Abstract
RNase T2 enzymes are conserved in most eukaryotic genomes, and expression patterns and phylogenetic analyses suggest that they may carry out an important housekeeping role. However, the nature of this role has been elusive. Here we show that RNS2, an intracellular RNase T2 from Arabidopsis thaliana, is essential for normal ribosomal RNA recycling. This enzyme is the main endoribonuclease activity in plant cells and localizes to the endoplasmic reticulum (ER), ER-derived structures, and vacuoles. Mutants lacking RNS2 activity accumulate RNA intracellularly, and rRNA in these mutants has a longer half-life. Normal rRNA turnover seems essential to maintain cell homeostasis because rns2 mutants display constitutive autophagy. We propose that RNS2 is part of a process that degrades rRNA to recycle its components. This process appears to be conserved in all eukaryotes.
Keywords: ribophagy
Ribonucleases (RNases) belonging to the RNase T2 family are acidic endonucleases without base specificity that are either extracellular or targeted to the secretory pathway (1). This family is conserved in the genome of almost all eukaryotic organisms so far analyzed, suggesting that it performs an important function that has been maintained throughout evolution (2, 3). Phylogenetic analyses have identified three subclasses present in plant genomes (3, 4). Class I proteins are highly diversified and show evidence of gene sorting, and their expression is commonly tissue-specific and/or regulated by biotic and abiotic stress (3). Class III includes mostly members of the S-RNases, enzymes involved in the process of gametophytic self-incompatibility in three plant families (5). Finally, class II includes proteins that are highly conserved in all plant genomes and are normally expressed at high levels in most plant tissues (3). On the basis of their conservation and gene expression characteristics, we proposed that class II RNases may have a housekeeping role in plant cells (3). Similar gene expression and phylogenetic studies in animals led us to the hypothesis that metazoan RNase T2 enzymes also have a housekeeping role and may be equivalent to class II enzymes from plants (2).
RNS2, one of five RNase T2 genes in the Arabidopsis genome, encodes the only class II protein in this model organism (3, 4). This RNase is present in all tissues at high levels (6, 7) and is localized in an intracellular compartment. RNS2 expression is increased even further during senescence and during inorganic phosphate (Pi) starvation (6, 8). Thus, it was hypothesized that RNS2 is part of a phosphate scavenging system that rescues plants that are under nutritional stress (9). However, because RNS2 and other class II RNase T2 proteins accumulate to high levels even under optimal growth conditions, this rescue function is unlikely to be the main role of these enzymes. Moreover, in vertebrates, in which RNase T2 enzymes are absolutely conserved (2), the mechanisms that control the response to phosphate starvation seem to be specific to the intestine and kidney (10), whereas RNase T2 genes are expressed constitutively in all tissues,(2, 3, 11). Therefore, the biological function that has led to the conservation of these enzymes in all eukaryotic organisms is still unknown.
Here we show that RNS2 is localized to the endoplasmic reticulum (ER) or ER-derived compartments and to the vacuole in Arabidopsis cells. Although a large fraction of the protein is present in vacuoles, the enzyme has a neutral pH optimum, suggesting that it may also function in the ER or another neutral pH compartment. We found that plants lacking RNS2 activity accumulate RNA intracellularly, most likely in the vacuole. Ribosomal RNA is degraded at a slower rate in mutant than in wild-type (WT) plants; thus, RNS2 is necessary for normal rRNA decay. In turn, deficient rRNA decay results in constitutive autophagy in mutant plants. Our results indicate that rRNA turnover is carried out by RNS2 in vacuoles or ER-derived compartments and that this process is necessary for normal cell homeostasis. A similar finding for an RNase T2 enzyme from zebrafish (12) suggests that this mechanism for rRNA recycling is conserved in all eukaryotes.
Results
RNS2 Localizes to ER and Vacuoles.
Previous work had shown that RNS2 is an intracellular protein, and the presence of a C-terminal extension suggested either a vacuolar or ER localization (7). To determine more definitively the localization of RNS2, we fused a cyan fluorescent protein (CFP) to the RNS2 polypeptide. RNS2 has an N-terminal secretion signal that targets the protein to the secretory pathway, in addition to the putative C-terminal extension, which could be an ER retention or vacuolar targeting signal. To avoid disrupting any potential localization signals, the CFP peptide was fused in frame after the N-terminal secretion signal (Fig. S1A). This construct was expressed in transgenic plants under the control of a strong constitutive promoter. The fusion protein is expressed well in plants and maintains RNase activity (Fig. S1B). Expression of CFP-RNS2 is also accompanied by an increase in RNS2 activity, suggesting that the CFP tag is removed from some of the proteins. Several new RNase bands appeared in the profile of plants expressing CFP-RNS2. These bands could be the result of different levels of glycosylation, a posttranslational modification common in RNase T2 enzymes. To test this hypothesis, plant extracts were treated with N-glycosydase F and then analyzed in the in gel assay (Fig. S2). We found that RNS2 is glycosylated and that the multiple bands in the CFP-RNS2 size range correspond to different glycosylation states of the protein.
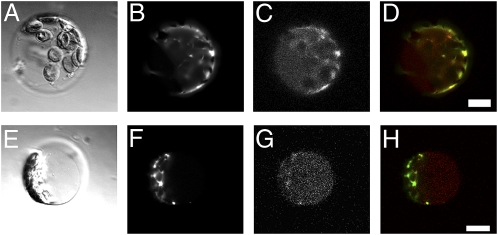
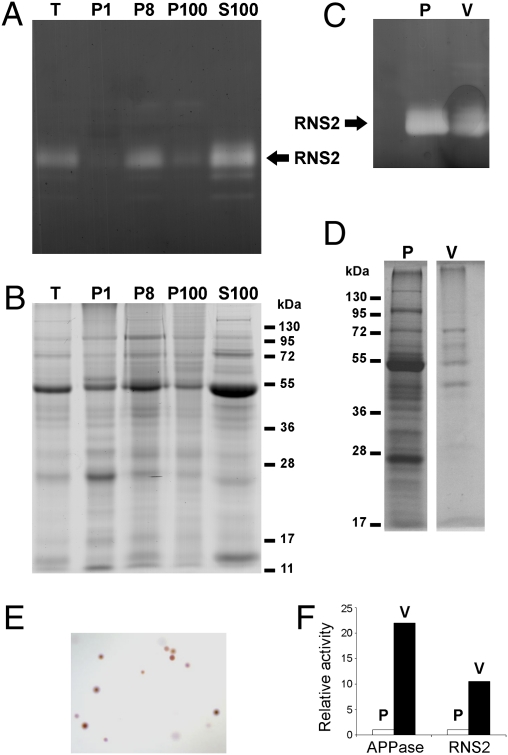
Confocal microscopy showed that the CFP-RNS2 fusion protein localizes to ER structures in leaf protoplasts, as determined by its colocalization with the ER marker YFP-HDEL, which contains a C-terminal HDEL tetrapeptide that promotes retrieval to the ER (13) (Fig. 1 and Fig. S3). Diffuse CFP-RNS2 signal is also observed in the protoplast vacuole. Confocal imaging of roots also revealed CFP-RNS2 fluorescence in ER bodies and vacuoles (Fig. S4). To confirm the microscopy results, we separated ER body-rich fractions (P1 and P8) and an ER network-rich fraction (P100) by subcellular fractionation (14) and analyzed the fractions using RNase activity in gel assays (Fig. 2A). We confirmed that RNS2 is present both in a fraction corresponding to small ER bodies (P8) and in the ER network (P100). However, a large fraction of RNS2 activity was found in the S100 that contains soluble cytoplasmic and vacuolar proteins. To confirm vacuolar localization, vacuoles were purified from leaf protoplasts (Fig. 2E) and then assayed for RNS2 activity (Fig. 2C). RNS2 activity was enriched in the vacuolar fraction, although not as much as the vacuolar marker acid phosphatase (Fig. 2F). Our results also confirm previous results by Carter et al. (15), who identified RNS2 as a vacuolar protein in a survey of vacuolar proteins through a proteomics approach.
Fig. 1.
Subcellular localization of CFP-RNS2. Plants expressing the CFP-RNS2 construct (#2 from Fig. S1) were transformed with YFP-HDEL, an ER marker. Protoplasts were prepared from adult leaves and imaged using confocal microscopy. A–D and E–H correspond to individual protoplasts. Protoplasts were analyzed in bright field (A and E), YFP conditions (B and F), and CFP conditions (C and G). (D and H) Merged CFP (red) and YFP (green) signals. Signal overlap is seen as yellow. (Scale bar, 10 μm.)
Fig. 2.
Subcellular localization of RNS2. (A) Protein extracts from wild-type plants were fractionated into P1 (enriched in large ER bodies), P8 (enriched in small ER bodies), P100 (enriched in microsomes), and S100 (enriched in soluble citoplasmic and vacuolar proteins) fractions and analyzed using an RNase activity in gel assay. “T” corresponds to the crude extract before fractionation. Each lane contains 10 μg of protein. The position of RNS2 is indicated. (B) SDS/PAGE stained with Coomassie Blue R-250 of samples as in A. (C) Vacuoles were purified from WT protoplasts. Total protoplast (P) proteins (30 μg) and purified vacuole (V) proteins (∼2–3 μg) were analyzed as in A. (D) SDS/PAGE stained with Coomassie Blue R-250 of samples as in C. (E) Purified vacuoles were stained with Neutral red and visualized under a light microscope to confirm purity and integrity. (F) Enzyme activities of RNS2 and the vacuole-specific enzyme marker acid phosphatase (APPase) were determined in protoplasts and vacuoles purified from the same protoplasts. The activities were normalized with respect to protoplasts.
RNS2 Is Not an Acidic RNase.
The presence of RNS2 in ER, ER-derived bodies, and vacuoles could mean that the enzyme functions in more than one cellular location. Alternatively, RNS2 could function only in the vacuole, and the high level of CFP signal in the ER network could be the result of overexpression of the fusion protein. Also, ER bodies are considered an alternate pathway for protein transport to the vacuole (16). The environment in the vacuole is different from that found in the ER. Although the vacuolar pH is acidic, the ER interior is maintained at a neutral pH (17, 18). Thus, determination of the pH preference of RNS2 activity may indicate whether its biological function is carried out in the ER or in the vacuole.
RNS2 was expressed in yeast cells using a system that facilitates collection of the protein and limits its potential toxic effect (19). Two class I RNase T2 genes from Arabidopsis, RNS1 and RNS3, were also expressed for comparison (Fig. 3A). The activities of the three RNases were tested over a range of pHs (Fig. 3B). We found that RNS1 and RNS3 have an acidic pH preference, as is common for most RNase T2 enzymes (1). In contrast, RNS2 has a pH preference near 7.5 and the activity at acidic pH is only a fraction of that at neutral pH. This finding suggests that RNS2 is more suited to carry out a biological function in the ER or ER-derived compartments than in the vacuole, although the high level of activity found in the vacuole suggests that RNS2 may function in both compartments. To confirm these results using native RNS2, plant extracts were analyzed using the RNase activity in gel assay at different pHs. Again, we found that RNS2 activity is higher at neutral pH (Fig. 3C), although the pH range of native RNS2 seems broader than that observed using yeast-expressed RNS2. On the other hand, RNS1 activity is detected only at acidic pH.
Fig. 3.
Characterization of the pH preference of RNS2. (A) Recombinant Arabidopsis RNases RNS1 (lane 1), RNS2 (lane 2), and RNS3 (lane 3) were obtained from a yeast expression system and analyzed by an RNase activity in gel assay. Each lane contains 1 μg of protein. (B) RNase activity of yeast-expressed RNS1 (●), RNS2 (■ and ◆), and RNS3 (▲) was assayed in vitro at different pHs as indicated. The activity of each enzyme relative to the maximum activity (at optimal pH) is reported. RNS2 activity (solid lines) near the pH optimum was assayed using two different buffers, Tris (◆) or phosphate (■), to discard any buffer artifact. (C) pH preference of native RNS2. Leaf protein extracts (10 μg) from WT and rns2-2 mutant plants were analyzed by an RNase activity in gel assay, with incubations at different pHs as indicated (below the panels). The position of RNS2 is indicated. At pH 6, it is possible to detect activity of RNS1 as an RNase band below the RNS2 activity.
Mutants Lacking RNS2 Activity Have Longer-Lived rRNA, and RNA Accumulates in Vacuoles.
A proposed function for RNS2 and other class II RNase T2 enzymes in plants is the scavenging of phosphate from bulk RNA during starvation. We hypothesized that the constitutive presence of this enzyme at high levels could indicate that RNS2 is involved in RNA recycling as part of normal cell homeostasis. Because the largest fraction of RNA in a cell is rRNA, we suspected that rRNA is a substrate for RNS2 activity.
We obtained an Arabidopsis line carrying a T-DNA insertion in the fifth intron of the RNS2 gene, truncating the encoded RNS2 protein before the second conserved active site motif (Fig. 4A and Fig. S5). Protein extracts from homozygous T-DNA insertion individuals were analyzed using RNase activity in gel assays (Fig. 4B). These mutants lack the main RNase activity found in Arabidopsis extracts. This RNase activity is present at high levels in all tissues of WT plants, and it is induced by Pi starvation and senescence, as described for the RNS2 transcript (7, 20). Thus, we identified this band as RNS2 and confirmed that the T-DNA insertion produced a null mutation. We named this mutant rns2-2.
Fig. 4.
Isolation of rns2-2, an Arabidopsis mutant that lacks RNS2 activity. (A) Schematic showing the organization of the RNS2 gene and the T-DNA insertion present in the rns2-2 mutant. Boxes represent exons, and lines represent introns (between exons). Start (arrow) and end (*) of the RNS2 ORF are indicated. (B) RNase activity in gel profile of protein extracts obtained from leaves of WT and rns2-2 mutant plants. A strong RNase activity in WT and absent in the rns2-2 mutant was identified as RNS2. Lanes contain 40 μg of protein. (C) SDS/PAGE stained with Coomassie Blue R-250 of samples as in B.
The rns2-2 plants did not show any obvious morphological phenotype, nor did they have any reproductive deficiency. We used an RNA-specific dye, SYTO-RNASelect, to test for changes in RNA accumulation in these plants. Comparison of WT and rns2-2 mutant roots showed a clear increase in fluorescence in mutant cells (Fig. 5 A–D). Quantification of the fluorescence signal from WT and mutant roots showed a roughly 10-fold increase in staining in mutant roots (Fig. 5E). This differential RNA accumulation seems to occur mainly in vacuoles, although in some cells the fluorescence is more evident around the contour of the vacuole, probably in the cytoplasm. A similar comparison was carried out using leaf protoplasts. About 10% of mutant protoplasts showed a large fluorescent body in the vacuole (Fig. 5 F–G); this body was never found in WT protoplasts.
Fig. 5.
Differential staining of mutant and WT cells using the RNA-specific dye SYTO RNASelect. Seven-day-old seedlings were transferred to liquid MS medium, stained with 5 μM SYTO RNASelect green, and observed using confocal microscopy. Green fluorescence (Left panels in A and B) and bright-field images (Right panels in A and B) were collected for WT (A) and rns2-2 (B) roots. A magnified image of the Inset boxed regions is shown in C and D. In all cases, the root tip lies on the right side of the image. (Scale bars, 75 μm.) Metamorph software was used to measure the mean fluorescence of six lateral lines across the elongation zone in the WT and rns2-2 roots stained with SYTO RNASelect (E). Leaf protoplasts were also stained with 5 μM SYTO RNASelect green and observed using confocal microscopy. Fluorescence images were collected for WT (F) and rns2-2 (G) protoplasts. Chlorophyll fluorescence (Left) and green fluorescence (Center) are shown. Merged images (Right) show chlorophyll in red and SYTO RNASelect in green. (Scale bar, 10 μm.)
We hypothesized that the substrate for RNS2 is rRNA. Lack of RNS2 would result in a longer half-life of rRNA. Because rRNA is long-lived, it is not possible to analyze its decay with the use of transcriptional inhibitors. Thus, we designed an in vivo labeling experiment to follow rRNA decay in mutant and WT plants. For this, 1-wk-old plants grown in liquid medium were incubated with [3H]-uridine for 30 min and then transferred to cold medium. Samples were taken 24, 48, and 96 h later, and the ratio of radioactive/total 28S rRNA was calculated. We found that 28S rRNA in the rns2-2 mutant plants decays at a significantly slower rate than in WT plants (Fig. 6A). We calculated the 28S rRNA half-life following standard methods described for mRNA (21) and found a significant difference (P < 0.05) between the 28S rRNA half-life in WT plants (38.0 ± 4.2 h) and in rns2-2 mutants (61.8 ± 15.2 h). The same decay analysis was carried out using a previously described (7) Arabidopsis line, which expresses an antisense RNS2 construct (AS-RNS2) that causes a decrease in RNS2 expression. As previously reported, these plants do show some RNS2 activity (7). Although the AS-RNS2 line does not fully phenocopy the rns2-2 mutation, both the antisense and rns2-2 lines show significant differences in 28S rRNA decay compared with that observed in WT plants (Fig. 6B). Similar analysis performed for 18S rRNA showed that the small ribosomal subunit is also more stable in rns2-2 mutants, with half-lives of 28.9 ± 1.7 h and 46.4 ± 4.4 h for WT and mutant, respectively (Fig. S6).
Fig. 6.
Decay of rRNA in wild-type plants and in lines with altered levels of RNS2 expression. One-week-old seedlings grown in liquid medium were incubated with [3H]-uridine for 30 min and then transferred to cold medium. At the indicated time points samples were extracted and total RNA was isolated. To determine the fraction of [3H]-RNA versus total RNA for the 28S rRNA, the samples were analyzed by agarose gel electrophoresis and transferred to nitrocellulose. Total 28S rRNA was determined by EtBr staining, then bands were excised and the tritium signal was quantified with a scintillation counter. (A) Comparison of the decay of 28S rRNA in WT and the rns2-2 mutant. (B) 28S rRNA decay in WT and in a RNS2 antisense line (AS-RNS2). Asterisks indicate statistically significant differences with respect to WT (P < 0.05).
Lack of RNS2 Activity Causes Constitutive Autophagy.
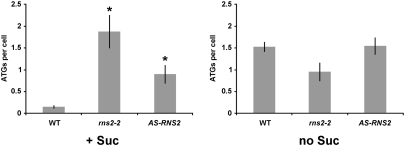
In yeast, ribosomes are selectively targeted for degradation by an autophagy-like process (termed ribophagy) in response to nutritional stress (22). RNS2 could be involved in a similar process in plant cells. Thus, we analyzed the autophagy process in WT, rns2-2, and AS-RNS2 plants. Autophagy is not commonly detected in WT cells of plants growing under normal nutrient conditions. However, when plants are subjected to nutritional stress, such as sucrose starvation, autophagosomes can be easily detected (Fig. 7 and Fig. S7). In contrast, rns2-2 and AS-RNS2 plants showed constitutive autophagy even under normal growth conditions, but the level of autophagy in these lines was not different from WT under extended sucrose starvation (Fig. 7 and Fig. S7). The number of autophagosomes per cell under normal growth conditions was almost 10-fold higher in rns2-2 than in WT plants.
Fig. 7.
Loss of RNS2 activity leads to constitutive autophagy. One-week-old WT, rns2-2, and AS-RNS2 seedlings were transferred to fresh solid MS medium with or without sucrose in the dark for 4 d, followed by staining with MDC. The number of autophagosomes per cell was quantified. Asterisks indicate statistically significant differences (P < 0.05) with respect to WT.
Discussion
We found that RNS2 is essential for normal degradation of ribosomes, as mutants lacking RNS2 activity have rRNA with an extended half-life, which accumulates intracellularly, probably in the vacuole. We found that rns2 mutants show constitutive autophagy, indicating that recycling of ribosomes is essential for maintaining cellular homeostasis. We propose that RNS2 is part of a housekeeping mechanism that recycles ribosomes throughout the life of a cell. This mechanism could be similar to ribophagy, the recycling of ribosomes observed under nutrient deficiency in yeast (22).
The mechanisms of ribosome degradation in eukaryotes are only now being investigated. Several reports have examined the fate of aberrant ribosomes with cis-acting mutations, in which rRNA sequences are mutated, or with trans-acting mutations, in which mutant proteins are absent from or fail to interact with the rRNA (23). These studies focus on the surveillance mechanisms involved in nonfunctional rRNA decay (NRD) and indicate that small and large ribosomal subunits are processed by independent decay pathways. Small ribosomal subunit NRD in yeast shares some components with the no-go mRNA decay pathway that targets mRNA in stalled ribosomes for degradation (24). 18S NRD involves the action of the exosome, and substrates for this process accumulate in P-bodies (24), conserved cytoplasmic granules that contain untranslated mRNA, a set of translational repressors, the mRNA decapping machinery, and the 5′-3′ exonuclease XRN1 (25). 25S NRD seems to use a different mechanism in which ubiquitylation is important (24, 26). Large subunit NRD substrates accumulate in the cytoplasm in a perinuclear compartment. Additionally, a quality control mechanism surveys the proper assembly of ribosomes in the nucleolus, and misfolded ribosomes are targeted for degradation by the TRAMP-exosome pathway (27, 28). All of these mechanisms target aberrant, short-lived rRNA; on the other hand, the decay mechanism for normal rRNA is not known.
Some clues regarding the degradation of ribosomes have come from studies of cells undergoing nutritional stress. When cells are starved, portions of the cytosol, including entire organelles, are recycled to obtain essential material for cell survival. The best-characterized process involved in this recycling is macroautophagy. Macroautophagy is nonselective and entails the formation of double-membrane vesicles in the cytoplasm called autophagosomes (29). In yeast, a unique type of selective autophagy occurs in parallel with nonselective autophagy. Yeast cells under nitrogen starvation specifically target ribosomes for degradation by ribophagy, which requires the action of Ubp3, a ubiquitin protease, and its cofactor Bre5 (22). Ribophagy also depends on some components of the basal, nonselective autophagy machinery. We hypothesize that RNS2 is an essential component of ribophagy, or a similar mechanism, and that this mechanism not only is active during nutritional stress, but also has a housekeeping role. This housekeeping role, missing in the absence of RNS2, is the degradation of effete ribosomes, probably to recycle essential nutrients such as nucleotides. Thus, we propose that the absence of RNS2 activity produces a nutrient imbalance in the cell that triggers constitutive production of autophagosomes in an attempt to restore the housekeeping function of the ribophagy-like process.
Our results suggest that RNS2 participates in a ribophagy-like mechanism rather than one of the mechanisms proposed for degradation of aberrant, short-lived RNA, such as NRD. Mutants lacking RNS2 activity accumulate rRNA mostly in the vacuole. This observation is consistent with a ribophagy-like process because ribophagosomes fuse with the vacuole; in contrast, NRD is associated with P-bodies or is cytoplasmic, depending on the rRNA subunit. A ribophagy-like role is also consistent with the observed localization of the RNS2 enzyme in the ER or ER-derived structures and the vacuole. However, the existence of ribophagy in plants has not been shown, and this hypothesis needs to be tested further.
A nutritional role for RNase T2 enzymes has been proposed previously. Expression of class II RNases, including RNS2, is normally high in most tissues (3, 8, 30, 31); however, these class II RNases are induced by senescence and phosphate starvation in plants (6, 30). It was proposed that these enzymes, along with several class I RNase T2 enzymes that are also induced by starvation, are part of a phosphate-scavenging system that rescues plants under nutritional stress (9). It is possible that a ribophagy-like mechanism also participates in this scavenging system.
Another argument in support of a housekeeping role for RNS2 and other class II RNases is the high degree of conservation and expression patterns of these enzymes among all higher eukaryotes (2, 3). This conservation suggests that the proposed rRNA decay mechanism in plants may also be conserved in other eukaryotes. This seems to be the case. Most vertebrates have only one RNaseT2 gene in their genomes, but zebrafish and other bony fishes have two (2). One of the two copies (RNase Dre1 in zebrafish) forms a fish-specific clade, whereas the other (RNase Dre2 in zebrafish) belongs to the same clade that is conserved in all vertebrates, and in zebrafish is expressed constitutively in all tissues (2). Haud et al. (12) show that zebrafish mutants lacking RNase Dre2 accumulate rRNA in the lysosome. They also propose that RNase Dre2 (or RNASET2, as it is called in humans) is part of the ribophagy machinery. Lack of RNASET2 results in engorged lysosomes that are particularly apparent within neurons of the brain, leading to white matter lesions. These lesions resemble neuropathologies that are associated with mutations in the RNASET2 gene in human infants (32).
Recently, another function was proposed for RNase T2 enzymes, in particular Rny1, the only RNase T2 enzyme present in Saccharomyces cerevisiae. Yeast cells under oxidative stress accumulate cleaved tRNAs (33). Rny1 is responsible for this cleavage after release of the enzyme from the vacuole into the cytosol (34). Rny1 also induces cell death in yeast in a process that is independent of its catalytic activity (34). Cleavage of tRNA is a conserved response to oxidative stress: accumulation of cleaved tRNAs was also observed in human and Arabidopsis (33). These results suggest that RNS2 could function in this process; thus, it is possible that the conservation of class II RNases in all eukaryotic organisms is related to this stress response. However, in humans the tRNA cleavage function is carried out by angiogenin (34–36), a member of the RNase A family, not by RNASET2. Evidently, even if RNase A enzymes can replace some of the functions of RNase T2 enzymes in animals, the biological roles retained by the RNase T2 family are important enough to justify its conservation throughout evolution of higher organisms.
This dual role for RNase T2 enzymes could explain the fact that whereas a large portion of RNS2 is vacuolar, the pH preference of the enzyme is neutral. The RNS2 lower efficiency at lower pH could be compensated by the high level of accumulation of this protein in vacuoles, while a role in the cytoplasm, similar to that performed by Rny1 or any ER-related function, could be efficiently performed by lower amounts of enzyme.
Our results, together with those of Haud et al. (12)show that RNS2 and other class II RNase T2 enzymes participate in a conserved mechanism that recycles rRNA and could have similarities to yeast ribophagy. Deficiencies in this mechanism result in constitutive autophagy in plants and enlarged lysosomes in animals, indicating that rRNA recycling is essential to maintain cellular homeostasis. We propose that this mechanism could explain the conservation of the RNase T2 family in most eukaryotes.
Materials and Methods
Plant Material.
Arabidopsis thaliana ecotype Columbia-0 was used for all of the experiments. The rns2-2 mutant (SALK_069588) was obtained from the Arabidopsis Biological Resource Center. The antisense RNS2 line (79c.6) was described before (7). Soil-grown plants were kept in chambers with 16 h light at 60% humidity and 21 °C. For seedling experiments, seeds were surface-sterilized and germinated on Arabidopsis growth medium as described (6).
Standard cloning techniques were used to build the CFP-RNS2 construct. The RNS2 cDNA (6) was obtained from Pamela Green (University of Delaware, Newark, DE). The CFP-coding region was amplified from pAVA321-CFP (37). The cauliflower mosaic virus 35S promoter was obtained from the pGD vector (38). After assembly, the 35Sp-CFP-RNS2 construct was inserted into the binary vector pCAMBIA 2301. The plasmid spYFP-HDEL (13) was provided by Chris Hawes (Oxford Brookes University, Oxford). Plants were transformed according to Bariola et al. (7). Mesophyll protoplasts were prepared from leaves using a modified version of the method described by Yoo et al. (39). See SI Materials and Methods for details.
RNA and Protein Preparation and Subcellular Fractionation.
RNA was prepared as previously described (40). Protein extracts were prepared as previously described (41), but without the addition of 2-mercaptoethanol. ER-enriched fractions were obtained following the method of Matsushima et al. (14). Vacuoles were purified from protoplasts according to Robert et al. (42). Quality of purified vacuoles was determined by Neutral Red staining and visualization with a bright-field microscope (42). Vacuole enrichment was measured by assaying the vacuolar marker enzyme acid phosphatase as described (43).
Ribonuclease Activity Assays.
RNase activity in gel assays were performed as described by Yen and Green (44), using high-molecular-weight purified torula yeast RNA (Sigma). In vitro RNase activity was assayed as described by McClure et al. (45), using the same substrate. For pH optima assays, acetate, phosphate, or Tris buffers at the indicated pH were used in place of the standard reaction buffers.
Analysis of rRNA Decay.
Approximately 200 Arabidopsis seeds were surface-sterilized, cold treated, and grown in 125-mL sterile flasks with 20 mL of Murashige and Skoog (MS) media (Phytotechnology) and 1% sucrose on an orbital shaker at 150 rpm at 22 °C under 16 h light. After 7 d, 0.5 mCi of [5-3H]-uridine (1 mCi/mL, 15–30 Ci/mmol; Moravek Biochemicals) were added to the medium. The flasks were subjected to vacuum for 1 min and shaken at 80 rpm for 30 min. The seedlings were then washed with sterile MS medium and resuspended in 20 mL of germination medium. Samples were taken 24, 48, and 96 h later. RNA was prepared using the Qiagen RNeasy Plant Mini Kit. Five micrograms of total RNA was separated by electrophoresis in the presence of ethidium bromide (EtBr) and blotted onto nylon membranes as previously described (46). Total 28S rRNA amount was determined by scanning the membrane on a Typhoon scanner using the fluorescence setting and adjusting the photomultiplier tube (PMT) setting to 550 to eliminate signal saturation. ImageQuant 5.2 software was used for quantification. Tritium labeling was measured by cutting out the fragment of nylon membrane corresponding to each ribosomal band. Background controls for each lane were used by cutting out a similar-sized membrane piece above the ribosomal bands. Membranes were analyzed on a liquid scintillation counter. The fraction (tritium signal/EtBr signal) of radioactive RNA remaining at each time was calculated. Half-life was calculated as described (21).
Autophagy Induction and Monodansylcadaverine Staining.
Autophagy was induced by sucrose starvation and monitored by monodansylcadaverine (MDC) staining as described (47). Seedling samples were harvested 4 d after transfer. The cells in the elongation zone were observed near the central focal plane of the root. The number of motile MDC-labeled autophagosomes in all cells visible in the focal plane was determined. This number was divided by the total number of cells visible in the focal plane and designated the number of autophagosomes per cell for that region. The data from all regions in a sample were averaged together to obtain a mean number of autophagosomes per cell. At least 150 cells were observed per sample.
Specific RNA Staining Using SYTO RNASelect.
Wild-type and rns2-2 seedlings were germinated and grown for 7 d on solid medium. Whole seedlings were transferred to liquid MS medium and stained with 5 μM SYTO RNASelect green (Invitrogen) for 3 h at room temperature. After staining, roots were washed three times with fresh MS and observed using confocal microscopy. Leaf protoplasts were transferred to W5 medium and stained with 5 μM SYTO RNASelect green for 12 h. After staining, cells were washed three times with fresh W5 and observed using confocal microscopy (microscopy details are in SI Materials and Methods).
Statistical Analysis and Experiment Repetitions.
All experiments were repeated at least three times. For gels, a representative sample is shown. Numerical data were analyzed with t test (for samples with an equal number of repetitions) or z-test (for samples with an unequal number of repetitions), and means were considered different when P < 0.05. Error bars in figures indicate SE.
Supplementary Material
Acknowledgments
The authors thank Margie Carter (Iowa State University) and Kirk Czymmek (University of Delaware) for their assistance with confocal microscopy, Curtis Mosher (Iowa State University) for assistance with the Metamorph Line Analysis, and Drena Dobbs for critical reading of the manuscript. We also thank Saralyn Ohanian and Ludmila Rizshsky for help with cloning and Justine Carroll for help with plant care. This work was funded in part by Roy J. Carver Charitable Trust Grant 06-2323 (to G.C.M.) and by Grant IOB-0515998 from the National Science Foundation (to D.C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.J.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009809108/-/DCSupplemental.
References
- 1.Irie M. Structure-function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol Ther. 1999;81:77–89. doi: 10.1016/s0163-7258(98)00035-7. [DOI] [PubMed] [Google Scholar]
- 2.Hillwig MS, et al. Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals. BMC Evol Biol. 2009;9:170. doi: 10.1186/1471-2148-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacIntosh GC, Hillwig MS, Meyer A, Flagel L. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol Genet Genomics. 2010;283:381–396. doi: 10.1007/s00438-010-0524-9. [DOI] [PubMed] [Google Scholar]
- 4.Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hua Z-H, Fields A, Kao TH. Biochemical models for S-RNase-based self-incompatibility. Mol Plant. 2008;1:575–585. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- 6.Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ. RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bariola PA, MacIntosh GC, Green PJ. Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bariola PA, et al. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- 9.Abel S, Ticconi CA, Delatorre CA. Phosphate sensing in higher plants. Physiol Plant. 2002;115:1–8. doi: 10.1034/j.1399-3054.2002.1150101.x. [DOI] [PubMed] [Google Scholar]
- 10.Berndt T, Kumar R. Novel mechanisms in the regulation of phosphorus homeostasis. Physiology (Bethesda, Md) 2009;24:17–25. doi: 10.1152/physiol.00034.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trubia M, Sessa L, Taramelli R. Mammalian Rh/T2/S-glycoprotein ribonuclease family genes: Cloning of a human member located in a region of chromosome 6 (6q27) frequently deleted in human malignancies. Genomics. 1997;42:342–344. doi: 10.1006/geno.1997.4679. [DOI] [PubMed] [Google Scholar]
- 12.Haud N, et al. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci USA. 2011;108:1099–1103. doi: 10.1073/pnas.1009811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irons SL, Evans DE, Brandizzi F. The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. J Exp Bot. 2003;54:943–950. doi: 10.1093/jxb/erg102. [DOI] [PubMed] [Google Scholar]
- 14.Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I. A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 2003;33:493–502. doi: 10.1046/j.1365-313x.2003.01636.x. [DOI] [PubMed] [Google Scholar]
- 15.Carter C, et al. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman E, Schmidt M. Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies provides an alternate pathway for protein transfer to the vacuole. Plant Physiol. 2004;136:3440–3446. doi: 10.1104/pp.104.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian H, Klämbt D, Jones AM. Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. J Biol Chem. 1995;270:26962–26969. doi: 10.1074/jbc.270.45.26962. [DOI] [PubMed] [Google Scholar]
- 18.Frigerio L, Hinz G, Robinson DG. Multiple vacuoles in plant cells: Rule or exception? Traffic. 2008;9:1564–1570. doi: 10.1111/j.1600-0854.2008.00776.x. [DOI] [PubMed] [Google Scholar]
- 19.delCardayré SB, et al. Engineering ribonuclease A: Production, purification and characterization of wild-type enzyme and mutants at Gln11. Protein Eng. 1995;8:261–273. doi: 10.1093/protein/8.3.261. [DOI] [PubMed] [Google Scholar]
- 20.Pérez-Amador MA, et al. Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 2000;122:169–180. doi: 10.1104/pp.122.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 23.Lafontaine DL. A ‘garbage can’ for ribosomes: How eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balagopal V, Parker R. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii K, Kitabatake M, Sakata T, Miyata A, Ohno M. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 2009;23:963–974. doi: 10.1101/gad.1775609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 28.Andersen KR, Jensen TH, Brodersen DE. Take the “A” tail: Quality control of ribosomal and transfer RNA. Biochim Biophys Acta. 2008;1779:532–537. doi: 10.1016/j.bbagrm.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Bassham DC. Function and regulation of macroautophagy in plants. Biochim Biophys Acta. 2009;1793:1397–1403. doi: 10.1016/j.bbamcr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Liang L, Lai Z, Ma W, Zhang Y, Xue Y. AhSL28, a senescence- and phosphate starvation-induced S-like RNase gene in Antirrhinum. Biochim Biophys Acta. 2002;1579:64–71. doi: 10.1016/s0167-4781(02)00507-9. [DOI] [PubMed] [Google Scholar]
- 31.Chai L, Ge X, Xu Q, Deng X. CgSL2, an S-like RNase gene in ‘Zigui shatian’ pummelo (Citrus grandis Osbeck), is involved in ovary senescence. Mol Biol Rep. 2011;38:1–8. doi: 10.1007/s11033-010-0070-x. [DOI] [PubMed] [Google Scholar]
- 32.Henneke M, et al. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nat Genet. 2009;41:773–775. doi: 10.1038/ng.398. [DOI] [PubMed] [Google Scholar]
- 33.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu H, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 36.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Assmann SM, Fedoroff NV. Characterization of the Arabidopsis heterotrimeric G protein. J Biol Chem. 2008;283:13913–13922. doi: 10.1074/jbc.M801376200. [DOI] [PubMed] [Google Scholar]
- 38.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 39.Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 40.Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacIntosh GC, Ulloa RM, Raices M, Tellez-Inon MT. Changes in calcium-dependent protein kinase activity during in vitro tuberization in potato. Plant Physiol. 1996;112:1541–1550. doi: 10.1104/pp.112.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert S, Zouhar J, Carter C, Raikhel N. Isolation of intact vacuoles from Arabidopsis rosette leaf-derived protoplasts. Nat Protoc. 2007;2:259–262. doi: 10.1038/nprot.2007.26. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed SU, et al. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol. 2000;149:1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yen Y, Green PJ. Identification and properties of the major ribonucleases of Arabidopsis thaliana. Plant Physiol. 1991;97:1487–1493. doi: 10.1104/pp.97.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClure BA, et al. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- 46.LeBrasseur ND, MacIntosh GC, Pérez-Amador MA, Saitoh M, Green PJ. Local and systemic wound-induction of RNase and nuclease activities in Arabidopsis: RNS1 as a marker for a JA-independent systemic signaling pathway. Plant J. 2002;29:393–403. doi: 10.1046/j.1365-313x.2002.01223.x. [DOI] [PubMed] [Google Scholar]
- 47.Contento AL, Xiong Y, Bassham DC. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 2005;42:598–608. doi: 10.1111/j.1365-313X.2005.02396.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.