Abstract
L-type prostaglandin synthase (L-PGDS) produces PGD2, a lipid mediator involved in neuromodulation and inflammation. Here, we show that L-PGDS and arrestin-3 (Arr3) interact directly and can be co-immunoprecipitated endogenously from MG-63 osteoblasts. Perinuclear L-PGDS/Arr3 co-localization is observed in PGD2-producing MG-63 cells and is induced by the addition of the L-PGDS substrate or co-expression of COX-2 in HEK293 cells. Inhibition of L-PGDS activity in MG-63 cells triggers redistribution of Arr3 and L-PGDS to the cytoplasm. Perinuclear localization of L-PGDS is detected in wild-type mouse embryonic fibroblasts (MEFs) but is more diffused in MEFs-arr-2−/−-arr-3−/−. Arrestin-3 promotes PGD2 production by L-PGDS in vitro. IL-1β-induced PGD2 production is significantly lower in MEFs-arr-2−/−-arr-3−/− than in wild-type MEFs but can be rescued by expressing Arr2 or Arr3. A peptide corresponding to amino acids 86–100 of arrestin-3 derived from its L-PGDS binding domain stimulates L-PGDS-mediated PGD2 production in vitro and in MG-63 cells. We report the first characterization of an interactor/modulator of a PGD2 synthase and the identification of a new function for arrestin, which may open new opportunities for improving therapies for the treatment of inflammatory diseases.
Keywords: Cyclooxygenase (COX) Pathway, Peptide Interactions, Prostaglandins, Protein Translocation, Protein-Protein Interactions, Arrestin, Prostaglandin Synthase
Introduction
Prostaglandins (PGs)5 are lipid mediators formed from arachidonic acid through the action of cyclooxygenases (COXs). COXs convert arachidonic acid released from the plasma membrane to an intermediate substrate, PGH2, which is metabolized by specific synthases to produce PGs like PGD2 (1, 2). PGD2 is involved in various physiological processes such as vasodilatation, bronchoconstriction (3), regulation of pain (4), and sleep (5) but is also implicated in inflammatory responses such as asthma (6) and atherosclerosis (7). PGD2 was shown to exhibit anti-inflammatory properties as well, as increased levels of PGD2 are observed during the resolution phase of inflammation (8–10). Recent work by our group showed that PGD2 displays anabolic properties in bone (11, 12).
There are two types of prostaglandin D2 synthase (PGDS). The hematopoietic PGDS (H-PGDS) is glutathione-requiring (13) and is expressed mainly in mast cells (14), megakaryocytes (15), and T-helper 2 lymphocytes (16). The lipocalin-type PGDS (L-PGDS), also called β-trace, is glutathione-independent and is expressed abundantly in the central nervous system (17, 18), the heart (19), the retina (20), and the genital organs (21). L-PGDS is also the only enzyme among the members of the lipocalin gene family and binds small lipophilic substances like retinoic acid (22), bilirubin (23), and gangliosides (24).
The arrestin family consists of ubiquitously expressed arrestin-2 and -3 (also known as β-arrestin-1 and -2) and two retinal arrestins (25). Arrestin-2 and -3 are multifunctional molecules in addition to their well known role in desensitization and internalization of G protein-coupled receptors (26). The identification of numerous non-receptor binding partners has expanded their functions to protein ubiquitination, chemotaxis, apoptosis, mitogen-activated protein kinases activation (27, 28), osteoclastogenesis inhibition (29), and regulation of the interleukin 1 (IL-1) pathway (30).
It is remarkable how very little is known about the interaction partners and the mechanisms regulating the activity of PG synthases considering their crucial physiological and pathological roles and the clinical problems associated with the long term use of COX inhibitors. Here we show that arrestin-3 interacts with L-PGDS and increases L-PGDS-mediated PGD2 production. An arrestin-3 peptide was identified as capable of inducing PGD2 production by L-PGDS. This is important because it shows that by identifying interacting partners of PG synthases we can not only further our understanding of the scarcely documented regulatory mechanisms of these critical enzymes but also provide approaches to identify potentially active small molecules that can modulate their activity. This could constitute an alternative approach for the development of modulators of PG synthases.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
The polyclonal anti-HA, polyclonal anti-myc, monoclonal anti-Arr3, and mouse anti-IgG antibodies as well as protein G-agarose were purchased from Santa Cruz Biotechnology. The anti-GST polyclonal antibody was from Bethyl Laboratories. The monoclonal anti-His antibody was from Cell Signaling Technology. The monoclonal anti-HA antibody was from Covance. The polyclonal and monoclonal L-PGDS antibodies, PGH2, HQL-79, and the prostaglandin D2 EIA kit were purchased from Cayman Chemical Co. The arrestin-3 antibody blocking peptide was from Santa Cruz. The HRP-conjugated anti-mouse and the HRP-conjugated anti-rabbit were from GE Healthcare. The Alexa Fluor 488 donkey anti-mouse, immunoglobulin G (IgG), Alexa Fluor 546 goat anti-mouse, and Alexa Fluor 546 goat anti-rabbit IgG antibodies were from Molecular Probes. Selenium tetrachloride (SeCl4) was from Sigma, whereas IL-1β was from Cedarlane.
Peptide Synthesis
Peptides arrestin-3 (Arr3) 56–70 (VTLTCAFRYGREDLD), 65–79 (GREDLDVLGLSFRKD), 76–90 (FRKDLFIANYQAFPP), and 86–100 (QAFPPTPNPPRPPTR) were synthesized by GenScript, with purity ∼80%. Peptides were acetylated at their N terminus to improve stability. The TAT-Arr3 89–103 (YGRKKRRQRRRGGGQAFPPTPNPPRPPTR) and scrambled TAT-Arr3 86–100 peptides (YGRKKRRQRRRGGGFPPRTRPQPANPTPP) were also synthesized by GenScript with purity ∼70%.
Yeast Two-hybrid Screen
A yeast two-hybrid screen was performed following the two-hybrid system standard protocol (31). Briefly, a plasmid containing the complete cDNA of Arr3 (pAS2–1-Arr3) was transformed into the yeast strain pJ69-4α according to the lithium yeast transformation protocol (32). This stably transformed clone was then transformed with a human HeLa MATCHMAKER cDNA Library or with the empty pGAD424 plasmid (Clontech). Positive clones were initially selected for growth in the absence of histidine. Clones showing positive interactions were then isolated, and these interactions were confirmed by growth on quadruple selective media (Trp−, Leu−, His−, and Ade−). pGADGH plasmids containing the library inserts from positive colonies were isolated and transformed into the DH5α bacterial strain. Plasmids were extracted from DH5α cells and transformed once more into yeast with either the bait (pAS2–1-Arr3) or the negative control (pAS2–1) and plated on quadruple selective medium (Trp−, Leu−, His−, and Ade−) to confirm the interaction. The selected plasmids were then sequenced by dideoxy sequencing, and the identities of the clones were determined by using the NCBI BLAST alignment tool.
Cell Culture and Transfections
Human embryonic kidney (HEK) 293 cells and the osteosarcoma MG-63 cell line were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere containing 5% CO2. Transient transfections of HEK293 cells grown to 50–70% confluence were performed using TransIT-LT1 Reagent (Mirus), whereas MG-63 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Mouse embryonic fibroblasts (MEFs) wt lacking Arr2, Arr3, or both arrestins (kindly provided by R. J. Lefkowitz) were maintained as described above and transfected with TransIT-20/20 Reagent (Mirus) according to the manufacturer's instructions.
Immunoprecipitations
The HEK293 cells were transiently transfected with the indicated constructs and maintained as described above for 48 h. Where indicated, cells were incubated for 15 min at 37 °C in the presence of 5 μm PGH2 before harvesting. The cells were then washed with ice-cold PBS and harvested in 300 μl of lysis buffer (150 mm NaCl, 50 mm Tris, pH 8, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 10 mm Na4P2O7, and 5 mm EDTA supplemented with protease inhibitors (9 nm pepstatin, 9 nm antipain, 10 nm leupeptin, and 10 nm chymostatin; Sigma)). After incubation in lysis buffer for 60 min at 4 °C, the lysates were clarified by centrifugation for 20 min at 14,000 × g at 4 °C. One μg of specific monoclonal antibodies was added to the supernatant. After 60 min of incubation at 4 °C, 40 μl of 50% protein G-agarose was added followed by 3 h of incubation at 4 °C. Samples were then centrifuged for 1 min in a microcentrifuge and washed three times with 1 ml of lysis buffer. Immunoprecipitated proteins were eluted by the addition of 50 μl of SDS sample buffer followed by 5 min in boiling water. Initial lysates and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotting using specific antibodies.
Recombinant Protein Production and Binding Assays
To produce His-tagged proteins, PCR fragments corresponding to the cDNA coding for full-length L-PGDS or Arr3 were inserted into the pRSETA expression vector (Invitrogen). These constructs were used to produce fusion proteins in OverExpressTM C41(DE3) Escherichia coli strain (Avidis) by following the manufacturer's instructions. The recombinant proteins were purified using nickel-nitrilotriacetic acid-agarose resin (Qiagen) as indicated by the manufacturer. The cDNA fragments coding for full-length or for different regions of Arr3 and for full-length L-PGDS were amplified by PCR and introduced into the pGEX-4-T1 vector (Amersham Biosciences) to produce the indicated glutathione S-transferase (GST)-tagged fusion proteins in the OverExpressTM C41(DE3) E. coli strain, which were purified using glutathione-SepharoseTM 4B (Amersham Biosciences) as indicated by the manufacturer. Purified recombinant proteins were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. Five μg of glutathione-Sepharose bound GST-tagged fusion protein was incubated with 5 mg of purified histidine protein in binding buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 10% glycerol, and 0.5% Igepal) supplemented with protease inhibitors (9 mm pepstatin, 9 mm antipain, 10 mm leupeptin, and 10 mm chymostatin) and 2 mm DTT overnight at 4 °C. The binding reactions were then washed 4 times with binding buffer. SDS sample buffer was added to the binding reactions, and the tubes were boiled for 5 min. The binding reactions were analyzed by SDS-PAGE, and immunoblotting was performed with the indicated specific antibodies.
Cell Fractionation
For isolation of cytoplasmic and nuclear fractions, MEF cells were plated in 6-well plates and transiently transfected with L-PGDS-HA to facilitate its detection. Plasma membranes were then disrupted in a buffer containing 10 mm HEPES, 10 mm KCl, 0.1 mm EDTA, 1% Igepal supplemented with protease inhibitors mixture. Centrifugation for 5 min to 14,000 × g at 4 °C allowed recovery of the cytoplasmic fraction. Pellets were resuspended with buffer containing 10 mm HEPES, 400 mm NaCl, 1 mm EDTA, DTT 1 mm, 10% glycerol, and protease inhibitors mixture and were incubated for 45 min at 4 °C. Samples were then centrifuged for 5 min at 14,000 × g at 4 °C to isolate the nuclear fraction in the supernatant. SDS sample buffer was added to the lysates, and samples were boiled for 5 min. Cytoplasmic (supernatant) and nuclear fractions were analyzed by SDS-PAGE, and immunoblotting was performed with the indicated specific antibodies.
In Vitro PGD2 Production Assays
His6-Arr3, His6-L-PGDS, or His6 alone were produced as described above and were incubated at a predetermined molar ratio in a buffer containing 1 m Tris-HCl, pH 8.0, 1 mm DTT, 0.5 m guanidine HCl (33), and 1 μg/ml IgG for 10 min at room temperature in 96-well plates. PGH2 to a final concentration of 0.5 μm was added to the wells, and the reaction was performed for 1 min and then stopped with 0.4 mg/ml SnCl2. PGD2 produced was then measured with the prostaglandin D2 EIA kit according to the manufacturer's instructions. For PGD2 production assays with peptides, the indicated peptides were added to the buffer with the same molar ratio, and PGD2 production was measured as described above.
PGD2 Production Assays in Cells
MEFs wt, Arr2 KO, Arr3 KO, or Arr (double knockout (dKO) were plated in 24-well plates and transiently transfected when needed with the indicated constructs. To eliminate PGD2 production by H-PGDS, cells were preincubated with HQL-79 (a specific H-PGDS inhibitor (34) to measure only L-PGDS-mediated PGD2 production) at 100 μm for 15 min at 37 °C. Where indicated, cells were incubated for 15 min at 37 °C in the presence of 5 μm PGH2 or starved with DMEM without FBS for 24 h and stimulated with 5 ng/ml IL-1β for 16 h. The cells were lysed as described above, and PGD2 in the lysates and in the media were measured with the prostaglandin D2 EIA kit according to the manufacturer's instructions. For PGD2 production assays with peptides, MG-63 cells were plated in 24-well plates, and 100 μm TAT peptide was added to the cells for 1 h at 37 °C (35). The assay was then performed as described above, and proteins in cell lysates were measured with a commercial kit (Bio-Rad) to normalize levels of PGD2.
Immunofluorescence Staining and Confocal Microscopy
For co-localization experiments, HEK293 or MEF cells were plated and transfected as described above with Arr3-GFP, COX-2, and/or L-PGDS-HA where indicated. Forty-eight hours later, 2 × 105 cells were transferred onto coverslips coated with 0.1 mg/ml poly-l-lysine (Sigma) and further grown overnight. Cells were washed with PBS, and where indicated, cells were incubated for 15 min at 37 °C in the presence of 5 μm PGH2. Cells were then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, washed again with PBS, permeabilized for 20 min with 0.1% Triton X-100 in PBS, and blocked with 0.1% Triton X-100 in PBS containing 5% nonfat dry milk for 30 min at room temperature. Cells were then incubated with primary antibodies diluted in blocking solution for 60 min at room temperature. Cells were washed twice with PBS, blocked again with 0.1% Triton X-100 in PBS containing 5% nonfat dry milk for 30 min at room temperature, and incubated with appropriate secondary antibodies diluted in blocking solution for 60 min at room temperature. The cells were washed twice with permeabilization buffer and twice with PBS, and coverslips were mounted using Vectashield Mounting Medium (Vector Laboratories). Confocal microscopy was performed using a scanning confocal microscope (FV1000; Olympus) coupled to an inverted microscope with a 63× oil immersion objective (Olympus), and images were processed with IMAGE-PRO PLUS 6.0. For inhibitory assays with SeCl4, MG-63 cells were directly transferred onto coverslips coated with 0.1 mg/ml poly-l-lysine (Sigma) and further grown overnight. Cells were washed with PBS, and where indicated, cells were incubated for 15 min at 37 °C in the presence of 100 μm SeCl4. Cells were then processed as described above.
Image Analysis
Co-localization was assessed by the examination of merged images that showed co-localized regions. In addition to the merged image, a pixel fluorogram was generated using IMAGE-PRO software. The fluorogram is a two-dimensional intensity histogram of the dual-color image, indicating the distribution of all pixels within the merged image as a scattergram. Pixel values of red and green components are displayed along the x axis and y axis, respectively, where dimmer pixels in the image are located toward the origin of the scatter plot, whereas brighter pixels are located farther out. A high degree of co-localization is revealed by a diagonal distribution (at 45°) of the dots on the fluorogram. A lack of co-localization is shown by two distinct populations with a minimal overlap of dots distributed toward the red and green axes, respectively (36).
Statistical Analysis
Statistical analyses were performed using PRISM v4.0 (GraphPad Software) using Student's t test. Data were considered significant when p values were <0.05 (*), <0.01 (**), or <0.001 (***).
RESULTS
L-PGDS Directly Interacts with Arrestin-3
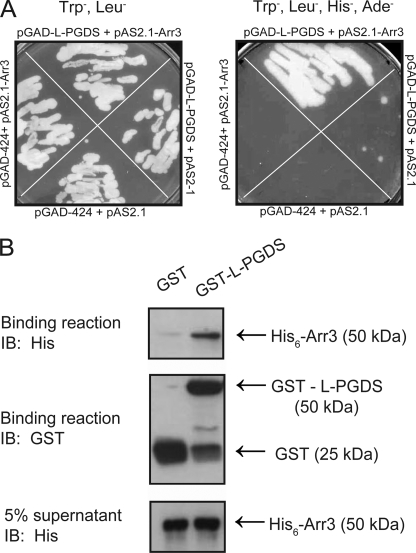
L-PGDS was identified as a potential Arr3 partner during a yeast two-hybrid screen of a human HeLa cell Matchmaker cDNA library using Arr3 as bait. As shown in Fig. 1A, strong growth on Trp−, Leu−, His−, and Ade− medium was present only in yeast strain pJ69-4α transformed with pAS2.1-Arr3 and pGADGH-L-PGDS constructs, whereas only minor growth was observed in yeast cells transformed with pAS2.1 and pGADGH-L-PGDS, indicating that L-PGDS is interacting with Arr3. No growth was observed in yeast cells transformed with pAS2.1-Arr3 and pGADGH-H-PGDS, the other PGD2 synthase, suggesting that Arr3 specifically interacts with L-PGDS (data not shown). Strong growth was also observed on Trp−, Leu−, His−, and Ade− medium in yeasts transformed with pAS2.1-Arr2 and pGADGH-L-PGDS (data not shown), indicating that L-PGDS also interacts with Arr2. To further confirm the interaction between L-PGDS and Arr3 and to determine whether this interaction was direct, we performed an in vitro binding assay using purified recombinant L-PGDS fused to GST and GST-L-PGDS along with purified recombinant Arr3 fused to His (His6-Arr3). The results presented in Fig. 1B illustrate that Arr3 binds to glutathione-Sepharose-bound GST-L-PGDS and not to glutathione-Sepharose-bound GST, showing that L-PGDS can interact directly with Arr3.
FIGURE 1.
The interaction between L-PGDS and Arr3 is direct. A, a yeast two-hybrid screen was performed using Arr3 as bait on a human HeLa MATCHMAKER cDNA Library. The interaction between L-PGDS and Arr3 was confirmed in pJ69-4α yeasts transformed with the indicated constructs grown on the selective yeast media Trp−, Leu− (left) or Trp−, Leu−, His− and Ade− (right), as detailed under “Experimental Procedures”. B, recombinant GST- and His-tagged proteins were produced in OverExpressTM C41(DE3) E. coli cells. GST pulldown assays were carried out using purified glutathione-Sepharose-bound GST (control) or GST- L-PGDS that were incubated with purified recombinant His6-Arr3 protein. The binding of Arr3 was detected by immunoblotting (IB) using a His6-specific monoclonal antibody, and the GST fusion proteins present in the binding reactions were detected using a GST-specific polyclonal antibody, as described under “Experimental Procedures.” Blots shown are representative of three independent experiments.
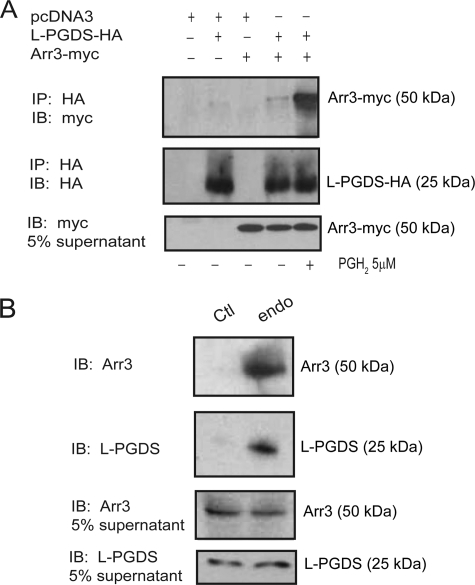
To investigate whether L-PGDS and Arr3 interact in a cellular context, we performed immunoprecipitation experiments in HEK293 cells transfected with pcDNA3-Arr3-myc and pcDNA3-L-PGDS-HA in the presence or absence of PGH2, the L-PGDS substrate. Cell lysates were incubated with a HA-specific monoclonal antibody, and co-immunoprecipitated Arr3 was detected by Western blot analysis with a myc polyclonal antibody. Our results demonstrate that Arr3-myc was modestly co-immunoprecipitated with L-PGDS in the absence of PGH2 (Fig. 2A). However, the L-PGDS/Arr3 co-immunoprecipitation was strongly increased by the addition of PGH2 (Fig. 2A). Co-immunoprecipitation experiments were also performed on endogenous proteins in the MG-63 osteoblast cell line, which produces PGD2 endogenously (11), contrary to HEK293 cells. Cell lysates were incubated with an L-PGDS-specific monoclonal antibody or with an isotypic normal mouse IgG as a negative control, and co-immunoprecipitated Arr3 was detected by Western blot analysis with an Arr3 polyclonal antibody. As shown in Fig. 2B, endogenous Arr3 co-immunoprecipitated with endogenous L-PGDS in MG-63 cells.
FIGURE 2.
Arr3 and L-PGDS interact in a cellular context. A, HEK293 cells were transiently transfected with HA-tagged L-PGDS and myc-tagged Arr3 constructs and were stimulated or not with 5 μm PGH2 for 15 min. Immunoprecipitations (IP) of L-PGDS were performed using an HA-specific monoclonal antibody, and immunoblotting (IB) was performed with HA- or myc-specific polyclonal antibodies. B, immunoprecipitation of endogenous L-PGDS in the MG-63 osteoblast cell line was performed using an L-PGDS-specific monoclonal antibody (endo), whereas a normal mouse anti-IgG isotype antibody was used as a control (Ctl). Immunoblotting was performed with L-PGDS- or Arr3-specific polyclonal antibodies. Immunoprecipitations and immunoblotting were performed as described under “Experimental Procedures.” Blots shown are representative of three independent experiments.
Co-localization of L-PGDS with Arr3
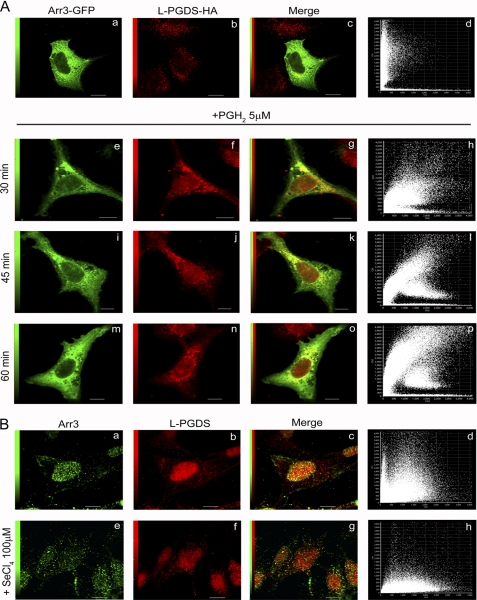
The possible co-localization of L-PGDS with Arr3 was studied by confocal microscopy first in HEK293 cells expressing Arr3-GFP and L-PGDS-HA. Arr3 and L-PGDS were both predominantly associated with vesicular structures in the cytoplasm, whereas Arr3 was also detected at the cell membrane (Fig. 3A). The scarcity of yellow pixels in the merged image (Fig. 3Ac) and analysis of the corresponding pixel fluorogram (Fig. 3Ad) show a low degree of co-localization between Arr3 and L-PGDS in basal conditions. Because PGH2 enhanced the L-PGDS/Arr3 interaction (Fig. 2A), co-localization of the two proteins was also studied after the addition of the L-PGDS substrate. Adding PGH2 significantly increased the number of co-localizing pixels between L-PGDS and Arr3, particularly in the perinuclear region (Fig. 3Ao) after 60 min of treatment.
FIGURE 3.
Co-localization analysis of Arr3 and L-PGDS by confocal immunofluorescence microscopy. A, HEK293 cells were transiently transfected with Arr3-GFP and L-PGDS-HA constructs and were treated with vehicle (upper panel) or 5 μm PGH2 (lower panel) for the indicated times to stimulate L-PGDS enzymatic activity. Cells were then fixed and prepared as described under “Experimental Procedures.” Arr3-GFP is shown in green, whereas L-PGDS was visualized using anti-HA monoclonal and Alexa Fluor 546-conjugated anti-mouse IgG antibodies (red). Overlays of staining patterns (c, g, k, and o) and the corresponding pixel fluorograms (d, l, h, p) are also shown (x axis, intensity of L-PGDS pixels; y axis, intensity of Arr3-GFP pixels). A high degree of co-localization is revealed by a diagonal distribution (at 45°) of the dots on the fluorogram, whereas a lack of co-localization is characterized by two distinct populations with a minimal overlap of dots distributed toward the x and y axes, respectively. Scale bars, 10 μm. B, MG-63 cells were treated with vehicle (upper panel) or 100 μm SeCl4 (lower panel) to inhibit PGDS enzymatic activity for 15 min. Cells were then fixed and prepared as described under “Experimental Procedures.” Arr3 was visualized using anti-Arr3 monoclonal antibody and Alexa Fluor 488-conjugated anti-mouse IgG antibodies (green, a and e), whereas L-PGDS was revealed with anti-L-PGDS polyclonal and Alexa Fluor 546-conjugated anti-rabbit IgG antibodies (red, b and f). Overlays of staining patterns (c and g) and the corresponding pixel fluorograms (d and h) are shown (x axis, intensity of L-PGDS pixels; y axis, intensity of Arr3 pixels). Scale bars, 10 μm.
Cellular distribution of endogenous Arr3 and L-PGDS was also examined in MG-63 osteoblasts. Intriguingly, localization of Arr3 and L-PGDS was predominant in the nuclear region in basal conditions in MG-63 cells (Fig. 3B, a and b) with a high degree of co-localization between the two proteins (Fig. 3B, c and d) that was not promoted by treatment with PGH2 (data not shown). The specificity of Arr3 detection in the nuclear region was demonstrated by using a blocking peptide (supplemental Fig. S1Ae) and by studying the localization of transfected Arr3-GFP in MG-63 cells (supplemental Fig. S1Ba). Contrary to HEK293 cells, MG-63 cells express COX enzymes and produce significant levels of PGH2 (37) and PGD2 (11) endogenously. Thus, we reasoned that inhibiting L-PGDS activity could perhaps modify the intracellular distribution of L-PGDS and Arr3 in MG-63 cells. Incubation of MG-63 cells with SeCl4, a PGDS inhibitor (38, 39), resulted in a marked redistribution of Arr3 out of the nuclear region to cytoplasmic punctates (Fig. 3Be). The change in L-PGDS distribution was less radical after SeCl4 treatment; L-PGDS remained largely in the perinuclear region but appeared more diffused (Fig. 3Bf). Consequently, co-localization of Arr3 with L-PGDS was drastically reduced in MG-63 cells when L-PGDS activity was inhibited (Fig. 3B, g and h).
The effect of COX-2 co-expression on the localization of L-PGDS was also studied in HEK293. Expression of COX-2 alone with L-PGDS did not alter the distribution of L-PGDS (compare Fig. 3Ab to Fig. 4Ab). However, when Arr3 was co-expressed, L-PGDS redistributed to the perinuclear region in presence of COX-2 (Fig. 4Af).
FIGURE 4.
Arr3 targets L-PGDS to the perinuclear region. A, HEK293 cells were transiently transfected with pcDNA3-L-PGDS-HA and pcDNA3-COX2-myc constructs (upper panel) or with pcDNA3-Arr3-GFP, pcDNA3-L-PGDS-HA, and pcDNA3-COX2 constructs (lower panel) for 48 h. The cells were then fixed and prepared as described under “Experimental Procedures.” Arr3-GFP is shown in green, whereas L-PGDS was visualized using anti-HA monoclonal and Alexa Fluor 546-conjugated anti-mouse IgG antibodies (red). COX-2 was visualized using anti-myc polyclonal and Alexa Fluor 633-conjugated anti-rabbit IgG antibodies (blue). Overlays of staining patterns (d and h) are also shown. Scale bars, 10 μm. B, wild-type MEFs (MEFs wt), MEFs Arr2 KO (k/o), MEFs Arr3 KO, or MEFs Arr dKO were transiently transfected with pcDNA3-L-PGDS-HA for 48 h. The cells were then fixed and prepared as described under “Experimental Procedures.” L-PGDS was visualized using anti-L-PGDS polyclonal and Alexa Fluor 546-conjugated anti-rabbit IgG antibodies (red), whereas the nucleus was stained with Hoechst and is shown in blue. Overlays of staining patterns (c, f, i, l) are also shown. Scale bars, 10 μm. C, lysates from wild-type MEFs or MEFs Arr dKO were separated into supernatant (S) and nuclear (N) fractions as described under “Experimental Procedures.” Immunoblotting (IB) was performed with L-PGDS, Shp2 (cytoplasmic control), or lamin A/C (nuclear control) specific polyclonal antibodies. Blots shown are representative of three independent experiments.
The role of Arr in L-PGDS distribution was also investigated in MEFs from wild-type mice and mice lacking Arr2, Arr3, or both. Confocal microscopy of wild-type MEFs that were transfected with an HA-tagged L-PGDS construct revealed prominent localization of L-PGDS in the perinuclear region and the nucleus (Fig. 4Bb). Interestingly, the bulk of L-PGDS-HA fluorescence was associated with intracellular vesicles scattered throughout the cell in MEFs-Arr double KO (Fig. 4Bk). Distribution of L-PGDS-HA in MEFs single Arr2 or Arr3 KO was intermediate to that observed for the wild-type MEFs and MEFs-Arr double KO (Fig. 4B, e and h). Moreover, L-PGDS was more abundant in the supernatant fraction depleted of nuclei in MEFs-Arr double KO than in wild-type MEFs, as assessed by Western blot (Fig. 4C).
L-PGDS-mediated PGD2 Production Is Modulated by Arr3
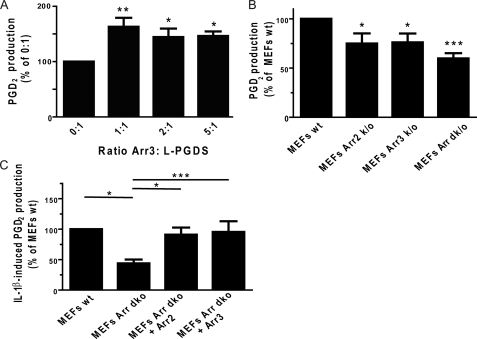
Next, we were interested in determining if Arr3 modulated PGD2 production by L-PGDS. We performed in vitro experiments using purified recombinant L-PGDS incubated with increasing amounts of purified Arr3 in the presence of the L-PGDS substrate PGH2, and PGD2 production was measured by competitive ELISA. As shown in Fig. 5A, incubation of L-PGDS with an equal molar ratio of Arr3 brought a 60% increase in PGD2 production when compared with L-PGDS alone, whereas a 2–5-fold excess of Arr3 failed to further increase it. We next explored whether Arr3 modulated L-PGDS-mediated PGD2 production in cells. Wild-type MEFs and MEFs Arr2 KO, Arr3 KO, or Arr double KO were incubated with PGH2. Fig. 5B shows that PGD2 production was decreased by ∼25% in MEFs lacking Arr2 or Arr3 and by 40% in MEFs lacking both arrestins compared with wild-type MEFs.
FIGURE 5.
Arr3 modulates L-PGDS-mediated PGD2 production. A, purified His6-L-PGDS was incubated with the indicated molar ratio of empty pRSETa vector control flow-through (0:1) or purified His6-Arr3, and PGD2 production was stimulated by the addition of 0.5 μm PGH2 (L-PGDS substrate) for 1 min. Reactions were stopped with 0.4 mg/ml SnCl2, and PGD2 was then measured with commercially available enzyme-linked immunoassays as described under “Experimental Procedures.” B, wild-type MEFs (MEFs wt) lacking Arr2 (MEFs Arr2 KO), Arr3 (MEFs Arr3 KO), or both arrestins (MEFs Arr dk/o) were preincubated with 100 μm HQL-79 for 15 min to inhibit H-PGDS (to measure L-PGDS activity specifically) and were stimulated with 5 μm PGH2 for 15 min. Supernatants were assessed for PGD2 production with commercially available enzyme-linked immunoassays as described under “Experimental Procedures.” C, wild-type MEFs lacking both arrestins (MEFs Arr dKO) or MEFs Arr dKO transfected with pcDNA3-Arr2 or -Arr3 for 24 h were incubated in DMEM without FBS for 24 h before stimulation with 5 ng/ml IL-1β for 16 h. Supernatants were assessed for PGD2 production with commercially available enzyme-linked immunoassays as described under “Experimental Procedures.” All values are the mean ± S.E. from at least three separate experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To put this new function in a physiologically relevant context, we stimulated MEFs with IL-1β, a pro-inflammatory cytokine that can stimulate prostanoid synthesis by stimulating the expression of COX-2 (40, 41). PGD2 production was decreased by roughly 50% in MEFs lacking both arrestins, in comparison with wild-type MEFs after stimulation with IL-1β (Fig. 5C). Expressing Arr2 or Arr3 in MEFs Arr double KO brought the level of PGD2 production back to that observed in wild-type MEFs (Fig. 5C), indicating that arrestins contribute to increased PGD2 production after stimulation with IL-1β in MEFs.
Determination of the L-PGDS Binding Domain on Arr3
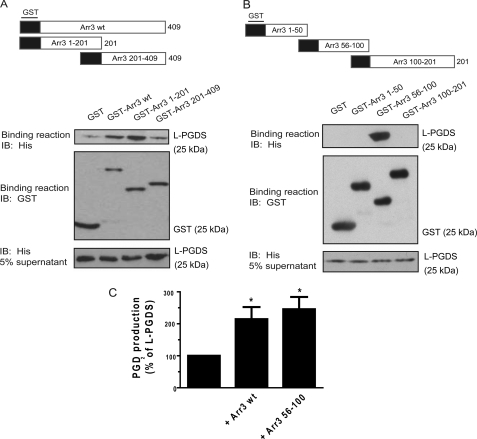
To identify the Arr3 domain involved in the interaction with L-PGDS, we generated truncated mutants of Arr3 that were introduced in the pGEX-4-T1 vector (Fig. 6A). These truncated mutants were then purified as GST fusion proteins and used in GST pulldown assays with purified His6-tagged L-PGDS. Protein complexes were analyzed by immunoblotting with a His-specific monoclonal antibody to detect the presence of L-PGDS. Specific interactions with L-PGDS were observed for the GST-Arr3 and GST-Arr3 1–201 constructs, indicating that L-PGDS binds to the N-terminal region of Arr3 (Fig. 6A). Very little specific L-PGDS binding to the GST-Arr3 201–409 construct was observed, close to what was observed for GST alone, throughout the different experiments that were performed. Thus, we decided to further characterize the Arr3 1–201 domain involved in L-PGDS binding by generating truncated GST mutants of this N-terminal region of Arr3 (Fig. 6B) that were used in the L-PGDS pulldown assays. Immunoblotting of the binding reactions revealed that only the region of Arr3 composed of amino acids 56–100 was able to bind to L-PGDS (Fig. 6B), suggesting that this domain is the major L-PGDS-interacting site on Arr3.
FIGURE 6.
Identification of the Arr3 domain involved in L-PGDS binding. The binding assays were carried out using purified glutathione-Sepharose-bound GST-truncated mutants of the full-length (A) or the N-terminal region (B) of Arr3 (represented schematically), which were incubated with purified recombinant His6-L-PGDS. The binding of L-PGDS was detected by immunoblotting (IB) using a His6-specific monoclonal antibody, and the GST fusion proteins present in the binding reaction were detected using a GST-specific polyclonal antibody as described under “Experimental Procedures.” Blots shown are representative of three independent experiments. C, purified His6-L-PGDS was incubated with pRSETa flow-through (control, 100%) or equivalent molar ratio of purified His6-Arr3 or His6-Arr3 amino acids 56–100, and PGD2 production was stimulated by the addition of 0.5 μm PGH2 (L-PGDS substrate) for 1 min. Reactions were stopped with 0.4 mg/ml SnCl2, and PGD2 was then measured with commercial enzyme-linked immunoassays as described under “Experimental Procedures.” All values are the mean ± S.E. from at least three separate experiments. *, p < 0.05.
Arr3 Peptides Increase PGD2 Production by L-PGDS
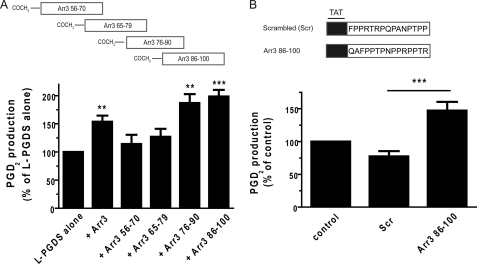
We then assessed whether amino acids 56–100 of Arr3 were sufficient to enhance PGD2 production by L-PGDS in vitro. L-PGDS-mediated PGD2 production was measured in the presence of purified His6-Arr3 or -Arr3 amino acids 56–100 proteins as described above. In these experiments, the addition of full-length Arr3 increased levels of PGD2 measured by 115%, in comparison with L-PGDS alone (Fig. 6C). Interestingly, incubation of L-PGDS with Arr3 amino acids 56–100 enhanced PGD2 production by 145%, demonstrating that amino acids 56–100 of Arr3 are sufficient to modulate PGD2 production by L-PGDS in vitro.
As schematically represented in Fig. 7A, 4 overlapping peptides of 15 amino acids corresponding to the region composed of residues 56–100 of Arr3 were synthesized. Their effects on L-PGDS-mediated PGD2 production was measured in vitro as above. Peptides Arr3 76–90 and Arr3 86–100 increased PGD2 production by L-PGDS by roughly 100% in vitro, whereas the effects of the Arr3 amino acids 56–70 and Arr3 amino acids 65–79 peptides were less significant (Fig. 7A). The Arr3 86–100 peptide and its scrambled sequence were then synthesized in fusion with the YGRKKRRQRRRGGG transduction peptide of the HIV TAT protein (Fig. 7B) to facilitate peptide entry into MG-63 cells. The cells were preincubated with the TAT peptides for 1 h, and PGD2 production was measured 15 min after the addition of PGH2. Quite interestingly, PGD2 production was roughly twice as high in MG-63 cells treated with the TAT-Arr3 86–100 peptide compared to cells treated with the control TAT-scrambled peptide (Fig. 7B).
FIGURE 7.
Arr3 peptides derived from the L-PGDS binding domain enhance PGD2 production by L-PGDS. A, shown is a schematic representation of peptides synthesized and used in PGD2 production in vitro assays. His6-L-PGDS was incubated with equivalent molar ratio of pRSETa, purified His6-Arr3, or indicated synthetic peptides, and PGD2 production was stimulated by the addition of 0.5 μm PGH2 (L-PGDS substrate) for 1 min. Reactions were stopped with 0.4 mg/ml SnCl2, and PGD2 was then measured with commercial enzyme-linked immunoassays. B, shown is a schematic representation of TAT peptides synthesized and used in PGD2 production in cells. MG-63 cells were preincubated with 100 μm TAT-Scrambled (Scr) or TAT-Arr3 amino acid 86–100 peptides for 1 h and with 100 μm HQL-79 (H-PGDS inhibitor, to specifically measure L-PGDS activity) for 15 min. Cells were then incubated with 5 μm PGH2 (L-PGDS substrate) for 15 min. Supernatants were assessed for PGD2 production by commercial enzyme-linked immunoassays as described under “Experimental Procedures.” All values are the mean ± S.E. from at least three separate experiments. **, p < 0.05; ***, p < 0.001.
DISCUSSION
Prostaglandin synthases recently garnered much interest because of their therapeutic potential, a fact that was reinforced by the clinical problems associated with the long term use of COX inhibitors. However, little is known about the regulation of these enzymes and their interacting partners. In the present study we have demonstrated that one of the two synthases that produce PGD2, L-PGDS, interacts with the scaffold protein Arr3.
A direct interaction between L-PGDS and Arr3 was suggested by our yeast two-hybrid screen and confirmed by the pulldown assays using purified recombinant proteins. Co-immunoprecipitation studies in HEK293 cells, which do not express COX enzymes (42), showed that L-PGDS and Arr3 interacted weakly in basal conditions. This interaction was, however, robustly promoted by the addition of the L-PGDS substrate, PGH2. Corroborating these results, confocal microscopy revealed weak co-localization between L-PGDS and Arr3 under basal conditions in HEK293 cells, but this co-localization was strongly increased in the perinuclear region after treatment with PGH2. Expression of COX-2 in HEK293 cells did not affect the intracellular distribution of L-PGDS. However, co-expression of Arr3 with COX-2 and L-PGDS resulted in localization of L-PGDS in the perinuclear region. Furthermore, confocal immunofluorescence microscopy and fractionation experiments showed that L-PGDS was more abundant in the cytoplasm of MEFs Arr double KO cells than in wild-type MEFs. Importantly, co-immunoprecipitation and co-localization between the two endogenous proteins was demonstrated in PGD2-producing MG-63 cells. Surprisingly, Arr3 demonstrated strong immunofluorescence staining in the nuclear region where it co-localized with L-PGDS in these cells. Inhibition of PGDS activity with SeCl4 (38, 39) resulted in a significant redistribution of Arr3 to the cytoplasm, whereas L-PGDS redistribution was subtle but still more diffused than in untreated cells. Taken altogether, our results suggest that 1) arrestin is involved in localizing L-PGDS in the perinuclear region when PGH2 is present, either when added exogenously or when produced by COX-2, and 2) co-localization between L-PGDS and Arr3 in the perinuclear region appears to be regulated by L-PGDS activity.
L-PGDS has been reported to localize to the rough endoplasmic reticulum, the outer nuclear membrane, the Golgi apparatus, lysosomes, multivesicular bodies, and endocytic vesicles of various cell types (43, 44). Arr3 is known to be distributed in the cytoplasm and associated to plasma and endosomal membranes. Arr3 is predominantly found out of the nucleus due to the presence of its two-leucine nuclear export signal (45). However, Neuhaus et al. (46) reported Arr3 to be localized in the nucleus of HEK293 cells and mature spermatozoa. COX-1 is located in the endoplasmic reticulum and perinuclear membranes, whereas COX-2 resides predominantly in the perinuclear envelope (47). Our results show co-localization of L-PGDS and Arr3 at the nuclear envelope and the perinuclear region (in the endoplasmic reticulum in HEK293 cells; data not shown). The limit of resolution of our confocal microscopy does not allow us to ascertain whether or not there is Arr3- or L-PGDS-associated immunofluorescence within the nucleus. The difference in Arr3 and L-PGDS localization between HEK293 and MG-63 cells under basal conditions could be due to the presence of endogenous COX, L-PGDS, and PGH2 in MG-63 cells. Interestingly, Arr3 co-expression caused perinuclear localization of L-PGDS when COX-2 was expressed in HEK293 cells. We propose that Arr3 participates in localizing L-PGDS in the vicinity of COX enzymes where it would get access to its COX-derived substrate, PGH2, resulting in greater production of PGD2.
A cellular role was demonstrated for Arr3 in L-PGDS-mediated PGD2 production in MEFs. MEFs lacking both Arr2 and Arr3 showed a ∼40% decrease in PGD2 production compared with wild-type MEFs after the addition of PGH2. Similarly, stimulation with the pro-inflammatory cytokine IL-1β resulted in ∼50% less PGD2 produced in MEF arrestin double KO cells than in wild-type MEFs. Transfection of Arr2 or Arr3 in MEF arrestin double KO cells brought the PGD2 production back to wild-type MEFs levels in response to IL-1β treatment. These results suggest that arrestin plays a significant, but not essential role in L-PGDS-mediated PGD2 production. Its function may reside in modulating PGD2 production in response to particular stimuli. PGD2 has been reported to be pro-inflammatory but also to be involved in the resolution of inflammation when produced in high concentrations (2, 8, 10). Further work will be necessary to determine whether arrestin-mediated increase in PGD2 production participates in the development and/or resolution of IL-1β-induced inflammation. Our data with IL-1β establish a new role for arrestin in linking inflammatory cytokine cell surface receptor signaling to intracellular prostaglandin synthesis.
Our study not only indicates that Arr3 could facilitate PGD2 production by bringing L-PGDS in the vicinity of COX enzymes and, thus, of its PGH2 substrate but that it can also increase L-PGDS enzymatic activity. Experiments performed with purified proteins in vitro showed that Arr3 stimulated L-PGDS to produce PGD2 optimally at a molar ratio of 1:1. Identification of residues 56–100 of Arr3 as the binding site for L-PGDS allowed us to generate two Arr3 peptides composed of amino acids 76–90 and 86–100 that nearly doubled PGD2 production by L-PGDS in vitro. Interestingly, this region of Arr3 is similar in Arr2, which also interacts with and regulates L-PGDS but differs in visual arrestins and is located for the most part on a loop region that is easily accessible for protein-protein interactions (48). It was shown that the proline-rich region 88–96 in non-visual arrestins is partly responsible for binding to the SH3 domain of Src family kinases such as c-Src (49) and Hck (50). This raises the interesting possibility that the L-PGDS-Arr3 functional interaction could be regulated by members of the Src family kinases and associated cell signaling pathways.
We were enthused when L-PGDS-mediated PGD2 production in MG-63 cells was increased almost 2-fold by the TAT-Arr3 86–100 fusion peptide compared with the control TAT-Arr3 scrambled peptide. One goal of our laboratory is to characterize the interacting partners of prostaglandin synthases to further our understanding of these critical enzymes but also to identify the interacting domains to derive peptides that could potentially modulate prostaglandin production. The data presented here show that this approach was successful for L-PGDS and should be applicable to other prostaglandin synthases. Increasing PGD2 levels could be beneficial in a number of conditions. For example, PGD2 is an endogenous sleep-promoting substance (51), and administration of selenium compounds markedly suppresses sleep (38, 52). Higher levels of PGD2 could help to resolve insomnia (53) and inflammation (54). The importance of PGD2 in the induction and maintenance of remission from ulcerative colitis was recently highlighted (10). Its best-studied metabolite, 15d-PGJ2, stimulates peroxisome proliferator-activated receptor-γ, which in turn mediates anti-inflammatory effects (55–59). Our group also demonstrated that PGD2 has a positive influence on bone anabolism (11, 12). Thus, modulating the interaction between arrestins and L-PGDS could turn out to have great therapeutic value toward inflammatory and other diseases.
In conclusion, we report an L-PGDS-Arr3 interaction that modulates PGD2 production. This is the first characterization of an interacting partner for L-PGDS and the identification of a new function for non-visual arrestins. Our findings also indicate that detailed analysis of the binding domains between a prostaglandin synthase and an interacting partner can lead to the generation of modulators of prostaglandin synthesis.
Supplementary Material
Acknowledgment
We thank Dr. R. J. Lefkowitz for kindly providing MEF cells.
This work was supported by a grant from Canadian Institutes of Health Research (to A. d. B. F. and J.-L. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PG
- prostaglandin
- Arr
- arrestin
- L-PGDS
- lipocalin-type prostaglandin D synthase
- H-PGDS
- hematopoietic prostaglandin D synthase
- MEF
- mouse embryonic fibroblast
- TAT
- transactivator of transcription
- dKO
- double knockout.
REFERENCES
- 1. Giles H., Leff P. (1988) Prostaglandins 35, 277–300 [DOI] [PubMed] [Google Scholar]
- 2. Ito S., Narumiya S., Hayaishi O. (1989) Prostaglandins Leukot. Essent. Fatty Acids 37, 219–234 [DOI] [PubMed] [Google Scholar]
- 3. Hardy C. C., Robinson C., Tattersfield A. E., Holgate S. T. (1984) N. Engl. J. Med. 311, 209–213 [DOI] [PubMed] [Google Scholar]
- 4. Eguchi N., Minami T., Shirafuji N., Kanaoka Y., Tanaka T., Nagata A., Yoshida N., Urade Y., Ito S., Hayaishi O. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 726–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueno R., Honda K., Inoué S., Hayaishi O. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1735–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matsuoka T., Hirata M., Tanaka H., Takahashi Y., Murata T., Kabashima K., Sugimoto Y., Kobayashi T., Ushikubi F., Aze Y., Eguchi N., Urade Y., Yoshida N., Kimura K., Mizoguchi A., Honda Y., Nagai H., Narumiya S. (2000) Science 287, 2013–2017 [DOI] [PubMed] [Google Scholar]
- 7. Ishizuka T., Matsui T., Okamoto Y., Ohta A., Shichijo M. (2004) Cardiovasc. Drug Rev. 22, 71–90 [DOI] [PubMed] [Google Scholar]
- 8. Gilroy D. W., Colville-Nash P. R., Willis D., Chivers J., Paul-Clark M. J., Willoughby D. A. (1999) Nat. Med. 5, 698–701 [DOI] [PubMed] [Google Scholar]
- 9. Ianaro A., Ialenti A., Maffia P., Pisano B., Di Rosa M. (2001) FEBS Lett. 508, 61–66 [DOI] [PubMed] [Google Scholar]
- 10. Vong L., Ferraz J. G., Panaccione R., Beck P. L., Wallace J. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12023–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallant M. A., Samadfam R., Hackett J. A., Antoniou J., Parent J. L., de Brum-Fernandes A. J. (2005) J. Bone Miner. Res. 20, 672–681 [DOI] [PubMed] [Google Scholar]
- 12. Gallant M. A., Chamoux E., Bisson M., Wolsen C., Parent J. L., Roux S., de Brum-Fernandes A. J. (2010) J. Rheumatol. 37, 644–649 [DOI] [PubMed] [Google Scholar]
- 13. Urade Y., Eguchi N. (2002) Prostaglandins Other Lipid Mediat. 68–69, 375–382 [DOI] [PubMed] [Google Scholar]
- 14. Urade Y., Ujihara M., Horiguchi Y., Igarashi M., Nagata A., Ikai K., Hayaishi O. (1990) J. Biol. Chem. 265, 371–375 [PubMed] [Google Scholar]
- 15. Fujimori K., Kanaoka Y., Sakaguchi Y., Urade Y. (2000) J. Biol. Chem. 275, 40511–40516 [DOI] [PubMed] [Google Scholar]
- 16. Tanaka K., Ogawa K., Sugamura K., Nakamura M., Takano S., Nagata K. (2000) J. Immunol. 164, 2277–2280 [DOI] [PubMed] [Google Scholar]
- 17. Urade Y., Fujimoto N., Hayaishi O. (1985) J. Biol. Chem. 260, 12410–12415 [PubMed] [Google Scholar]
- 18. Blödorn B., Mäder M., Urade Y., Hayaishi O., Felgenhauer K., Brück W. (1996) Neurosci. Lett. 209, 117–120 [DOI] [PubMed] [Google Scholar]
- 19. Eguchi Y., Eguchi N., Oda H., Seiki K., Kijima Y., Matsu-ura Y., Urade Y., Hayaishi O. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14689–14694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beuckmann C. T., Gordon W. C., Kanaoka Y., Eguchi N., Marcheselli V. L., Gerashchenko D. Y., Urade Y., Hayaishi O., Bazan N. G. (1996) J. Neurosci. 16, 6119–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerena R. L., Eguchi N., Urade Y., Killian G. J. (2000) J. Androl. 21, 848–854 [PubMed] [Google Scholar]
- 22. Tanaka T., Urade Y., Kimura H., Eguchi N., Nishikawa A., Hayaishi O. (1997) J. Biol. Chem. 272, 15789–15795 [DOI] [PubMed] [Google Scholar]
- 23. Beuckmann C. T., Aoyagi M., Okazaki I., Hiroike T., Toh H., Hayaishi O., Urade Y. (1999) Biochemistry 38, 8006–8013 [DOI] [PubMed] [Google Scholar]
- 24. Mohri I., Taniike M., Okazaki I., Kagitani-Shimono K., Aritake K., Kanekiyo T., Yagi T., Takikita S., Kim H. S., Urade Y., Suzuki K. (2006) J. Neurochem. 97, 641–651 [DOI] [PubMed] [Google Scholar]
- 25. Lefkowitz R. J., Whalen E. J. (2004) Curr. Opin. Cell Biol. 16, 162–168 [DOI] [PubMed] [Google Scholar]
- 26. Gurevich V. V., Gurevich E. V. (2004) Trends Pharmacol. Sci. 25, 105–111 [DOI] [PubMed] [Google Scholar]
- 27. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 28. Song X., Coffa S., Fu H., Gurevich V. V. (2009) J. Biol. Chem. 284, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pierroz D. D., Rufo A., Bianchi E. N., Glatt V., Capulli M., Rucci N., Cavat F., Rizzoli R., Teti A., Bouxsein M. L., Ferrari S. L. (2009) J. Bone Miner. Res. 24, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y., Tang Y., Teng L., Wu Y., Zhao X., Pei G. (2006) Nat. Immunol. 7, 139–147 [DOI] [PubMed] [Google Scholar]
- 31. Gietz R. D., Woods R. A. (2002) Methods Mol. Biol. 185, 471–486 [DOI] [PubMed] [Google Scholar]
- 32. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 33. Inui T., Ohkubo T., Emi M., Irikura D., Hayaishi O., Urade Y. (2003) J. Biol. Chem. 278, 2845–2852 [DOI] [PubMed] [Google Scholar]
- 34. Aritake K., Kado Y., Inoue T., Miyano M., Urade Y. (2006) J. Biol. Chem. 281, 15277–15286 [DOI] [PubMed] [Google Scholar]
- 35. Farrell C. J., Lee J. M., Shin E. C., Cebrat M., Cole P. A., Hayward S. D. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4625–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amirand C., Viari A., Ballini J. P., Rezaei H., Beaujean N., Jullien D., Käs E., Debey P. (1998) J. Cell Sci. 111, 3551–3561 [DOI] [PubMed] [Google Scholar]
- 37. Li L., Pettit A. R., Gregory L. S., Forwood M. R. (2006) Cytokine Growth Factor Rev. 17, 203–216 [DOI] [PubMed] [Google Scholar]
- 38. Matsumura H., Takahata R., Hayaishi O. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9046–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qu W. M., Huang Z. L., Xu X. H., Aritake K., Eguchi N., Nambu F., Narumiya S., Urade Y., Hayaishi O. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17949–17954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mifflin R. C., Saada J. I., Di Mari J. F., Adegboyega P. A., Valentich J. D., Powell D. W. (2002) Am. J. Physiol. Cell. Physiol. 282, C824–34 [DOI] [PubMed] [Google Scholar]
- 41. Molina-Holgado E., Ortiz S., Molina-Holgado F., Guaza C. (2000) Br. J. Pharmacol. 131, 152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murakami M., Kambe T., Shimbara S., Yamamoto S., Kuwata H., Kudo I. (1999) J. Biol. Chem. 274, 29927–29936 [DOI] [PubMed] [Google Scholar]
- 43. Nagasaka T., Hiraide M., Sugimoto T., Shindo K., Shiozawa Z., Yokota S. (2004) Histochem. Cell Biol. 121, 483–491 [DOI] [PubMed] [Google Scholar]
- 44. Urade Y., Fujimoto N., Kaneko T., Konishi A., Mizuno N., Hayaishi O. (1987) J. Biol. Chem. 262, 15132–15136 [PubMed] [Google Scholar]
- 45. Scott M. G., Le Rouzic E., Périanin A., Pierotti V., Enslen H., Benichou S., Marullo S., Benmerah A. (2002) J. Biol. Chem. 277, 37693–37701 [DOI] [PubMed] [Google Scholar]
- 46. Neuhaus E. M., Mashukova A., Barbour J., Wolters D., Hatt H. (2006) J. Cell Sci. 119, 3047–3056 [DOI] [PubMed] [Google Scholar]
- 47. Morita I., Schindler M., Regier M. K., Otto J. C., Hori T., DeWitt D. L., Smith W. L. (1995) J. Biol. Chem. 270, 10902–10908 [DOI] [PubMed] [Google Scholar]
- 48. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 49. Luttrell L. M., Ferguson S. S., Daaka Y., Miller W. E., Maudsley S., Della Rocca G. J., Lin F., Kawakatsu H., Owada K., Luttrell D. K., Caron M. G., Lefkowitz R. J. (1999) Science 283, 655–661 [DOI] [PubMed] [Google Scholar]
- 50. Barlic J., Andrews J. D., Kelvin A. A., Bosinger S. E., DeVries M. E., Xu L., Dobransky T., Feldman R. D., Ferguson S. S., Kelvin D. J. (2000) Nat. Immunol. 1, 227–233 [DOI] [PubMed] [Google Scholar]
- 51. Ram A., Pandey H. P., Matsumura H., Kasahara-Orita K., Nakajima T., Takahata R., Satoh S., Terao A., Hayaishi O. (1997) Brain Res. 751, 81–89 [DOI] [PubMed] [Google Scholar]
- 52. Takahata R., Matsumura H., Kantha S. S., Kubo E., Kawase K., Sakai T., Hayaishi O. (1993) Brain Res. 623, 65–71 [DOI] [PubMed] [Google Scholar]
- 53. Urade Y., Hayaishi O. (2000) Vitam. Horm. 58, 89–120 [DOI] [PubMed] [Google Scholar]
- 54. Gilroy D. W., Colville-Nash P. R., McMaster S., Sawatzky D. A., Willoughby D. A., Lawrence T. (2003) FASEB J. 17, 2269–2271 [DOI] [PubMed] [Google Scholar]
- 55. Bell-Parikh L. C., Ide T., Lawson J. A., McNamara P., Reilly M., FitzGerald G. A. (2003) J. Clin. Invest. 112, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bishop-Bailey D., Hla T. (1999) J. Biol. Chem. 274, 17042–17048 [DOI] [PubMed] [Google Scholar]
- 57. Ide T., Egan K., Bell-Parikh L. C., FitzGerald G. A. (2003) Thromb. Res. 110, 311–315 [DOI] [PubMed] [Google Scholar]
- 58. Jiang C., Ting A. T., Seed B. (1998) Nature 391, 82–86 [DOI] [PubMed] [Google Scholar]
- 59. Ricote M., Li A. C., Willson T. M., Kelly C. J., Glass C. K. (1998) Nature 391, 79–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.