadzu LC-10Ai LC system equipped with an SPD-20A ultraviolet-visible (UV) detector using a 4.6 × 250 mm Waters Spherisorb S5 SAX column. A two-segment gradient elution was achieved using mobile phase A (water, pH 3.5, adjusted with HCl) and mobile phase B (2.0 M NaCl, pH 3.5, adjusted with HCl) at a flow rate of 1.0 mL/min. The seven heparin lyase-digested heparin samples were loaded onto the column in a concentration of 10 μg/10 μL and washed with 0% to 60% B over 60 min. The elution was monitored at 232 nm of wavelength. Analyses were all performed in triplicate.
Preparation and Characterization of Oligosaccharide Standards
Heparin (400 mg in 50 mM, pH 7.4, sodium phosphate buffer) was exhaustively digested by heparin lyase 1 (20 IU, activity against heparin) in the same conditions described above. Buffer salts and disaccharide components within the product mixture were removed by chromatography on a 100 × 5.0 cm Bio-Gel P10 column eluted at 1.2 mL/min with 0.2 M NaCl in distilled water. The remaining oligosaccharide mixture was desalted on a 100 × 2.0 cm P2 column and lyophilized. The resulting mixture was fractionated on a 20 × 250 mm semi-preparative strong anion exchange (SAX)-high performance liquid chromatography (HPLC) column (Waters Spherisorb S5) eluted with a salt gradient (see below) over 60 min at a flow rate of 4.0 mL/min with absorbance detected at 232 nm. Individual peaks were desalted on a Bio-Gel P2 column and were further purified by repeated separation on the same SAX-HPLC column. Oligosaccharide standards produced by heparin lyase 2 digestion were prepared in an identical manner. The purity of each of the oligosaccharides prepared from HPLC was determined to be >95% pure by analytical SAX-HPLC and PAGE analysis. The structures of these standards were characterized by HPLC-ESI-MS, and one-dimensional (1D) and two-dimensional (2D) NMR experiments.18 High resolution mass spectra were obtained for all new compounds using ESI-Fourier transform-mass spectrometry (FT-MS) on an LTQ XL Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA).29
Specificity of Heparin Lyase 3
Each heparin substrate (20 μg in 100 μL of pH 7.4, 50 mM sodium phosphate buffer) was incubated with an excessive amount of heparin lyase 3. The reaction was incubated in a 30ºC water bath for 12 h. The reaction was quenched by heating at 100ºC and the product was lyophilized before further analysis. The oligosaccharide products were then redissolved and analyzed on a 22% PAGE gel at 200 V for 70 min, and were subsequently subjected to analytical SAX-HPLC (see above) using the oligosaccharide standards for composition assignment.
Action Patterns of Heparin Lyase 1 and Heparin Lyase 2
The action patterns of heparin lyases 1 and 2 were examined using substrates of heparin-derived oligosaccharides with a size range from degree of polymerization (dp) 8 to dp16 that have been prepared by heparin lyase 1 partial digestion of porcine intestinal mucosa heparin.18 In these experiments, 50 μg of each oligosaccharide substrate was dissolved in 200 μL of pH 7.4, 50 mM sodium phosphate buffer, and 1.0 μL of heparin lyase 1 or heparin lyase 2 (0.01 mU/μL) was added to each reaction. The reactions were incubated in a 30ºC water bath and 40 μL aliquots were removed at time points of 10 min, 20 min, 30 min, 1 h and 12 h each. Aliquots were immediately heated in a 100ºC water bath to stop the reaction, and were then dried by lyophilization and redissolved in 20 μL of de-ionized distilled water. A 2 μL portion of each sample was analyzed by electrophoresis on a 22% PAGE gel. The remaining 18 μL of each sample was subjected to analytical SAX-HPLC. Undigested oligosaccharides were used as 0 min control.
LMWH Generated by Heparin lyase 3 Digestion
Heparin sodium (20 mg from Celsus Laboratories) was incubated with 10 IU (activity against heparan sulfate) heparin lyase 3 for 12 h at 25ºC. A second 15 IU heparin lyase 3 was then added for another 12 h incubation to exhaustively digest the substrates. The reaction was terminated in 100ºC water bath for 10 min and the precipitant was removed by centrifugation at 10,000 × g for 30 min. The product was reconstituted in 16% (w/w) NaCl and precipitated by 80% (w/w) methanol.30 Residual sodium chloride was removed using a 3000 molecular weight cut off (MWCO) membrane centrifuge concentrator. The sample was then dried by lyophilization.
The amount of LMWH in the lyophilized sample was determined by micro carbazole assay.31 The LMWH was analyzed by electrophoresis on a 15% PAGE gel at 200 V for 30 min. The gel was fixed and stained with 0.5% (w/v) alcian blue in 2% (v/v) acetic acid and de-stained with distilled water. The gel was scanned, digitized, and analyzed by the software UN-scan-it gel (Silk Scientific, Orem, UT). The molecular weight of the resulting LMWH was calculated based on the heparin oligosaccharide standards and was compared to that of the intact heparin and a commercial LMWH from Pfizer.32,33 1D NMR experiments were performed at 800 MHz (Bruker Avance 800) for digestion completion evaluation. Negative control was made by adding thermally inactivated heparin lyase 3 to the same amount of heparin substrate. A coagulation analyzer (ACL 8000, Beckman Coulter, Fullerton, CA) was used to determine anti-factor Xa, anti-factor IIa (thrombin), and activated partial thromboplastin time (aPTT) activity following previously published protocols.34
Results and Discussion
Heparin Mapping
Heparin oligosaccharides mixtures were prepared by exhaustive digestion using heparin lyases 1, 2 and 3, respectively. PAGE and SAX-HPLC analyses of these oligosaccharide mixtures shows that heparin lyase 2 digestion resulted in the most complete digestion (95%) of the glycosidic linkages to uronic acid cleaved (Table S1). In contrast, heparin lyase 3 resulted in the least complete (5%) digestion (Table S1). Most of the oligosaccharides contained in the heparin lyase 3 digestion product mixture were in a size larger than degree of polymerization (dp16) (Figure S1), suggesting that only a small percentage of heparin linkages were cleaved. Heparin lyase 1 afforded an intermediate level of heparin digestion (75%) leaving some (13% in molar percentage) undigested tetrasaccharides and a small quantity of undigested hexasaccharides within the products (Table S1).
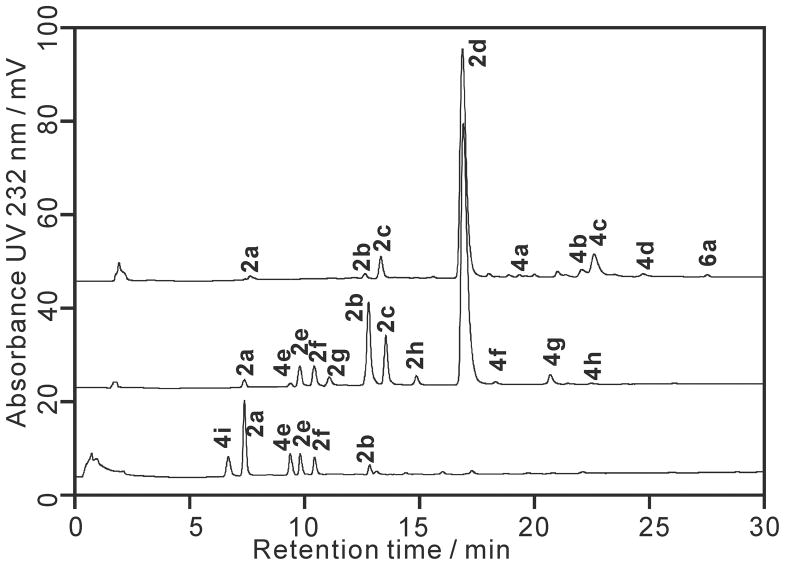
Disaccharides have a relatively small size and are only slightly charged, causing an inadequate fixation by the staining solution alcian blue and hence difficultly in observing them on the PAGE gel (Figure S1).35 Therefore, the oligosaccharide mixtures prepared using individual lyase were further analyzed by SAX-HPLC and the major oligosaccharide peaks in the chromatogram were assigned using heparin oligosaccharide standards (Figure 2). Reference standards used to characterize the heparin lyase 1 digestion products were prepared directly from a large-scale heparin lyase 1 digestion of heparin (from Celsus). The most prominent peak in the chromatogram of heparin lyase 1 digested heparin was the trisulfated disaccharide 2d (ΔUA2S-GlcNS6S). Tetrasaccharide 4d (ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S), which is composed of two units of 2d and is known to be cleavable to heparin lyase 1, was also present in the “completely” digested product mixture. Compared with previous studies,23 the percentage of 4d was greatly reduced due to the large excess of heparin lyase 1 used in the current study. Smaller substrates such as tetrasaccharides bearing heparin lyase 1 sensitive linkages are reportedly more difficult to digest.36 Evaluation of the digestion completeness was made by monitoring the UV absorbance at 232 nm versus time. This is apparently too imprecise a measure to detect the small change in UV absorbance occurring when minor resistant oligosaccharides are digested. 4c (ΔUA2S-GlcNS6S-GlcA-GlcNS6S) also showed resistance to heparin lyase 1 digestion and was the major tetrasaccharide product. 6a (ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S), a hexasaccharide with 3-O-sulfo glucosamine residue at its reducing end, was the major hexasaccharide product.
Figure 2.
Chromatograms of heparin lyases produced oligosaccharides for heparin mapping. SAX-HPLC chromatograms are: a. heparin lyase 1 treated heparin; b. heparin lyase 2-treated heparin; and c. heparin lyase 3 treated heparin. The structures determined for each peak are: 2a ΔUA-GlcNAc, 2b ΔUA-GlcNS6S, 2c ΔUA2S-GlcNS, 2d ΔUA2S-GlcNS6S, 2e ΔUA-GlcNS, 2f ΔUA-GlcNAc6S, 2g ΔUA2S-GlcNAc, 2h ΔUA2S-GlcNAc6S, 4a ΔUA2S-GlcNS-IdoA2S-GlcNS, 4b ΔUA2S-GlcNS6S-IdoA2S-GlcNS, 4c ΔUA2S-GlcNS6S-GlcA-GlcNS6S, 4d ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S, 4e ΔUA-Gal-Gal-Xyl-O-CH2CONHCH2COOH, 4f ΔUA-GlcNAc6S-GlcA-GlcNS3S, 4g ΔUA-GlcNAc6S-GlcA-GlcNS3S6S, 4h ΔUA-GlcNS6S-GlcA-GlcNS3S6S, 4i ΔUA-Gal-Gal-Xyl-O-Ser, 6a ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S.
Heparin lyase 2 digestion afforded a greater number of disaccharide products than did heparin lyase 1, cleaving both tetrasaccharides 4c and 4d. Interestingly, the three tetrasaccharide products, 4f (ΔUA-GlcNAc6S-GlcA-GlcNS3S), 4g (ΔUA-GlcNAc6S-GlcA-GlcNS3S6S) and 4h (ΔUA-GlcNS6S-GlcA-GlcNS3S6S), observed in the heparin lyase 2 product mixture, each had a 3-O-sulfo group containing glucosamine residue at their reducing end. These 3-O-sulfo group containing tetrasaccharides are absent in heparin lyase 1 digestion chromatogram, demonstrating that heparin lyase 1 cannot act on higher oligosaccharides, such as 6a to convert it into 4g and 2d.18 These observations are consistent with our previous understanding of the differences in the specificity of heparin lyase 1 and 2.10,11
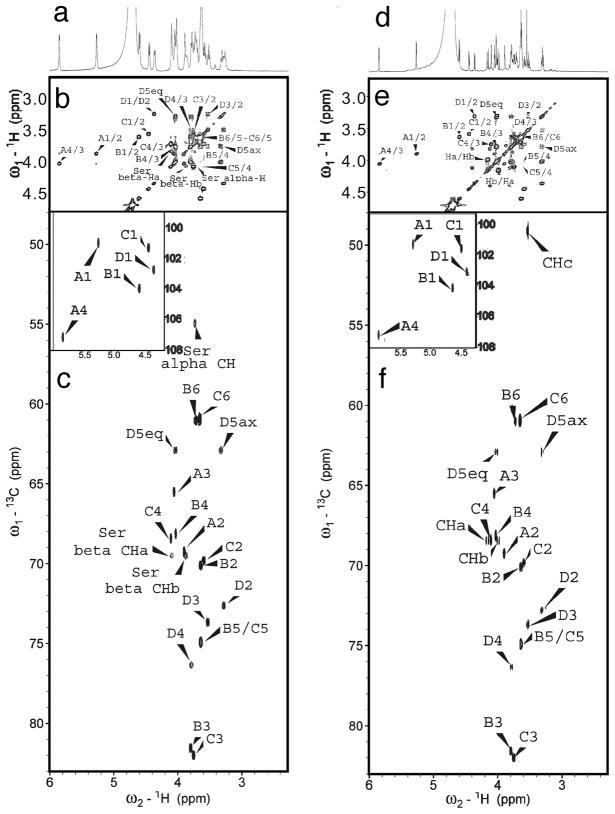
Heparin lyase 3 digested heparin released only under sulfated disaccharides containing 0, 1 or 2 sulfo groups with no 2-O-sulfo group in the unsaturated uronic acid residues and tetrasaccharides containing residues derived from the under sulfated core protein linkage region (Figure 2). One of the heparin lyase 3 tetrasaccharide products, 4i (ΔUA-Gal-Gal-Xyl-O-Ser) has a structure associated with the core protein linkage region and has been previously reported by our group and other groups.37–39 1D and 2D NMR spectra (Figure 3, Table S2) indicated that 4e (ΔUA-Gal-Gal-Xyl-O-CH2CONHCH2COOH) had a structure similar to 4i but is distinct from any previously reported structure. The anomeric protons A1 (5.264 ppm), B1 (4.603 ppm), C1 (4.453 ppm) and D1 (4.353 ppm) in 4e had the same chemical shifts as 4i. Differences in the HMQC spectra originated from two different amino residues attached to these tetrasaccharides. 4i had the expected serine amino acid residue attached to the reducing end of the core tetrasaccharide. The alpha proton and the beta protons in the serine residue could be easily assigned in the HMQC spectrum. However, the same cross-peaks were absent in the HMQC spectrum of 4e. Instead, the resonances of Ha (4.179 ppm), Hb (3.984 ppm) and Hc (3.533 ppm) in the HMQC spectrum for 4e indicated an O-glycolylglycine residue. Assignment of the structures of 4e and 4i are shown in Figure S4. The structures of these two compounds were further confirmed by high-resolution mass spectrometry with the molecular ion peaks [M-H]− for 4e and 4i detected at 746.19 and 718.20, respectively (Figure S4).
Figure 3.
1D and 2D NMR spectra of the linkage core tetrasaccharides. Spectra for 4i are: a. 1D 1H-NMR; b. 1H, 1H-COSY; and c. 1H, 13C-HMQC (insert shows the anomeric signals). Spectra for 4e are: d. 1D 1H-NMR; e. 1H, 1H-COSY; and f. 1H, 13C-HMQC (insert shows the anomeric signals). The structures and labeled positions can be found in Figure S4.
The seven commercial heparin samples were digested by the three heparin lyases individually and were analyzed in triplicate by SAX-HPLC (Figure S2). The assigned peaks were integrated with the relative peak areas shown in Table S1. The average micromolar concentrations of the oligosaccharide compositions are presented in Figure 4. Each heparin lyase afforded a unique chromatogram containing different disaccharides and oligosaccharides, providing more structural information for each individual heparin sample than would a simple disaccharide analysis. The relative quantities of certain oligosaccharides varied significantly between the different commercial heparin samples (Figure 4 and Table S1) suggesting such analyses might be useful for quality evaluation within the heparin manufacturing process. The manufacturing process may also modify the structure of the reducing end of intact heparin resulting in a reduction in the formation of 4i and an increase in the formation of the new structure of 4e. The relative amount of 4e to 4i is on average, 1 to 1.3 (Table S1).
Figure 4.
Heparin oligosaccharide mapping results based on the quantification of SAX-HPLC data. Heparin (MW 12 000 Da) at 83 μM was treated with: a. heparin lyase 1; b. heparin lyase 2; and c. heparin lyase 3. The micromolar concentrations of each product are shown with standard deviations. The structures of these oligosaccharides can be found in Figure 2.
Specificity of Heparin Lyase 3
The specificity of heparin lyase 3 has been previously studied in our group using heparin derived tetrasaccharides and hexasaccharides as primary substrates.10 Heparin lyase 3 cleaves the glucosamine (1→4) iduronic acid / glucuronic acid linkage, where the uronic acid residue contains no 2-O-sulfo group. Furthermore, heparin lyase 3 acts on either heparan sulfate polysaccharide11,17 or heparan sulfate precursor-derived oligosaccharides in a random endolytic action pattern.40 The current study examines the heparin lyase 3 sensitivity of some previously prepared heparin-derived oligosaccharides.18 Oligosaccharide substrates 4c (ΔUA2S-GlcNS6S-GlcA-GlcNS6S) and 6b (ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-GlcA-GlcNS6S) having the same internal GlcNS6S (1→4) GlcA linkage gave different results when treated with heparin lyase 3 under identical reaction conditions. Tetrasaccharide 4c was cleaved, while hexasaccharide 6b was not a substrate. Similarly, hexasaccharide 6a (ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S) was converted by heparin lyase 3 into a tetrasaccharide and a disaccharide product, whereas octasaccharide 8a (ΔUA2S-GlcNS6S-IdoA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS3S6S) bearing the same potentially cleavable linkage failed to serve as a substrate (Figure 1, Figure S3). Hexasaccharides 6c (ΔUA2S-GlcNS6S-IdoA2S-GlcNAc-GlcA-GlcNS6S) and 6d (ΔUA2S-GlcNS6S-IdoA-GlcNAc6S-GlcA-GlcNS6S) were both digested by heparin lyase 3 (Figure S3), this result is consistent with our previous report.10 These experiments suggest that heparin lyase 3 can cut an oligosaccharide at a linkage between glucosamine (1→4) unsulfated iduronic acid / glucuronic acid only when that linkage is two saccharide units away from the non-reducing end. It is likely that extra fully sulfated saccharide residues flanking the cleavage site from the non-reducing end may prevent the action of heparin lyase 3.
Action Patterns of Heparin Lyase 1 and Heparin Lyase 2
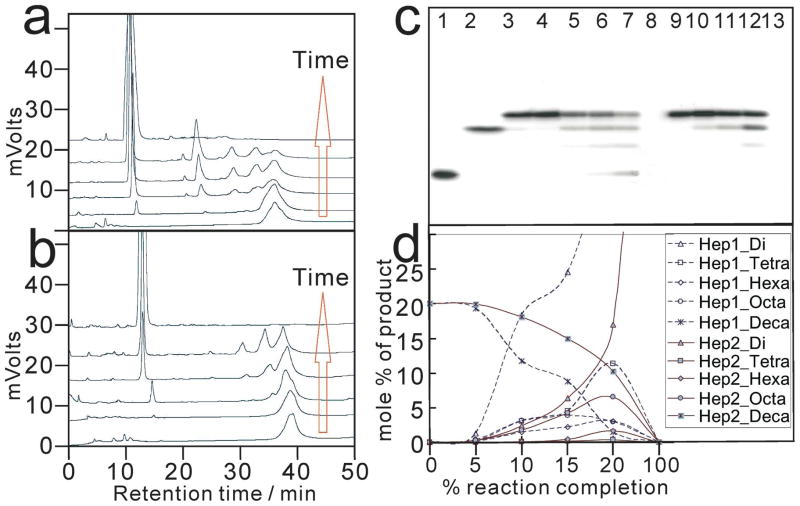
At a time-point corresponding to 20% completion of heparin lyase 1 or 2 respective digestion on a decasaccharide 10a (ΔUA2S-[-GlcNS6S-IdoA2S-]4-GlcNS6S), exolytic products were 3-fold more prominent than endolytic products (Figure 5). Thus, for oligosaccharide substrates, heparin lyase 1 and 2 showed an exolytic bias in their action patterns. This observation is in contrast to the endolytic action patterns of both lyases on polysaccharide substrates.17,40 Heparin lyase 1 has also been reported to act predominantly by an exolytic, processive mechanism, depolymerizing its substrate by cleaving linkages starting from the non-reducing end of an octasaccharide.42 Under similar protocol, heparin lyase 2 showed an endolytic, non-random action pattern.36 However, the current study shows that both heparin lyase 1 and heparin lyase 2 have a similar exolytic preference on small oligosaccharide substrates although their substrate specificities are different. These observations suggest a reason for the controversy surrounding the action pattern of these important enzymes, which needs to be further clarified by additional studies.
Figure 5.
Heparin lyase 1 and heparin lyase 2 action pattern studies using decasaccharide 10a as a model substrate. 10a ΔUA2S-[-GlcNS6S-IdoA2S-]4-GlcNS6S. SAX-HPLC chromatograms of a digestion time course are shown for a. heparin lyase 1 and b. for heparin lyase 2. The red arrows indicate increasing digestion time. PAGE (22% total acrylamide) analyses of 10a treated by heparin lyase 1 and heparin lyase 2 are shown in c. where lane 1 is 4d; lane 2 is 8b ΔUA2S-[-GlcNS6S-IdoA2S-]3-GlcNS6S; lane 3 through lane 8 are 10a incubated with heparin lyase 1 from time-point 0 to time-point 5; lane 9 through lane 13 are 10a treated by heparin lyase 2 from time-point 0 to time-point 5; lane 8 and lane 13 are 10a exhaustively digested by heparin lyase 1 and heparin lyase 2, respectively. The mole percent of each size product (indicated with different symbols) is plotted as a function of percent reaction completion in d. for heparin lyase 1 (dotted lines) and heparin lyase 2 (solid lines).
LMWH Produced by Heparin Lyase 3
A low molecular weight heparin was prepared by the complete heparin lyase 3 digestion of heparin. This new LMWH closely resembled the LMWHs previously reported in our laboratory.43 Unlike heparin lyase 1 and 2, heparin lyase 3 is incapable of cleaving linkages within heparin’s AT-III binding site.15 This property of heparin lyase 3 should make it an useful tool for the development of novel anticoagulant agents. This new LMWH had a number-averaged molecular weight (MN ) of 5300 Da, a weight-averaged molecular weight (MW ) of 7700 Da, and a polydispersity (PD) of 1.45, as determined by PAGE analysis on a 15% gel (Table S3). The reaction completion of heparin lyase 3 on heparin was estimated by 1D 1H-NMR spectroscopy, which clearly showed the H-4 signals in unsaturated uronic acid residues (Figure S5) corresponding to disaccharide and other oligosaccharides produced. The digestion completion was less than 5% ([cleaved linkages/linkages to uronic acid] × 100%) based on the integration of the H-4 signals in unsaturated uronic acid residues and all anomeric proton signals observed. The aPTT activity of this novel LMWH was comparable to a commercially available LMWH but showed a lower anti-factor Xa/anti-factor IIa ratio (Table S3).
Under sulfated domains in heparin chains have previously been reported.44–46 In the above specificity study of heparin lyase 3, we found that the linkage of glucosamine (1→4) iduronic acid / glucuronic acid, where the uronic acid residue has no 2-O-sulfo group, could not be acted on by the enzyme when the adjacent tetrasaccharide residues were fully sulfated (6b and 8a in Figure 1). Nevertheless, the result of PAGE analysis showed that heparin was partially digested by heparin lyase 3 (Figure S5). This suggests that the pharmaceutical heparin chains have internal under sulfated domains that can be cleaved by heparin lyase 3 (Figure 1). In this case, one intact heparin chain (MW = 12000 Da, Table S3) could be cut into two shorter chains (MW = 7700 Da). Based on the weight average molecular weight of the resulting LMWH product, one intact heparin chain approximately has only one internal under sulfated region that is susceptible to heparin lyase 3.
Conclusions
Pharmaceutical heparins from a variety of commercial sources were examined using heparin lyases digestion followed by HPLC analysis. The heparin mapping chart based on the quantification by HPLC is another new type of fingerprint of pharmaceutical heparins in addition to the regular NMR spectra and disaccharide analysis chromatogram. The mapping methodology may also serve as a way of evaluating and monitoring heparin manufacturing processes. The specificity of heparin lyase 3 and the action patterns of heparin lyase 1 and heparin lyase 2 have also been determined in current study. Heparin lyase 3 has been shown unexpectedly to cleave heparin at linkages of glucosamine (1→4) iduronic acid / glucuronic acid with no 2-O-sulfo group on the uronic acid. We believe that the use of large amounts of recombinant heparin lyase 3 and sensitive analytical methods explain why we were the first to report heparin sensitivity to heparin lyase 3. The susceptibility of linkages to heparin lyase 3 is also affected by the structure and length of the adjacent saccharide residues. By using heparin lyase 3 we observe that most of the pharmaceutical heparin chains have only one internal under sulfated domain and one under sulfated domain adjacent to the core protein linkage region. Heparin lyase 3 is incapable of cleaving the AT binding site present in an intact heparin chain. Cleavage of heparin’s under sulfated domains affords a novel LMWH with similar average molecular weight as other current available LMWHs.
Supplementary Material
Acknowledgments
The authors acknowledge support from the US National Institutes of Health Grants HL101721, HL096972 and GM38060. Bo Yang and Tatiana N. Laremore for the assistance with mass spectrometry, Fuming Zhang for collecting the pharmaceutical heparins, and Dr. Madje and Dr. Mousa at Pharmaceutical Research Institute, Albany College of Pharmacy, for measurement of anticoagulant activities. Z. Xiao is a recipient of a Chinese scholarship from the State Scholarship Fund to pursue his study in the US as a joint Ph.D. student.
Footnotes
Abbreviations: Ac, acetyl; APS, ammonium persulfate; aPTT, activated partial thromboplastin time; AT, antithrombin III; ΔUA, 4-deoxy-α-L-threo-hex-4-eno-pyranosyluronic acid; dp, degree of polymerization; EDTA, ethylenediamine tetracetic acid; ESI, eletrospray ionization; FT, Fourier transform; Gal, galactopyranose; GlcA, β-D-glucopyranosyl uronic acid; GlcN, 2-deoxy-2-amino-β-D-glucopyranose; HMQC, heteronuclear multiple-quantum coherence; HPLC, high performance liquid chromatography; IdoA, α-L-iduropyranosyl uronic acid; LMWH, Low molecular weight heparin; MS, mass spectrometry; NMR, nuclear magnetic resonance; 1D, one dimensional; PAGE, polyacrylamide gel electrophoresis; S, sulfo; SAX, strong anion exchange; Ser, serine; TEMED, N,N,N’,N’-tetramethylenediamine; 2D, two dimensional; UV, ultra violet; Xyl, xylopyranose.
Supporting Information Available: Quantification of oligosaccharide compositions (Table S1), NMR data for oligosaccharide standards (Table S2), molecular weights (Table S3), PAGE analysis for digested heparins (Figure S1), chromatogram of digested heparins (Figure S2), analysis of heparin lyase 3 treated oligosaccharides (Figure S3), structures and mass spectra of 4e and 4i (Figure S4), PAGE and NMR analysis of LMWH (Figure S5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rabenstein DL. Heparin and heparan sulfate: structure and function. Nat Prod Rep. 2002;19:312–331. doi: 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- 2.Linhardt RJ. Perspective: 2003 Claude S. Hudson Award Address in Carbohydrate Chemistry. Heparin: Structure and Activity. J Med Chem. 2003;46:2551–2564. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- 3.Toida T, Linhardt RJ. Structure and bioactivity of sulfated polysaccharides. Trends Glycosci Glycobiol. 2003;15:29–46. [Google Scholar]
- 4.Petitou M, Casu B, Lindahl U. 1976–1983, a critical period in the history of heparin: the discovery of the antithrombin binding site. Biochimie. 2003;85:83–89. doi: 10.1016/s0300-9084(03)00078-6. [DOI] [PubMed] [Google Scholar]
- 5.Capila I, Linhardt RJ. Heparin-protein interactions. Angew Chemie Int Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang VC, Linhardt RJ, Bernstein H, Cooney CL, Langer R. Purification and characterization of heparinase from Flavobacterium heparinum. J Biol Chem. 1985;260:1849–1857. [PubMed] [Google Scholar]
- 8.Lohse DL, Linhardt RJ. Purification and characterization of heparin lyases from Flavobacterium heparinum. J Biol Chem. 1992;267:24347–24355. [PubMed] [Google Scholar]
- 9.Linhardt RJ, Galliher PM, Cooney CL. Polysaccharide lyases. Appl Biochem Biotech. 1986;12:135–177. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- 10.Desai UR, Wang H, Linhardt RJ. Substrate specificity of the heparin lyases from Flavobacterium heparinum. Arch Biochem Biophys. 1993;306:461–468. doi: 10.1006/abbi.1993.1538. [DOI] [PubMed] [Google Scholar]
- 11.Desai UR, Wang HM, Linhardt RJ. Specificity studies of heparin lyases from Flavobacterium heparinum. Biochemistry. 1993;32:8140–8145. doi: 10.1021/bi00083a012. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich CP, Silva ME, Michelacci YM. Sequential Degradation of Heparin in Flavobacterium heparinum: purification and properties of five enzymes involved in heparin degradation. J Biol Chem. 1973;248:6408–6415. [PubMed] [Google Scholar]
- 13.Nader HB, Porcionatto MA, Tersariol IL, Pinhal MA, Oliveira FW, Moraes CT, Dietrich CP. Purification and substrate specificity of heparitinase I and heparitinase II from Flavobacterium heparinum. Analyses of the heparin and heparan sulfate degradation products by 13C NMR spectroscopy. J Biol Chem. 1990;265:16807–16813. [PubMed] [Google Scholar]
- 14.Shaya D, Zhao W, Garron ML, Xiao Z, Cui Q, Zhang F, Sulea J, Linhardt RJ, Cygler M. Catalytic mechanism of heparinase II revealed by site-directed multagenesis and the crystal structure with its substrate. J Biol Chem. 2010;285:20051–20061. doi: 10.1074/jbc.M110.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shriver Z, Sundaram M, Venkataraman G, Fareed J, Linhardt RJ, Biemann K, Sasisekharan R. Cleavage of the antithrombin III binding site in heparin by heparinases and its implication in the generation of low molecular weight heparin. Proc Natl Acad Sci USA. 2000;97:10365–10370. doi: 10.1073/pnas.97.19.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu G, LeBrun L, Gunay NS, Hoppensteadt D, Walenga J, Fareed J, Linhardt RJ. Heparinase 1 acts on a synthetic heparin pentasaccharide corresponding to the antithrombin III binding site. Thrombos Res. 2000;100:549–556. doi: 10.1016/s0049-3848(00)00368-6. [DOI] [PubMed] [Google Scholar]
- 17.Jandik KA, Gu K, Linhardt RJ. Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Z, Zhao W, Yang B, Zhang Z, Guan H, Linhardt RJ. Heparinase 1 selectivity for the 3,6-di-O-sulfo-2-deoxy-2-sulfamido-α-D-glucopyranose (1,4) 2-O-sulfo-α-L-idopyranosyluronic acid (GlcNS3S6SIdoA2S) linkages. Glycobiology. 2010 doi: 10.1093/glycob/cwq123. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbull JE, Hopwood JJ, Gallagher JT. A strategy for rapid sequencing of heparan sulfate and heparin saccharides. Proc Natl Acad Sci USA. 1999;96:2698–2703. doi: 10.1073/pnas.96.6.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shriver Z, Raman R, Venkataraman G, Drummond K, Turnbull J, Toida T, Linhardt RJ, Biemann K, Sasisekharan R. Sequencing of 3-O-sulfated containing heparin decasaccharides with a partial antithrombin III binding site. Proc Natl Acad Sci USA. 2000;97:10359–10364. doi: 10.1073/pnas.97.19.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korir AK, Limtiaco FK, Gutierrez SM, Larive CK. Ultra performance ion-pair liquid chromatography coupled to electrospray Time-of-flight mass spectrometry for compositional profiling and quantification of heparin and heparan sulfate. Anal Chem. 2008;80:1297–1306. doi: 10.1021/ac702235u. [DOI] [PubMed] [Google Scholar]
- 22.Guerrini M, Naggi A, Guglieri S, Santarsiero R, Torri G. Complex glycosaminoglycans: profiling substitution patterns by two-dimensional nuclear magnetic resonance spectroscopy. Anal Biochem. 2005;337:35–47. doi: 10.1016/j.ab.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Linhardt RJ, Rice KG, Kim YS, Lohse DL, Wang HM, Loganathan D. Mapping and quantification of the major oligosaccharide components of heparin. Biochem J. 1988;254:781–787. doi: 10.1042/bj2540781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbull JE. Oligosaccharide mapping and sequence analysis of glycosaminoglycans. In: Graham JM, Higgins JA, editors. Methods in Molecular Biology Biomembrane Protocols I Isolation and Analysis. Vol. 19. Humana Press Inc; Totowa, NJ: 1993. pp. 253–267. [DOI] [PubMed] [Google Scholar]
- 25.Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: Recent progress and future challenges. Omics. 2010;14:389–399. doi: 10.1089/omi.2009.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasisekharan R, Bulmer M, Moremen KW, Cooney CL, Langer R. Cloning and expression of heparinase I gene from Flavobacterium heparinum. Proc Natl Acad Sci USA. 1993;90:3660–3664. doi: 10.1073/pnas.90.8.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaya D, Tocilj A, Li Y, Myette J, Venkataraman G, Sasisekharan R, Cygler M. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J Biol Chem. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- 28.Godavarti R, Davis M, Venkataraman G, Cooney C, Langer R, Sasisekharan R. Heparinase III from Flavobacterium heparinum: cloning and recombinant expression in Escherichia coli. Biochem Biophys Res Commun. 1996;225:751–758. doi: 10.1006/bbrc.1996.1246. [DOI] [PubMed] [Google Scholar]
- 29.Laremore TN, Ly MK, Zagorevski DV, Linhardt RJ. High-resolution preparative separation of glycosaminoglycan oligosaccharides by polyacrylamide gel electrophoresis. Anal Biochem. 2010;401:236–241. doi: 10.1016/j.ab.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, Zhang Z, Thistle R, McKeen L, Hosoyama S, Toida T, Linhardt RJ, Page-McCaw P. Structural characterization of glycosaminoglycans from zebrafish in different ages. Glycoconjugate J. 2009;26:211–218. doi: 10.1007/s10719-008-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesaretti M, Luppi E, Maccari F, Volpi N. A 96-well assay for uronic acid carbazole reaction. Carbohydr Polym. 2003;54:59–61. [Google Scholar]
- 32.Edens RE, Al-Hakim A, Weiler JM, Rethwisch DG, Fareed J, Linhardt RJ. Gradient polyacrylamide gel electrophoresis for determination of molecular weights of heparin preparations and low-molecular-weight heparin derivatives. J Pharm Sci. 1992;81:823–827. doi: 10.1002/jps.2600810821. [DOI] [PubMed] [Google Scholar]
- 33.Ly M, Wang Z, Laremore TN, Zhang F, Zhong W, Pu D, Zagorevski DV, Dordick JS, Linhardt RJ. Analysis of E. coli K5 capsular polysaccharide heparosan. Anal Bioanal Chem. 2010 doi: 10.1007/s00216-010-3679-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mousa SA, Zhang F, Aljada A, Chaturvedi S, Takieddin M, Zhang H, Chi L, Castelli MC, Friedman K, Goldberg MM, Linhardt RJ. Pharmacokinetics and pharmacodynamics of oral heparin solid dosage form in healthy human subjects. J Clin Pharmacol. 2007;47:1508–1520. doi: 10.1177/0091270007307242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice KG, Rottink MK, Linhardt RJ. Fractionation of heparin-derived oligosaccharides by gradient polyacrylamide-gel electrophoresis. Biochem J. 1987;244:515–522. doi: 10.1042/bj2440515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhomberg AJ, Shriver Z, Biemann K, Sasisekharan R. Mass spectrometric evidence for the enzymatic mechanism of the depolymerization of heparin-like glycosaminoglycans by heparinase II. Proc Natl Acad Sci USA. 1998;95:12232–12237. doi: 10.1073/pnas.95.21.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Desai UR, Han X, Toida T, Linhardt RJ. Strategy for the sequence analysis of heparin. Glycobiology. 1995;5:765–774. doi: 10.1093/glycob/5.8.765. [DOI] [PubMed] [Google Scholar]
- 38.Shibata S, Midura RJ, Hascall VC. Structural analysis of the linkage region oligosaccharides and unsaturated disaccharides from chondroitin sulfate using CarboPac PA1. J Biol Chem. 1992;267:6548–6555. [PubMed] [Google Scholar]
- 39.Sugahara K, Tsuda H, Yoshida K, Yamada S, Beer T, Vliegenthart JFG. Structure determination of the octa- and decasaccharide sequences isolated from the carbohydrate-protein linkage region of porcine intestinal heparin. J Biol Chem. 1995;270:22914–22923. doi: 10.1074/jbc.270.39.22914. [DOI] [PubMed] [Google Scholar]
- 40.Kuberan B, Babu P. Fluorescent-tagged heparan sulfate precursor oligosaccharides to probe the enzymatic action of heparintinase I. Anal Biochem. 2010;396:124–132. doi: 10.1016/j.ab.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linhardt RJ, Fitzgerald GL, Cooney CL, Langer R. Mode of action of heparin lyase on heparin. Biochim Biophys Acta. 1982;702:197–203. doi: 10.1016/0167-4838(82)90503-9. [DOI] [PubMed] [Google Scholar]
- 42.Ernst S, Rhomberg AJ, Biemann K, Sasisekharan R. Direct evidence for a predominantly exolytic processive mechanism for depolymerization of heparin-like glycosaminoglycans by heparinase I. Proc Natl Acad Sci USA. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linhardt RJ, Loganathan D, Hakim AA, Wang H, Walenga JM, Hoppensteadt D, Fareed J. Oligosaccharide mapping of low molecular weight heparins: structure and activity differences. J Med Chem. 1990;33:1639–1645. doi: 10.1021/jm00168a017. [DOI] [PubMed] [Google Scholar]
- 44.Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 45.Yamada S, Yamane Y, Tsuda H, Yoshida K, Sugahara K. A major common trisulfated hexasaccharide core sequence, hexuronic acid (2-sulfate)-glucosamine (N-sulfate)-iduronic acid -N-acetylglucosamine-glucuronic acid-glucosamine (N-sulfate), isolated from the low sulfated irregular region of porcine intestinal heparin. J Biol Chem. 1998;273:1863–1871. doi: 10.1074/jbc.273.4.1863. [DOI] [PubMed] [Google Scholar]
- 46.Yamada S, Sakamoto K, Tsuda H, Yoshida K, Sugiura M, Sugahara K. Structural studies of octasaccharides derived from the low-sulfated repeating disaccharide region and octasaccharide serines derived from the protein linkage region of porcine intestinal heparin. Biochemistry. 1999;38:838–847. doi: 10.1021/bi981889n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.