Abstract
In Diptera (Insecta), alternatively spliced male-specific and female-specific products of the doublesex (dsx) gene play key role in regulating development of the adult genital structures from the genital disc. Analysis of the pattern of nucleotide substitution of different domains of the dsx gene in 29 dipteran species showed that, over short evolutionary times, purifying selection predominated on the domain common to both sexes, the female-specific exons, and the and male-specific exon. However, over longer the evolutionary time frames represented by between-family comparisons, the male-specific exon accumulated nonsynonymous substitutions at a much more rapid rate than either the common domain or the female-specific exon. Overall, the accumulation of nonsynonymous substitutions in the male-specific exon occurred at a significantly greater than linear rate relative to the common domain, whereas the accumulation of nonsynonymous substitutions in the female-specific exon occurred at less than linear rate relative to the common domain. The evolution of the male-specific exon of dsx thus shows a pattern reminiscent of that seen in the “runaway” evolution of male secondary sexual characters at the morphological level, consistent with the hypothesis that female choice is an important factor in the morphological diversification of insect male genitalia.
Keywords: Diptera, doublesex, genital development, sex-specific exon, sexual selection
1. Introduction
The great morphological variety of male genitalia in insects has been known for over a century and a half and has been exploited as a source of characters for species-level taxonomy of many insect groups. For many years, the most widely held explanation of this surprising diversity was the “lock-and-key” hypothesis, first advanced by the pre-Darwinian entomologist Dufour (1944), whereby different genital morphologies are hypothesized to pose a barrier to between-species copulation (Shapiro and Porter 1989). The lock-and-key hypothesis was subsequently “Darwinized” (in the terminology of Shapiro and Porter 1989) as an evolutionary hypothesis, whereby the evolution of distinctive male genital morphologies was seen as a form of pre-mating isolating mechanism, which arose by natural selection as a mechanism to prevent gametic wastage in non-viable interspecific copulation attempts (Jordan 1896). However, numerous objections to the lock-and-key hypothesis have been raised, including both (1) the observation that the male genitalia generally vary much more markedly among taxa than do female genitalia; and (2) data from several groups showing the lack of any obvious covariation between male and female genital morphology (Eberhard 1985; Eberhard and Ramirez 2004; Shapiro and Porter 1989).
More recently, hypotheses explaining the variety of male insect genital morphology as the result of sexual selection have become increasingly popular (Eberhard 1985; Arnqvist 1998; Hosken and Stockley 2004). As with many aspects of sexual selection, the exact mechanism that might have given rise to the diversity of insect male genitalia has so far proved elusive (Hosken and Stockley 2004). Eberhard (1985) favored a classic Fisherian (Fisher 1930) runaway process, while others have argued for a Zahavian mechanism (Zahavi 1975) whereby genital morphology provides a reliable indicator of male genetic quality (Arnqvist and Thornhill 1998).
An alternative to hypotheses proposing direct selection on genital structures themselves suggests that differences in genital morphology arose as pleiotropic effects of alleles selected for their effects on other characters (Mayr 1963). An objection to this hypothesis has been that it does not explain why such pleiotropic effects would impact the male genitalia specifically, rather than other morphological structures (Eberhard 1985). On the other hand, although some theoretical models have suggested a role of genetic drift in the evolution of traits used in mate choice (Lande 1981), the literature on insect genital morphology has largely ignored the possibility that interspecies differences in these structures have arisen through fixation of selectively neutral mutations by random genetic drift.
In Drosophila melanogaster (Diptera: Drosophilidae), the sex-determination gene doublesex (dsx) plays a key role in regulating development of the adult genital structures from the genital disc (Sánchez and Guerrero 2001; Vincent et al. 2001). By alternative splicing, the dsx gene encodes the female-specific transcription factor DsxF (exons 1, 2, and 3) and the male-specific transcription factor DsxM (exons 1, 2, and 4). In addition to their role in the development of the genitalia, these transcription factors play roles in the nervous system in regulating the development of sex-specific behaviors (Rideout et al. 2007; Siwicki et al. 2009; Waterbury et al. 1999). In transgenic Drosophila melanogaster with dsx genes from Anastrepha obliqua (Diptera: Tephiritidae), normal genital morphologies did not develop; this result demonstrates that amino acid changes in DsxF and DsxM can effect genital morphology (Alvarez et al. 2009).
Previous results have suggested that purifying selection acts on the amino acid sequences encoded by both the female-specific exon and the male-specific exon of dipteran dsx (Ruiz et al. 2007). Here I extend these analyses in order to compare the patterns of evolutionary diversification in functionally different regions of the dsx gene. Specifically, I test whether the pattern of nonsynonymous nucleotide substitutions in the male-specific exon of dsx is consistent with the hypothesis that such substitutions play a role to the diversification of male genitalia of Diptera. Moreover, I address functional constraints on dsx at different evolutionary time scales in order to gain insight into potential evolutionary mechanisms underlying male genital diversification. For example, if sexual selection has repeatedly favored novel male genital morphologies, rapid evolution of nonsynonymous sites in the male-specific exon might be predicted. Conversely, on the lock-and-key hypothesis, one might predict both covariation of male-specific and female-specific exons and rapid diversification between closely related species as a result of selection for isolating mechanisms.
2. Methods
Coding sequences of dsx from 29 species of Diptera were analyzed (Table 1). Anastrepha fraterculus is a species complex including 4 recognized cryptic species, designated Sp1–Sp4 (Ruiz et al. 2007), for each of which sequence data were available (Table 1). The data included 11 species in the genus Anastrepha (family Tephritidae); 4 species in the genus Bactrocera (Tephiritidae) and 8 species in the genus Drosophila (Drosophilidae). Sequences for the common domain (exons 1–2), the female-specific domain (exon 3), and the male-specific domain were retrieved from the NCBI sequence database by homology search. In the case of Drosophila species, when predicted mRNAs in the Refseq database did not include all of these exons, they were located by homology search in genomic shotgun sequences (Table 1).
Table 1.
Sequences used in analyses.
| Family | Species | Sequences |
|---|---|---|
| Tephritidae | Anastrepha fraterculus Sp1 | DQ494334, DQ494344 |
| Anastrepha fraterculus Sp2 | DQ494325, DQ494335 | |
| Anastrepha fraterculus Sp3 | DQ494326, DQ494336 | |
| Anastrepha fraterculus Sp4 | DQ494327, DQ494343 | |
| Anastrepha amita | DQ494333, DQ494342 | |
| Anastrepha serpentina | DQ494338, DQ494339 | |
| Anastrepha obliqua | AY948420, DQ948421 | |
| Anastrepha bistrigata | DQ494332, DQ494341 | |
| Anastrepha striata | DQ494340, DQ494341 | |
| Anastrepha sororcula | DQ494330, DQ494339 | |
| Anastrepha grandis | DQ494328, DQ494337 | |
| Bactrocera tryoni | AF029675, AF029676 | |
| Bactrocera dorsalis | AY669317, FJ176944 | |
| Bactrocera correcta | FJ185165, FJ185166 | |
| Bactrocera oleae | AJ547621, AJ547622 | |
| Ceratitis capitata | AF434087, AF434935 | |
| Drosophilidae | Drosophila melanogaster | NM_169202, NM_169203 |
| Drosophila erecta | XM_001979206, NW_001956552 | |
| Drosophila sechellia | XM_002038714, NW_001999695 | |
| Drosophila pseudoobscura | XM_001358983, NC_009005 | |
| Drosophila persimilis | XM_002013310, NW_001985953 | |
| Drosophila yakuba | XM_002086742, NT_167065 | |
| Drosophila simulans | XM_002102506, NT_167061 | |
| Drosophila ananassae | XM_001954765, NW_001939291 | |
| Muscidae | Musca domestica | AY461853, AY461854 |
| Calliphoridae | Lucilia cuprina | GU784833, GU784834 |
| Phoridae | Megaselia scalaris | AF283695, AF283696 |
| Culicidae | Anopheles gambiae | XM_309601, XM560052 |
| Aedes aegypti | DQ440532, DQ440534 |
Using the MEGA 4.1 program (Tamura et al. 2007), sequences were aligned at the amino acid level, and the alignment imposed on the DNA sequence (Supplementary Figure S1). In pairwise comparisons of sequences within and between genera, any site at which the alignment postulated a gap in any sequence was excluded from all comparisons (“complete deletion”). However, when all available sequences were compared pairwise, partial deletion (i.e., deletion only of sites with gaps between the two sequences compared) was used; this was done to increase the number of sites compared in the case of the male-specific exon, for which the alignment postulated numerous gaps between distantly related sequences. However, in preliminary analyses, the results using complete and partial deletion showed the same trends (not shown).
A phylogenetic tree of the common domain was reconstructed using the neighbor-joining method (Saitou and Nei 1986), based on the maximum composite likelihood (MCL) distance (Tamura et al. 2007). The reliability of clustering patterns in the tree was assessed by bootstrapping (Felsenstein 1985); 100 boostrap samples were used. The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by Nei and Gojobori’s (1986) method. In preliminary analyses, other methods (Li 1993; Yang and Nielsen 2000) yielded similar estimates; therefore, only the results using the Nei and Gojobori (1986) method are presented, because that method is expected to have a lower variance than the others (Nei and Kumar 2000). The variance of mean dS and dN was estimated by the bootstrap method (Nei and Kumar 2000).
After computing dN values for all pairwise comparisons separately for each domain (common, female-specific, and male-specific), I estimated the correlation coefficient between the dN values for each domain and those for each other domain. Because the pairwise dN values for a given domain are not independent of each other, randomization tests were used to test the significance of the correlation coefficients. This test involved creating 1000 random populations of paired dN values by sampling with replacement from the observed dN values.
In order to test for the linearity of the relationship of dN in different domains, I used the technique of allometric regression, which involves regression of log-transformed values (Sokal and Rohlf 1971). In the present case, since dN was often equal to zero in closely related comparisons, I used the natural logarithm of dN + 1 (Sokal and Rohlf 1971). In significance tests regarding the slope of the allometric regression line, the standard error of the slope estimator was estimated from the randomization procedure described above.
3. Results
3.1. Sister Pair Comparisons
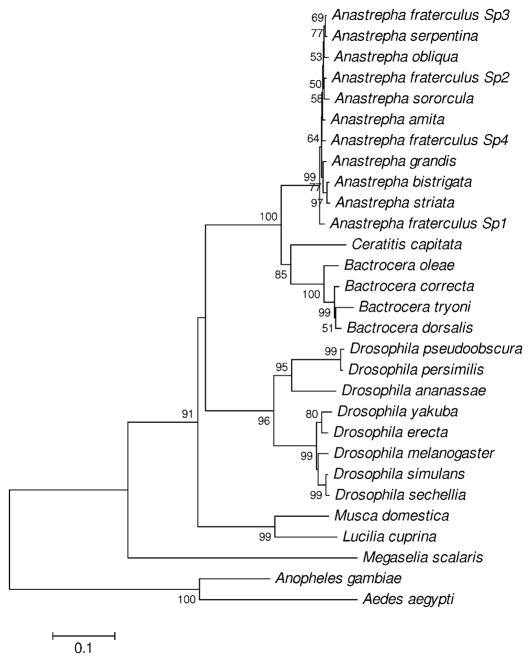
The NJ tree of common domain sequences (Figure 1) showed a topology very similar to that obtained in previous analyses (Ruiz et al. 2007). On the basis of this tree, seven “sister” pairs of sequences were chosen; and dS and dN in the common domain, the female-specific exon, and the male-specific exon were estimated between these seven phylogenetically independent pairs of sequences (Table 2). In the common domain, mean dS for the seven comparisons was significantly greater than mean dN (P < 0.05; Table 2). Likewise, in the male-specific exon, mean dS was significantly greater than mean dN (P < 0.05; Table 2). In the female-specific exon, mean dS and mean dN did not differ significantly (Table 2). However, the latter result did not occur because of elevated dN in the female-specific exon, but rather because there were no synonymous differences in the female specific exon between any of the seven pairs (Table 2). As a consequence, mean dS in the female-specific exon was significantly lower than mean dS in the common domain (P < 0.05; Table 2). However, there were no significant differences between mean dN in the common domain and mean dN in either of the two sex-specific exons (Table 2). Thus the sister-pair comparisons showed evidence of purifying selection on the common domain and both sex-specific exons, with no evident differences among these three regions with regard to the strength of purifying selection.
Figure 1.
NJ tree of the common domain of dsx from 29 species of Diptera, based on the maximum composite likelihood distance. Numbers on the branches represent the percentages of 1000 bootstrap samples supporting the branch; only values ≥ 50% are shown.
Table 2.
Numbers of synonymous (dS) nucleotide substitutions per synonymous site and nonsynonymous (dN) nucleotide substitutions per nonsynonymous site between sister pairs of dsx genes of Diptera.
| Species comparison | Common | Female-specific | Male-specific | |||
|---|---|---|---|---|---|---|
| dS ± S.E. | dN ± S.E. | dS ± S.E. | dN ± S.E. | dS ± S.E. | dN ± S.E. | |
| Anastrapha fraterculus Sp3 vs. A. Serpentina | 0.0000 ± 0.0000 | 0.0031 ± 0.0031 | 0.0000 ± 0.0000 | 0.0147 ± 0.0148 | 0.0818 ± 0.0314 | 0.0085 ± 0.0060 |
| Anastrepha fraterculus Sp2 vs. A. Sororcula | 0.0256 ± 0.0068 | 0.0077 ± 0.0035 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0223 ± 0.0158 | 0.0042 ± 0.0043 |
| Anastrepha striata vs. A Bistrgiata. | 0.0144 ± 0.0083 | 0.0031 ± 0.0022 | 0.0000 ± 0.0000 | 0.0146 ± 0.0146 | 0.0225 ± 0.0160 | 0.0042 ± 0.0042 |
| Batrocera tryoni vs. B. Dorsalis | 0.1249 ± 0.0261 | 0.0070 ± 0.0034 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0458 ± 0.0231 | 0.0042 ± 0.0042 |
| Drosophila pseudoobscura vs. D. Persimilis | 0.0343 ± 0.0109 | 0.0011 ± 0.0011 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0256 ± 0.0149 | 0.0031 ± 0.0031 |
| Drosophila yakuba vs. D. erecta | 0.1330 ± 0.0229 | 0.0163 ± 0.0043 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.1482 ± 0.0511 | 0.0090 ± 0.0052 |
| Drosophila simulans vs. D. sechellia | 0.0351 ± 0.0112 | 0.0034 ± 0.0019 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0491 ± 0.0202 | 0.0030 ± 0.0030 |
| Mean | 0.0523 ± 0.0000 | 0.0060 ± 0.0020a | 0.0000 ± 0.0000b | 0.0042 ± 0.0027 | 0.0565 ± 0.0174 | 0.0052 ± 0.0010a |
Paired t-tests of the hypothesis that mean dS equals mean dN:
P < 0.05.
Paired t-tests of the hypothesis that mean dS or dN equals the corresponding value for the common domain:
P < 0.05.
3.2. Comparisons within and between Genera
There were two genera of Tephritidae (Anastrepha and Bactrocera) and one of Drosophilidae (Drosophila) represented by at least 4 species in the data set. In order to examine the patterns of nucleotide substitution over a somewhat longer evolutionary time frame than that represented by the sister-pair comparisons, I estimated mean dS and dN in the different domains of dsx for all pairwise comparisons within and between these three genera (Table 3). Within each genus, the observed pattern was generally similar to that seen in the sister-pair comparisons. In the common domain and in the male-specific exon, mean dS was significantly greater than mean dN (P < 0.001 in every case; Table 3). On the other hand, in the female-specific exon, mean dS and mean dN did not differ significantly; but this pattern was due to greatly reduced dS rather than elevated dN (Table 3). In fact, not a single synonymous substitution was observed in the female-specific exon in any of the pairwise comparisons within the tephritid genera Anastrepha and Bactrocera (Table 3). In all three genera, mean dS in the female-specific exon was significantly lower than that in the common domain (P < 0.001 in every case; Table 3).
Table 3.
Mean numbers of synonymous (dS) nucleotide substitutions per synonymous site and nonsynonymous (dN) nucleotide substitutions per nonsynonymous site in dsx genes in comparisons within and between genera of Tephritidae and Drosophilidae.
| Comparison | Common | Female-specific | Male-specific | |||
|---|---|---|---|---|---|---|
| dS ± S.E. | dN ± S.E. | dS ± S.E. | dN ± S.E. | dS ± S.E. | dN ± S.E. | |
| Within genera | ||||||
| Anastrepha | 0.0428 ± 0.0083 | 0.0067 ± 0.0016d | 0.0000 ± 0.0000g | 0.0053 ± 0.0039 | 0.4361 ± 0.0797 | 0.0549 ± 0.0116d |
| Bactrocera | 0.1622 ± 0.0210 | 0.0088 ± 0.0028d | 0.0000 ± 0.0000g | 0.0074 ± 0.0073 | 0.1276 ± 0.0314 | 0.0148 ± 0.0058d |
| Drosophila | 0.5891 ± 0.0375 | 0.0375 ± 0.0069d | 0.0786 ± 0.0433g | 0.0000 ± 0.0000g | 0.4914 ± 0.0812 | 0.0300 ± 0.0085d |
| Between genera | ||||||
| Anastrepha vs. Bactrocera | 0.9708 ± 0.1312 | 0.0351 ± 0.0208d | 0.2817 ± 0.1713f | 0.0055 ± 0.0042 | 0.7984 ± 0.1807 | 0.1052 ± 0.0237d, e |
| Anastrepha vs. Drosophila | --a | 0.1692 ± 0.0208 | 0.8113 ± 0.4083 | 0.0481± 0.0253g | 2.4335 ± 0.6452 | 0.5684 ± 0.0833c, g |
| Bactrocera vs. Drosophila | --a | 0.1807 ± 0.0216 | 1.2927 ± 0.5395 | 0.0338 ± 0.0213b,g | 2.4357 ± 0.5142 | 0.5582 ± 0.0795d, g |
The quantity could not be estimated because of saturation of synonymous sites.
Z-tests of the hypothesis that mean dS equals mean dN:
P < 0.05;
P < 0.01;
P < 0.001.
Z-tests of the hypothesis that mean dS or dN equals the corresponding value for the common domain:
P < 0.05.;
P < 0.01;
P < 0.001.
In Drosophila, there were no nonsynonymous differences in all pairwise comparisons of the female-specific exon; and mean dN was significantly lower in the latter exon than in the common domain (P < 0.001; Table 3). Mean dN in the male-specific exon was significantly greater than that in the common domain in Anastrepha (P < 0.001) but not in the two other genera (Table 3). Thus, the within-genus comparisons showed purifying selection on all domains. In Drosophila, there was evidence that purifying selection on the female-specific exon was stronger than that on the common domain, while in Anastrepha there was evidence that purifying selection on the male-specific exon was more relaxed than that on the common domain.
In comparisons between Drosophila and the two tephritid genera, dS in the common domain could not be estimated because of saturation of synonymous sites; and in the sex-specific exons, dS estimates were very high and close to saturation (Table 3). However, in the comparison between Anastrepha and Bactrocera, mean dS was significantly greater than mean dN in the common domain (P < 0.001; Table 3). In all between-genus comparisons, mean dS was significantly greater than mean dN in the male-specific exon (Table 3). And in the comparison between Bactrocera and Drosophila, mean dS was significantly greater than mean dN in the female-specific exon (Table 3).
In all three between-genus comparisons, mean dN in the male-specific exon was significantly greater than that in the common domain (Table 3). In the comparisons between both tephritid genera and Drosophila, mean dN in the female-specific exon was significantly lower than that in the common domain; and in the comparison between Anastrepha and Bactrocera, mean dS in the female-specific exon was significantly lower than that in the common domain (Table 3). Thus, the comparisons between genera again showed evidence of purifying selection on all domains, but there was a clear indication in these more distant comparisons that purifying selection on the male-specific exon was more relaxed than that on the common domain. By contrast, the female-specific exon showed evidence of stronger purifying selection and thus greater functional constraint than the common domain.
In order to examine possible causes of the reduced dS in the female-specific exon of all three genera, I examined nucleotide usage in this exon. Mean % G+C at third codon positions in the female-specific exon was 48.1% in Anastrepha, 45.2% in Bactrocera, and 55.8% in Drosophila. These values did not show a consistent pattern of difference from those observed in the common domain (45.6%, 39.9%, and 70.5%, respectively) or the male-specific exon (46.0%, 41.9%, and 76.2% respectively). Thus, it did not appear that a biased nucleotide content was responsible for the reduced dS. However, of 90 nucleotide sites in this exon, 69 were completely conserved in all species of the three genera in the data set. The conserved sites fell mainly in 7 blocks of 4 or more consecutive sites that were conserved in all three genera (Supplementary Figure S1). Most notable among these were two blocks each consisting of 11 consecutive nucleotides conserved in all three genera (sites 22–32 and 80–90; Supplementary Figure S1). These extensive conserved blocks included both synonymous and nonsynonymous sites, accounting for the the reduced dS in the female-specific exon. On the other hand, these blocks were not conserved in comparisons with species outside Tephritidae and Drosophilidae.
3.2. All Pairwise Comparisons
In all pairwise comparisons among 29 dipteran species, mean dN in the male-specific exon (0.7039 ± 0.0496) was significantly greater than that in the common domain (0.2096 ± 0.0143) or that in the female-specific domain (0.0515 ± 0.0174; Z-tests; P < 0.001 in each case). Likewise, mean dN in the common domain was significantly greater than that in the female-specific domain (Z-test; P < 0.001). Thus, these comparisons showed that purifying selection was strongest on the female-specific exon and most relaxed on the male-specific exon. Overall, 18 of 30 (60%) of amino acid sites encoded in the female-specific exon were conserved in all 29 species. By contrast, there was only one amino acid residue, a proline (at position 69 of 152 encoded by the male-specific exon of D. melanogaster), that was conserved in all 29 species.
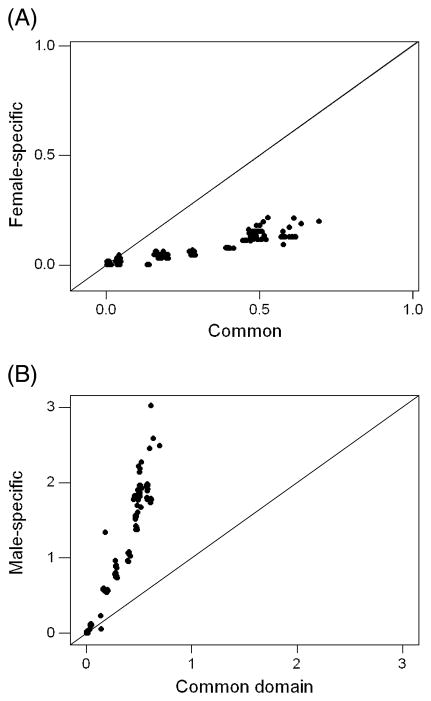
When the pairwise dN values in the female-specific exon were plotted against those in the common domain, there was a strong positive correlation (r = 0.935; P < 0.001; randomization test; Figure 2A). Likewise dN values in the male-specific exon were positively correlated with those in the common domain (r = 0.948; P < 0.001; randomization test; Figure 2B). However, inspection of the plots showed that dN in the male-specific exon increased much more rapidly as a function of dN in the common domain than did dN in the female-specific exon (Figure 2). When the method of allometric regression was applied to the relationship between dN in the male-specific exon and dN in the common domain, a slope of 2.562 ± 0.132 was obtained. This slope was significantly greater than zero (P < 0.001; Z-test) and significantly greater than 1.0 (P < 0.001; Z-test), indicating that dN in the male-specific exon increased as a function of dN in the common domain at a greater than linear rate. By contrast, the slope of the allometric regression of dN in the female-specific exon on dN in the common domain 0.294 ± 0.016. The latter value was significantly greater than zero (P < 0.001; Z-test) but significant less than 1.0 (P < 0.001; Z-test). Thus, dN in the female-specific exon increased as a function of dN in the common domain at a less than linear rate.
Figure 2.
The number of nonsynonymous substitutions per nonsynonymous site (dN) in (A) the female-specific exon and (B) the male-specific exon (r = 0.948; P < 0.001; randomization test) plotted against dN in the common domain for all pairwise comparisons among 29 dipteran species.
Discussion
Analysis of the pattern of nucleotide substitution of different domains of the dipteran dsx gene showed that over short evolutionary times purifying selection predominated on the domain common to both sexes, the female-specific exons, and the and male-specific exon. These patterns were seen in comparisons between closely related species pairs and indeed all comparisons within the families Tephritidae and Drosophilidae. However, over longer evolutionary time frames represented by between-family comparisons, the male-specific exon accumulated nonsynonymous substitutions at a much more rapid rate than either the common domain or the female-specific exon. Overall, the accumulation of nonsynonymous substitutions in the male-specific exon occurred at a significantly greater than linear rate relative to the common domain, whereas the accumulation of nonsynonymous substitutions in the female-specific exon occurred at less than linear rate relative to the common domain. The evolution of the male-specific exon of dsx thus shows a pattern reminiscent of that seen in the “runaway” evolution of male secondary sexual characters at the morphological level (Eberhard 1985). Assuming that amino acid changes in the sex-specific exons of dsx play a role in the evolution of genital morphology, the present results are consistent with the hypothesis that female choice is an important factor in the evolution of insect male genitalia.
The fact that the male-specific exon and the female-specific exon of dsx evolve at very different rates is consistent with the fact that female genital morphology is evolutionarily more conservative than male genital morphology, an observation often cited as evidence against the lock-and-key hypothesis (Eberhard 1985). On the other hand, the fact that dsx plays a role not only in the development of the genitals but also a role in the development of sex-specific behaviors (Rideout et al. 2007; Siwicki et al. 2009; Waterbury et al. 1999) is intriguing in the light of Mayr’s (1983) pleiotropy hypothesis for male genital evolution. Because of the dual role of dsx, changes in male genital structures, but not other morphological traits, might occur as a pleiotropic consequence of selection for changes in male sexual behavior, answering a major objection to Mayr’s theory (Eberhard 1985). However, it is not clear what would be the basis for selection specifically focused on the behavioral role of dsx but not on its role in genital development. The observed evolutionary pattern is suggestive of a role for sexual selection in the evolution of the male-specific exon of dsx, whether as a consequence of its role in the genitalia, or its role in the nervous system, or both.
Most discussions of the evolution of male traits by sexual selection have assumed that positive, directional selection drives the process (Fisher 1930; Kirkpatrick and Ryan 1991; Pomiankowski and Isawa 1998). On the other hand, Lande (1981) presented a quantitative genetic model in which a runaway process could be started if female preferences were subject to genetic drift. Here I describe a simple model (Figure 3) to illustrate how changes in the focus of female choice might drive a runaway process in the absence of positive selection. This model is consistent with the hypothesis of Nei (2007) that phenotypic divergences can result from the random fixation of neutral or nearly neutral mutations, in combination with changes over time in the focus of functional constraints.
Figure 3.
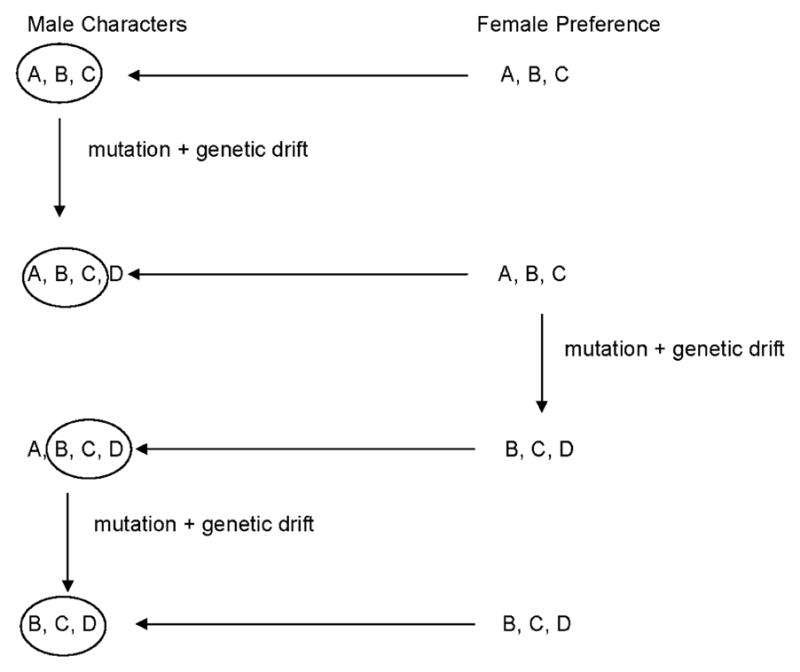
Schematic illustration of a simple model whereby runaway evolution of traits used in mate choice by females can occur through a process of mutation and genetic drift, in the absence of positive Darwinian selection.
Assume that females choose mates on the basis of certain traits (A, B, and C) of a given male structure (such as the genitalia), and that these traits are not essential to the biological function of the structure (Figure 3). Female choice focusing on A, B, and C will result in stabilizing selection at the phenotypic level and purifying selection on the genetic basis of A, B, and C. Certain other features of the structure in question that are neither necessary for its function nor used by the female in mate choice will be free to vary at random. As a result, mutations causing changes to these traits will not be deleterious, and some of them may be fixed by drift. Suppose that one such mutation is fixed so that the structure in question now has the properties A, B, C, and D in nearly all males of the species (Figure 3). Under these circumstances, a mutation that causes the female to choose on the basis of B, C, and D rather than A, B, and C will confer no fitness disadvantage and may become fixed by drift. If the latter happens, females will choose on the basis of B, C, and D; and the genetic basis of A will no longer be subject to purifying selection. A mutation that eliminates A will no longer be selected against and may be fixed by genetic drift (Figure 3). Thus, by this process the properties of a male structure used by the female for mate choice will be subject to runaway evolution in the total absence of positive selection, simply due to changes in the focus of the purifying selection resulting from female choice.
Contrary to a widespread impression, there is no unambiguous “signature” of positive Darwinian selection at the molecular level that can detect cases where an individual nonsynonymous substitution was selectively favored (Hughes 2007). Thus it is possible that some of the nonsynonymous substitutions occurring in the male-specific exon of dsx were fixed by positive selection. Nonetheless, the overall pattern of nucleotide substitution in the dsx gene suggests that positive selection has not played an important role in the evolutionary diversification of this gene. The pattern of nonsynonymous substitution in the male-specific exon of dsx suggests strong purifying selection over a short to intermediate term. However, over longer periods, a relaxation of purifying selection is detectable, giving rise to the positively allometric relationship between dN in the male-specific exon and dN in the common domain. Such a pattern is most consistent with the hypothesis that the male-specific exon of dsx is subject to purifying selection, but that the set of residues subject to strong purifying selection changes over evolutionary time. On this hypothesis, it is the shift over time in the focus of purifying selection that gives rise to the runaway evolution of the male-specific exon, consistent with the model outlined above.
Although there is evidence that amino acid changes in dsxM can cause changes in male genital morphology (Alvarez et al. 2009), the mechanistic basis of this effect is not known. Thus, it remains to be determined whether the evolution of dipteran male genital structures is driven by the runaway evolution of the male-specific exon of dsx. However, the hypothesis of a non-Darwinian runaway process presents an attractive explanation for the evolution of male genitalia in insects and many other groups of animals. This hypothesis is consistent with evidence that selection on insect male genital morphology in contemporary populations of insects and spiders is largely stabilizing (Eberhard et al. 1998); with the observation that certain structures of male insect genitalia are not needed for successful mating (Scudder 1971); and with the apparent arbitrariness of male traits used in female choice (Eberhard 1985). Moreover, this hypothesis provides an attractive alternative model for the evolution of sexually selected traits in general that does not require the problematic assumption of a genetic correlation between female preference and male trait postulated by Fisher’s (1930) model and its successors (Kirkpatrick and Ryan 1991). In the future, formal population genetics models as well as computer simulations may shed further light on the proposed mechanisms outlined in the above verbal model.
In contrast to the male-specific exon of dsx, the female-specific exon shows extraordinary conservation. Of the 30 amino acid positions in the female-specific exon, 60% were conserved in all 29 dipteran sequences analyzed here. Moreover, the conservation of this exon was not confined to the amino acid sequences encoded; rather, there were blocks of sequence, including both synonymous and nonsynonymous sites, there were conserved in all sequences from Tephritidae and Drosophilidae. This conservation suggests that the female-specific exon may have functions beyond protein-coding, perhaps a regulatory function in splicing of the dsx pre-mRNA. Indeed, it has been shown that insertion mutations in this exon can affect sex-specific splicing in Drosophila melanogaster (Nagoshi and Baker 1990). The strong conservation of the female-specific exons contrasts with the evolution of the male-specific exon, consistent with the distinct biological roles of DsxF and DsxM proteins (Sánchez and Guerrero 2001).
Supplementary Material
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez M, Ruiz MF, Sánchez L. Effect of the gene doublesex of Anastrepha on the somatic sexual development of Drosophila. PLoS One. 2009;4(4):e5141. doi: 10.1371/journal.pone.0005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnqvist G. Comparative evidence for the evolution of genitalia by sexual selection. Nature. 1998;393:784–786. [Google Scholar]
- Arnqvist G, Thornhill R. Evolution of animal genitalia: patterns of phenotypic and genotypic variation and condition dependence of genital and non-genital morphology in water strider (Heteroptera: Gerridae: Insecta) Genet. Res. 1998;71:193–212. [Google Scholar]
- Dufour L. Anatomie générale des diptères. Ann Sci Nat. 1844;1:244–264. [Google Scholar]
- Eberhard WG. Sexual Selection and Animal Genitalia. Harvard University Press; Cambridge MA: 1985. [Google Scholar]
- Eberhard WG, Huber BA, Rodriguez RL, Briceño RD, Salas I, Rodriguez V. One size fits all? Relationships between the size and degree of variation in genitalia and other body parts in twenty species of insects and spiders. Evolution. 1998;52:415–431. doi: 10.1111/j.1558-5646.1998.tb01642.x. [DOI] [PubMed] [Google Scholar]
- Eberhard WG, Ramirez N. Functional morphology of the male genitalia of four species of Drosophila: failure to confirm both lock and key and male-female conflict predictions. Ann Entomol Soc Amer. 2004;97:1007–1017. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; Oxford: 1930. [Google Scholar]
- Hosken DJ, Stockley P. Sexual selection and genital evolution. Trends Ecol Evol. 2004;19:87–93. doi: 10.1016/j.tree.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Jordan K. On mechanical isolation and other problems. Novi Zool. 1896;3:426–525. [Google Scholar]
- Kirkpatrick M, Ryan MJ. The evolution of mating preferences and the paradox of the lek. Nature. 1991;350:33–38. [Google Scholar]
- Li WH. Unbiased estimates of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Harvard University press; Cambridge MA: 1963. [Google Scholar]
- Nagoshi RN, Baker BS. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Pomiankowski A, Isawa Y. Runaway ornament diversity caused by Fisherian sexual selection. Proc Nat Acad Sci USA. 1998;95:5106–5111. doi: 10.1073/pnas.95.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Billeter JC, Goodwin SF. The sex determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr Biol. 2007;17:1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MF, Eirín-López JM, Stefani RM, Perondini AL, Selivon D, Sánchez L. The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insetcs. Dev Genes Evol. 2007;217:725–731. doi: 10.1007/s00427-007-0178-8. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sánchez L, Guerrero I. The development of the Drosophila genital disc. BioEssays. 2001;23:698–707. doi: 10.1002/bies.1099. [DOI] [PubMed] [Google Scholar]
- Scudder GG. Comparative morphology of insect genitalia. Ann Rev Entomol. 1971;16:379–406. [Google Scholar]
- Shapiro AM, Porter AH. The lock-and-key hypothesis: evolutionary and biosystematic interpretation of insect genitalia. Annu Rev Entomol. 1989;34:231–245. [Google Scholar]
- Siwicki KK, Kravitz EA. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr Opin Neurobiol. 2009;19:200–206. doi: 10.1016/j.conb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2. W.H. Freeman; San Francisco: 1971. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vincent S, Perkins LA, Perrimon N. Doublesex surprises. Cell. 2001;106:399–402. doi: 10.1016/s0092-8674(01)00468-8. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zahavi A. Mate selection – a selection for a handicap. J Theoret Biol. 1975;53:205–214. doi: 10.1016/0022-5193(75)90111-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.