Abstract
In the absence of x-ray structures of sodium and calcium channels their homology models are used to rationalize experimental data and design new experiments. A challenge is to model the outer-pore region that folds differently from potassium channels. Here we report a new model of the outer-pore region of the NaV1.4 channel, which suggests roles of highly conserved residues around the selectivity filter. The model takes from our previous study (Tikhonov, D. B., and Zhorov, B. S. (2005) Biophys. J. 88, 184–197) the general disposition of the P-helices, selectivity filter residues, and the outer carboxylates, but proposes new intra- and inter-domain contacts that support structural stability of the outer pore. Glycine residues downstream from the selectivity filter are proposed to participate in knob-into-hole contacts with the P-helices and S6s. These contacts explain the adapted tetrodotoxin resistance of snakes that feed on toxic prey through valine substitution of isoleucine in the P-helix of repeat IV. Polar residues five positions upstream from the selectivity filter residues form H-bonds with the ascending-limb backbones. Exceptionally conserved tryptophans are engaged in inter-repeat H-bonds to form a ring whose π-electrons would facilitate passage of ions from the outer carboxylates to the selectivity filter. The outer-pore model of CaV1.2 derived from the NaV1.4 model is also stabilized by the ring of exceptionally conservative tryptophans and H-bonds between the P-helices and ascending limbs. In this model, the exceptionally conserved aspartate downstream from the selectivity-filter glutamate in repeat II facilitates passage of calcium ions to the selectivity-filter ring through the tryptophan ring. Available experimental data are discussed in view of the models.
Keywords: Biophysics, Calcium Channels, Computer Modeling, Ion Channels, Sodium Channels
Introduction
Voltage-gated ion channels are involved in the control of many physiological functions. Upon membrane depolarization, channels rapidly transit from the resting to the open state, then close to inactivated state(s), and return to the resting state upon membrane repolarization. The human genome encodes nine voltage-gated sodium (NaV1.1-NaV1.9) and ten voltage-gated calcium channels categorized as L (CaV1.1-CaV1.4), P/Q (CaV2.1), N (CaV2.2), R (CaV2.3), and T (CaV3.1-CaV3.3) types. The pore-forming α1-subunit of calcium and sodium channels folds from a single polypeptide chain of four homologous repeats (Fig. 1A). Repeats I to IV are arranged clockwise when viewed extracellularly (1). Each repeat contains a voltage-sensing domain S1-S4, an outer helix S5, an inner helix S6, and a membrane-diving P-loop between S5 and S6. The P-loop includes a P-helix, a P-turn, and an ascending limb. Four ascending limbs, which line the outer pore, contain selectivity-filter residues in positions p50 (see Footnote 2 for residue labels).2 Repeats I, II, III, and IV contain selectivity-filter glutamates in calcium channels (the EEEE ring) and Asp, Glu, Lys, and Ala residues in sodium channels (the DEKA ring). The ascending limbs also contain the outer carboxylates three or four positions downstream from the selectivity-filter residues.
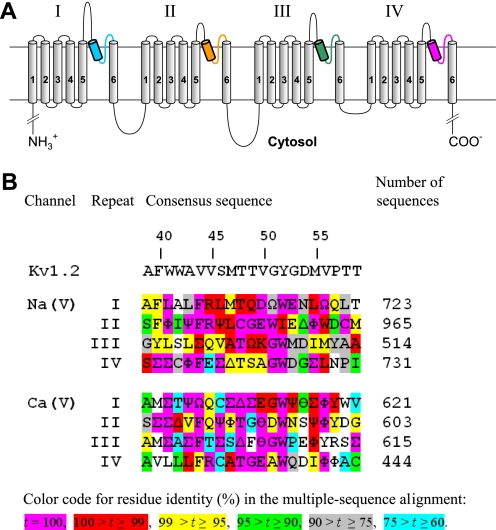
FIGURE 1.
A, transmembrane topology of the pore-forming α1-subunit in calcium and sodium channels. The α1 subunit has four repeats. Each repeat contains six transmembrane helices (S1-S6), and a P-loop between S5 and S6 helices. The homology models in this study are composed of highlighted P-helices and ascending limbs, which contribute to the P-loops. P-helices and ascending limbs are colored as in the NaV1.4 models (Figs. 2–5). B, residue conservation in P-loops of sodium and calcium channels. Each position shows the dominant residue in the multiple sequence alignments obtained with query sequences CAC1C_Human (CaV1.2) and SCN4A_HUMAN (NaV1.4) for sodium and calcium channels, respectively. Greek letters encode residues of similar physico-chemical properties: ϴ = E/D (acidic), Ω = Y/F (aromatic), Ψ = V/I (β-branched, hydrophobic), Φ = M/L (long hydrophobic), Σ = V/I/L/M/F (hydrophobic), Δ = S/T (hydroxy-substituted Cβ). The rightmost column shows number of aligned sequences from the translated nucleotide database (nucleotide collection) found by tblasn with the respective query sequence. Relative numbers of residues in P-loops (3) are shown above the Kv1.2 sequence.
Voltage-gated potassium, sodium, and calcium channels are believed to share a generally similar folding of transmembrane helices and P-helices (2, 3). This justifies homology modeling of sodium and calcium channels based on available x-ray structures of potassium channels. The homology models are used to predict structural peculiarities of sodium and calcium channels, propose mechanisms of ion permeation, and explain data on ligand actions. The most difficult region for homology modeling is the outer pore, whose folding in sodium and calcium channels is different from that in potassium channels.
Tetrodotoxin (TTX)3 and saxitoxin, the hallmark outer-pore blockers of NaV1.4, were used to probe the outer-pore region. These data have been accommodated in competing models of NaV1.4 with the toxins (4, 5). Despite the fact that similar experimental data sets underlie the models, they differ significantly in terms of mutual disposition and orientation of the pore helices and conformations of the ascending limbs. An explanation for this difference is flexibility of side chains in the ascending limbs that according to experimental data interact with specific toxin moieties. This flexibility can compensate a substantial backbone difference between the two models. Thus, the toxin-channel distance constraints, which are derived from experiments, are insufficient for elaboration of an unambiguous model of the outer pore.
In previous models, the P-loop domains of sodium and calcium channels (4–7) had been built without serious considerations of the outer-pore stability. This stability can be supported by specific inter- and intra-repeat contacts including the ascending limbs that line the outer pore. The contacts, which stabilize the outer pore of potassium channels, are known from the crystallographic data. In sodium and calcium channels such contacts are unknown.
Multiple-sequence alignment of 137 voltage-gated calcium channel proteins shows exceptionally conserved residues in the P-domains, including the signature-sequence TXEXW with the selectivity filter glutamates (8). In the last decade, hundreds of new sequences of sodium and calcium channel proteins in various organisms have become available. Because the pore-forming α1-subunits of voltage-gated calcium and sodium channels contain four homologous repeats, local sequence alignments of segments from individual repeats can be used to reveal conserved residues in each repeat. Considering highly conserved residues beyond the selectivity filter and their interaction patterns in homology models may suggest functional roles for these residues and new inter-residue interactions. These can be used as distance constraints to improve the homology models.
In this work we have generated and explored multiple sequence alignments of P-loops in individual repeats of hundreds of calcium and sodium channel proteins. Analysis of these alignments allowed us to propose a network of hydrophobic, hydrogen bonding, and knob-into-the-hole contacts involving conserved residues, which stabilize the outer-pore region. Using available experimental data and the above contacts as distance constraints, we elaborated our homology models (5, 6, 9). The new inter-repeat contacts suggest new important roles for some of the exceptionally conserved residues. The new models allow us to propose details of the ion permeation mechanisms in sodium and calcium channels and provide structural rationale for recent experimental data on sequence peculiarities of sodium channels in garter snakes that feed on toxic pray.
MATERIALS AND METHODS
To reveal conserved residues in the P-loops of sodium and calcium channels we aligned many sequences from the translated nucleotide database using NCBI BLAST program tblastn. Sequences of 21-amino acid segments, which include P-helices, P-turns, and ascending limbs in four repeats of NaV1.4 (SCN4A_HUMAN) and four repeats of CaV1.2 (CAC1C_HUMAN) were used as queries. Default parameters were used for the tblasn algorithm, except for the maximal number of aligned sequences, which was set to 5,000. The search was limited by the ENTREZ queries (“calcium channel” OR “Ca2+ channel”) and (“sodium channel” OR “Na+ channel”). A total of ∼6,000 sequences were returned. The sequence alignments for 21-amino acid segments of Ca2+ and Na+ channels returned by tblastn were unambiguous. To minimize the influence of possible errors in the data base, repeat entries that contained atypical residue types (less than four occurrences in a matching position) were excluded (8); about 3% of the sequences were thus excluded. We also manually removed sequences with insertions/deletions at the N or C termini. Unequal numbers of sequences returned for individual repeats may be due to the fact that most calcium and sodium channels are hetero-tetramers and in some cases sequences of specific repeats, which are too different from the query sequence, were not returned by tblastn. Detailed analysis of this possibility, which would require specific bioinformatics tools, is beyond the scope of this study.
The returned sequences were analyzed and manually condensed in Fig. 1 where different colors encode different levels of conservation. For some positions, we use Greek letters to encode residues with similar physico-chemical properties (see legend to Fig. 1). An amino acid may be attributed to different categories. For example, when leucine and methionine align in a matching position with other hydrophobic residues, we categorize all residues in this position as hydrophobic. However, when most of the residues in a matching position are leucine or methionine, we categorize them as long hydrophobic residues. Such categorization highlights conserved structural features (aromaticity, charge, length, β-branched atom), which will be useful for future elaboration of the models.
Homology models of NaV1.4 and CaV1.2 channels were built from the P-loop and S6 segments using the sequence alignments with potassium channels proposed before (7). The backbones of the ascending limbs were initially folded as in Ref. 5, 6. All calculations were performed using the ZMM program. The nonbonded energy was calculated using the AMBER force field (10, 11) with a cut-off distance of 8 Å. The hydration energy was calculated using the implicit solvent method (12). Electrostatic interactions were calculated using the distance-dependent dielectric function. The atomic charges of TTX have been calculated by the semi-empirical method AM1 (13) using MOPAC. The Monte Carlo-minimization method (14) was used to optimize the models. During energy minimizations, α carbons of the P-helices were constrained to their corresponding positions in the template using pins. A pin is a flat-bottom energy function, which allows an atom to deviate penalty-free up to 1 Å from the template and imposes a penalty of 10 kcal mol-1 Å-1 for deviations >1 Å.
Each model was MC-minimized until 2,000 consecutive minimizations did not decrease the energy of the apparent global minimum. The multi-MCM protocol (9) was used to dock TTX and impose specific distance constraints. No specific energy terms were used for cation-π interactions, which were accounted for due to partial negative charges at the aromatic carbons (15). Further details of methodology can be found elsewhere (9).
RESULTS
Analysis of Multiple Sequence Alignments
The multiple sequence alignments are summarized in Fig. 1B where residues are colored according to their conservation in individual repeats. As expected, most of the selectivity filter residues in positions p50 (the DEKA ring in sodium channels and EEEE ring in calcium channels) are exceptionally (100%) conserved. The selectivity-filter alanine A4p50 in the sodium channels is highly (97.8%) conserved, and 2.2% of the sequences contain G4p50 or S4p50 residues. Also expected was a high conservation of the outer-carboxylate residues in the sodium channels. Thus, repeats I and II contain exceptionally conserved E1p53 and highly (99.0%) conserved E2p53 (Q2p53 in 1% of the sequences). Residues D3p54, E3p54, and Q3p54 are found respectively, in 71.2, 12.8, and 9.9% of repeat III sequences. Residues D4p53 and N4p53 are found, respectively, in 83.6 and 11.6% of repeat IV sequences (the remaining 4.8% of residues in this position are designated “B”, i.e. either D or N). Three outer-carboxylates in positions p53 are the TTX-sensing residues that were proposed to face the outer pore and interact with the TTX molecule (4, 5). In contrast, carboxylate 3p54 contributes to binding of μ-conotoxin (1) suggesting that it faces the extracellular media. In calcium channels the outer carboxylates in positions 1p54, 3p54, and 4p54 are moderately conserved suggesting that they are not critical for calcium permeation. A notable exception is a highly conserved D2p51 next to the selectivity filter glutamate E2p50. Residue D2p51 was proposed by us to play an important role in calcium permeation (6).
The ascending limbs contain tryptophan residues, which are exceptionally conserved in all repeats of calcium and sodium channels. In all four repeats of calcium channels, these tryptophans are in the matching positions p52. Repeats I, III, and IV of Na+ channels have tryptophans in position p52, whereas repeat II contains tryptophan W2p51. Noteworthy, the matching position in calcium channels contains either highly conserved aspartate D2p51 (98%) or asparagine (2%). Sodium channels evolved from calcium channels (16). The exceptional conservation of the tryptophans and replacement of the calcium channel D2p51 with sodium channel W2p51 strongly suggests that these residues are important for key channel function(s). These are likely ion permeation and/or the outer-pore folding and stability.
Main peculiarities of the tryptophan side chain are the large planar heterocyclic ring with π-electrons, which may be engaged in π-cation interactions with permeating ions, and the NH group. Besides, the tryptophan side chain has the largest nonpolar surface area among naturally occurring amino acids. Examples of structural roles of tryptophans in proteins are shown in supplemental Fig. S1. In potassium channels the conserved tryptophans in the pore helix (positions p41 and p42) participate in intra- and inter-subunit H-bonds, providing stabilization for the outer-pore structure (supplemental Fig. S1A). In some proteins, tryptophans form box-like structures, which accommodate cationic ligands (supplemental Fig. S1, B and C) with π-cations apparently contributing to the ligand-protein interactions.
Conserved Tryptophans in the Outer Pore of Sodium Channels
The high conservation of tryptophans in the ascending limbs of sodium and calcium channels has long been known (8) and all these tryptophans have been previously mutated in sodium channels. Cysteine substitutes of the ascending-limb tryptophans are accessible by externally applied methane thiosulfonate reagents (17). Furthermore, cysteine substitutions of W4p52, W3p52, and W1p51 form disulfide bonds involving engineered cysteines in the neighboring ascending limbs (18). These data strongly indicate that the side chains of the conserved tryptophans are not buried in the protein, but face the outer pore. Alanine substitutions of W4p52 alter the ionic selectivity of sodium channels (19). Cysteine substitution of W4p52 abolishes the NaV1.4 block by lidocaine (20), a local anesthetic that binds in the inner pore (15). W4p52 was suggested to directly interact with the local anesthetic bound in the inner pore (20), but in view of our models of sodium channels, direct contact of W4p52 with the local anesthetic molecule is unlikely, suggesting that the mutation W4p52C affects the drug binding allosterically. W1p51 was proposed to contribute to the pore of the sodium channel (21).
Even though the above studies imply importance of the outer-pore tryptophans, their structural and functional roles, which underlie the exceptional conservation, remain unclear. How could one arrange the pore-facing side chains of the conserved tryptophans? The tryptophans are unlikely to face the pore lumen with their edges because the side-chain NH group and aromatic hydrogens would retard the cation permeation. The more likely possibility is that the tryptophans face the pore by their π-electron rich aromatic faces that would favorably interact with the permeating cations. Box-like structures involving tryptophans are seen in some proteins (supplemental Fig. S1, B and C). With this hypothesis in mind, we revised our previous model (5). In the previous model, the side chain of W3p52 is partially exposed to the pore and its NH hydrogen is 2.5–2.8 Å away from the backbone oxygen of W4p52. This distance is ∼1 Å larger than a typical H-bond length. Here we hypothesized that the side chain NH groups of the conserved tryptophans are engaged in inter-repeat H-bonds that stabilize the outer-pore structure. We biased the inter-repeat H-bond for W4p52 with a distance constraint and used analogous constraints to impose inter-repeat H-bonds involving the side chains of W1p52 and W4p52. The side chain of W2p51 projects toward repeat I rather than to repeat III. Therefore, we imposed an H-bond between the side chain NH group of W2p51 and the backbone carbonyl of Y1p51. We have used our previous model (5) as the starting structure, MC-minimized the model with the above constraints, and then refined it by unconstrained MC-minimizations. The proposed set of H-bonds was readily formed with only modest deformations of the ascending-limb backbones in the starting model (5).
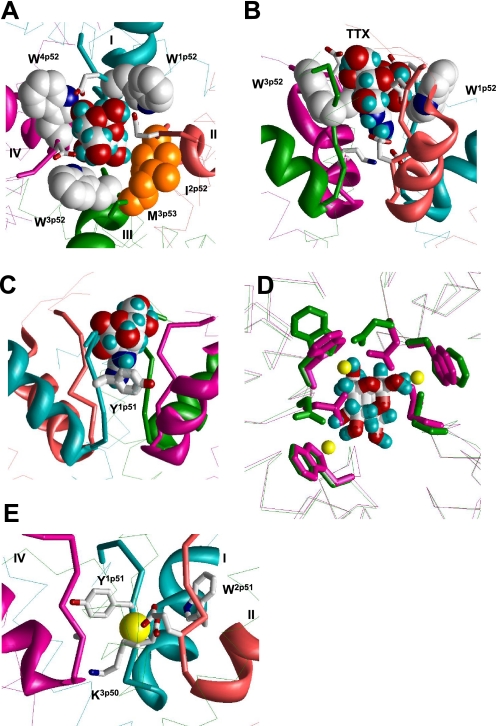
The critical test for the new model was to explore whether the inter-repeat H-bonds agree with experimental data on TTX binding. Therefore, we energy-optimized the model with the TTX molecule bound to the outer pore as described before (5). To minimize distortions of the TTX receptor upon the ascending-limb deformations caused by new inter-repeat constraints, we used distance constraint to keep specific TTX interactions with the DEKA ring and the outer carboxylates as described before (5). More recent experimental data show that the aromatic ring of Y1p51 is engaged in π-cation interactions with TTX, which stabilize TTX binding in TTX-sensitive sodium channels (22). This non-conservative aromatic residue is a well-known determinant of TTX sensitivity (23–25). For example, NaV1.2 and NaV1.4, which have, respectively, F1p51 and Y1p51, are TTX-sensitive, whereas the NaV1.5 channel, which has C1p51, is low-sensitive to TTX. In our previous model Y1p51 faces the pore and approaches the TTX molecule, but it is not engaged in π-cation interaction with TTX (5). To satisfy the new experimental data (22), we imposed a distance constraint between the guanidinium group of TTX and the aromatic ring of Y1p51. In the MC-minimized model, TTX fits perfectly in the outer pore, which is lined by tryptophans in positions p52, the highly conserved I2p52, and a hydrophobically conserved residue in position 3p53 (Fig. 2). The outer carboxylate side chains E1p53, E2p53, and D4p53 protruded through the gaps between the tryptophans to make specific contacts with the TTX molecule (Fig. 2A) whose guanidinium group reached the selectivity-filter residues D1p50 and E2p50 (Fig. 2B) and was also engaged in cation-π interaction with Y1p51 (Fig. 2C). The position and orientation of the TTX molecule did not change substantially versus our previous model (5), while the ascending-limb backbones underwent only moderate changes. The TTX binding appeared invariant to these changes because TTX interacted with flexible side chains.
FIGURE 2.
Binding of TTX and Ca2+ in the revised model of NaV1.4. A and B, top and side views of TTX-bound channel. Backbones in repeats I, II, III, and IV are cyan, opaque, green, and magenta, respectively. P-helices are shown as ribbons, ascending limbs as thick α-tracings, and S6s as thin α-tracings. TTX fits perfectly in the outer pore and interacts with known TTX-sensing residues. C, cation-π interaction between Y1p51 and guanidine group of TTX (22). This interaction is achieved by only a modest deformation of our previous model (5), in which Y1p51 faces the pore and interacts with TTX by the aromatic ring edge rather than the aromatic plane. D, superposition of α tracings of the current model, which is MC-minimized either in the presence of Na+ ions (yellow Na+ ions, magenta side chains and backbones) or TTX (green side chains and backbones). Side chains of W1p52, W3p52, W4p52, and TTX-sensing outer carboxylates E1p53, E2p53, and D4p53 are shown as sticks. The root mean square deviation between α carbons in these models is as small as 0.2 Å. Note that hydroxyl groups of the TTX molecule are very close to positions of Na+ ions. E, possible binding of a Ca2+ ion in the Na+ channel. The ion binds to acidic residues D1p50 and E2p50 of the DEKA locus and displaces the ammonium group of K3p50 toward the inner pore. The aromatic side chains of Y1p51 and W2p51 are close enough to the Ca2+ ion to interact with it through water molecules in the first hydration shell. The complex likely corresponds to the blocked channel state. In the absence of another cation-binding site in the narrowest part of the outer pore, an incoming ion cannot displace the bound Ca2+ ion.
TTX-sensing Residues in the Pore Helices
Aside from well-known TTX-sensing residues in the ascending limbs, mutations of several other residues affect TTX binding. Intriguing examples are provided by the analysis of NaV1.4 sequences from garter snakes that feed on the tetrodotoxic newt (26). The single mutation I4p46V increases TTX resistance of the Warrenton population of garter snakes by more than 4-fold (26). This effect can hardly be explained by direct interaction of the TTX molecule with the hydrophobic residue I4p46, which is located in the P-helix and does not protrude in the outer pore. In our model the ascending limbs of all four domains closely approach the same-domain P-helices and tightly pack against them. In particular, the conserved isoleucine I4p46 forms a knob-into-the-hole contact with the exceptionally conserved glycine G4p51 (Fig. 3, A and B). Mutation I4p46V creates a void of a methyl group size in the tight intra-repeat contact between the P-helix and the ascending limb. P-helices in our models are practically immobile due to several strong contacts with S6 and S5 helices (15). To fill the void, glycine G4p51 would move toward valine V4p46 and pull the ascending limb of repeat IV away from the pore axis. The shifted ascending limb would widen the outer pore and thus deform the TTX receptor.
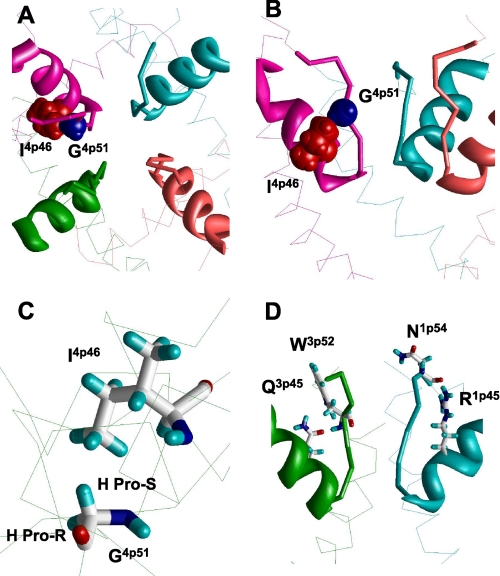
FIGURE 3.
Contacts between P-helices and ascending limbs contribute to the outer- pore stability. A and B, top and side views at the outer pore of NaV1.4 in which space-filled G4p51 and I4p46 form a tight knob-into-the-hole contact in our model. Mutation I4p46V reduces TTX sensitivity of garter snakes that adapted to the tetrodotoxic newt pray (26). It is hardly possible to explain this fact by direct interaction of I4p46 with TTX. In view of our model, the I4p46V mutation would result in a deformation of the outer pore due to shift of G4p51, which would move toward V4p46 to retain the knob-into-the-hole contact. This shift would move the ascending limb in domain IV. The ascending limb accommodates a TTX-sensing residue D4p53, which would move away from the pore axis thus decreasing the TTX sensitivity. A native knob-into-the-hole contact in repeat III involves G3p51 and V3p46 (not shown). C, G4p51 is involved in the hole-into-the-knob contact by its pro-S hydrogen, which is not substituted by the methyl group in the G4p51A mutant. D, conserved polar residues in positions p45 may stabilize the outer pore by forming H-bonds with ascending limbs. In our model the side chain of R1p45 interacts with the backbone carbonyl group of N1p54, whereas Q3p45 interacts with the backbone NH group of W3p52. These contacts may explain why cysteine substitutions of R1p45 and R2p45 destabilize binding of the positively charged TTX (27).
In the Brenton populations of garter snakes, which are about two times less sensitive to TTX than the Warrenton population, the sodium channel has two mutations in the P-loop region: I4p46V and G4p51A (26). At first glance, the methyl group of A4p51 would fill the void at the V4p46 side chain without causing any shift of the ascending limb of repeat IV. However, closer inspection of our model shows that G4p51 is involved in a hole-into-the-knob contact by its pro-S hydrogen, whereas it is the pro-R hydrogen that is substituted by the methyl group in the G4p51A mutant (Fig. 3C). In our model, the pro-R hydrogen approaches the ascending limb of domain III suggesting that mutations G4p51A and I4p46V concertedly cause the outer-pore widening, which decreases the TTX sensitivity of the Brenton populations of garter snakes.
The lowest sensitivity to TTX is observed in the Willow-Creek population of garter snakes, whose NaV1.4 channels, besides the above-described mutation L4p46V, have three additional mutations in the P-loop region. These include mutation D4p53N, which changes a residue that directly interacts with TTX (Fig. 2A). Thus, our model explains the intriguing data on the TTX resistance of garter snakes by the influence of mutations I4p46V and G4p51A on the outer-pore geometry and by direct influence of mutation D4p53N on TTX binding.
Other TTX-sensing residues in the P-helix, which do not directly interact with the TTX molecule in our model, can also affect TTX binding by modifying the outer-pore structure. Indeed, the bundle of four ascending limbs contains intrinsically flexible strands, which are not stabilized by H-bonds of a regular secondary structure. Therefore, the geometry of the bundle should be stabilized by specific contacts. Mutations, which affect such stabilizing contacts, would affect the outer-pore structure and therefore the TTX sensitivity of the channel. Intriguingly, cysteine substitutions of highly conserved R1p45 and exceptionally conserved R2p45 decrease the sensitivity of NaV1.4 to TTX by ∼10- and ∼1000-fold, respectively (27). These data seem paradoxical because the cationic TTX molecule should electrostatically repel from the cationic arginines and their replacement by cysteines is expected to eliminate the repulsion and hence increase the TTX potency. The fact that the TTX potency is decreased can be explained as follows. In the KcsA K+ channel, position p45 is occupied by the glutamate residue whose side chain projects toward the extracellular space, parallel to the ascending limb (supplemental Fig. S2). These data prompt us to suggest that mutations R1p45C and R2p45C eliminate contributions of the arginines in the outer-pore stability and thus affect TTX binding indirectly. Intensive MC-minimizations of our current model from multiple starting orientations of the R1p45 and R2p45 side chains yielded low-energy conformations in which these side chains donate intra-repeat H-bonds to the backbone carbonyls in positions p53 (Fig. 3D). Noteworthy, positions 1p53 and 2p53 are occupied by TTX-sensing glutamates from the outer-carboxylates ring. In view of our model, the cysteine substitution of R1p45 and R2p45 would deform the structure of the outer-carboxylate ring and thus its interactions with TTX. The exceptionally conserved Q3p45 in the homologous position can also form specific intra-repeat contacts with the ascending-limb backbone of repeat III. Glutamine Q4p45 is moderately conserved (67.4.5%), but 28.4% of the sequences contain glutamate E4p45, which can also be engaged in an intra-repeat H-bond with the ascending limb backbone. One such possibility is shown in Fig. 3D where the side chain carbonyl of Q3p45 accepts an H-bond from the backbone NH group of W3p52. It should be noted however that mutations Q4p45C and Q3p45C have only a small effect on the TTX potency (27). Other specific interactions involving side chains of long H-bonding residues in positions p45 are also possible.
Na+ Binding Sites
Previously we used our TTX-based model of the sodium channel (5) to predict putative binding sites for Na+ ions and propose a scheme of ion permeation. The scheme involves six Na+ ions: four in the outer-carboxylates ring, one in the selectivity filter ring, and one between the two rings (9). To explore whether this scheme is consistent with our present model, we loaded the outer pore with six Na+ ions as proposed before (9). The first ion was constrained to D3p54, the second ion between D3p54 and D4p53, the third ion between D4p53 and E1p53, the fourth ion between E1p53 and E2p53, the fifth ion between E2p53 and E2p50, and the sixth ion to D1p50. The selectivity-filter lysine K3p50 was constrained between E2p50 and D1p50. MC-minimization yielded a structure (Fig. 4A) in which four Na+ ions were engaged in cation-π contacts with the conserved tryptophans. The scheme of the Na+ binding sites is shown in Fig. 4E.
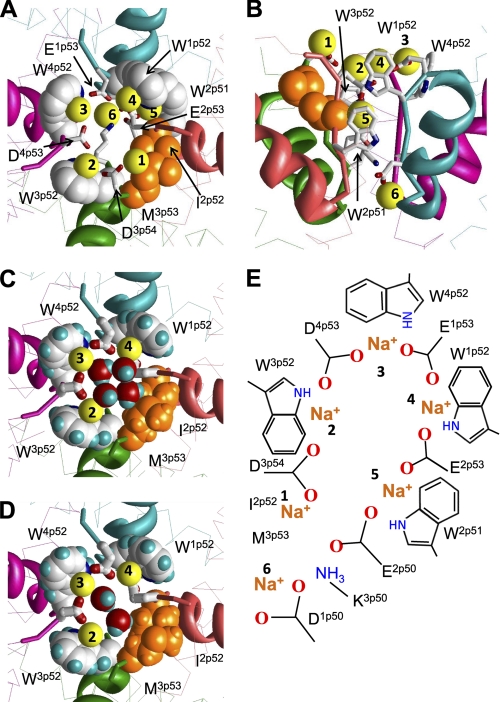
FIGURE 4.
Homology model of the NaV1.4 channel illustrating possible role of conserved tryptophans in ion permeation. A and B, extracellular and side views. The side chains of W1p52, W3p52, and W4p52 (space-filled with white carbons and blue nitrogens) along with I2p52 and M3p53 (space-filled side chains with orange heavy atoms) form a ring between the outer carboxylates (sticks) and the selectivity filter DEKA locus (sticks). Na+ ions are numbered as in scheme E. Ions 1 to 5 interact with the outer carboxylates (9) and ions 2 to 4 are simultaneously engaged in cation-π interactions with the conserved tryptophans in positions p52. Exceptionally conserved W2p51 provides a cation-attractive transient site between the outer carboxylates and the selectivity filter. A Na+ ion bound to W2p51 can simultaneously interact with an outer carboxylate E2p53 and an acidic residue in the DEKA locus (9). C and D, extracellular views show four and two water molecules, respectively, which fill the gap between Na+ ions at the level of the conserved tryptophans, providing additional coordination bonds for the first solvation shell of these ions. E, scheme of Na+ binding sites in the outer pore of the Na+ channel.
The first Na+ ion in the most extracellular position interacts with D3p54. This ion would not move deeper into the outer pore along the pore axis due to unfavorable interactions with the hydrophobic corner (Fig. 4B). The latter is formed by the highly conserved I2p52 and hydrophobically conserved residues in position 3p53 where Met, Ile, Thr, and Leu residues are found, respectively, in 78.0, 15.1, 5.0, and 1.8% of the sequences. The second, third, and fourth Na+ ions are engaged in cation-π interactions with tryptophans W3p52, W4p52, and W1p52, respectively. The fifth Na+ ion is engaged in cation-π interactions with W2p51 below the hydrophobic corner and this ion simultaneously interacts with the outer carboxylate E2p53 and the selectivity-filter glutamate E2p50. Importantly, the exceptionally conserved W2p51 is located deeper in the outer pore than tryptophans in positions p52. According to our model, W2p51 would stabilize the Na+ ion moving from the outer-carboxylates to the selectivity filter. The sixth Na+ ion is coordinated at the selectivity filter. Our model implies that Na+ ions move through the outer pore not linearly, but along a spiral path, consecutively occupying positions 1 through 6.
The space between Na+ ions in our model at the level of the conserved tryptophans is filled with water molecules. Asymmetry of the pore at this level (three tryptophans and the hydrophobic corner) dictates such orientation of the water dipoles, in which partial positive charges are directed toward the hydrophobic corner. Fig. 4, C and D show models with four and two water molecules at the level of the conserved tryptophans. The water molecules are linked to each other by H-bonds and a water molecule forms H-bonds with M3p53. It should be noted that structures in Fig. 4, C and D are just possible examples of Na+ hydration in the outer pore.
Thus, our current model agrees with our previous models in terms of TTX (5) and ion binding (9), but it proposes a new structural role for the exceptionally conserved tryptophans, whose π-electrons are predicted to face the ion permeation pathway and thus participate in ion permeation. The structures obtained by MC-minimizing the P-loops domain with TTX on one hand and with Na+ ions on the other hand are very similar (Fig. 2D). The root mean square deviation the ascending-limbs backbone atoms (positions p50–p54) between the two structures is as small as 0.4 Å. Na+ ions, which interact with the conserved tryptophans, are close to the positions of the hydroxyl hydrogens of a bound TTX molecule.
The Outer-pore Model for Ca2+ Channels
Modeling the outer pore of calcium channels is challenging due to deficiency of experimental data on binding specific toxins as small as TTX. Recently we developed a model of Ca2+ binding in the calcium channel selectivity filter, which predicts multiple Ca2+ chelation patterns involving long flexible side chains in the EEEE ring (6). Our present model of the sodium channel suggests the importance of the inter-repeat H-bonds involving conserved tryptophans for outer-pore stability and ion permeation. Sodium channels evolved from calcium channels (16). Many P-loop residues in sodium channels, including selectivity-filter residues, have changed during the evolution, but tryptophans W1p52, W3p52, and W4p52, did not change. The calcium channel tryptophan, W2p52 did not disappear during the evolution, but apparently shifted one position in the N-terminal direction to become exceptionally conserved W2p51 (Fig. 1B). These data allow us to suggest that the exceptionally conserved tryptophans in calcium channels play roles similar to those that we proposed for sodium channels.
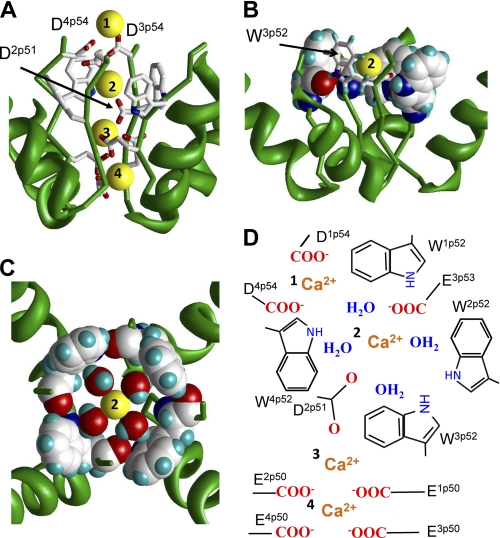
To model the tryptophan ring in the calcium channel, we used distance constraints to impose inter-repeat H-bonds involving the side chains and backbone carbonyls of the four Wp52s, MC-minimized the structure, and arrived at the model shown in Fig. 5. How could Ca2+ ions permeate through the tryptophan ring? Unlike Na+ ions, which can be directly engaged in cation-π interactions with tryptophans, strongly hydrated Ca2+ ions should interact with the aromatic moieties through water molecules (28). We placed a Ca2+ ion with four waters in the Wp52 ring, constrained D2p51 and E2p53 to the ion as proposed before (6), MC-minimized the structure with the constraints, and refined it without constraints. In the resulting structure (Fig. 5, B and C) four waters and the Ca2+ ion are in the same plane. The hydrated Ca2+ fits perfectly in the tryptophan ring. Each water molecule interacts with the tryptophan π-electrons by a hydrogen atom. The inner “diameter” of the H-bonded Wp52 ring is model-independent. The perfect match of the inner diameter with the outer “diameter” of the Ca2+ ion surrounded by four waters is hardly a coincidence. The Ca2+ ion in the tryptophan ring is hexa-coordinated (four waters, E2p53, and D2p51) and occupies the center of the octahedral bipyramid. This coordination geometry is found for calcium in many x-ray structures (29) and is predicted to be the most populated in water (30).
FIGURE 5.
Possible role of exceptionally conserved tryptophans in positions p52 of calcium channels. A, side view of the predicted pattern of Ca2+ binding in the CaV1.2 model. The most extracellular Ca2+ ion is coordinated by the outer carboxylates D1p54 and D4p54. The outer carboxylate E3p53 and highly conserved D2p51 coordinate the hydrated Ca2+ ion in the Wp52 ring. Two Ca2+ ions at lower levels of the outer pore are coordinated by the selectivity filter glutamates. B, side view at a Ca2+ ion in the ring of W52s. Residues W1p52, W2p52, and W4p52 are space-filled, and W3p52 is shown by sticks. C, top view at ring of four Wp52s, which is stabilized by inter-repeat H-bonds. Each H-bond involves the side chain NH group in the given repeat and the backbone carbonyl in the next repeat. In this arrangement, the tryptophan side chains form a cation-attractive path by maximizing exposure of π-electrons to the pore lumen. The size of the tryptophan-ring inner surface matches the dimension of the Ca2+ ion with the first hydration shell. Four water molecules and the Ca2+ ion lie in a plane, which is normal to the pore axis. D3p54 and highly conserved D2p51 provide two axial coordination bonds (not shown) to complete the hexa-coordinated geometry, which is seen in x-ray structures and predicted by molecular dynamics simulations (see text). The water oxygens interact with the Ca2+ ion, while water hydrogens interact with π-electrons of the tryptophans. D, schematic view of model A. Calcium ions are labeled with numbers.
Combining the current outer-pore model with our recent modeling study of Ca2+ binding to the selectivity-filter glutamates (6), we propose the following permeation scheme (Fig. 5, A and D). It involves two Ca2+ ions bound to the selectivity filter glutamates, a Ca2+ ion at the level of tryptophan ring and a Ca2+ ion bound to the outer ring of residues in positions p54. The outer carboxylates are not conserved in calcium channels (Fig. 1). In some channels the outer carboxylates may attract Ca2+ ions from the extracellular media to the outer pore. Positions p54 are occupied by residues whose side chains contain at least one electronegative atom (99.0, 41.1, 77.3, and 97.9% in repeats I, II, III, and IV, respectively). These residues would contribute to the first solvation shell of the incoming Ca2+ ion. The incoming Ca2+ ion would exchange its waters to the water molecules in the tryptophan ring, which along with the highly conserved D2p51 would coordinate the ion. Vacancies in the first coordination spheres of other Ca2+ ions would be also filled by waters (6). Thus, our model suggests that the tryptophan ring participates in the permeation process by stabilizing the conducting conformation of the outer pore and by helping to deliver the external Ca2+ ion to the selectivity filter. The water molecules may not move with Ca2+ and remain H-bonded to π-electrons of the tryptophans, suggesting a mechanism of Ca2+ dehydration.
Comparing Calcium Binding in the Selectivity Filters of Ca2+ and Na+ Channels
The selectivity filter region in the Ca2+ channel has a net charge of −5 proton charge units (the EEEE locus and D2p51), while this region in the Na+ channel has only −1 proton charge units (the DEKA locus and neutral W2p51). Na+ channels are blocked by divalent cations at the selectivity filter level (28). To reveal possible structural determinants of the block, we docked a Ca2+ ion into the outer-pore region of the Na+ channel from multiple starting positions and MC-minimized the complex using an approach described elsewhere (31). In the energetically optimal structures, the Ca2+ ion strongly bound to D1p50 and E2p50 in the DEKA locus and destroyed contacts of these residues with the ammonium group of K3p50 that moved toward the inner pore (Fig. 2E). In some of the MC-minimized structures, the Ca2+ ion bound to the outer carboxylates without destroying the salt bridge between D1p50 and K3p50. The conserved tryptophans W1p52, W3p52, and W4p52, which are located externally to the selectivity filter, were too far from the Ca2+ ion bound to the DEKA locus. However, the aromatic ring of Y1p51, which is 4–5 Å from the Ca2+ ion in our model, would interact with the ion through a water molecule in agreement with the proposition that interaction of Y1p51 and the channel-blocking Ca2+ ion is mediated by a water molecule (28). The proximity of Y1p51 to the Ca2+ ion is supported by the data that NaV1.5, which contains C1p51, is less sensitive to the Ca2+ block than NaV1.4 (32). The conserved W2p51 is at a similar distance from the Ca2+ ion and could also interact with it through a water molecule.
In the Ca2+ channel, up to five negative charges at the selectivity filter allow simultaneous binding of two or three calcium ions (6). The repulsion between the ions weakens the binding and facilitates the ion permeation (6). In the sodium channel, binding of the Ca2+ ion to D1p50 and E2p50 is very strong because no additional binding sites for a divalent cation are within 6 Å from the DEKA locus. The lack of neighboring divalent cations likely explains why Ca2+ ions do not permeate through the sodium channel, but block it.
DISCUSSION
In the present work we aligned many sequences of the P-loop segments of sodium and calcium channels and revealed exceptionally and highly conserved residues whose roles were not explained before. We suggest that the evolutionary conservation of these residues is due to their participation in maintaining the outer-pore structure and their involvement in the ion permeation. Our models provide a structural explanation for the conservation of the residues and relate it to functional peculiarities of calcium and sodium channels. The proposed tryptophan ring is symmetrical in calcium channels. A single Ca2+ ion surrounded by four water molecules fits snugly into the ring. The conserved D2p51 would deliver the ion to the selectivity-filter glutamates. The selectivity-filter glutamates along with D2p51 may bind three Ca2+ ions (6). A system of three divalent ions and five ionized carboxylates has an excessive positive charge of one proton-charge unit that may facilitate the ion permeation (6). In sodium channels, the positive charge of one proton-charge unit in the fully ionized DEKA ring is counterbalanced by a Na+ ion. If aspartate D2p51, which is present in all Ca2+ channels, were to remain in Na+ channels to deliver a monovalent cation to the DEKA ring, the net negative charge of the amino acids at the selectivity filter (−2 proton charge units) would attract a Ca+ ion and thus decrease the channel selectivity to Na+. Evolution of Na+ channels from Ca2+ channels involved substitution of the negatively charged aspartate D2p51 with the electroneutral (but cationophilic) W2p51. The substitution, which may have resulted from deletion of D2p51 in the Ca2+ channel sequence, likely helped to reduce affinity for Ca2+ ions.
The exact role of residues in position 2p51 needs further experimental analysis. It is well known that mutations in the EEEE locus in calcium channels and the DEKA locus in sodium channels, which did not change residues in position 2p51, alter the Na+ and Ca2+ selectivity (33, 34). On the other hand, the observation that the prokaryotic homotetrameric channel NaChBac has the EEEE locus initially led to categorize it as a Ca2+-selective channel (8). Subsequently expressed NaChBac is found to be a Na+-selective channel (35). Additional acidic residues in the ascending limbs, in particular in position p51, are required to render the NaChBac channel Ca2+-selective (36).
Here we proposed that the exceptionally conserved tryptophans in the ascending limbs of calcium and sodium channels participate in stabilizing the outer-pore structure by H-bonds, which involve the side chain NH groups. The ascending limbs contain free backbone carbonyls, which can accept H-bonds. In our model of CaV1.2, the side chain of Wp52 in a given repeat donates an H-bond to the backbone carbonyl of the neighboring-repeat Wp52 (Fig. 5). A similar network of H-bonds involving tryptophans Wp52 is proposed for the sodium channels. Importantly, the tryptophan rings do not preclude the selectivity filter and outer carboxylates residues from exposing their side chains into the pore lumen and thus participate in the ion permeation process. Due to the network of inter-repeat H-bonds, the outer-pore lumen at the level of p52 is rather narrow, but in our model it still accommodates a TTX molecule. It will be interesting to further explore whether our narrow outer-pore model is consistent with intriguing data on synergistic and antagonistic interactions between tetrodotoxin and mu-conotoxin in blocking voltage-gated sodium channels (37).
Although Ca2+ and Na+ channels are believed to have similar architecture of the outer pore, the ion permeation mechanisms in these channels are significantly different. Thus, strong ion coupling in the Ca2+ channels seems critical for the anomalous mole fraction effect, while the narrow part of the Na+ channel pore is predominantly single-occupied (16). Our computational methodology does not allow a direct detailed simulation of the flux coupling. However, our models predict that the equilibrium binding sites for permeating ions are separated by 5–6 Å along the pore axis. At such distances, the divalent ions in the Ca2+ channel repel each other more strongly than monovalent ions in the Na+ channel. This repulsion compensates the strong binding of divalent ions to their binding sites.
Limitations of our model should be spelled out. Our computational methodology allows one to reproduce structural details of known complexes of proteins with organic ligands (38) and calcium ions (31). However a homology model, which includes only a part of a large transmembrane protein and lacks membrane lipids, is not expected to correspond to the global energy minimum. Therefore, we initially preserved the structural stability of the models by pin constraints imposed to alpha carbons of the P-helices (general folding) and specific H-bonding constrains involving conserved tryptophans and polar residues in positions p45. When metal ions and a TTX molecule (in Na+ channel) were added, the models did not change significantly even after intensive MC-minimizations without constraints because permeating cations counterbalanced excessive negative charges of the pore-exposed residues, while TTX stabilized the model by H-bonds and other interactions. (The importance of permeating cations for supporting the native structure of ion channels is well known.) Thus, our modeling approach allowed us to propose specific contacts, which stabilize the ascending limbs in sodium and calcium channels. Imposing these contacts as constraints in MC minimizations resulted in the models that can explain various experimental data.
Another limitation is related to the proposed permeation mechanism. We did not model the permeation process, but predicted transient binding sites for permeant ions. Therefore, the proposed details of the permeation mechanisms should be considered as hypotheses derived from static snapshots. The MCM method used in the present work is not appropriate for realistic modeling of the permeation process because it involves large random transitions between sampled structures. The intensive sampling of the conformational space allows predicting stable structures, but MCM neglects local energy barriers, which are important to simulate the ion permeation. The proposed details of the ion permeation mechanism could be tested by computing the free energy profile for the ion permeating through the outer pore, but such simulations, which would require different computational methodologies, are beyond the scope of our study. Our models lay the groundwork for the dynamic calculations that will be needed to address specific effects such as ion coupling in the pore.
The outer pore of ion channels governs selectivity, permeation, slow inactivation, and binding of toxins. Many data indicate similar dispositions of S6, S5, and P-helices in voltage-gated potassium, calcium, and sodium channels. However, the ascending limbs in calcium and sodium channels are generally believed to fold not like in potassium channels. Here we elaborated our previous models (5, 6) to explain roles of exceptionally conserved residues, which were revealed in multiple-sequence alignments, and explained recently published experimental data. The fact that the P-helices and S6s in our homology models are built using a potassium channel template further supports the notion that the folding of these segments in potassium, sodium, and calcium channels is generally similar.
Supplementary Material
Acknowledgments
We thank Ludmila Milova for generating multiple sequence alignments and preparing Fig. 1. We thank Daniel Garden for reading the manuscript. Computations were made possible by the facilities of the Shared Hierarchical Academic Research Computing Network.
This study was supported by grants from Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research (MOP-53229) (to B. S. Z.), and by a grant from the RAS Program Molecular and Cell Biology (to D. B. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
We designate residues using labels that are universal for P-loop channels (Fig. 1). A residue label includes repeat number, which is omitted if all repeats are referred to, segment index (i, inner helix; p, P-loop; o, outer helix), and position in the segment.
- TTX
- tetrodotoxin
- DEKA
- selectivity-filter ring of Asp, Glu, Lys, and Ala residues from the four P-loop domains of sodium channels
- EEEE
- selectivity-filter ring of glutamate residues from the four P-loop domains of calcium channels
- MC
- Monte Carlo
- MCM
- Monte Carlo minimization.
REFERENCES
- 1. Dudley S. C., Jr., Chang N., Hall J., Lipkind G., Fozzard H. A., French R. J. (2000) J. Gen. Physiol. 116, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruhova I., Zhorov B. S. (2010) J. Gen. Physiol. 135, 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhorov B. S., Tikhonov D. B. (2004) J. Neurochem. 88, 782–799 [DOI] [PubMed] [Google Scholar]
- 4. Lipkind G. M., Fozzard H. A. (1994) Biophys. J. 66, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tikhonov D. B., Zhorov B. S. (2005) Biophys. J. 88, 184–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng R. C., Tikhonov D. B., Zhorov B. S. (2010) Eur. Biophys J. 39, 839–853 [DOI] [PubMed] [Google Scholar]
- 7. Zhorov B. S., Folkman E. V., Ananthanarayanan V. S. (2001) Arch. Biochem. Biophys. 393, 22–41 [DOI] [PubMed] [Google Scholar]
- 8. Durell S. R., Guy H. R. (2001) Biochem. Biophys. Res. Commun. 281, 741–746 [DOI] [PubMed] [Google Scholar]
- 9. Tikhonov D. B., Zhorov B. S. (2007) Biophys. J. 93, 1557–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiner S. J., Kollman P. A., Case D. A., Singh U. C., Ghio C., Alagona G., Profeta S., Weiner P. (1984) J. Am. Chem. Soc. 106, 765–784 [Google Scholar]
- 11. Weiner S. J., Kollman P. A., Nguyen D. T., Case D. A. (1986) J. Comput. Chem. 7, 230–252 [DOI] [PubMed] [Google Scholar]
- 12. Lazaridis T., Karplus M. (1999) Proteins 35, 133–152 [DOI] [PubMed] [Google Scholar]
- 13. Dewar M. J., Zoebisch E. G., Healy E. F., Stewart J. J. (1985) J. Amer. Chem. Soc. 107, 3902–3909 [Google Scholar]
- 14. Li Z., Scheraga H. A. (1987) Proc. Natl. Acad. Sci. U. S. A. 84, 6611–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bruhova I., Tikhonov D. B., Zhorov B. S. (2008) Mol. Pharmacol. 74, 1033–1045 [DOI] [PubMed] [Google Scholar]
- 16. Hille B. (2001) Ion Channels of Excitable Membranes, Sinauer Associates Inc., Sunderland, MA [Google Scholar]
- 17. Struyk A. F., Cannon S. C. (2002) J. Gen. Physiol. 120, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong W., Li R. A., Tian Y., Tomaselli G. F. (2003) J. Gen. Physiol. 122, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsushima R. G., Li R. A., Backx P. H. (1997) J. Gen. Physiol. 109, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsang S. Y., Tsushima R. G., Tomaselli G. F., Li R. A., Backx P. H. (2005) Mol. Pharmacol. 67, 424–434 [DOI] [PubMed] [Google Scholar]
- 21. Carbonneau E., Vijayaragavan K., Chahine M. (2002) Pflugers Arch. 445, 18–24 [DOI] [PubMed] [Google Scholar]
- 22. Santarelli V. P., Eastwood A. L., Dougherty D. A., Horn R., Ahern C. A. (2007) J. Biol. Chem. 282, 8044–8051 [DOI] [PubMed] [Google Scholar]
- 23. Chen L. Q., Chahine M., Kallen R. G., Barchi R. L., Horn R. (1992) FEBS Letts. 309, 253–257 [DOI] [PubMed] [Google Scholar]
- 24. Backx P. H., Yue D. T., Lawrence J. H., Marban E., Tomaselli G. F. (1992) Science 257, 248–251 [DOI] [PubMed] [Google Scholar]
- 25. Satin J., Kyle J. W., Chen M., Bell P., Cribbs L. L., Fozzard H. A., Rogart R. B. (1992) Science 256, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 26. Geffeney S. L., Fujimoto E., Brodie E. D., 3rd, Brodie E. D., Jr., Ruben P. C. (2005) Nature 434, 759–763 [DOI] [PubMed] [Google Scholar]
- 27. Yamagishi T., Li R. A., Hsu K., Marbán E., Tomaselli G. F. (2001) J. Gen. Physiol. 118, 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santarelli V. P., Eastwood A. L., Dougherty D. A., Ahern C. A., Horn R. (2007) Biophys. J. 93, 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McPhalen C. A., Strynadka N. C., James M. N. (1991) Adv. Protein Chem. 42, 77–144 [DOI] [PubMed] [Google Scholar]
- 30. Ikeda T., Boero M., Terakura K. (2007) J. Chem. Physics 127, 074503-1–074503-8 [DOI] [PubMed] [Google Scholar]
- 31. Cheng R. C., Zhorov B. S. (2010) Eur. Biophys. J. 39, 825–838 [DOI] [PubMed] [Google Scholar]
- 32. Chahine M., Chen L. Q., Kallen R. G., Barchi R. L., Horn R. (1992) Biophys. J. 62, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heinemann S. H., Terlau H., Stühmer W., Imoto K., Numa S. (1992) Nature 356, 441–443 [DOI] [PubMed] [Google Scholar]
- 34. Tang S., Mikala G., Bahinski A., Yatani A., Varadi G., Schwartz A. (1993) J. Biol. Chem. 268, 13026–13029 [PubMed] [Google Scholar]
- 35. Ren D., Navarro B., Xu H., Yue L., Shi Q., Clapham D. E. (2001) Science 294, 2372–2375 [DOI] [PubMed] [Google Scholar]
- 36. Yue L., Navarro B., Ren D., Ramos A., Clapham D. E. (2002) J. Gen. Physiol. 120, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang M. M., McArthur J. R., Azam L., Bulaj G., Olivera B. M., French R. J., Yoshikami D. (2009) Channels 3, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garden D. P., Zhorov B. S. (2010) J. Computer-aided Mol. Design 24, 91–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.