Abstract
Ribosome biogenesis is required for normal cell function, and aberrant ribosome biogenesis can lead to p53 activation. However, how p53 is activated by defects of ribosome biogenesis remains to be determined. Here, we identified human UTP14a as an SSU processome component by showing that hUTP14a is nucleolar, associated with U3 snoRNA and involved in 18 S rRNA processing. Interestingly, ectopic expression of hUTP14a resulted in a decrease and knockdown of hUTP14a led to an increase of p53 protein levels. We showed that hUTP14a physically interacts with p53 and functionally promotes p53 turn-over, and that hUTP14a promotion of p53 destabilization is sensitive to a proteasome inhibitor but independent of ubiquitination. Significantly, knockdown of hUTP14a led to cell cycle arrest and apoptosis. Our data identified a novel pathway for p53 activation through a defect in rRNA processing and suggest that a ribosome biogenesis factor itself could act as a sensor for nucleolar stress to regulate p53.
Keywords: Cell Cycle, p53, Proteasome, Protein Degradation, rRNA Processing, SSU Processome, hUTP14a, Ribosomal Stress
Introduction
Ribosome biogenesis is required for normal cell function, and aberrant ribosome biogenesis can lead to p53 activation. In eukaryotes ribosome biogenesis including transcription of ribosome RNA genes (rDNA),3 rRNA processing, and assembly of 40 S and 60 S ribosome subunits takes place in the nucleolus. A 47 S rRNA precursor (pre-rRNA) containing the sequences for the mature rRNAs (18 S, 5.8 S, and 28 S rRNA), two external transcribed spacers (ETS) and two internal transcribed spacers (ITS) is transcribed by RNA polymerase I (pol I). After chemical modification at numerous sites, the 47 S pre-rRNA is cleaved to produce 18 S, 5.8 S, and 28 S rRNAs. The 18 S rRNA is incorporated in the 40 S small subunit (SSU) with small subunit ribosomal proteins. The 5.8 S rRNA and 28 S rRNA are incorporated into the 60 S large subunit with 5 S rRNA, which is independently transcribed by RNA polymerase III elsewhere. Both the large and small subunits are assembled in the nucleolus and transferred to the cytoplasm for protein synthesis.
Polymerase I transcription and pre-rRNA processing can be disrupted by serum starvation, depletion of nucleotides, chemical reagents, and malfunction of nucleolar proteins. All of these nucleolar functional disruptions have been shown to result in nucleolar stress signaling to p53. For example, actinomycin D selectively inhibits RNA pol I-dependent transcription at low concentrations (5–10 nm) (1, 2) and stimulates stabilization of p53. The chemotherapeutic agent 5-fluorouracil (5-FU), which blocks pre-rRNA processing by incorporating newly synthesized rRNA activates p53. Functional disruption of nucleolar proteins, which are required for rRNA processing, also activates p53. Bop1 is an essential factor in 28 S rRNA processing and its dominant negative mutant inhibits ribosomal biogenesis and elicits p53 activation (3). For 18 S rRNA processing, the U3 snoRNA base pairs with the 47 S pre-rRNA at A0, A1, and A2 sites (4, 5) and mediates 18 S rRNA maturation (6). If a protein is nucleolar, associated with U3 snoRNA and required for 18 S rRNA processing, it is identified as UTP (U3 protein). UTPs are also known as SSU processome as they play important roles in 40 S subunit biogenesis (7, 8). Recently, disruption of human UTP18 was found to induce p53 activation (9). In addition, inhibition of nucleophosmin (B23) activity by ARF (10) and reduction of ribosomal protein S6 (11) also activates p53.
All of these nucleolar or ribosomal stresses activate p53, which induces cell cycle arrest and/or apoptosis. The in vivo significance of this p53 activation has been determined from a number of mouse models. For example, A mouse model with the juvenile spermatogonial depletion phenotype (jsd) showed that utp14b is required for spermatogenesis in mice (12, 13). A p53-dependent pathway has been found to mediate apoptosis in spermatogonial differentiation in utp14bjsd mice (14). However, the mechanisms by which nucleolar disruptions direct p53 activation are largely undefined.
In unstressed cells, the p53 protein level remains low through regulation of its protein stability by a number of negative regulators. MDM2 serves as a key negative feedback regulator for p53 and various stresses activate distinct cellular signaling pathways leading to the suppression of MDM2 activity and activation of p53 (15, 16). Thus, the p53-MDM2 feedback loop plays an essential role in response to a multitude of genotoxic and cytotoxic stressors. It has been found that 5 S rRNA and ribosomal protein (RP) of the large subunit RPL5 interact with MDM2 (17) and RPL5 participates in MDM2 nuclear export (18). It was thought that p53-MDM2 might “hitch a ride” on the ribosome for cytoplasmic degradation (19). Therefore, nucleolar stress was thought to induce p53 accumulation due to a failure in nucleolus-dependent export and degradation of p53 in the cytoplasm (20). Later studies found that treating cells with either a lower dose of actinomycin D or serum starvation inhibits ribosome assembly and consequently releases free ribosomal proteins from the nucleolus to the nucleoplasm (21). Moreover, it has been found that several ribosomal 60 S proteins including RPL11 (22, 23), RPL23 (24, 25), and RPL5 (23) interact with MDM2. This binding inhibits the MDM2 E3 ligase function, resulting in p53 accumulation and activation. A small ribosomal subunit protein RPS7 (27, 28) has also been shown to interact with MDM2. In addition, RPL26 was found to increase the translational rate of p53 mRNA by binding to its 5′-untranslated region (29). All these findings identify p53 as a molecule which is critical in sensing nucleolar stress, and suggest that RPs may play a pivotal role in the p53 response to nucleolar stress. A recent study revealed that defects in 18 S and 28 S rRNA processing activate p53 by an RPL11-dependent pathway (9). However, the specific linkage between pre-rRNA processing and p53 activation has not been determined.
Recently, mutations in human UTP14C have been found in 3 of 234 nonobstructive and azoospermic/severely oligospermic males, suggesting that the human UTP14C gene is associated with human spermatogenesis and fertility, and raising the possibility UTP14C may be functionally equivalent to mouse utp14b (30). More importantly, expression of UTP14C mRNA has also been found to be associated with human ovarian cancer (31). Mouse utp14b and human UTP14C are retrogenes of the X-linked utp14a gene and UTP14A, respectively. Thus, functional study of human UTP14a will determine the fundamental molecular mechanisms by which UTP14c functions in spermatogenesis deficiency and ovarian cancer. Therefore, we set out to investigate the function of human UTP14a (hUTP14a) and explore the mechanism by which nucleolar stress activates the p53 pathway. In this study, we identified hUTP14a as the mammalian homolog of yeast Utp14 which functions in 18S rRNA processing, and found that hUTP14a itself acts as a nucleolar stress sensor, which signals to p53.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
The expression plasmids coding pCI-neo-Flag-hUTP14a and series deletion mutants Del-1 (amino acids 1–267), Del-2 (amino acids 268–645), and Del-3 (amino acids 646–771) were obtained by RT-PCR-cloning using total RNA extracted from HeLa cells as a template. The plasmids were verified by DNA sequencing. Plasmids coding pEGFP-hUTP14a and its series deletion mutants were constructed by inserting hUTP14a cDNA fragments from pCI-neo-Flag-hUTP14a into the pEGFP plasmid. Plasmids coding GST-hUTP14a and its series deletion mutants were constructed by inserting hUTP14a cDNA fragments from pCI-neo-Flag-hUTP14a into the pGEX-4T1 plasmid. Plasmids coding MDM2, p53 and p53 deletion mutants were kindly provided by Dr. Yongfeng Shang (Peking University Health Science Center). Plasmid coding HA-Ub was constructed by inserting an ubiquitin cDNA fragment into a pCMV-HA plasmid. Antibodies directed against p53 or MDM2 were purchased from Santa Cruz Biotechnology, p53 phosphorylation antibodies including anti-Ser15, anti-Thr18, anti-Ser20, anti-Ser33, and anti-Ser37 are from Cell Signaling. Anti-p21Waf1/Cip1, anti-nucleolin, anti-PARP, and anti-β-actin were from Santa Cruz Biotechnology, and anti-HA and anti-Flag monoclonal antibody M2 were from Sigma. Anti-1A6/DRIM monoclonal antibody was raised in our laboratory. TRITC-conjugated goat anti-mouse IgG and FITC-conjugated goat anti-rabbit IgG were from the Zhongshan Goldenbridge Biotechnology Co., Ltd.
Generation of an Anti-hUTP14a Antibody
A rabbit polyclonal antibody was generated against peptides derived from the C-terminal 18 amino acids (amino acids 751–768: QRNPKRITTRHKKQLKKC) of the hUTP14a protein and used after affinity purification.
Cell Culture and Transfections
Cell lines were grown in DMEM or RPMI 1640 medium supplemented with 10% FCS according to the instructions of the ATCC (American Type Culture Collection). For knockdown of hUTP14a expression, three hUTP14a-specific siRNAs (small interference RNA) (A-1: 5′-CAGGAAGAACUAGCGGAUUtt-3′; A-2: 5′- GACGCCGGUUUCUCAUUAAtt-3′; A-3: 5′- GAGAUUGAACGGAUCCACAtt-3′) together with an unrelated control siRNA (siNC: 5′-ACUACCGUUGUUAUAGGUG-3′) were chemically synthesized (Shanghai GenePharma Co., Ltd). These synthesized siRNAs were transfected into cells at a concentration of 100 nm. Transfections were carried out with LipofectamineTM 2000 (Invitrogen) according to the manufacturer's instructions.
Immunofluorescence Staining
Cells were plated on coverslips in 6-well plates 1 day before harvest. 1A6/DRIM, hUTP14a, Flag-hUTP14a, and nucleolin were detected, respectively, with the monoclonal anti-1A6/DRIM antibody 6D9, the polyclonal antibody against hUTP14a, and the monoclonal antibody against Flag or nucleolin-specific polyclonal antibody after cells were fixed with methanol/acetone. The 1A6/DRIM- or Flag-hUTP14a-specific immunocomplexes were identified with TRITC-conjugated goat anti-mouse IgG. The hUTP14a and nucleolin specific immunocomplexes were detected with FITC-conjugated goat anti-rabbit IgG. Immunofluorescence signals were recorded with confocal laser scanning microscopy (Leica TCS-ST2).
Newly Synthesized rRNA Analysis
Pulse-chase labeling was performed as described previously (32). In brief, 72 h after transfection of chemically synthesized siRNA, HeLa cells were labeled with l-[methyl-3H]methionine (PerkinElmer Life Sciences) for 30 min. RNA was evaluated at various time points. Total RNA was isolated with Trizol reagent (Invitrogen) and resolved on a 1% agarose-glyoxal gel (Ambion). The RNA was transferred to a nylon membrane and the newly synthesized rRNAs were detected by fluorography.
Immunoprecipitation Analyses of Protein-RNA Association
Immunoprecipitation was performed as described previously (33). Briefly, HeLa cell lysates were prepared in buffer A (25 mm Tris-Cl, pH 7.5, 100 mm KCl, 1 mm dithioerythreitol, 2 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 0.05% Nonidet P-40, 1 unit/ml RNasin) and used directly for immunoprecipitation. Antibody was coupled with a 50% suspension of protein A-Sepharose beads (Amersham Biosciences) in IPP500 (500 mm NaCl, 10 mm Tris-Cl, pH 8.0, 0.05% Nonidet P-40). Coupled beads were incubated with cellular extracts for 2 h at 4 °C. After washes, precipitated proteins were analyzed by Western blotting and co-precipitated RNA was isolated with Trizol® reagent (Invitrogen).
Immunoblot
Proteins from cellular fractions were separated on SDS-PAGE and transferred onto PVDF membrane. Membranes were probed with corresponding primary antibodies after blocking with 5% milk in PBS/T (0.5% Tween-20 in PBS buffer). After extensive washing with PBS/T, the membrane was incubated with HRP-conjugated secondary antibody followed by detection using the ECL-Kit (Amersham Biosciences).
Northern Blot
To visualize U3 snoRNA, Northern blot was performed as described previously (34) with minor modifications. In brief, the RNA sample was loaded onto 7% polyacrylamide-8.3 m urea gel and blotted onto a BrightStar®-PLUS positively charged Nylon membrane (Ambion) by electroblotting (Bio-Rad). The U3 snoRNA-specific RNA probe was labeled with biotin-UTP using the Riboprobe® System (Promega) with SalI-linearized pGEM-T-U3 (34). Hybridization was carried out for 16 h at 65 °C after 3 h of prehybridization. Detection was carried out with the BrightStar® BioDetectTM Kit (Ambion) according to the manufacturer's instructions.
Reverse Transcription PCR
For U3 snoRNA amplification, RT-PCR was performed as previously described (34).
GST Pull-down
GST or GST fusion proteins were expressed in E. coli and crosslinked to glutathione-Sepharose beads (BD Product). In vitro transcription/translation was performed with a rabbit reticular lysis-coupled transcription/translation kit (Promega). GST pull-down was carried out by incubating the GST fusion proteins with in vitro transcribed/translated proteins. The GST fusion protein interacting proteins were detected by Western blotting probed with appropriate antibodies.
Flow Cytometric Analysis
For DNA content analysis, cells were trypsinized, washed with PBS, and fixed in 75% ice-cold ethanol at 4 °C overnight. Cells were rehydrated in PBS on the second day. Following RNase digestion, cells were stained with propidium iodide (PI). Flow cytometry analysis was performed using red (PI) emission (at 630 nm). Data from 104 cells were collected and analyzed by using Cellquest software (Becton Dickinson). For apoptosis analysis, cells were double-stained with annexin V and PI and subjected to flow cytometric analysis.
BrdU Incorporation Assay
The cell population in S-phase was determined with a BD PharmingenTM BrdU Flow kit according to the manufacturer's instruction. Briefly, cells were pulsed for 16 h with bromodeoxyuridine (BrdU), and were then stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU and 7-amino-actinomycin D (7-AAD). Positive BrdU staining and 7-AAD staining were analyzed by FACScan (Becton Dickinson). Data from 104 cells were analyzed by using Cellquest software (Becton Dickinson). Cells in S-phase were quantitated by comparing FITC (DNA incorporation) versus 7-AAD (total DNA) staining.
Growth Curve
A growth curve was plotted with the Cell Counting Kit-8 (CCK-8, Dojindo) according to the manufacturer's instructions. In brief, cells were seeded in 96-well plates. WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] was added to these cells and incubated for 4 h at different time points. Absorbance at 450 nm was measured using a microplate reader. The experiment was repeated three times in duplicate. Growth curves were plotted using the mean ± S.D. of absorbance at 450 nm versus the time points.
RESULTS
Expression Profile of hUTP14a in Different Human Cell Lines
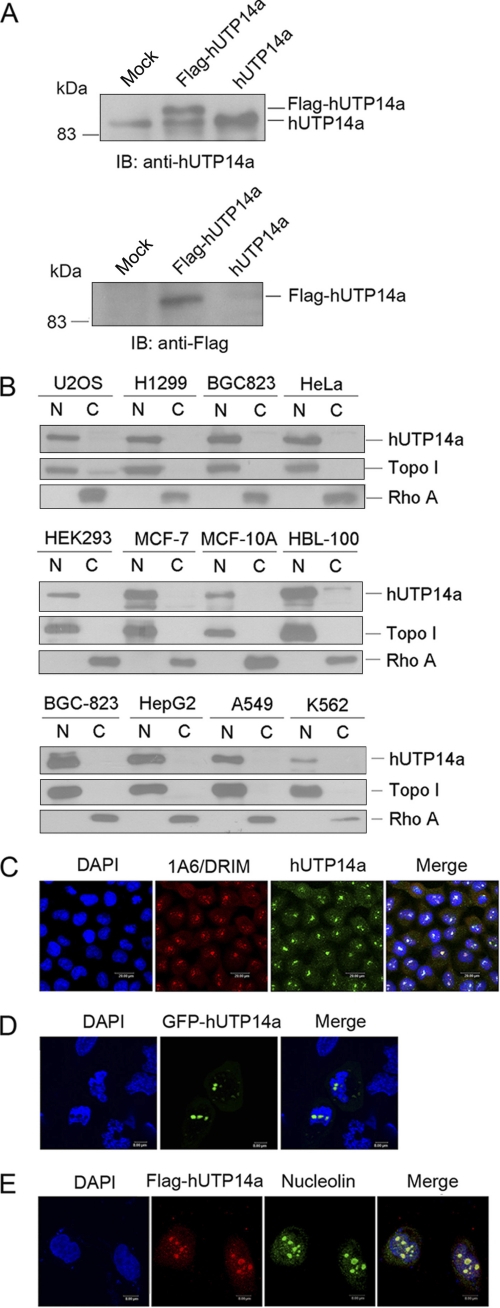
Sequence alignment showed that hUTP14a is conserved among Saccharomyces cerevisiae, mice, and humans (supplemental Fig. S1). To evaluate the function of hUTP14a protein and to detect endogenous hUTP14a expression, we raised a rabbit polyclonal antibody against hUTP14a and confirmed its specificity by Western blotting utilizing whole cell lysates of Flag-hUTP14a or hUTP14a-transfected HeLa cells (Fig. 1A). A band at the expected molecular mass for hUTP14a of 88 kDa was observed in HeLa cells and hUTP14a-transfected cells. Flag-tagged hUTP14a was recognized by both anti-hUTP14a antibody and anti-Flag antibody M2 as a band of the same size. In addition, we analyzed the expression of hUTP14a in various cell lines. Cellular fractions were prepared and proteins from the fractions were separated on SDS-PAGE, and transferred onto a PVDF membrane. The blot was probed with anti-hUTP14a. Fig. 1B shows that hUTP14a was ubiquitously expressed in the nuclear extracts of the cell lines under evaluation. Subcellular localization of endogenous hUTP14a was determined by indirect immunofluorescence performed with the polyclonal anti-hUTP14a antibody and an anti-1A6/DRIM monoclonal antibody was used as a nucleolar protein marker. As shown in Fig. 1C, hUTP14a was predominantly localized in the nucleolus and co-localized with 1A6/DRIM (also known as human UTP20). To investigate the localization of ectopically expressed hUTP14a, GFP-hUTP14a was transfected into U2OS cells and GFP fluorescence showed the same localization pattern with endogenous hUTP14a (Fig. 1D). Flag-hUTP14a was transfected into U2OS cells and double indirect immunofluorescence staining was performed with anti-Flag antibody and anti-nucleolin antibody. The confocal image showed Flag-hUTP14a co-localized with nucleolin in the nucleolus (Fig. 1E).
FIGURE 1.
Expression profile of hUTP14a in different human cell lines. A, specificity of anti-hUTP14a antibody was verified by Western blotting. Whole cell extracts were prepared from HeLa cells and Flag-hUTP14a- or hUTP14a-transfected HeLa cells. Equal amount of proteins from the extracts were separated by SDS-PAGE, and transferred onto PVDF membranes. Blots were probed with anti-hUTP14a antibody. Endogenous and exogenous hUTP14a are indicated (upper panel). Blots as described above were stripped and reprobed with anti-Flag antibody M2 (lower panel). B, expression of hUTP14a in various human cell lines. Cytosolic and nuclear extracts were fractionated from multiple cell lines. The same amount of protein was separated on SDS-PAGE and transferred onto a PVDF membrane. Blots were probed with anti-hUTP14a antibody. Cellular fractionation was controlled by using nuclear protein topoisomerase I (Topo I) as a nuclear marker and cytoplasmic protein RhoA as a cytosolic marker. N represents nuclear extract and C represents cytosolic extract. Cell lines are indicated at the top of the blots. C, endogenous hUTP14a is localized to the nucleolus. Immunofluorescence was performed with 1A6/DRIM-specific monoclonal antibody and anti-hUTP14a polyclonal antibody on U2OS cells. 1A6/DRIM immuno-signals were detected with TRITC-conjugated goat anti-mouse IgG and hUTP14a immuno-complexes were recognized with FITC-conjugated goat anti-rabbit IgG. Nucleus was stained with DAPI. D, GFP-hUTP14a displayed the same localization pattern as endogenous hUTP14a. Plasmid coding GFP-hUTP14a was transfected into U2OS cells. Localization of GFP protein was observed and recorded with confocal microscopy. E, ectopic Flag-hUTP14a is localized to the nucleolus. U2OS cells were transfected with Flag-hUTP14a. Immunofluorescence was performed using anti-Flag monoclonal antibody and anti-nucleolin polyclonal antibody. Flag-hUTP14a immuno-signals were detected with TRITC-conjugated goat anti-mouse IgG and nucleolin immuno-complexes were recognized with FITC-conjugated goat anti-rabbit antibody. The nuclei were stained with DAPI.
hUTP14a Is an SSU Processome Component
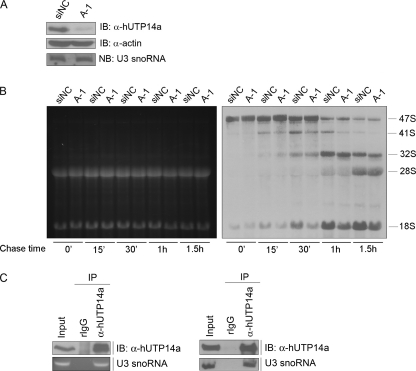
To investigate whether hUTP14a is required for pre-rRNA processing, hUTP14a-specific siRNA was transfected into HeLa cells, and pulse chase labeling was performed to analyze newly synthesized rRNA 72-h post-transfection. Efficiency of hUTP14a silencing was determined by Western blotting using anti-hUTP14a antibody (Fig. 2A). The pulse chase results showed that at the chase time points 15 min, 30 min, 1 h, and 1.5 h, the level of 41 S rRNA decreased (Fig. 2B, right). In addition, the 18 S rRNA level decreased markedly at chase time points 15 min, 30 min, 1 h, and 1.5 h in hUTP14a-deficient cells. To determine whether hUTP14a is associated with U3 snoRNA, immunoprecipitation was performed using anti-hUTP14a antibody. Protein and RNA from the precipitates were analyzed in parallel. Proteins extracted from one-half of the precipitate were subjected to Western blotting to analyze for hUTP14a. RNA was extracted from the remaining half of the precipitate and was used for RT-PCR to analyze for U3 snoRNA. As shown in Fig. 2C (left panel), U3 snoRNA was amplified from the hUTP14a-specific immunoprecipitates. To confirm this observation, RNA was extracted from the hUTP14a-specific immunoprecipitates and subjected to Northern blotting to analyze for U3 snoRNA. The results showed that U3 snoRNA was present in the hUTP14a-specific immunoprecipitates (Fig. 2C, right panel) demonstrating in vivo association between hUTP14a and U3 snoRNA. In addition, Northern blotting showed that U3 snoRNA levels were not affected by silencing of hUTP14a expression (Fig. 2A, lower panel). These results demonstrated that depletion of hUTP14a inhibited 18 S rRNA processing. Taken these results together, we identified hUTP14a as an SSU processome component.
FIGURE 2.
hUTP14a is an SSU processome component. A, HeLa cells were transfected with a hUTP14a-specific siRNA (A-1) or a control siRNA (siNC). Cell lysates were prepared 72-h post-transfection and subjected to Western blotting. Blots were probed with anti-hUTP14a antibody. β-Actin was used as a loading control. RNA extracted from the cells described above was resolved on a 1% agarose-glyoxal gel and transferred onto a nylon membrane. The blot was hybridized with biotin-labeled U3 snoRNA probe. B, cells transfected with siRNAs as described in A were pulse labeled with [3H]methionine and analyzed at different time points 72-h post-transfection. Total RNA was extracted and resolved on a 1% agarose gel. After staining with ethidium bromide and photographing the gel (left panel), RNA was transferred from the agarose gel to a nylon membrane. The [3H]methionine-labeled RNA was detected with radioautography (right panel). C, immunoprecipitation was performed with anti-hUTP14a antibody or pre-immune rabbit IgG (rIgG) on whole cell extracts of HeLa cells. Proteins from the precipitates were separated with SDS-PAGE and transferred onto PVDF membranes followed by immunoprobing with anti-hUTP14a antibody. Ten percent of the cell extract was loaded as input control (left upper panel). RNA extracted from precipitates as described above was subjected to RT-PCR to amplify U3 snoRNA. The PCR products were resolved on a 2% agarose gel and stained with ethidium bromide (left lower). Immunoprecipitation was repeated as described above, and proteins from immunoprecipitates were subjected to Western blotting to analyze hUTP14a (right upper). RNA was extracted from the immunoprecipitates and transferred onto a nylon membrane. Northern blotting analysis was performed with a biotin-labeled U3 snoRNA probe. 2.5% of the total RNA was employed as input control (right lower panel).
hUTP14a Promotes p53 Protein Turnover via a Proteasome-dependent Pathway
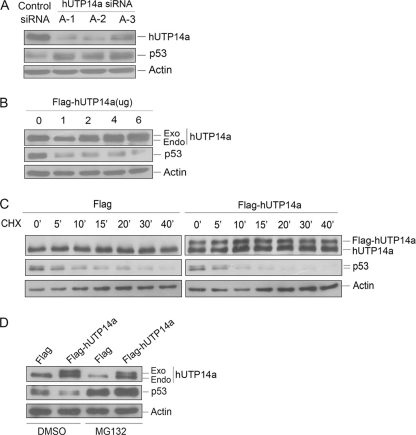
We first examined p53 protein levels after silencing endogenous hUTP14a expression. Three siRNAs targeting hUTP14a were transfected into U2OS cells together with a control siRNA. Whole cell extracts were prepared 72 h post-transfection. Proteins from the cell extracts were subjected to Western blotting to evaluate protein levels of hUTP14a and p53. As shown in Fig. 3A, knockdown of hUTP14a resulted in elevated levels of p53. These experiments were repeated in the breast cancer cell line MCF-7, which expresses wild-type p53 and the same result was obtained (data not shown). Accordingly, a plasmid coding Flag-hUTP14a was transfected into U2OS cells, and protein levels of p53 were determined by Western blotting. As shown in Fig. 3B, ectopic expression of Flag-hUTP14a resulted in decreased p53 levels. We next asked if hUTP14a causes decrease in p53 levels by affecting the stability of p53. U2OS cells were transfected with Flag-hUTP14a. At 16-h post-transfection, cells were treated with cycloheximide for different time periods before harvest. Protein levels of p53 were determined by Western blotting. As shown in Fig. 3C, the half-life of p53 in the Flag vector transfected cells was about 20 min. However, the p53 half-life was shortened to less than 10 min when Flag-hUTP14a was ectopically expressed. Thus, ectopic expression of hUTP14a destabilized p53 protein. To determine whether this hUTP14a-mediated p53 destabilization was related to the proteasome pathway, Flag-hUTP14a was transfected into U2OS cells. Cells were treated with either a protein inhibitor, MG132 or DMSO, which was used as a vehicle for MG132. Cell lysates were extracted, and proteins from the extracts were subjected to Western blotting. The results showed that the hUTP14a-induced p53 decrease was blocked by MG132 (Fig. 3D). These results demonstrated that hUTP14a-induced p53 degradation is proteasome-dependent.
FIGURE 3.
hUTP14a promotes p53 protein turnover in a proteasome-dependent manner. A, U2OS cells were transfected with three hUTP14a-specific siRNAs (A-1, A-2, and A-3) or a control siRNA, respectively. Whole cell lysates were prepared 72-h post-transfection. Proteins from lysates were separated on SDS-PAGE and transferred onto a PVDF membrane. The upper part of the blot was probed with anti-hUTP14a antibody, and the lower part was probed with anti-p53 antibody. β-Actin (Actin) was evaluated as a loading control. B, U2OS cells were transfected with increasing amounts of Flag-hUTP14a plasmid. Cells were harvested 24-h post-transfection. Proteins from cell lysates were separated on SDS-PAGE and transferred onto a PVDF membrane. The blot was probed with anti-hUTP14a or anti-p53 antibody as in A. β-Actin was evaluated as a loading control. C, U2OS cells were transfected with either Flag-hUTP14a or Flag vector plasmid. Cells were treated with 15 μg/ml cycloheximide for 4 h at 16-h post-transfection. Cells were harvested at indicated time points, and cell lysates were prepared and subjected to Western blotting analysis. Expression of Flag-hUTP14a and endogenous hUTP14a was detected with anti-hUTP14a antibody. The lower part of the blot was probed with anti-p53 antibody. β-Actin was used as a loading control. D, Flag-hUTP14a or Flag vector plasmid was transfected into U2OS cells. Cells were treated with DMSO or MG132 before harvesting 20 h post-transfection. Protein levels were evaluated by Western blotting as described above.
UTP14a Interacts Directly with p53
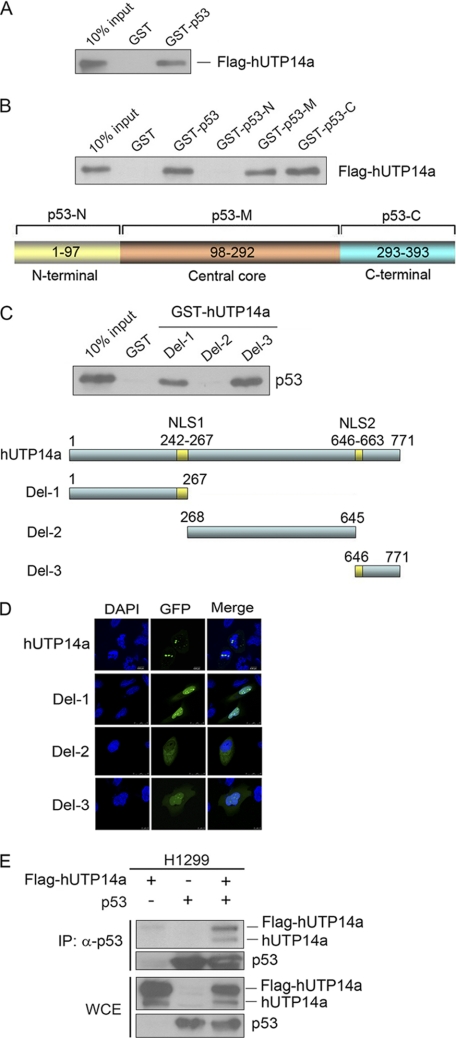
To investigate whether hUTP14a physically interacts with p53, GST pull-down experiments were performed with E. coli expressed GST-p53 fusion protein or GST protein and in vitro transcribed/translated Flag-hUTP14a. The GST fusion protein-bound Flag-hUTP14a was determined by Western blotting using anti-UTP14a antibody. These results showed that hUTP14a specifically interacted with p53 (Fig. 4A). To map the interaction domain in p53, GST pull-down experiments were performed with in vitro transcribed/translated Flag-hUTP14a and GST-p53 deletion mutants. Fig. 4B shows that hUTP14a interacted with the central core region and C terminus of p53. Conversely, the interacting domain in hUTP14a was mapped using in vitro translated p53 and GST-hUTP14a deletion mutants with GST pull-down experiments. Fig. 4C shows that both the N and C terminus of hUTP14a were required for interaction with p53. Further, the cellular localization of hUTP14a deletion mutants was examined. The plasmids coding GFP-hUTP14a deletion mutants were transfected into U2OS cells separately. The localization of GFP proteins was observed under confocal microscopy after nuclear staining with DAPI. The results showed that both p53-interacting domains in hUTP14a were localized in the nucleus (Fig. 4D). To confirm the interaction between hUTP14a and p53, Flag-hUTP14a and p53 plasmids were transfected into a p53-null cell line H1299. Immunoprecipitation was performed with anti-p53 antibody, and proteins from the precipitates were separated with SDS-PAGE and transferred onto a PVDF membrane. The blot was hybridized with anti-hUTP14a antibody and anti-p53 antibody. Fig. 4E shows that both Flag-hUTP14a and endogenous hUTP14a were present in the p53-specific immunoprecipitates, confirming that hUTP14a was associated with p53 in cells.
FIGURE 4.
hUTP14a specifically interacts with p53 in vitro and in cells. A, GST pull-down was performed with E. coli expressed GST-p53 fusion protein or GST and in vitro transcribed/translated Flag-hUTP14a. The GST fusion protein-bound Flag-hUTP14a proteins were analyzed by Western blot probed with anti-hUTP14a antibody. Ten percent of the Flag-hUTP14a protein was loaded as an input control. B, GST pull-down was performed with GST or GST-p53 deletion mutant fusion proteins and in vitro translated Flag-hUTP14a. The GST-p53 bound Flag-hUTP14a proteins were analyzed by Western blot probed with anti-hUTP14a antibody. Ten percent of the Flag-hUTP14a protein was loaded as an input control. A schematic structure of p53 deletion mutants is shown in the lower panel. C, GST pull-down was performed with GST or GST-hUTP14a deletion mutant fusion proteins and in vitro translated p53. The GST-hUTP14a-bound p53 proteins were analyzed by Western blot probed with anti-p53 antibody. Ten percent of the p53 protein was loaded as an input control. A schematic structure of hUTP14a deletion mutants is shown in the lower panel. D, plasmids coding GFP-hUTP14a deletion mutants were transfected into U2OS cells, and GFP proteins were observed under confocal microscopy after the nucleus was stained with DAPI. E, Flag-hUTP14a and p53 plasmids were transfected into H1299 cells. Cell lysates were extracted 24 h post-transfection. Immunoprecipitation was performed with anti-p53 antibody, and proteins from the immunoprecipitates were subjected to Western blotting. The upper part of the blot was probed with anti-hUTP14a antibody, and the lower part was hybridized with anti-p53 antibody. The expression level of hUTP14a and p53 in whole cell extracts (WCE) was determined by Western blotting.
hUTP14a Promotes p53 Degradation Possibly through a Ubiquitin-independent Pathway
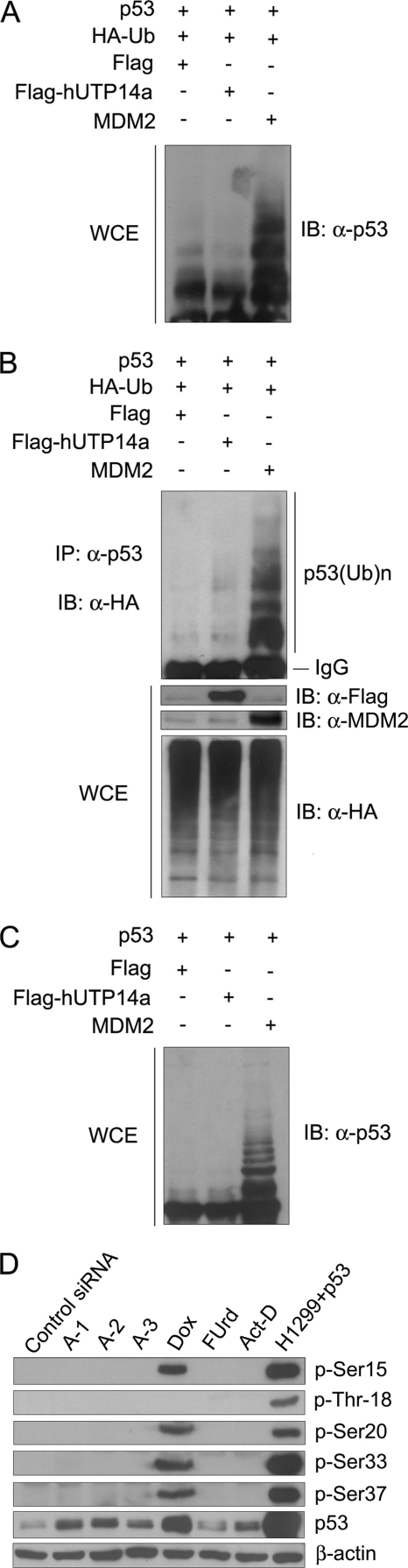
The next question raised was whether hUTP14a-induced p53 degradation depends on ubiquitination. To this end, H1299 cells were co-transfected with p53 and HA-Ub plasmids in the presence of either Flag-hUTP14a or Flag vector or MDM2, which is known to ubiquitinate p53 protein. Cells were treated with MG132 before harvest. Polyubiquitination of p53 was identified by Western blotting. As shown in Fig. 5A, polyubiquitination of p53 was observed when p53 was co-transfected with HA-Ub in the presence of MDM2. However, co-expression of Flag-hUTP14a with HA-Ub did not show any effect on p53 polyubiquitination. To confirm that hUTP14a induces p53 degradation independent of ubiquitination, H1299 cells were co-transfected with p53 and Flag-hUTP14a or MDM2 expression plasmids in the presence of HA-Ub. Immunoprecipitation experiments were performed with anti-p53 antibody and Western blotting was performed with anti-HA antibody. As shown in Fig. 5B, p53 polyubiquitination was enhanced when p53 was co-transfected with HA-Ub and MDM2. In contrast, Flag-hUTP14a showed no effect on p53 polyubiquitination. We further determined that hUTP14a had no effect on p53 polyubiquitination in the absence of ubiquitin overexpression. H1299 cells were co-transfected with p53 and Flag-hUTP14a or MDM2 expression plasmids. Cells were treated with MG132 before harvest. Polyubiquitination of p53 was examined by Western blotting. As shown in Fig. 5C, p53 is polyubiquinated by MDM2 but not by Flag-hUTP14a. These results suggested that Flag-hUTP14a may induce p53 degradation in an ubiquitin-independent manner.
FIGURE 5.
hUTP14a promotes p53 degradation possibly through an ubiquitin-independent pathway. A, H1299 cells were co-transfected with p53 and Flag-hUTP14a or MDM2 or Flag vector plasmid in the presence of HA-Ub. Cells were treated with 10 μm MG132 for 4 h before harvest. Equal amount of protein of whole cell extracts were subjected to Western blotting for detection of p53. B, H1299 cells were co-transfected with p53 plasmid and HA-Ub with either Flag-hUTP14a or MDM2 or Flag vector plasmid. Cells were treated with 10 μm MG132 for 4 h before harvesting at 20 h post-transfection. Immunoprecipitation was performed with anti-p53 antibody. The proteins from the precipitates were separated on SDS-PAGE and transferred to a PVDF membrane. The blot was probed with anti-HA antibody (upper panel). Expression of hUTP14a or MDM2 was detected by immunoblot with anti-Flag antibody or anti-MDM2 antibody (middle panels). Equal amounts of protein from whole cell extracts (WCE) were subjected to Western blotting and probed for HA-Ub with anti-HA antibody (lower panel). C, H1299 cells were co-transfected with p53 and Flag-hUTP14a or MDM2 expression plasmid or Flag vector. Cells were treated with 10 μm MG132 for 4 h before harvest. Equal amount of proteins of whole cell extracts were subjected to Western blotting for detection of p53. D, U2OS cells were transfected with hUTP14a specific siRNAs (A-1, A-1, or A-3) or a control siRNA. Cells were harvested and whole cell extracts were prepared 72-h post-transfection. Proteins from the cell extracts were separated on SDS-PAGE and transferred onto a PVDF membrane. Blots were probed with p-Ser15, p-Thr18, p-Ser20, p-Ser33, or p-Ser37 p53 antibody. Proteins extracted from U2OS cells treated with 5 μm of doxorubicin for 16 h, 5 μm of FUrd for 15 min, or 5 nm of actinomycin D for 4 h or from p53-transfected H1299 cells were loaded as controls. Total p53 levels were determined by Western blotting and probing with anti-p53. β-Actin was used as a loading control.
To determine whether knockdown of hUTP14a can stabilize p53 by affecting p53 phosphorylation, p53 phosphorylation at Ser-15, Thr-18, Ser-20, Ser-33, and Ser-37 was evaluated after silencing hUTP14a. Cell lysates from p53-transfected H1299 cells were used as positive control. As shown in Fig. 5D, DNA damage reagent doxorubicin induced p53 phosphorylation at Ser-15, Ser-20, Ser-33, and Ser-37 sites, while no detectable p53 phosphorylation at these phosphorylation sites was induced by hUTP14a depletion. Moreover, treatment of cells with actinomycin D or 5-FU (FUrd) did not show detectable phosphorylation of these phosphorylation sites.
Knockdown of hUTP14a Inhabits Cell Growth and Induces Apoptosis
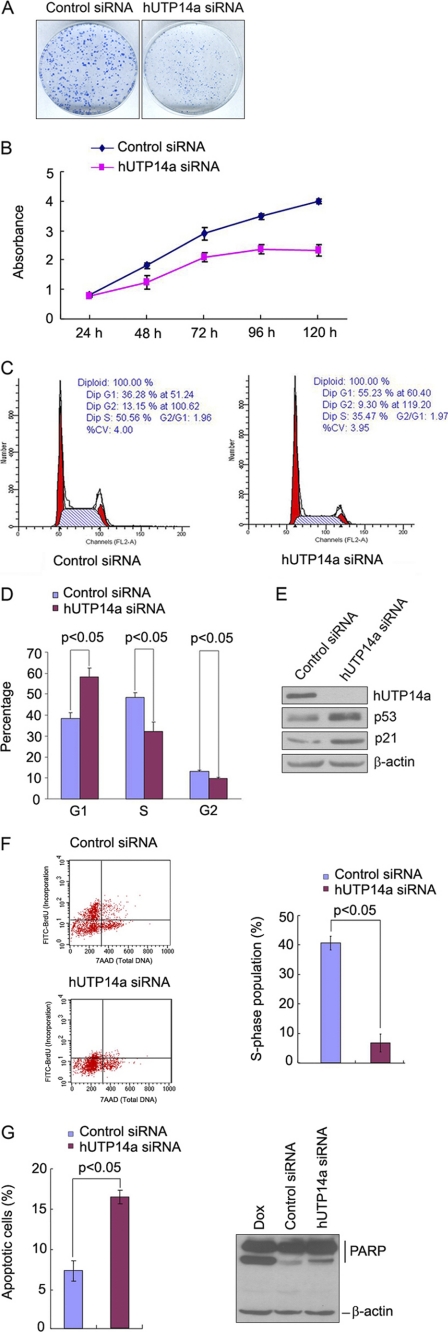
We then asked whether hUTP14a modulates p53 activity. As hUTP14a is ubiquitously expressed in various human cell lines, most of these experiments were conducted with knockdown of hUTP14a. First, we examined cell growth when hUTP14a was silenced in U2OS cells. Colony formation was performed with U2OS cells following knockdown of hUTP14a by siRNA. As shown in Fig. 6A, a significant reduction in both the size and the number of colonies was observed in hUTP14a-deficient cells. To confirm the effect of hUTP14a knockdown on cell growth, a growth curve was plotted after hUTP14a was depleted by siRNA in U2OS cells. As shown in Fig. 6B, cell growth was significantly inhibited in the hUTP14a-deficient cells. Accordingly, the cell cycle was analyzed with flow cytometry when hUTP14a was silenced in U2OS cells. The result showed that silencing of hUTP14a results in cell cycle arrest at G1. To verify this phenomenon, the same experiments were performed in another p53-expressing cell line MCF-7 and identical results were obtained (Fig. 6C). Cell cycle analyses after hUTP14a knockdown were repeated three times in duplicate in MCF-7 cells, and the results are summarized in Fig. 6D. The percentage of the cells was 58.3% in G1, 32.0% in S, and 9.7% in G2/M in hUTP14a-depleted cells, while it was 38.5% in G1, 48.4% in S, and 13.1% in G2/M in control siRNA-transfected cells. Knockdown of hUTP14a resulted in a significant accumulation of cells in G1 and concomitantly fewer cells in S and G2/M (p < 0.05), indicating that knockdown of hUTP14a arrested cells in G1. In accordance with these cell cycle analysis results, Western blotting showed that knockdown of hUTP14a caused increased levels of p53 and consequently elevated p21 expression (Fig. 6E). To confirm that depletion of hUTP14a caused G1 arrest, BrdU incorporation was determined with the BrdU Flow kit (BD Product). Cells were pulsed with BrdU for 16 h after transfection with hUTP14a-specific siRNA. Cells were stained with FITC-conjugated anti-BrdU to evaluate for DNA incorporation during the pulse and with 7-AAD to determine total DNA content. As shown in Fig. 6F (left panel), the cell number of FITC-BrdU-staining positive in hUTP14a-depleted cells decreased dramatically compared with that in control siRNA-treated cells. The S-phase population was quantitated as the percentage of cells staining FITC (BrdU) positive versus 7-AAD stained cells. Data from three independent experiments in duplicate are shown in Fig. 6F (right panel). Cells transfected with hUTP14a siRNA had about 6.8% of cells in S-phase while cells treated with control siRNA had 40.6% of cells in S-phase. The S-phase population in hUTP14a-deficient cells was significantly reduced compared with the control siRNA-treated cells (p < 0.05). This experiment further indicated that knockdown of hUTP14a arrested cells in G1. Taking these findings together, we conclude that knockdown of hUTP14a stabilized p53 and inhibited cell growth by arresting cells in G1. We then examined the effect of hUTP14a knockdown on cell apoptosis. U2OS cells were transfected with hUTP14a-specific siRNA, and cells were then double stained with annexin V and propidium iodide and subjected to flow cytometric analysis 72-h post-transfection. As shown in Fig. 6G (left panel), hUTP14a knockdown caused 16.4% of the cells to undergo apoptosis, while control siRNA-transfected cells only resulted in 7.2% apoptotic cells, indicating that knockdown of hUTP14a significantly increased cell apoptosis. To further confirm hUTP14a-induced apoptosis, PARP cleavage was determined by Western blotting after hUTP14a was depleted. As shown in Fig. 6G (right panel), knockdown of hUTP14a enhanced PARP cleavage, while control siRNA resulted in a modest level of cleaved PARP and p53. Thus, we demonstrate that knockdown of hUTP14a-induced apoptosis.
FIGURE 6.
Knockdown of hUTP14a inhibits cell growth and induces apoptosis. A, U2OS cells were transfected with hUTP14a siRNA (A-1). Two thousand cells were seeded in 6-mm plates 72-h post-transfection. Cells were grown for 10 days, and cell colonies were stained with Coomassie Bright Blue after fixation with ethanol/acetic acid. B, U2OS cells were transfected with hUTP14a siRNA (A-1), and cells were seeded into 96-well plates 72-h post-transfection. WST-8 was added to cells at different time points, and absorbance at 450 nm was measured. This experiment was repeated three times in duplicate, and growth curves were plotted with the mean ± S.D. of absorbance at 450 nm versus time points. C–E, MCF-7 cells were transfected with hUTP14a siRNA (A-1). Cell cycle was analyzed by flow cytometry 72-h post-transfection. A representative result is shown in C and a summary of results from three independent experiments in duplicate is shown in D. Whole cell extracts were prepared from the siRNA-transfected cells. Proteins from the extracts were separated by SDS-PAGE and transferred onto a PVDF membrane. The blot was probed with antibodies directed against hUTP14a, p53, or p21. β-Actin was used as a loading control (E). F, U2OS cells were transfected with hUTP14a siRNA (A-1) or a control siRNA. BrdU incorporation was determined with a BrdU Flow kit 72-h post-transfection as described under “Experimental Procedures.” A representative result of cell number of positive BrdU staining and 7-AAD staining is shown (left panel). The S-phase population was plotted by comparing BrdU staining versus 7-AAD-stained cells. Data are presented as mean ± S.D. from three independent experiments in duplicate (right panel). G, U2OS cells were transfected with hUTP14a siRNA (A-1) or a control siRNA and cells were then double-stained with annexin V and PI and cell apoptosis was determined by flow cytometry 72-h post-transfection. A summary of the results from three independent experiments in duplicate is shown. Bars represent the mean ± S.D. Whole cell extracts were prepared from hUTP14a siRNA-transfected cells 72-h post-transfection. Proteins from whole cell extracts were separated on SDS-PAGE and transferred onto a PVDF membrane. The upper part of the blot was probed with anti-PARP antibody, and the lower part was probed with anti-β-actin antibody (right panel). Statistical analyses were performed with one-tailed unpaired t test.
DISCUSSION
It has previously been found that nucleolar stress causes stabilization of p53. The mechanism of nucleolar stress-induced p53 activation has focused on the MDM2 interacting RPs including RPL5, RPL11, and RPL23. On one hand, these ribosomal proteins bind MDM2 and inhibit the E3 ubiquitin ligase activity resulting in stabilization of p53. On the other hand, RPL5 and RPL26 bind p53 mRNA to enhance p53 synthesis. The significance of multiple MDM2 interacting RPs in the p53 response to nucleolar stress has been discussed by Zhang and Lu (35). It was thought that multiple RPs might sense different growth inhibitory or ribosomal stresses so that the various steps of ribosome biogenesis can be effectively monitored. These findings suggest that nucleolar stress-induced p53 activation is far more complex than previously thought. This prompted us to consider the possibility that defects in ribosome biogenesis may trigger p53 activation through other mechanisms.
We first identified hUTP14a as an SSU processome component by demonstrating that hUTP14a is nucleolar, associated with U3 snoRNA, and that knockdown of hUTP14a inhibits 18 S rRNA processing. Upon further investigation we found that knockdown of hUTP14a simultaneously caused increase in p53 levels and ectopic expression of hUTP14a induced decrease in p53 levels. Interestingly, ectopic expression of hUTP14a shortened the half-life of p53 from 20 min to less than 10 min. In addition, hUTP14a-induced p53 decrease was inhibited by the proteasome inhibitor MG132. These findings indicate that hUTP14a induces p53 degradation through a proteasome-dependent pathway. GST pull-down experiments showed that hUTP14a interacts directly with p53. This interaction was further confirmed by immunoprecipitation performed with p53-specific antibody on cell lysates from cells transfected with Flag-hUTP14a and p53. Thus, we provide the first evidence for a pre-rRNA processing factor binding p53 directly and promoting p53 degradation.
In proteasomes, proteins can be degraded either via ubiquitin-dependent or ubiquitin-independent pathways (36). In ubiquitin-dependent protein degradation, the substrate is polyubiquitinated by specific E3 ubiquitin ligases, marking it for degradation by 26 S proteasomes. p53 protein has been found to be degraded in ubiquitin-dependent and ubiquitin-independent pathways (37). For example, the human papilloma virus (HPV) E6 protein interacts with the central DNA binding region and the C terminus region of p53 (38, 39). Binding of E6 to the DNA binding region enhances p53 degradation through the ubiquitin pathway in the presence of E6-associated protein (E6AP), whereas binding to the C terminus enhances ubiquitin-independent degradation (40). Our in vivo ubiquitination experiments demonstrated that hUTP14a had no effect on p53 ubiquitination, whereas p53 ubiquitination was promoted by MDM2. This raises the possibility of an ubiquitin-independent pathway for hUTP14a-induced p53 degradation. Phosphorylation of p53 usually modulates its stability (41). Because p53 phosphorylation at Ser-15, Thr-18, Ser-20, Ser-33, and Ser-37 stabilizes p53 (42–44), we therefore evaluated p53 phosphorylation at Ser-15, Thr-18, Ser-20, Ser-33, and Ser-37 when hUTP14a was depleted. However, no p53 phosphorylation was found at these sites in the absence of hUTP14a. Moreover, treatment of cells with actinomycin D or 5-FU failed to induce p53 phosphorylation at Ser-15, Thr-18, Ser-20, Ser-33, and Ser-37 under our experimental conditions. It is therefore unlikely that these nucleolar stresses cause p53 stabilization through modification of p53 phosphorylation at these DNA damage-induced phosphorylation sites. In the present study, the biologic result of hUTP14a deficiency-induced p53 activation was arrest of cells at G1 and induction of apoptosis. Acetylation of p53 is induced in response to stress, and p53 acetylation causes p53 activation and stabilization (for review, see Ref. 45). An acetylation-defective p53–8KR mutant is completely unable to induce cell cycle and apoptotic regulators. Therefore, whether depletion of hUTP14a can induce p53 acetylation needs to be determined.
In the present study, protein binding experiments showed that association of hUTP14a and p53 requires the central DNA binding domain and the C terminus of p53, the N terminus domain (amino acids 1–267) and the C terminus (amino acids 646–771) of hUTP14a. It is as yet unclear how the two p53-interaction domains of hUTP14a function in a coordinated manner to induce p53 degradation. A model of dual-site regulation of MDM2 E3-ubiquitin ligase activity has been proposed by Wallace et al. (46). In this model, the interaction between the p53-BOX-I domain and the N terminus of MDM2 promotes conformational changes in MDM2. These conformational changes stabilize the interaction of the MDM2 acid domain and the DNA binding domain of p53 and further facilitate MDM2-mediated p53 ubiquitination. The C terminus of p53 has been demonstrated to mediate ubiquitin-independent p53 degradation by E6 (40). We would like to raise the possibility that the interaction between hUTP14a and the DNA binding region of p53 might induce a conformational change to facilitate the binding of hUTP14a to the C terminus of p53 and thereby mediate ubiquitin-independent degradation of p53. Our GST pull-down experiments showed that hUTP14a also interacted with MDM2 in vitro (data not shown). However, we failed to find hUTP14a/MDM2/p53 complexes in in vivo experiments. Because RPs bind and target MDM2 for degradation in response to nucleolar stress, we speculate that hUTP14a as a novel nucleolar stress sensor, may interact with MDM2 and affect MDM2 level and activity under nucleolar stress. Whether hUTP14a affects MDM2-p53 interaction is currently under investigation.
Our demonstration of direct p53 targeting by a ribosome biogenesis factor is a novel finding. We propose that hUTP14a is required for 18 S rRNA processing and that it contributes to keeping p53 at low levels in unstressed cells. When hUTP14a is deficient, 18 S rRNA processing is inhibited, and hUTP14a-induced p53 degradation is also simultaneously inhibited. Thus, hUTP14a itself functions as a nucleolar stress sensor in addition to its function in ribosome biogenesis. Our findings provide a novel mechanism for p53 activation in which a ribosome biogenesis factor itself functions as a nucleolar stress sensor serving to signal stress directly to p53.
An RPL11-dependent pathway has recently been demonstrated to mediate p53 degradation by defects in 18 S and 28 S rRNA processing. Moreover, it has been shown that RPL26 binds the 5′-UTR of p53 mRNA and activates p53 translation after DNA damage (29). We therefore cannot rule out the possibility that other alternative pathways also function in p53 activation caused by depletion of hUTP14a. We also speculate that p53 acetylation may be involved in the mechanisms by which cells survey ribosome biogenesis and activate p53 to ensure normal cell proliferation.
Supplementary Material
Acknowledgments
We thank Dr. Michael A McNutt for editing the English in this manuscript. We would also like to thank Dr. Qihua He and Dr. Hounan Wu for assistance with the confocal microscopy and flow cytometry analysis.
This work was supported by grants from the National Natural Science Foundation of China (Grants No. 30571038 and 30771224), the Key Construction Program of the National “985” Project (Grant No. 985-2-016-24), Grants from the National High Technology Research and Development Program of China (863 Program) (Grant No. 2008AA02Z131 and 2006AA02A402), and from the National Basic Research Program of China (973 Program) (Grant No. 2010CB529303).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- rDNA
- ribosome RNA genes
- SSU
- small subunit
- snoRNA
- small nucleolar RNA
- hUTP
- human U3 protein.
REFERENCES
- 1. Iapalucci-Espinoza S., Franze-Fernández M. T. (1979) FEBS Lett. 107, 281–284 [DOI] [PubMed] [Google Scholar]
- 2. Perry R. P., Cheng T. Y., Freed J. J., Greenberg J. R., Kelley D. E., Tartof K. D. (1970) Proc. Natl. Acad. Sci. U.S.A. 65, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pestov D. G., Strezoska Z., Lau L. F. (2001) Mol. Cell. Biol. 21, 4246–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kass S., Tyc K., Steitz J. A., Sollner-Webb B. (1990) Cell 60, 897–908 [DOI] [PubMed] [Google Scholar]
- 5. Hughes J. M., Ares M., Jr. (1991) EMBO J. 10, 4231–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fournier M. J., Maxwell E. S. (1993) Trends Biochem. Sci. 18, 131–135 [DOI] [PubMed] [Google Scholar]
- 8. Venema J., Tollervey D. (1995) Yeast 11, 1629–1650 [DOI] [PubMed] [Google Scholar]
- 9. Hölzel M., Orban M., Hochstatter J., Rohrmoser M., Harasim T., Malamoussi A., Kremmer E., Längst G., Eick D. (2010) J. Biol. Chem. 285, 6364–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. (2003) Mol. Cell 12, 1151–1164 [DOI] [PubMed] [Google Scholar]
- 11. Volarevic S., Stewart M. J., Ledermann B., Zilberman F., Terracciano L., Montini E., Grompe M., Kozma S. C., Thomas G. (2000) Science 288, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 12. Bradley J., Baltus A., Skaletsky H., Royce-Tolland M., Dewar K., Page D. C. (2004) Nat. Genet. 36, 872–876 [DOI] [PubMed] [Google Scholar]
- 13. Rohozinski J., Bishop C. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11695–11700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shetty G., Shao S. H., Weng C. C. (2008) Endocrinology 149, 2773–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vogelstein B., Lane D., Levine A. J. (2000) Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 16. Vousden K. H., Lu X. (2002) Nat Rev Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 17. Marechal V., Elenbaas B., Piette J., Nicolas J. C., Levine A. J. (1994) Mol. Cell. Biol. 14, 7414–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roth J., Dobbelstein M., Freedman D. A., Shenk T., Levine A. J. (1998) EMBO J. 17, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sherr C. J., Weber J. D. (2000) Curr. Opin. Genet. Dev. 10, 94–99 [DOI] [PubMed] [Google Scholar]
- 20. Rubbi C. P., Milner J. (2003) EMBO J. 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scheer U., Hock R. (1999) Curr. Opin Cell Biol. 11, 385–390 [DOI] [PubMed] [Google Scholar]
- 22. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Mol. Cell. Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai M. S., Lu H. (2004) J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 27. Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 28. Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Mol. Cell 35, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 30. Rohozinski J., Lamb D. J., Bishop C. E. (2006) Biol. Reprod. 74, 644–651 [DOI] [PubMed] [Google Scholar]
- 31. Rohozinski J., Anderson M. L., Broaddus R. E., Edwards C. L., Bishop C. E. (2009) PLoS One 4, e5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strezoska Z., Pestov D. G., Lau L. F. (2002) J. Biol. Chem. 277, 29617–29625 [DOI] [PubMed] [Google Scholar]
- 33. Pluk H., Soffner J., Lührmann R., van Venrooij W. J. (1998) Mol. Cell. Biol. 18, 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Y., Liu J., Zhao H., Lü W., Zhao J., Yang L., Li N., Du X., Ke Y. (2007) Biochim. Biophys. Acta 1773, 863–868 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y., Lu H. (2009) Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asher G., Tsvetkov P., Kahana C., Shaul Y. (2005) Genes Dev. 19, 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asher G., Shaul Y. (2005) Cell Cycle 4, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 38. Lechner M. S., Laimins L. A. (1994) J. Virol. 68, 4262–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Coffino P. (1996) J. Virol. 70, 4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camus S., Menéndez S., Cheok C. F., Stevenson L. F., Laín S., Lane D. P. (2007) Oncogene 26, 4059–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 42. Shieh S. Y., Ikeda M., Taya Y., Prives C. (1997) Cell 91, 325–334 [DOI] [PubMed] [Google Scholar]
- 43. Sakaguchi K., Saito S., Higashimoto Y., Roy S., Anderson C. W., Appella E. (2000) J. Biol. Chem. 275, 9278–9283 [DOI] [PubMed] [Google Scholar]
- 44. Unger T., Juven-Gershon T., Moallem E., Berger M., Vogt Sionov R., Lozano G., Oren M., Haupt Y. (1999) EMBO J. 18, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace M., Worrall E., Pettersson S., Hupp T. R., Ball K. L. (2006) Mol. Cell 23, 251–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.