Abstract
The zymogen activation mechanism and physiological functions of the most ancient and highly conserved basic amino acid-specific proprotein convertase 7 (PC7) are not known. Herein, we characterized the biosynthesis, subcellular localization, and trafficking of the membrane-bound full-length rat and human PC7. The prosegment of PC7 is primarily secreted alone as a non-inhibitory protein via the conventional, Golgi-dependent, secretory pathway. Mature PC7 is partially sulfated and thus reaches the cell surface via the conventional route. However, a fraction of PC7 reaches the cell surface through a brefeldin A- and COPII-independent unconventional secretory pathway. The latter trafficking may explain the rapid (<10 min) transit of a fraction of PC7 from the ER to the cell surface. Electron microscopy further confirmed the localization of PC7 to the cell surface of HEK293 cells. Within the cytosolic tail, only two cysteines (Cys699 and Cys704) are palmitoylated, but this modification does not affect the choice of trafficking pathway. Swapping the transmembrane-cytosolic tail (TMCT) sequences of the convertases Furin and PC7 revealed that PC7TMCT-Furin is much more sulfated and hence traffics more efficiently through the conventional secretory pathway. In contrast, the FurinTMCT-PC7 is no longer sulfated and thus reaches the cell surface by the unconventional pathway. Because trafficking of PC7CT-Furin and FurinCT-PC7 resemble their wild type counterparts, we deduce that the transmembrane domain of PC7 regulates the sorting of PC7 toward the unconventional secretory pathway. In conclusion, PC7 is distinct from other proprotein convertases in its zymogen activation, subcellular localization, and trafficking.
Keywords: Cell Surface Protein, Protease, Protein Palmitoylation, Protein Processing, Trafficking, Endoproteolysis, Furin, Proprotein Convertase PC7, Unconventional Secretory Pathway, Zymogen Activation
Introduction
Mammalian genomes encode nine secretory proprotein convertases (PCs)2 related to bacterial subtilisin and yeast kexin (1, 2). Seven of them, PC1/3, PC2, Furin, PC4, PC5/6, PACE4, and PC7, cleave precursors at either single or pairs of basic amino acids (aa) within the motif Arg/Lys-[X]n-Arg↓ (n = 2 or 4 aa), and the last two convertases, SKI-1/S1P and PCSK9, cleave at nonbasic sites. The basic aa-specific PCs are involved in the processing of multiple protein precursors, including polypeptide hormones, proteases, receptors, and growth factors (3).
Whereas the physiological functions of most PCs are now better understood (2, 4), the unique functional roles of PC7, the most ancestral and conserved mammalian member of the family of basic aa-specific convertases (5, 6), are barely explored. Northern blot analyses revealed a wide expression of PC7 mRNA in all rat tissues and cell lines analyzed (5). Quantitative real time-PCR (qPCR) analysis of PC7 expression in adult mouse tissues showed that colon, kidney, duodenum, and heart are the richest sources of PC7 mRNA (supplemental Fig. S1). These data suggest that PC7 may have multiple physiological functions, some of which may be redundant with other convertases.
Biosynthetic analyses of rat PC7 (r-PC7) or human PC7 (h-PC7) revealed that the protease is first synthesized as a proPC7 zymogen, which within the endoplasmic reticulum (ER) rapidly undergoes an autocatalytic cleavage at KRAKR140↓ (rat) (5) or RRAKR141↓ (human) (7). PC7 also undergoes a number of post-translational modifications, including N-glycosylation (5) and cytosolic tail Cys-palmitoylation (8). However, the zymogen activation mechanism is still unknown, as this requires exit of the prosegment-PC7 complex from the ER and/or separation of the inhibitory prosegment from mature PC7. Mature PC7 is the active protease that cleaves substrates in trans at basic aa.
A number of investigations aimed at defining the sequence recognition of PC7 and its redundancy with other convertases, suggested that although less efficient than Furin, PC7 specifically cleaves overexpressed substrates at Arg↓ residues both in vitro (9–16) and in cell lines (17–26). Thus, although Furin and PC7 have been proposed as the major gp160 processing convertases, rat liver microsomal gp160 processing activity was essentially resolved from Furin and only partially overlapped with PC7. Density gradient studies revealed that PC7 resides in lighter subcellular fractions than Furin (27). Interestingly, whereas overexpression of the prosegments of Furin, PC5, and PC7 resulted in potent inhibitors of substrate cellular processing (22, 28), only the prosegment of PC7 is secreted into the medium (10, 22, 29). Finally, the C-terminal KRAKR140 motif in the prosegment of r-PC7 is critical for its convertase inhibitory activity (30). Altogether, these data point out the particularities of PC7 in its zymogen activation and subcellular localization.
We herein characterize the zymogen activation, subcellular localization and secretory pathways of PC7. Our data show that the active convertase reaches the cell surface by a conventional, but also by an unconventional secretory pathway, while the prosegment traffics through the regular Golgi-dependent route and is secreted alone. Our data also suggest that the transmembrane domain of PC7, but not that of Furin, contains critical elements controlling its trafficking through the unconventional pathway.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
r-PC7, h-PC7, rat, or human soluble PC7 (r-sPC7; h-sPC7), human Furin, and human low density lipoprotein receptor (LDLR)-V5 were cloned in pIRES-2-EGFP vector as previously described (10, 18, 31). PC7 and Furin mutant cDNAs were generated and cloned into pIRES2-EGFP (Clontech). All of the oligonucleotides used in the various PC7 and Furin constructions are listed in supplemental Table S1. The cDNA of Sar1p-(H79G), kindly provided by Dr Y. Wang, was subcloned by digestion into pIRES2-EGFP vector. GFP-Flotillin-1 was kindly provided by Dr. B. Nichols. Brefeldin A (BFA) was purchased from Calbiochem.
Quantitative Real-time PCR
qPCR of RNA was performed as previously described (32). Briefly, each cDNA sample was submitted to two PCR amplifications each performed in triplicate: one for the control ribosomal S16 gene used for normalizing, and another to measure mouse PC7 mRNA levels. The oligonucleotides used for both genes are listed in supplemental Table S2. The Mx3500P system from Stratagene was used to perform and analyze the qPCRs.
Cell Culture and Transfections
HEK293 and COS-1 cells (American Type Culture Collection) were routinely cultivated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and were maintained at 37 °C under 5% CO2. At 80–90% confluence, HEK293 cells were transfected with a total of 0.6 μg of cDNAs with Effectene (Qiagen) and COS-1 cells with a total of 4 μg of cDNAs in Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, the cells were washed and incubated in serum-free medium for an additional 16 h before cell lysis.
Immunoprecipitation and Western Blot Analyses
Cells were lysed in ice-cold precipitation assay buffer (50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing a mixture of protease inhibitors (Roche Applied Science). Proteins were analyzed by 8% SDS-PAGE. For immunoprecipitation, cell lysates were incubated overnight at 4 °C with streptavidin (SA)-agarose (Fluka) and washed 5× with cold lysis buffer. Following addition of reducing Laemmli sample, solubilized proteins were separated by 8% SDS-PAGE. Proteins were visualized using primary antibodies (Ab): Ab:PC7, which recognizes both the prosegment and mature PC7 (1:10,000, 22), anti-Furin Ab (1:2000, MON148, Alexis), or anti-LDLR Ab (1:1000 R&D Systems). Bound primary antibodies were detected with corresponding species-specific HRP-labeled secondary antibodies and revealed by enhanced chemiluminescence. Quantitation of band intensity was done with Scion image software from the Scion Corporation (Frederick, MD). All experiments were performed at least in triplicate. Quantitations are normalized to PC7 without treatment or co-transfected with an empty vector (pIRES) and considered as 100%.
Biosynthetic Analyses and Immunoprecipitation
HEK293 or COS-1 cells were transiently transfected in 60-mm dishes as described above. Biosynthesis were performed 2 days post-transfection and the cells were washed and pulsed-labeled in Cys/Met-free or Leu-free RPMI 1640 medium containing 0.2% BSA for either (PerkinElmer Life Sciences) either 2 h with 250 μCi/ml of [35S]Cys/Met, 4 h with 500 μCi/ml of [3H]Leu for the radiolabeled of the prosegment of h-PC7, 2 h with 500 μCi/ml of Na235SO4 for sulfation of PC7, Furin and their chimeras, or 2 h with 700 μCi/ml of [3H]palmitate for the palmitoylation of PC7. After the pulse, the media were recovered, and the cells lysed as mentioned previously (33). The cells lysates were immunoprecipitated with Ab:PC7, a prosegment-specific antibody (Ab:pPC7) (22) or anti-Furin Ab. The immunoprecipitated proteins were resolved by SDS-PAGE on 8% Tricine gels, dried and autoradiographed, as described (33). In some cases, during the 30 min pre-incubation times for pulse analysis, the cells were pretreated with BFA (2.5 μg/ml), and this treatment was continued during the 2 h pulse period.
Enzymatic Digestion of Carbohydrates
Proteins from scratched cells were lysed in ice cold precipitation assay buffer (see above) and then incubated with endoglycosidase H (endoH) or N-glycosidase F (PGNase F) for 1h at 37 °C (NEB). Deglycosylated proteins were separated by 8% SDS-PAGE and revealed by immunoblotting. To identify the nature of PC7 glycosylation, HEK293 cells were pulsed-labeled with Na235SO4 or [35S]Cys/Met. Cells lysates in assay buffer were immunoprecipitated with Ab:PC7, eluted and denatured with 1× glycoprotein denaturing buffer (NEB) at 100 °C for 10 min. Immunoprecipitates were incubated overnight with PGNase F at 37 °C, boiled 10 min in Tricine sample buffer + β-mercaptoethanol (Bio-Rad) and resolved by SDS-PAGE on 8% Tricine gels, dried, and autoradiographed.
Cell Surface Biotinylation
For biochemical detection of cell surface PC7 by Western blot analysis, Furin or LDLR at the plasma membrane, HEK293 or COS-1 cells were transiently transfected with r-PC7, h-PC7, Furin, their chimeras, or LDLR. Cells were washed with ice-cold phosphate-buffered saline (PBS) adjusted to pH 8.0, and biotinylated with 0.2 mg of sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-(biotin-amido) hexanoate) (Pierce) for 30 min at 4 °C. After washing with 100 mm glycine in PBS, pH 8.0, to quench the reaction, cell lysates are immunoprecipitated with SA agarose (Fluka), and immunoprecipitates are then treated as described above. For the detection of cell surface PC7 by biosynthesis analysis, cells were biotinylated with 1 mg of sulfo-NHS-LC-(sulfosuccinimidyl-6-(biotin-amido) hexanoate) (Pierce) and immunoprecipitated with Ab:PC7. After elution in Tricine sample buffer (Bio-Rad), 30 μl of immunoprecipitates were incubated with 500 μl of DMEM + 0.1% BSA, 500 μl of precipitation assay buffer, and 50 μl of neutravidin (NA)-agarose (Pierce). The immunoprecipitates were resolved by SDS-PAGE on 8% Tricine gels, dried, and autoradiographed. Note that a fraction (1/3) of the lysates was kept before NA precipitation in order to normalize the quantity of cell surface PC7 to that of total PC7.
Immunofluorescence
HEK293 cells were plated on collagen-coated culture dishes (MatTek) whereas COS-1 cells were plated on uncoated culture dishes. The following day, cells were transfected. Twenty-four hours post-transfection, the cells were fixed 15 min with 4% paraformaldehyde for PC7 cell surface labeling or 5 min at −20 °C with 100% methanol for PC7 intracellular staining. Cell fixed with paraformaldehyde were incubated 5 min in 150 mm glycine to stabilize the aldehydes. Cells are then incubated overnight at 4 °C with primary antibodies: Ab:PC7 (1:1000), anti-LDLR Ab (1:100 R&D Systems), anti-calnexin Ab (1:500 Abcam), anti-calreticulin Ab (1:250 Abcam), anti-Golgin 97 Ab (1:500, Santa Cruz Biotechnology), anti-EEA1 Ab (1:100, Abcam), anti-mannose 6-phosphate Ab (mannose-6P, 1/500, Abcam), and anti-LAMP-1 Ab (1/30, DSHB). Antigen antibody complexes were revealed by 1-h incubation with corresponding species-specific Alexa fluor (488, 555, or 647)-tagged antibodies (Molecular Probes), and the nuclei were stained by Hoescht 33258 for 1 min (100 μg/ml, Sigma-Aldrich). Cells were covered with DABCO (Sigma-Aldrich) in PBS/90% glycerol, and immunofluorescence analyses were performed with a confocal microscope (Zeiss LSM-710).
Electron Microscopy
HEK293 cells in 100 mm dishes were transiently transfected with 1 μg h-PC7 or pIRES-2-EGFP cDNA using Effectene (Qiagen). Twenty-four hours post-transfection, the cells were collected in 1 ml of PBS and pelleted by centrifugation for 5 min at 1,200 rpm. The cell pellet was then fixed in 5% paraformaldehyde + 0.5% gluteraldehyde in 0.1 m PBS for 15 min at 4 °C. The pellet was centrifuged again and then stored for 1 h at 4 °C in fixative. Immunolabeling of cryosections was done according to the method of Tokuyasu et al. (34) at the Facility for Electron Microscopy Research at McGill University. Primary antibodies were diluted to 1:10 in the case of PC7 and 1:5 for Golgin 97, calreticulin, and calnexin. PC7 is labeled with the Ab:PC7 conjugated to 10 nm colloidal gold particles and cell compartment markers are visualized by secondary antibodies conjugated to 18 nm colloidal gold particles. Analyses were performed with a Tecnai 12 transmission electron microscope (FEI Company).
Reverse Transcription PCR (RT-PCR) Analysis of Xbp-1 Splicing
HEK293 cells were lysed, and total RNA was collected (TRIzol®, Invitrogen), as recommended by the manufacturer. Typically, 250 ng of total RNA were used for cDNA synthesis in a total volume of 20 μl using SuperScript II reverse transcriptase, 25 μg/ml oligo(dT)12–18, 0.5 mm 2′-deoxynucleoside 5′-triphosphates, and 40 units of RNaseOUT, all products from Invitrogen, and used according to the recommendations of the manufacturer. Primers used to amplify the Xbp-1 cDNA bearing the intron target of IRE1α ribonuclease activity and PCR conditions were previously described (35). A 289-bp amplicon was generated from unspliced Xbp-1; a 263-bp amplicon was generated from spliced Xbp-1. 4-h treatment with 5 μg/ml tunicamycin was used as control for ER stress.
RESULTS
Zymogen Activation and Biosynthesis of PC7
In an effort to understand the physiological functions of PC7 and because it is highly expressed in adult kidney (supplemental Fig. S1), we first characterized its zymogen processing in kidney-derived HEK293. Accordingly, HEK293 cells overexpressing r-PC7 or h-PC7 were labeled for 4 h with [3H]Leu (Fig. 1). Cell lysates and media were then immunoprecipitated with a polyclonal antibody, which recognizes both the prosegment and mature PC7 (Ab:PC7), or a prosegment-specific antibody (Ab:pPC7) (22), and the proteins were resolved by SDS-PAGE. Our data show that proPC7 is processed into PC7 intracellularly, likely in the ER (Fig. 1A), as previously reported (5). The membrane-bound form of mature PC7 remains in the cell and is not shed into the medium (Fig. 1A), as originally demonstrated (5, 7). Similar results were obtained in COS-1 cells (not shown).
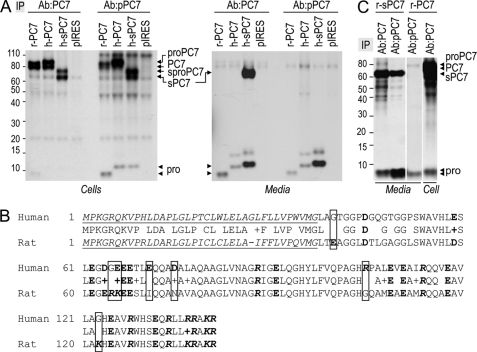
FIGURE 1.
Zymogen activation of PC7. A, autoradiographs of labeled PC7 and its prosegment in cell lysates (left panel) and media (right panel) of HEK293 cells expressing r-PC7, h-PC7, or h-sPC7 and pulse-labeled with [3H]Leu for 4 h. Proteins were immunoprecipitated with the Ab:PC7 (recognizing both the prosegment and mature PC7) or Ab:pPC7 (recognizing only the prosegment and the zymogen form of PC7) and analyzed by SDS-PAGE separation. B, alignment of the prosegment of h-PC7 and r-PC7. The signal peptide appears in italic and is underlined; bold residues emphasize negative charges, and bold and italic residues, the positive charges. Framed boxes emphasize differences in charge between the two species. C, autoradiographs of labeled PC7 and its prosegment in the media and cell lysates of HEK293 cells expressing soluble or full-length rat PC7 (r-sPC7, r-PC7) and pulse-labeled with [3H]Leu for 4 h. Proteins were immunoprecipitated with either the Ab:PC7 or Ab:pPC7 and analyzed by SDS-PAGE separation.
Interestingly, using Ab:pPC7, we also noticed that the cellular mature full-length PC7 partially co-immunoprecipitates with its prosegment, but that the latter is secreted alone from HEK293 cells (Fig. 1A). The migration position of the rat prosegment (apparent molecular mass ∼9 kDa) is smaller than the corresponding human (∼12 kDa). We believe the difference between the ∼9 and ∼12 kDa forms may be related to the overall presence of 6 net negative charges in the human prosegment versus 2 in the rat one (Fig. 1B). The human immunoreactive media protein that migrates at ∼15 kDa (Fig. 1A) is not reproducibly seen, and hence was not further investigated.
We repeated the same experiments using a soluble form of h-PC7 (h-sPC7) (10) (Fig. 1A). Using the Ab:pPC7, we noticed that the prosegment form did not co-immunoprecipitate with mature sPC7 in the media (Fig. 1A). A similar result was also obtained in COS-1 cells (not shown). Altogether, the data suggest that in the media of cells expressing human full-length PC7 or sPC7, the prosegment is not bound to the mature enzyme. Indeed, sPC7 secreted into the medium is catalytically active on many substrates (9–16).
Using the Ab:pPC7, we noted that in contrast to the h-prosegment, the r-prosegment co-immunoprecipitated with mature r-sPC7 in the media (Fig. 1C). We also noticed that using the Ab:pPC7, but not the Ab:PC7, two forms of the r-prosegment appeared in the media. The lower form appeared in the media of both full-length and r-sPC7 using the Ab:pPC7 (Fig. 1C). It may correspond to the free r-prosegment not bound to mature PC7. It seems that the Ab:pPC7 is better at detecting the latter form than the Ab:PC7. We presume that this form lost the dibasic residues LysArg141 at the C terminus of the r-prosegment, likely due to a basic carboxypeptidase trimming e.g. CPD (36). The separation of the prosegment from the mature enzyme requires the action of a basic carboxypeptidase, because the C terminus Lys-Arg141 of the prosegment is critical for its binding to the enzyme and its inhibition (30).
We therefore conclude that (1) h-prosegment dissociated more efficiently from mature PC7 than the r-prosegment; (2) contrary to other PCs, the prosegment of either species does not undergo a secondary internal cleavage; (3) the prosegment is secreted as an independent 102-aa polypeptide, which may have its own biological function.
The Subcellular Localization of PC7
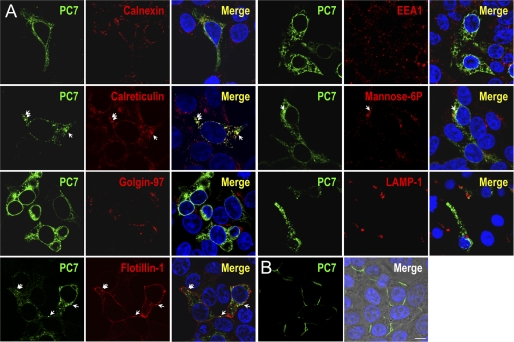
Under cell permeabilization conditions (Fig. 2A), PC7 significantly colocalizes with the chaperone calreticulin, a soluble 60 kDa KDEL-protein marker of the ER, but not with calnexin, a membrane-bound ER-resident protein. Our data suggest that, at least partially, PC7 is present in a sub-compartment of the ER co-localizing with calreticulin. Some PC7 was also found to colocalize with GFP-flotillin-1, a known marker of clathrin-independent endocytosis (37), suggesting that PC7 is internalized by clathrin-independent routes. In rare occasions, we observed a late endosomal localization together with the mannose-6-phosphate receptor (Fig. 2A). In contrast, we do not observe any colocalization with markers of the trans Golgi network (TGN; Golgin-97), early endosomes (EEA1), or lysosomes (Lamp1). Finally, under non-permeabilizing conditions, PC7 clearly localizes to the cell surface (Fig. 2B).
FIGURE 2.
Subcellular localization of PC7. A, immunofluorescence of PC7 (green labeling) on permeabilized HEK293 cells expressing r-PC7. Cell compartment markers are labeled in red. PC7 co-localizes with calreticulin in the ER, but not with calnexin, another ER marker. There is also some co-localization in clathrin-independent vesicles with GFP-Flotillin-1 and in late endosomes with the mannose-6-phosphate. In contrast, no co-localization is observed in the TGN with Golgin 97, in early endosomes (EEA1), or in lysosomes (LAMP-1). B, PC7 cell surface immunofluorescence of non-permeabilized HEK293 cells expressing r-PC7 shows that PC7 is present at the cell surface (green labeling). Cell nuclei are marked by Hoescht 33258 staining (blue labeling). Bar, 10 μm.
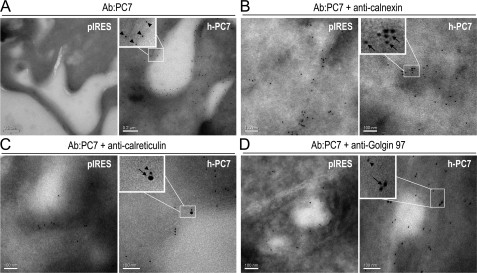
Further confirmation of the cell surface localization of h-PC7 in HEK293 cells was achieved by electron microscopy (Fig. 3A). Whereas h-PC7 was concentrated at and below the cell surface, it was also found to colocalize closely with calreticulin (Fig. 3C) and to a lesser extent with Golgin-97 (Fig. 3D). Similar to the immunofluorescence data, no co-localization was observed with calnexin (Fig. 3B). We therefore conclude that overexpressed h-PC7 mostly localizes to the ER and at the cell surface of HEK293 cells.
FIGURE 3.
Electron microscopy. Immunolabeling of cryo-sections of HEK293 cells transfected with pIRES-2-EGFP or h-PC7. PC7 is labeled with the Ab:PC7 conjugated to 10 nm colloidal gold particles (arrowheads). Cell compartment markers are visualized by secondary antibodies conjugated to 18 nm colloidal gold particles (arrows). A, PC7 labeling was abundant at and below the cell surface of cells expressing h-PC7. Co-localization was observed with calreticulin (C), but not with calnexin (B) and to a lesser extent with Golgin 97 (D). PC7 labeling was absent in cells transfected with the control empty vector (pIRES). Bars, 200 nm (A), 100 nm (B–D).
PC7 Reaches the Cell Surface via Golgi-dependent and -independent Secretory Pathways
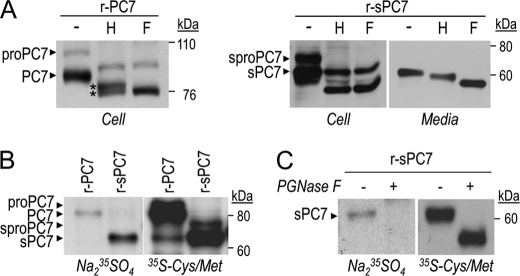
To characterize the trafficking of active PC7 and its inhibitory prosegment (30), we incubated cell extracts of HEK293 cells expressing r-PC7 with endoH or PGNase F (Fig. 4A). Western blot analysis showed that PGNase F treatment shifted the apparent molecular masses of proPC7 and PC7 from ∼97 kDa and ∼85 kDa to their predicted molecular masses of ∼88 kDa and ∼75 kDa, respectively. EndoH digestion of proPC7 resulted in a similarly sized ∼88-kDa form as observed with PGNase F (Fig. 4A), suggesting that the majority of proPC7 is in the ER. In contrast, endoH digestion of mature PC7 resulted in a protein doublet, with apparent molecular masses of ∼77 and ∼75 kDa (Fig. 4A, asterisk). This suggests that in the ∼77 kDa form one or more of the four potential N-glycosylation sites of PC7 (5) is/are endoH-resistant. This is not the case for the ∼75 kDa PC7 form, which is generated by either endoH or PGNase F. Thus, a fraction of the PC7 pool (∼75 kDa) remains completely endoH-sensitive, suggesting that it is still localized in the ER.
FIGURE 4.
PC7 reaches the cell surface by an ER/Golgi-dependent secretory pathway. A and B, post-translational modifications of PC7. Western blot analysis of PC7 in cell lysates or media from HEK293 cells expressing r-PC7 or r-sPC7. A, treatment with endoH (H) and PGNase F (F) of PC7 isolated from cell lysates and media showed that PC7 is sensitive and partially resistant to endoH treatment. Stars correspond to two PC7 forms: the endoH-sensitive one and the form partially resistant to endoH digestion. B, autoradiographs of labeled PC7 in cell lysates of HEK293 cells expressing r-PC7 or r-sPC7 and pulse-labeled with Na235SO4 and [35S]Cys/Met for 2 h demonstrate that a fraction of PC7 traffics through the conventional secretory pathway. C, r-sPC7 samples similar to those obtained in panel B were digested (+) or not (−) with PGnase F.
EndoH digestion of the secreted soluble form of PC7 (sPC7) in the media, results in a single intermediate-sized product, which is of higher molecular mass than the PGNase F-treated form (Fig. 4A, right panel). This observation agrees with the results of full-length PC7, whereby some of its N-glycosylation sites are resistant to endoH. This is the typical behavior of a protein trafficking through the conventional secretory pathway, whereby it exits the ER and reaches the cell surface via the Golgi apparatus, acquiring endoH resistance along the way. Similar results were obtained with hPC7 (not shown).
To confirm the different behaviors of PC7 and its soluble form, we investigated whether PC7 is sulfated, a post-translational modification that takes place in the Golgi apparatus (38, 39). Incubation of HEK293 cells expressing r-PC7, r-sPC7, h-PC7, or h-sPC7 with Na235SO4 for 2 h revealed that proPC7 is not sulfated, agreeing with its ER localization (Fig. 4B; see also Fig. 7C). In contrast, its mature form is sulfated to a much lower extent in full-length PC7 as compared with that of sPC7, whereas their expression in cells is similar, as shown by [35S]Cys/Met labeling (Fig. 4B). This modification seems to affect an N-glycosylation site, since it is eliminated upon treatment of the 35SO4-labeled rPC7 with PGNase F (Fig. 4C). These data suggest that a fraction of full-length PC7 reaches the TGN via the conventional secretory pathway and is sulfated at one or more glycosyl moieties.
FIGURE 7.
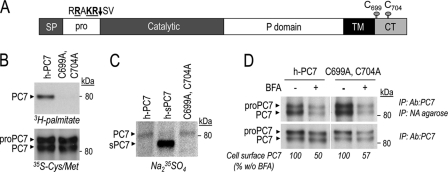
Role of PC7 palmitoylation. A, schematic diagram depicting proPC7, in which the cleavage site of the prosegment and cysteines involved in palmitoylation are emphasized. SP, signal peptide; Pro, prosegment; TM, transmembrane domain; CT, cytosolic tail. B, cells expressing wild type and mutant PC7-(C699A, C704A) were pulse-labeled with [3H]palmitic acid or [35S]Cys/Met for 2 h, immunoprecipitated with Ab:PC7, and then analyzed following SDS-PAGE separation. C, autoradiograph of labeled PC7 in lysates from HEK293 cells expressing WT PC7 or PC7-(C699A, C704A), pulse-labeled with Na235SO4 for 2 h and then immunoprecipitated with Ab:PC7. D, cell surface biotinylation of WT PC7 and PC7-(C699A, C704A) incubated or not with BFA (2.5 μg/ml). Autoradiographs of labeled PC7 in lysates from HEK293 cells expressing either WT PC7 or PC7-(C699A, C704A), pulse-labeled with [35S]Cys/Met for 2 h, biotinylated, and immunoprecipitated with Ab:PC7, eluted, and then immunoprecipitated with NA-agarose. One-third of lysates were kept before NA-agarose precipitation to normalize the quantity of cell surface PC7. The percent of cell surface PC7/total PC7, estimated by Scion image analysis and normalized to that obtained without BFA treatment (value = 100), showed that PC7 Cys-palmitoylation has no effect on PC7 trafficking. Notice that the autoradiogram of proteins immunoprecipitated with Ab:PC7 and NA was obtained after 4 days of exposure whereas that of proteins immunoprecipitated with Ab:PC7 was obtained after 2 h of exposure.
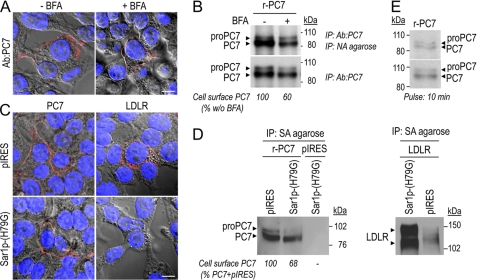
We next wished to assess whether the fraction of the mature PC7 pool, which is sensitive to endoH, remains in the ER or can exit this compartment and reach the cell surface by an unconventional secretory pathway, bypassing the TGN (40). Accordingly, HEK293 cells expressing PC7 were treated with BFA, which inhibits the transport of proteins from the ER to the Golgi (41). Under non-permeabilizing conditions, immunostaining of transfected HEK293 cells (Fig. 5A), or COS-1 cells (supplemental Fig. S2) revealed that 6 h treatment of cells with BFA does not prevent the cell surface localization of PC7. However, the same BFA treatment prevented the cell surface localization of the LDLR (supplemental Fig. S2), which traffics through the TGN pathway (42). Cell surface biotinylation of newly synthesized [35S]Cys/Met-labeled PC7 confirmed that a significant portion of PC7 (50–60% of untreated cells) can reach the cell surface in the presence of BFA (Fig. 5B). Similar results were obtained with human PC7 (Fig. 7D). These data suggest that a significant fraction of PC7 traffics to the cell surface via an unconventional route.
FIGURE 5.
PC7 reaches the cell surface by a non-conventional secretory pathway through a BFA- and COPII-coated vesicles independent way. A, immunostaining of non-permeabilized HEK293 cells expressing r-PC7 (red labeling) treated (+) or no (−) with BFA (5 μg/ml) for 6 h. Nuclei of transfected cells are marked by Hoescht 33258 staining (blue labeling). Bar, 10 μm. B, cell surface biotinylation of PC7. Autoradiograph of labeled PC7 in lysates of transfected HEK293 cells, incubated or not with BFA (2.5 μg/ml) and pulse-labeled with [35S]Cys/Met for 2 h (B) or 10 min (E). Cells were biotinylated, immunoprecipitated with the Ab:PC7, eluted and then immunoprecipitated with NA-agarose. One-third of lysates were kept before NA-agarose precipitation to normalize the quantity of cell surface PC7. The percent of cell surface PC7/total PC7, estimated by Scion image analysis and normalized to that obtained without BFA treatment (value = 100), showed that despite BFA treatment, ∼60% of PC7 reaches the cell surface. Notice that the autoradiogram of proteins immunoprecipitated with Ab:PC7 and NA was obtained after 4 days of exposure whereas that of proteins immunoprecipitated with Ab:PC7 was obtained after 2 h of exposure. C, immunostaining of PC7 and LDLR (red labeling) in cells expressing r-PC7 and either Sar1p-(H79G) or empty vector (pIRES) or as control human LDLR and either Sar1p-(H79G) or pIRES. Nuclei of transfected cells are marked by Hoescht 33258 staining (blue labeling). Bar, 10 μm. D, cell surface biotinylation of PC7 and a positive control, the LDLR, in the presence of the dominant-negative Sar1p-(H79G). Western blot analysis of PC7 or LDLR on lysates from HEK293 cells co-expressing r-PC7 or LDLR with either Sar1p-(H79G) or pIRES, biotinylated and immunoprecipitated with streptavidin (SA)-agarose. The percent of cell surface PC7, estimated by Scion image analysis and normalized to that obtained with PC7 + pIRES transfection (value = 100), showed that the dominant-negative Sar1p-(H79G) prevents the cell surface localization of LDLR but not that of PC7.
To further support the observation that some PC7 reaches the cell surface by an unconventional secretory route, we tested whether COPII-coated vesicles, usually needed for the conventional secretory pathway, are critical for PC7 trafficking. Thus, we co-transfected HEK293 cells with or without the dominant-negative Sar1p-(H79G) form of Sar1p, which prevents the formation of COPII vesicles (43). Immunocytochemistry (Fig. 5C) and cell surface biotinylation (Fig. 5D) revealed that PC7 reaches the cell surface in a COPII-independent manner. Cell surface PC7 in HEK293 cells co-expressing PC7 and Sar1p-(H79G) was almost similar to cells co-expressing PC7 and an empty vector (pIRES). In contrast, the dominant-negative Sar1p-(H79G) completely prevents the control LDLR cell surface localization (Fig. 5, C and D). All these data strongly support the notion that a significant fraction of the zymogen-cleaved PC7 pool bypasses the TGN, and reaches the cell surface by a COPII-independent unconventional secretory pathway. Finally, using a short 10 min pulse with [35S]Cys/Met, it was observed by cell surface biotinylation that some r-PC7 already reached the cell surface within this short interval (Fig. 5E).
We also showed by immunoprecipitation that BFA treatment or co-transfection of PC7 with Sar1p-(H79G) completely prevents secretion of the prosegment (Fig. 6). Altogether, these data demonstrate that in contrast to mature PC7, the prosegment of PC7, is primarily secreted via the conventional secretory pathway.
FIGURE 6.
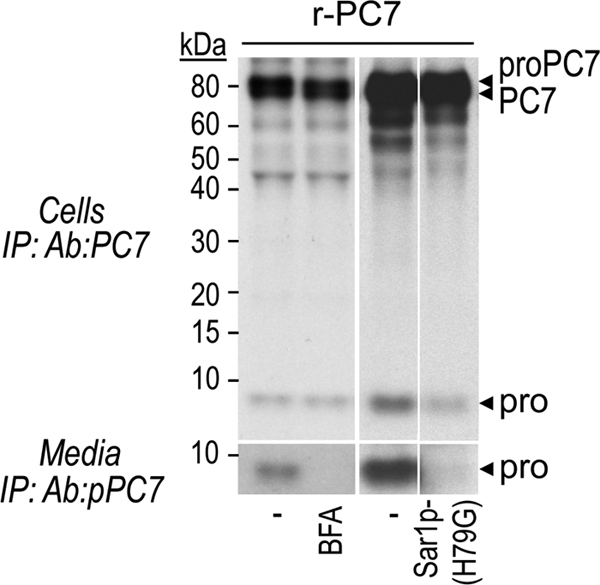
The prosegment of PC7 mainly traffics through the conventional secretory pathway. Autoradiographs of labeled PC7 and its prosegment in lysates and media of transfected HEK293 cells, pulse-labeled with [35S]Cys/Met for 2 h and incubated or not with BFA (2.5 μg/ml) or co-transfected with either the dominant-negative Sar1p-(H79G) or empty vector (pIRES).
The Transmembrane Domain of PC7 Regulates Its Trafficking to the Unconventional Pathway
To define which PC7 domain is required for its trafficking through the unconventional secretory pathway, we first tested the role of PC7 Cys-palmitoylation since it is unique within the PC family (8). We note that both human and rat cytosolic tails (CT) contain 5 and 8 Cys residues, respectively. Earlier work (8) indicated that Cys699 and Cys704 within the CT of h-PC7 are palmitoylated (Fig. 7A). The authors showed the presence of a residual palmitoylation and suspected an artifactual Ser palmitoylation (44). We herein generated the double C699A, C704A in WT human PC7 sequence. [3H]palmitate incorporation revealed that indeed these Cys are palmitoylated, while the double mutant is not (Fig. 7B) and no residual Cys-palmitoylation is present. Thus, in HEK293 cells, Cys699 and Cys704 are the only palmitoylation sites in h-PC7.
To determine its involvement in PC7 trafficking, we assessed whether the sulfation of the PC7-(C699A, Cys704) mutant is modulated. We clearly showed that it is sulfated to the same extent as WT PC7 (Fig. 7C). We also observed this mutant at the cell surface like WT PC7 (Fig. 7D). In addition, the absence of palmitoylation does not direct PC7 through the conventional secretory pathway since the same proportion of PC7-(C699A, Cys704) (∼60%) reaches the cell surface in presence of BFA as compared with WT PC7 (Fig. 7D). Altogether, these data demonstrate that PC7 Cys-plamitoylation does not modulate its trafficking.
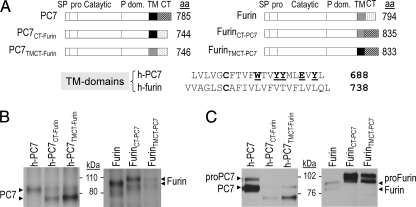
To determine whether specific sequences within the transmembrane (TM) or TMCT domains of PC7 are critical for its trafficking through the conventional or unconventional secretory pathways, we swapped these domains in h-PC7 with the corresponding ones of h-Furin (PC7CT-Furin; PC7TMCT-Furin) and vice versa (FurinCT-PC7 and FurinTMCT-PC7) (Fig. 8A). We then compared the Na235SO4 incorporation into PC7 and Furin versus that of their chimeras with swapped CT and TMCT domains (Fig. 8B). We first noted that while Furin and FurinCT-PC7 were sulfated, the FurinTMCT-PC7 chimera is largely unsulfated. This is not due to the retention of FurinTMCT-PC7 in early secretory compartments, because cell-surface biotinylation of FurinCT-PC7 and FurinTMCT-PC7 were similar and even at much higher than that of Furin (Fig. 8C), as the latter is well known to be rapidly retrieved to the TGN via its CT (45). This may also explain the lower cell surface levels of PC7CT-Furin and PC7TMCT-Furin chimeras as compared with PC7 (Fig. 8C). In contrast, while less sulfated than sPC7, PC7TMCT-Furin, is at least 3-fold more sulfated than wild type PC7. It thus seems that the critical information resides in the TM domain of PC7, since PC7CT-Furin and FurinCT-PC7 are as well sulfated as their wild-type counterparts (Fig. 8B). We conclude that the TM domain of PC7 (Fig. 8A) may contain critical sorting information that may regulate its entry into the unconventional secretory pathway, and as a corollary the TM domain of Furin (Fig. 8A) favors its trafficking through the conventional secretory pathway.
FIGURE 8.
The transmembrane domain of PC7 is required for its trafficking via an unconventional pathway. A, schematic diagram depicting chimeras of h-PC7 and human Furin. SP, signal peptide; Pro, prosegment; Catalytic, catalytic domain; P dom., P domain; TM, transmembrane domain; CT, cytosolic tail. We also show the alignment of the TM domains of h-PC7 and h-Furin. B, autoradiographs of labeled PC7 and Furin in lysates from HEK293 cells expressing h-PC7, Furin, or different PC7 and Furin fused constructions, pulse-labeled with Na235SO4 for 2 h and then immunoprecipitated with Ab:PC7 or anti-Furin Ab. C, cell surface biotinylation of PC7 and Furin from HEK293 cells expressing h-PC7, Furin, or their chimeras. After biotinylation, proteins were immunoprecipitated with SA-agarose and revealed by Western blot using Ab-PC7 or anti-Furin Ab.
DISCUSSION
Processing sites compatible with selective PC-cleavage specificities are found in multiple polypeptides and protein precursors, including hormonal peptides, proteases, receptors, viral envelope glycoproteins, and growth factors (3). The proprotein convertase PC7, the most ancient and conserved PC-like enzyme, is the least studied member of the mammalian basic aa-specific PCs. When it was first discovered in 1996 (5, 7), it was thought to have similar properties to other members of the family in terms of its zymogen activation, cellular trafficking and cleavage specificity (10, 19, 29). While Furin knock-out mice exhibited early embryonic death during development with multiple endothelial and heart defects (46), PC7 knock-out mice were viable and did not exhibit visible anatomical differences (47). Furthermore, other differences between PC7 and Furin began to surface upon analysis of the subcellular localization of PC7, which suggested that it concentrates in less dense compartments than Furin (27). Therefore, it was of interest to identify specific PC7 properties, which would differentiate this enzyme from the other PCs and define its non-redundant physiological roles.
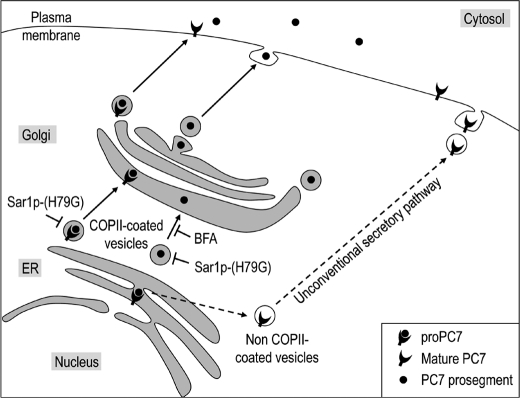
In the present study, we demonstrated that like most PCs, proPC7 undergoes its autocatalytic zymogen cleavage into PC7 within the ER (2), as this process is insensitive to BFA treatment (Fig. 5B). However, different from the other PCs, no secondary cleavage of the prosegment of PC7 takes place, and the latter is secreted as a 9–12 kDa protein not bound to PC7 (Fig. 1). Our studies suggest that the prosegment of PC7 traffics by the conventional secretory pathway before reaching the cell surface and/or media (Fig. 6), and this model is schematized in Fig. 9. Our data also suggest that a basic carboxypeptidase digested the LysArg140 of r-prosegment since free prosegment has a lower molecular mass in HEK293 cells expressing full-length PC7 compared with the prosegment bound to mature s-PC7 in media and the secreted enzyme is active on substrates in vitro (9–16). This new mechanism implies that the prosegment, which is a potent inhibitor of PC7 (22, 30), detaches from the mature enzyme without the need of a secondary cleavage, which usually occurs in other PCs (45).
FIGURE 9.
Schematic diagram of PC7 trafficking. The prosegment of PC7 traffics through the conventional ER/TGN secretory pathway whereas the full-length and mature PC7 traffics through both conventional secretion and an unconventional secretory pathway that is insensitive to BFA treatment and independent of COPII-coated vesicle formation.
The ability of a convertase to efficiently cleave a given precursor depends on their tissue co-expression during embryonic development and/or in the adult, the subcellular localization of the active form of the convertase, and the contact time with the substrate. Thus, active PC1/3 and PC2 are localized in dense core secretory granules (48) and hence are mostly responsible for the processing of neural and endocrine precursors sorted to the regulated secretory pathway (2, 49–51). In contrast, Furin, PC5/6, and PACE4 are localized to the TGN, cell surface, and/or endosomes, and are responsible for the processing of multiple precursors trafficking through the conventional constitutive secretory pathway (2, 45). According to the literature, the intracellular localization of PC7 remains unclear. Thus, PC7 has been reported to localize within subcellular compartments distal to the TGN and can be stored in undefined vesicular stacks close to the Golgi apparatus (19, 52). Herein, we show that PC7 is present in the ER and is endocytosed by a clathrin-independent manner (Fig. 2A). The lack of localization of PC7 within lysosomes is consistent with the work of Wouters et al. (27). In addition, we could not detect PC7 immunoreactivity in the TGN of HEK293 cells (Fig. 2A), probably because of its fast transit through this compartment. However, by electron microscopy, PC7 was found to loosely localize with the TGN-marker Golgin 97 but at separate spatial positions (Fig. 3D), as previously described (19, 52). This finding supports our biochemical data, as well as the previous electron microscopy observations of van de Loo et al. (19). We also clearly demonstrated by three different techniques that PC7 is present at the cell surface of HEK293 cells and also in COS-1 cells by immunocytochemistry, contrary to other studies, which showed that PC7 is not, or only occasionally, present at the cell surface of CHO cells (19) or the lymphoblastoid T2 cells (52). It is thus conceivable that the preferred subcellular localization of PC7 may be cell-type dependent, requiring more detailed localization analyses in various cells and tissues.
To better define the trafficking of PC7, we first tested its endoH sensitivity and sulfation (Fig. 4, A and B). Thus, although PC7 can be sulfated, likely by a Golgi-associated sulfotransferase (38, 39) at an N-glycosylation site(s) (Fig. 4C), one or more of its predicted four N-glycosylation sites (5) remain PGnase F-sensitive. This suggests that like its prosegment, PC7 reaches the cell surface through a Golgi-dependent secretory pathway. While cellular forms of PC7 are only partially resistant to endoH (Fig. 4A, left panel), the secreted form of r-sPC7 exhibits some glycosyl moieties that are completely resistant to endoH resulting in an intermediate-sized form between the PGnase F treated and the non-digested form (Fig. 4A, right panel). This suggests that the cellular form of both r-sproPC7 and r-sPC7 that are completely digested by endoH are localized to the ER. Only the intermediate-sized form of r-sPC7 that is in part sensitive to endoH is the one that exited the ER and could be secreted.
BFA- and/or COPII-coated vesicle-independent secretory pathways were discovered in the last decade (40), suggesting that not all proteins traffic through the conventional TGN-dependent secretory pathway. To our surprise, incubation of HEK293 or COS-1 cells with BFA, although drastically reducing the level of cell surface LDLR (supplemental Fig. S2), had little effect on those of cell surface PC7, as observed by immunocytochemistry (Fig. 5A) or by surface biotinylation of newly biosynthesized [35S]Cys/Met-labeled PC7 (Fig. 5B). Furthermore, overexpression of the dominant-negative Sar1p-(H79G), which inhibits COPII-dependant vesicle budding from the ER (43), only slightly reduced the level of cell surface PC7 (Fig. 5, C and D). Overall, these data suggest that a significant fraction of PC7 (∼50–60%) reaches the cell surface by an unconventional, Golgi-independent, secretory pathway that is insensitive to BFA or COPII inhibition (Fig. 9). This traffic is not the result of an ER stress due to PC7 overexpression, as would be generated upon incubation of cells with the N-glycosylation inhibitor tunicamycin (supplemental Fig. S3).
The molecular mechanisms of the unconventional secretory pathway are not well defined. The ER has been described to be closely associated with essentially all other organelles in the cell including the plasma membrane (53). Contact sites between domains of the ER and the plasma membrane are involved in numerous processes including lipid transfer, signaling, and coupled transport of molecules through both membranes (54). Whether PC7 reaches the cell surface in the presence of BFA through fusion between ER-derived and plasma membranes, as reported for yeast Ist2 (55), or through a vesicle-mediated transport mechanism, as observed during phagosome formation (56) needs further exploration. In addition, the role of this unconventional secretory pathway is not well known. First, this pathway may be a reminiscent of the yeast secretory pathway that is conserved in high eukaryotes, or it allows a faster supply of newly synthesized proteins and lipids to the plasma membrane (57), as reported for a fraction of CD45 (58). In our model, we observed that a fraction of PC7 rapidly reaches the cell surface in less than 10 min after synthesis (Fig. 5E), suggesting that PC7 may use the unconventional secretory pathway as a means to rapidly get to the cell surface.
To define the domain(s) in PC7 that is/are critical for its trafficking through the unconventional secretory pathway, we first mutated the PC7 Cys-palmitoylation sites. Previously, Cys-palmitoylation of PC7 was reported to stabilize the enzyme without affecting its subcellular localization (8). Here, we demonstrated that cell surface PC7 localization is not affected by the absence of Cys-palmitoylation (Fig. 7D). We also demonstrate that it does not affect PC7 trafficking since sulfation of PC7-(C699A, C704A) is unchanged compared with WT PC7 (Fig. 7C) and BFA treatment does not modulate the proportion of cell surface WT PC7 versus its non-palmitoylated mutant (Fig. 7D).
To test whether the transmembrane or cytosolic domains of PC7 are required for its choice of trafficking, we swapped its TMCT or CT domains with those of Furin and vice versa. We noticed that the sulfation of PC7TMCT-Furin, but not that of PC7CT-Furin, is enhanced as compared with WT PC7 (Fig. 8B). In contrast, the sulfation of FurinTMCT-PC7, but not that of FurinCT-PC7, is highly decreased as compared with WT Furin (Fig. 8B). This suggests that a motif within the TM of PC7, which remains to be defined, is critical for the trafficking of this enzyme. Alignment of the TM of h-PC7 and h-Furin revealed that while both of them contain a free Cys, only PC7 exhibits the unique presence of the aa Trp678, Tyr681, 682, 687, and Glu685 (Fig. 8A). Whether these aa contribute to the sorting of PC7 to the unconventional pathway is yet to be determined.
Furin mostly localizes to the TGN, it cycles between the cell surface and TGN via endosomes, and part of it is shed into the medium (45). PC7 seems to localize at the cell surface (Figs. 2B and 3A), and is not shed into the medium (Fig. 1). We surmise that the difference in endocytosis may be related to the CT of each enzyme. Accordingly, swapping of the CT or TMCT of PC7 with those of Furin decreased the PC7 cell surface localization, and the reverse is true for the opposite swap (Fig. 8C). Finally, while no PC7 shedding was observed in any construct, Furin shedding remained unabated in all constructs (not shown), suggesting that this cleavage only depends on the presence of a favorable cleavage motif within the extracellular domain, a site found in Furin (59), but not in PC7.
In conclusion, our present data show the uniqueness of PC7 zymogen activation, its subcellular localization and trafficking. The data also suggest that these unique properties of PC7 may allow this enzyme to specifically process certain precursors in a manner different from any other convertase.
Supplementary Material
Acknowledgments
We thank Marie-Claude Asselin and Ann Chamberland for excellent technical assistance. We are also thankful to Dr. Y. Wang for Sar1p-(H79G) plasmid, Dr. B. Nichols for GFP-Flotillin-1 plasmid, Dr. A. Prat, and Dr. D. Constam for their precious advice. Many thanks to all the members of the Seidah laboratory for helpful discussions and to Brigitte Mary for efficacious editorial assistance.
This research was supported by CIHR Grants MOP-44363, a Strauss Foundation grant, and a Canada Chair Grant 216684.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- PCs
- proprotein convertases
- BFA
- brefeldin A
- CT
- cytosolic tail
- endoH
- endonuclease H
- ER
- endoplasmic reticulum
- LDLR
- low density lipoprotein receptor
- mannose-6P
- mannose-6 phosphate
- NA
- neutravidin
- PBS
- phosphate-buffered saline
- PC7
- proprotein convertase 7
- PGNase F
- N-glycosidase F
- pPC7
- prosegment of PC7
- qPCR
- quantitative real time-PCR
- RT-PCR
- reverse transcription-PCR
- SA
- streptavidin
- TGN
- trans Golgi network
- TM
- transmembrane domain
- aa
- amino acids.
REFERENCES
- 1. Seidah N. G., Prat A. (2007) J. Mol. Med. 85, 685–696 [DOI] [PubMed] [Google Scholar]
- 2. Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008) Int. J. Biochem. Cell Biol. 40, 1111–1125 [DOI] [PubMed] [Google Scholar]
- 3. Seidah N. G., Chrétien M. (1999) Brain Res. 848, 45–62 [DOI] [PubMed] [Google Scholar]
- 4. Creemers J. W., Khatib A. M. (2008) Front Biosci. 13, 4960–4971 [DOI] [PubMed] [Google Scholar]
- 5. Seidah N. G., Hamelin J., Mamarbachi M., Dong W., Tardos H., Mbikay M., Chretien M., Day R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3388–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siezen R. J., Leunissen J. A. (1997) Protein Sci. 6, 501–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meerabux J., Yaspo M. L., Roebroek A. J., Van de Ven W. J., Lister T. A., Young B. D. (1996) Cancer Res. 56, 448–451 [PubMed] [Google Scholar]
- 8. Van de Loo J. W., Teuchert M., Pauli I., Plets E., Van de Ven W. J., Creemers J. W. (2000) Biochem. J. 352, 827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Basak A., Zhong M., Munzer J. S., Chrétien M., Seidah N. G. (2001) Biochem. J. 353, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Munzer J. S., Basak A., Zhong M., Mamarbachi A., Hamelin J., Savaria D., Lazure C., Hendy G. N., Benjannet S., Chrétien M., Seidah N. G. (1997) J. Biol. Chem. 272, 19672–19681 [DOI] [PubMed] [Google Scholar]
- 11. Touré B. B., Munzer J. S., Basak A., Benjannet S., Rochemont J., Lazure C., Chrétien M., Seidah N. G. (2000) J. Biol. Chem. 275, 2349–2358 [DOI] [PubMed] [Google Scholar]
- 12. Blais V., Fugère M., Denault J. B., Klarskov K., Day R., Leduc R. (2002) FEBS Lett. 524, 43–48 [DOI] [PubMed] [Google Scholar]
- 13. Siegfried G., Basak A., Cromlish J. A., Benjannet S., Marcinkiewicz J., Chrétien M., Seidah N. G., Khatib A. M. (2003) J. Clin. Invest. 111, 1723–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Basak S., Chrétien M., Mbikay M., Basak A. (2004) Biochem. J. 380, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scamuffa N., Basak A., Lalou C., Wargnier A., Marcinkiewicz J., Siegfried G., Chrétien M., Calvo F., Seidah N. G., Khatib A. M. (2008) Gut 57, 1573–1582 [DOI] [PubMed] [Google Scholar]
- 16. Fugere M., Appel J., Houghten R. A., Lindberg I., Day R. (2007) Mol. Pharmacol. 71, 323–332 [DOI] [PubMed] [Google Scholar]
- 17. Decroly E., Wouters S., Di Bello C., Lazure C., Ruysschaert J. M., Seidah N. G. (1996) J. Biol. Chem. 271, 30442–30450 [DOI] [PubMed] [Google Scholar]
- 18. Decroly E., Benjannet S., Savaria D., Seidah N. G. (1997) FEBS Lett. 405, 68–72 [DOI] [PubMed] [Google Scholar]
- 19. Van de Loo J. W., Creemers J. W., Bright N. A., Young B. D., Roebroek A. J., Van de Ven W. J. (1997) J. Biol. Chem. 272, 27116–27123 [DOI] [PubMed] [Google Scholar]
- 20. Canaff L., Bennett H. P., Hou Y., Seidah N. G., Hendy G. N. (1999) Endocrinology 140, 3633–3642 [DOI] [PubMed] [Google Scholar]
- 21. Lopez-Perez E., Seidah N. G., Checler F. (1999) J. Neurochem. 73, 2056–2062 [PubMed] [Google Scholar]
- 22. Zhong M., Munzer J. S., Basak A., Benjannet S., Mowla S. J., Decroly E., Chrétien M., Seidah N. G. (1999) J. Biol. Chem. 274, 33913–33920 [DOI] [PubMed] [Google Scholar]
- 23. McColl B. K., Paavonen K., Karnezis T., Harris N. C., Davydova N., Rothacker J., Nice E. C., Harder K. W., Roufail S., Hibbs M. L., Rogers P. A., Alitalo K., Stacker S. A., Achen M. G. (2007) FASEB J. 21, 1088–1098 [DOI] [PubMed] [Google Scholar]
- 24. Longpré J. M., McCulloch D. R., Koo B. H., Alexander J. P., Apte S. S., Leduc R. (2009) Int. J Biochem. Cell Biol. 41, 1116–1126 [DOI] [PubMed] [Google Scholar]
- 25. Nelsen S. M., Christian J. L. (2009) J. Biol. Chem. 284, 27157–27166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khatib A. M., Lahlil R., Scamuffa N., Akimenko M. A., Ernest S., Lomri A., Lalou C., Seidah N. G., Villoutreix B. O., Calvo F., Siegfried G. (2010) PLoS. ONE 5, e11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wouters S., Decroly E., Vandenbranden M., Shober D., Fuchs R., Morel V., Leruth M., Seidah N. G., Courtoy P. J., Ruysschaert J. M. (1999) FEBS Lett. 456, 97–102 [DOI] [PubMed] [Google Scholar]
- 28. Nour N., Basak A., Chrétien M., Seidah N. G. (2003) J. Biol. Chem. 278, 2886–2895 [DOI] [PubMed] [Google Scholar]
- 29. Seidah N. G. (2011) Annals of the New York Academy of Sciences, in press [Google Scholar]
- 30. Bhattacharjya S., Xu P., Zhong M., Chrétien M., Seidah N. G., Ni F. (2000) Biochemistry 39, 2868–2877 [DOI] [PubMed] [Google Scholar]
- 31. Ozden S., Lucas-Hourani M., Ceccaldi P. E., Basak A., Valentine M., Benjannet S., Hamelin J., Jacob Y., Mamchaoui K., Mouly V., Desprès P., Gessain A., Butler-Browne G., Chrétien M., Tangy F., Vidalain P. O., Seidah N. G. (2008) J. Biol. Chem. 283, 21899–21908 [DOI] [PubMed] [Google Scholar]
- 32. Zaid A., Roubtsova A., Essalmani R., Marcinkiewicz J., Chamberland A., Hamelin J., Tremblay M., Jacques H., Jin W., Davignon J., Seidah N. G., Prat A. (2008) Hepatology 48, 646–654 [DOI] [PubMed] [Google Scholar]
- 33. Benjannet S., Elagoz A., Wickham L., Mamarbachi M., Munzer J. S., Basak A., Lazure C., Cromlish J. A., Sisodia S., Checler F., Chrétien M., Seidah N. G. (2001) J. Biol. Chem. 276, 10879–10887 [DOI] [PubMed] [Google Scholar]
- 34. Tokuyasu K. T. (1980) Histochem. J. 12, 381–403 [DOI] [PubMed] [Google Scholar]
- 35. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arolas J. L., Vendrell J., Aviles F. X., Fricker L. D. (2007) Curr. Pharm. Des 13, 349–366 [DOI] [PubMed] [Google Scholar]
- 37. Glebov O. O., Bright N. A., Nichols B. J. (2006) Nat. Cell Biol. 8, 46–54 [DOI] [PubMed] [Google Scholar]
- 38. Ong E., Yeh J. C., Ding Y., Hindsgaul O., Fukuda M. (1998) J. Biol. Chem. 273, 5190–5195 [DOI] [PubMed] [Google Scholar]
- 39. Honke K., Taniguchi N. (2002) Med. Res. Rev. 22, 637–654 [DOI] [PubMed] [Google Scholar]
- 40. Nickel W., Rabouille C. (2009) Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 41. Lippincott-Schwartz J., Yuan L., Tipper C., Amherdt M., Orci L., Klausner R. D. (1991) Cell 67, 601–616 [DOI] [PubMed] [Google Scholar]
- 42. Bos C. R., Shank S. L., Snider M. D. (1995) J. Biol. Chem. 270, 665–671 [DOI] [PubMed] [Google Scholar]
- 43. Aridor M., Bannykh S. I., Rowe T., Balch W. E. (1995) J. Cell Biol. 131, 875–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alvarez E., Gironès N., Davis R. J. (1990) J. Biol. Chem. 265, 16644–16655 [PubMed] [Google Scholar]
- 45. Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roebroek A. J., Umans L., Pauli I. G., Robertson E. J., van Leuven F., Van de Ven W. J., Constam D. B. (1998) Development 125, 4863–4876 [DOI] [PubMed] [Google Scholar]
- 47. Villeneuve P., Feliciangeli S., Croissandeau G., Seidah N. G., Mbikay M., Kitabgi P., Beaudet A. (2002) J. Neurochem. 82, 783–793 [DOI] [PubMed] [Google Scholar]
- 48. Malide D., Seidah N. G., Chrétien M., Bendayan M. (1995) J. Histochem. Cytochem. 43, 11–19 [DOI] [PubMed] [Google Scholar]
- 49. Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 [DOI] [PubMed] [Google Scholar]
- 50. Benjannet S., Rondeau N., Day R., Chrétien M., Seidah N. G. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3564–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marcinkiewicz M., Ramla D., Seidah N. G., Chrétien M. (1994) Endocrinology 135, 1651–1660 [DOI] [PubMed] [Google Scholar]
- 52. Leonhardt R. M., Fiegl D., Rufer E., Karger A., Bettin B., Knittler M. R. (2010) J. Immunol. 184, 2985–2998 [DOI] [PubMed] [Google Scholar]
- 53. Staehelin L. A. (1997) Plant J. 11, 1151–1165 [DOI] [PubMed] [Google Scholar]
- 54. Levine T., Loewen C. (2006) Curr. Opin. Cell Biol. 18, 371–378 [DOI] [PubMed] [Google Scholar]
- 55. Nickel W., Seedorf M. (2008) Annu. Rev. Cell Dev. Biol. 24, 287–308 [DOI] [PubMed] [Google Scholar]
- 56. Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P. H., Steele-Mortimer O., Paiement J., Bergeron J. J., Desjardins M. (2002) Cell 110, 119–131 [DOI] [PubMed] [Google Scholar]
- 57. Marie M., Sannerud R., Avsnes D. H., Saraste J. (2008) Cell Mol. Life Sci. 65, 2859–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baldwin T. A., Ostergaard H. L. (2002) J. Biol. Chem. 277, 50333–50340 [DOI] [PubMed] [Google Scholar]
- 59. Plaimauer B., Mohr G., Wernhart W., Himmelspach M., Dorner F., Schlokat U. (2001) Biochem. J. 354, 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.