Abstract
Background
Cancer biomarkers are the backbone for the implementation of individualized approaches to bladder cancer (BCa). Hyaluronic acid (HA) and all seven members of the HA-family i.e., HA-synthases (1,2,3), HYAL-1 hyaluronidase and HA-receptors (CD44s, CD44v and RHAMM) function in tumor growth and progression. However, the diagnostic and prognostic potential of these seven HA family members has not been compared simultaneously in any cancer. We evaluated the diagnostic and prognostic potential of HA-family members in BCa.
Methods
Using quantitative-PCR and immunohistochemistry, the expression of HA family members was evaluated in prospectively collected bladder tissues (n=72); mean and median follow-up: 29.6 ± 5.3; and 24 months, respectively. Transcript levels were also measured in exfoliated urothelial cells from urine specimens (n=148).
Results
Among HA family members, HA-synthase(s), HYAL-1, CD44v and RHAMM transcript levels were 4–16-fold elevated in BCa tissues when compared to normal tissues (P<0.0001), however, CD44s levels were lower. In univariate and multivariate analyses, tumor-stage (P=0.003), lymph node invasion (P=0.033), HYAL-1 (P=0.019) and HAS1 (P=0.027) transcript levels and HYAL-1 staining (P=0.021) independently associated with metastasis. Tumor-stage (p=0.019) and HYAL-1 (p=0.046) transcript levels also associated with disease specific mortality. While HA-synthase and HYAL-1 transcript levels were elevated in exfoliated urothelial cells from BCa patients, the combined HAS2-HYAL-1 expression detected BCa with overall 85.4% sensitivity and 79.5% specificity and predicted BCa recurrence within 6-months (P=0.004; RR=6.7).
Conclusion
HYAL-1 and HAS1 expression predicted BCa metastasis and HYAL-1 expression also predicted disease-specific survival. Furthermore, the combined HAS2-HYAL-1 biomarker detected BCa and significantly predicted its recurrence.
Keywords: Prognostic makers, hyaluronic acid, HA-synthase, HYAL-1, HA-receptors, hyaluronidase, diagnosis, recurrence
INTRODUCTION
Clinical and pathological parameters, such as tumor grade, stage, and lymph node invasion provide important prognostic information but have limited ability to predict development of metastases, or survival among BCa patients (1). Hyaluronic acid, HA receptors, and HA-degrading enzyme or hyaluronoglucosaminidase (HAase) have been implicated in tumor growth, angiogenesis, and metastasis (2–4). HA is a non-sulfated glycosaminoglycan involved in many physiological functions (2). Some of the molecules in HA signaling pathway function in tumor growth and progression and are useful biomarkers for cancer diagnosis and prognosis (2–6). Elevated HA level in urine is an accurate diagnostic marker (HA test) for detecting BCa, regardless of tumor grade and stage (7–12). HA is synthesized by HA-synthases: HAS1, HAS2 and HAS3 (13). Two studies reported that HAS1 expression is elevated in BCa and promotes tumor growth, infiltration and angiogenesis (14,15). Similarly, HAS2 and HAS3 also promote tumor growth and metastasis (16,17.
CD44 is a well characterized HA receptor (2). CD44 mRNA is frequently alternatively spliced and these mRNA splice variants generate different CD44 isoforms. In BCa, contradictory findings have been reported regarding the correlation of CD44 expression with tumor grade and disease-free survival (18–21). Miyake et al reported that the ratio of the CD44 variant isoform (v8–10) and CD44s (CD44-standard isoform) correlates with disease-free survival (20). RHAMM is another well characterized HA receptor involved in cell migration and motility and compensates for certain CD44 functions (22). Only one study has been reported regarding RHAMM expression in BCa and it showed that RHAMM levels, examined by immunohistochemistry, are increased in BCa tissues (18).
HYAL-1 type HAase is expressed by tumor cells and promotes tumor growth, infiltration, and angiogenesis (3,23). HYAL-1 expression in prostate cancer specimens is an independent predictor of biochemical recurrence following surgery (24, 25). HAase levels are elevated in the urine of high-grade BCa patients (HAase test; ref 10) and together with HA (HA-HAase test) detect BCa with high accuracy (10,11). Eissa et al have shown that HYAL-1 mRNA levels measured by semi-quantitative PCR are a marker for BCa (26). Recently we showed that HYAL-1 expression is elevated in bladder tumor tissues and is an independent predictor of muscle invasion (27).
In this study we examined the expression of seven HA family members in bladder tissues and urine specimens, by quantifying mRNA and protein levels to compare their diagnostic and prognostic accuracy alone and as a biomarker profile. We chose all of the molecules from the same biological pathway, rather than biomarkers in different functional pathways relevant to the malignant phenotype, because we hypothesized that due to their functional synergy, the combination of some of the HA-family molecules may have better diagnostic and prognostic potential than the individual molecules.
MATERIALS AND METHODS
Tissue specimens and patients: Tissue specimens
All specimens were obtained based on the availability of the specimen for research purpose and under a protocol approved by University of Miami’s Institutional Review Board. Normal bladder tissues (NBL; n=28) were obtained either from organ donors or from patients who underwent cystectomy. A portion of each BCa (n=44) and NBL tissue was paraffin embedded and the other was flash frozen. Total RNA was isolated from frozen bladder tissues (~ 30 mg) using the RNeasy Mini kit (Qiagen, Valencia, CA). The characteristics of all bladder specimens are presented in Table 1.
Table 1. Specimen and patient characteristics.
Characteristics of bladder tissue and urine specimens are shown.
| Tissue specimens n= 72 (NBL = 28) Organ donors (NBL-O) = 18 BCa patients undergoing cystectomy (NBL-B) = 10 |
Urine Specimens (n = 137) |
||
| BCa = 44; Transurethral resection (TURBT) = 11 Cystectomy = 33 |
NormalA | n = 29 Group 1, n = 18B Age: 38.3 ±13.9; Median: 30.5 yrs Gender: Female = 6; Male = 12 Group 2: n = 11 Age: 63.2 ± 10.1; Median: 61 yrs Gender: Female = 6; Male = 5 |
|
| Gender | Female n = 9 Male, n = 35 |
BCa | n = 48 LG = 14 Age: 68.4 ± 8.5; Median = 70.5 yrs Gender: Female = 8; Male = 6 HG = 34 Ta = 14; T1 = 13 T2 = 13 T3 = 7 T4 = 1 Age: 68.3 ± 10.1; Median = 68.5 yrs Gender: Female = 10; Male = 23 |
| Smoker | +) = 25 (−): 2 Unknown: 17 |
BGU | n = 34 Urinary tract infection = 6 Benign prostatic hyperplasia = 9 Urethral strictures = 4 Chronic prostatitis = 4 Urolithiasis = 3 Renal cyst = 2 Dysuria = 1 Benign adrenal mass = 2 History of cervical cancer = 2 Hydrocele = 1 Age: 57 ± 12.3; Median = 59 Gender: Female = 10; Male = 24 |
| Grade | LG = 7 (all, stage Ta) HG = 37 |
HXBCa | n = 31 Age: 64.8 ± 7.5; Median = 63 Gender: Female = 8; Male = 23 |
| Stage | Ta = 8; T1 = 3 T2 = 12; T3 = 16; T4 = 5 Concomitant CIS = 4 |
Prostate cancer | n = 6; Age: 66.6 ± 5.7; Median = 65 |
| LN | (+) = 11 (−) = 26 Unknown =7 |
||
| Metastasis (all BCa patients) Metastasis in patients with stage ≥ T2 tumor |
(−) = 19 (+) = 16 Unknown = 9 (−) = 12 (+) = 16 Unknown = 5 |
||
| Age | Metastasis (+): 63.7 ± 11yrs Median: 60 yrs Metastasis (−): 65 ± 12.3 yrs Median 68 yrs |
||
| Neoadjuvant Chemotherapy | (+) = 9 (−) = 28 Unknown = 7 |
||
| Adjuvant Chemotherapy |
(+) = 11 (−) = 34 Unknown = 7 |
||
| Radiation | (+) = 6 (−) = 29 Unknown = 9 |
||
| Death | (−) = 23 (+) = 19; BCa specific = 16 Unknown = 2 |
||
| Mean Follow-up For patients with stage ≥ T2 tumor |
29.6 ± 5.3; median = 24 (11–43 months) 28.7 ± 6.1; median = 20 (11–27) |
||
Normal: Normal individuals were healthy volunteers who did not have chronic and/or acute illness and were not taking any prescription medication, at the time specimen collection.
The normal individuals in Group 1 were included in all figures and calculations. Group 2 individuals were age-matched individuals with respected to BCa patients. Biomarkers were assayed among Group 2 individuals to determine whether the biomarker levels vary with respect to age. In the BGU category, 12 patients had microscopic or gross hematuria and underwent cystoscopy to rule out BCa. All HXBCa patients underwent cystoscopy.
Urine specimens
Urine specimens (n=148) were prospectively collected from healthy individuals, and patients with BCa, benign genitourinary (BGU) conditions or a history of BCa (HXBCa; Table 1). Clinical follow-up was collected on patients with HXBCa. All urine specimens were brought to the laboratory within two hours of collection and processed for total RNA isolation using the ZR urine isolation kit™ (Zymo Research Corp. Orange, CA). Briefly, urine specimens (100 – 150 ml) were passed through a syringe filter provided in the kit and the exfoliated cells captured on the filter were lysed in an RNA lysis buffer. RNA was then purified as per the manufacturer’s instructions. No fixation or isolation of exfoliated urothelial cells is necessary when using this kit.
Quantitative RT-PCR (Q-PCR)
Total RNA isolated from tissues or exfoliated cells was subjected to Q-PCR using the iQ real time PCR system (BioRad, Hercules, CA) and primers and probes specific for each transcript (Table 2; ref, 14,28). Each cDNA sample was simultaneously subjected to β-actin Q-PCR. The normalized transcript levels for each gene were calculated as (1/2Δct × 100); ΔCt = Ct (transcript) – Ct (β-actin). The variance of the PCR assay was examined by performing the Q-PCR RNA isolated from 15 specimens in quadruplicate for each marker and then computing the intra-class correlation. The intra-class coefficient for all markers varied between 0.955 and 0.994 with P < 0.001. This indicated that the replicate values within a sample were highly correlated or that the variance was low.
Table 2. Sequences of the primers and probes used in Q-PCR assays.
For CD44v, the primer and probe sequences were designed in exon 12, which is alternatively spliced in CD44s but is present in all variant isoforms. Therefore, CD44v primers and probes will amplify any and all alternatively spliced variants.
| Gene | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| HAS1 | 5-GGTGGGGACGTGC GGATC-3 | 5-ATGCAGGATACACAGTGGAAGTAG-3 | FAM 5-CCCGCTCCACATTGAAGGCTACCCAG-3BHQ |
| HAS2 | 5-TGAACAAAACAGTTGCCCTTT-3 | 5-TTCCCATCTATGACCATGACAA-3 | FAM 5-ATCGCTGCCTATCAAGAAGATCCAGAC-3 BHQ1 |
| HAS3 | 5-CTCTACTCCCTCCTCTATATGTC-3 | 5-AACTGCCACCCAGATGGA-3 | FAM 5-AATGAGGCCAAT GAAGTTCACCACAAT-3BHQ1 |
| CD44s | 5-CTGTACACCCCATCCCAGAC-3 | 5-TGTGTCTTGGTCTCTGGTAGC-3 | FAM5-TGGATCACCGACAG CACAGAC AGAAT-3BHQ |
| CD44v | 5-CAGGTGGAAGAAGAGACCCAA-3 | 5-GCTGAGGTCACTGGGATGAA-3 | FAN5-ACCCACACACGAAGGAAAGCAGGACC-3BHQ |
| RHAMM | 5-CAGCTGGAAGATGAAGAAGGA-3 | 5-GCATGTAGTTGTAGCTGAAAAGG-3 | FAM5-TGAAGAAATTAACAAGTGGCGTCT-3BHQ |
| HYAL-1 | 5-AGCCAGGGTAGCATCGACA-3 | 5-AAGCCCTCCTCCTCCTTAACC-3 | FAM5-CAGGCACAGATGGCTGTGGAGTT-3BHQ1 |
Immunohistochemistry
Five-micron sections of paraffin-fixed bladder tissues were placed on positively charged slides. The slides were sequentially deparaffinized, rehydrated and subjected to antigen retrieval by heating the slides at 95°C for 25 minutes in the Target-Retrieval Solution (DakoCytomation, Carpentaria, CA). The slides were incubated at 4°C for 16 hours, with the following primary reagents: 1. biotinylated HA binding protein (1 μg/ml; for HA staining); 2. anti-HYAL-1 IgG (1 μg/ml); 3. anti-HAS1 IgG (1.3-μg/ml); 4. anti-HAS2 IgG (0.5 μg/ml); 5. anti-HAS3 IgG (1.5 μg/ml). The slides were developed using the Dako LSAB kit and 3,3′-diaminobenzidine staining. The same batch of antibodies and commercial reagents were used in all experiments.
The specificity and validation of the HA-binding protein, and of rabbit polyclonal antibodies for HAS1, HAS2 and HYAL-1 have been reported previously (14,1523-25,27, 28). The affinity purified anti-HAS3 antibody was custom synthesized against a c-terminal sequence in HAS3 protein (ARRCGKKPEQYSLA) by (Genscript Corp; Piscataway, NJ). The specificity of anti-HAS3 antibody was established by down regulating HAS3 expression in BCa cell lines by HAS3siRNA transfection, followed by immunoblotting (data not shown). As outlined in the reviews by Bordeaux et al and Bonner et al (29, 30), for validation purposes, specimens slides were incubated with anti-HAS1, HAS2, HAS3, HYAL-1 antibodies in the presence of peptides (against which the respective antibodies were generated); for the biotinylated HA-binding protein, the slides were incubated with biotinylated HA-binding protein and HA (1 mg/ml). As a control for biotin-streptavidin conjugated-link solution in LSAB kit, IHC was performed by eliminating the primary antibody.
Stained slides were graded by two individuals in a blinded fashion. To account for the heterogeneity in staining, each specimen was graded for staining intensity (0 to 3 +) and then multiplied by the area in the specimen staining, with that intensity (e.g., 25% × 0 = 0; 50% × 1+ = 50 and 25% × 2+ = 50). The intensity scores in all areas were added to obtain the staining score for the entire specimen (e.g., 0 + 50 + 50 = 100). Therefore, each specimen received a staining score between 0 and 300 (27). The intensity scores of the two readers then were averaged to obtain the final score. The slides were also evaluated using the IP Image Analysis software and the results were comparable to the readers’ scores. There was significant correlation between staining scores of the two readers (Spearman r=0.852; 95% CI: 0.773 – 0.967; P=< 0.001) and between the average scores of the two readers and the IP image analysis scores (Spearman r=0.863; 95% CI: 0.804 – 0.915; P=0.003).
Statistical analyses
Differences in biomarker levels among bladder tissues (e.g., NBL versus low-grade, NBL versus high-grade) were compared using the Mann-Whitney U-test, because the data showed a non-normal distribution. Similar analysis was conducted when comparing the biomarker levels in various categories of urine specimens (e.g., BGU versus tumor, etc). All of the P-values reported in this study are two-tailed. Logistic regression single-parameter model (i.e., univariate analysis) was used to determine: 1. the association of clinical parameters, and biomarker levels (i.e., transcript levels or staining scores) with metastasis and disease-specific survival; 2. association of urinary biomarker levels with BCa. Cox-proportional hazards model (i.e., multivariate analysis) was used to determine which of the pre- and post-operative parameters and/or tissue biomarkers predict metastasis and disease-specific survival.
The levels of the combined biomarkers (e.g., HAS2-HYAL-1) for each study subject were calculated as follows: [intercept +(α × (HAS2)1) + (β × (HYAL-1)1)]; α and β: HAS2 and HYAL-1 coefficients, respectively and (HAS2)1 and (HYAL-1)1: HAS2 and HYAL-1 levels in subject # 1, respectively. The intercept and coefficients for each marker were computed by simultaneously analyzing the two variables (e.g., (HAS2 and HYAL-1) in the logistic regression model (i.e., bivariate analysis).
Receiver operating characteristic (ROC) curves were generated to determine the association between tissue biomarker levels and metastasis or disease-specific survival and of the various urine biomarkers (both single and the combined) with BCa. Cut-off values were selected from the ROC curve data by a statistical program (JMP®6 Software from SAS) for calculating sensitivity and specificity of each biomarker. A biomarker level that yielded the highest efficacy (i.e., sensitivity – (1-specificity)) was selected by the program as the cut-off limit. Cross-validation using boot-strap modeling (specific sampling rate = 0.5; re-sampling = 104) was performed to obtain the mean ± SD and 95% CI for the sensitivity, specificity and accuracy of each biomarker. Statistical analyses were carried out using the JMP® Software Program (version 6.0; SAS Institute, Cary, NC).
RESULTS
HA-synthase, HYAL-1, CD44v, RHAMM expression increases but CD44s levels decrease in BCa tissues
Transcript expression
We measured the levels of HA-synthases, HYAL-1, and HA receptor transcripts in 72 bladder tissues by Q-PCR. Figure 1 shows that when compared to NBL tissues, HAS1, HAS2, HAS3 and HYAL-1 levels were 4–16-fold elevated in both low-grade (P<0.0001 for all markers) and high-grade (P<0.0001) BCa tissues. However, the differences in the transcript levels between low- and high-grade tumor tissues were not statistically significant. Among HA receptors, while CD44s mRNA levels decreased 3–6-fold in BCa tissues (P<0.0001), CD44v levels were ~ 5-fold higher in BCa tissues (P<0.0001; Figure 1). The CDD4v/CD44s ratio was 14.6-fold (111.4 ± 69.2; P < 0.0001) and 37.3-fold (283.4 ± 60.8; P < 0.0001) higher in low-and high-grade BCa tissues, respectively, when compared to NBL tissues (7.6 ± 15.6). RHAMM expression was also significantly elevated in low-grade (P=0.007) and high-grade (P<0.0001) tissues (Figure 1). In this study NBL tissues were obtained from organ donors (NBL-O) and BCa patients (NBL-B). As shown in Figure 1, the levels of each HA-family member was very similar among NBL-O and NBL-B tissues (P > 0.05 for each marker). The sensitivity and specificity of HA-family molecules for distinguishing NBL and BCa tissues were as follows: HAS1: 72%, 75%; HAS2: 81.4%, 100%; HAS3: 72.1%, 89.3%; HYAL-1: 81.4%, 82.1%; CD44s: 73.2%, 81.1%; CD44v: 78.1%; 85.7%).
Figure 1. Scatter diagrams of HA-synthase and HYAL-1 mRNA levels in bladder tissues.
mRNA levels of each of the seven HA-family molecules in bladder specimen are shown. The mean ± SD scores for each biomarker are indicated. NBL: normal bladder; NBL-O: NBL tissue obtained from organ donors; NBL-B: NBL tissue obtained from BCa patients at the time of cystectomy. LG: low-grade BCa; HG: high-grade BCa.
Protein expression
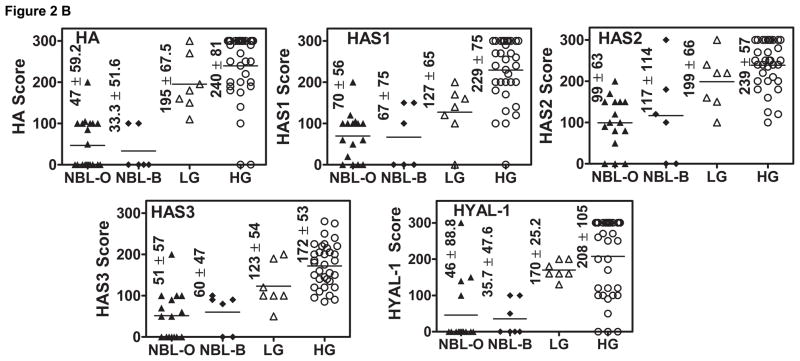
Since HAS (1, 2, 3) and HYAL-1 transcript levels were significantly elevated in BCa tissues, we performed IHC in the same set of tissues to determine whether the observed differences in the transcript levels among normal and tumor specimens were accompanied by similar changes in the protein expression. Figure 2A shows that both the tumor associated stroma and tumor cells expressed HA, HAS1, HAS2 and HAS3; HYAL-1 expression was observed only in tumor cells. As shown in Figure 2B, for HA and HAS2 staining, the difference between low-grade tumors and NBL and between high-grade tumors and NBL were statistically significant (P < 0.0001; Figure 2B). For HAS1, HAS3 and HYAL-1 staining only the differences between high-grade and NBL were statistically significant (P<0.0001). For HYAL-1, the differences between low-grade and high-grade BCa tissues were also statistically significant (P=0.002). The overall sensitivity and specificity of HA-family proteins to distinguish between NBL and tumor tissues was as follows: HA: 95%, 95.2%; HAS1: 77.5; 85.7%; HAS2: 77.5%, 90.5%; HAS3: 67.4%, 100%; HYAL-1: 80%, 86.4%. No consistent staining pattern was observed among NBL and BCa tissues for RHAMM and CD44 (data not shown).
Figure 2. Analyses of HA-synthase(s) and HYAL-1 expression in bladder tissues.
A: HA, HAS1, HAS2, HAS3 and HYAL-1 were localized in normal bladder, low-grade (LG) and high-grade (HG) BCa tissues by IHC. Representative specimens from each category are shown. B: Scatter diagrams of staining scores of HA, HAS1, HAS2, HAS3 and HYAL-1 in bladder specimens are shown. Five NBL specimens could not be stained due to poor fixation resulting in the loss of tissue from the slides during staining.
Association of HA-family members with metastasis
In this study, the majority of the patients had high-grade (n=37) and muscle invasive BCa (n=33) BCa. Among patients with high-grade BCa who later experienced metastasis, the mean and median transcript levels of HAS1 (5.1± 5.5; 2.4) and HYAL-1 (18±12.5; 13.2) were significantly higher when compared to those who did not develop metastasis (HAS1: 1.4±1.1; 1.1; HYAL-1 5.5± 5.5; 4.0). Similarly, the mean and median transcript levels of HAS1 (5.7± 5.1; 2.1) and HYAL-1 (16.3± 9.2; 10.7) were higher among patients who died of the disease when compared to those who did not die (HAS1: 2.0± 1.7; 1.6; HYAL-1 8.3± 6.8; 5.9). Among all markers, only the mean and median staining inferences for only HYAL-1 were higher among patients who had metastasis (271.8± 62.3; 300) or died from BCa (242± 102; 300), when compared to those who did not experience metastasis (HYAL-1: 126.8± 58; 120) or die of BCa (162± 86; 160).
Univariate analysis showed that stage, lymph node status, HYAL-1 and HAS1 transcript levels, and HYAL-1 staining inferences significantly associated with metastasis and disease specific mortality (Table 3A). In the multivariate model, stage, lymph node, HYAL-1 and HAS1 mRNA levels and HYAL-1 staining independently associated with metastasis (Table 3B). For disease specific mortality, only stage and HYAL-1 mRNA levels were significant predictors (Table 3B).
Table 3. Determination of the association between metastasis/disease specific mortality and clinical parameters and HA-family members.
The pre- and post-operative parameters included age, gender, tumor grade, stage, lymph node status and concomitant presence of CIS.
| Table 3A: Univariate analysis | ||||||
|---|---|---|---|---|---|---|
| Parameter | Chi-square | P-vale | Odds Ratio | Chi-square | P-Value | Odds Ratio |
| Metastasis | Disease-specific mortality | |||||
| Stage | 8.64 | 0.003* | 5.4; 2.2 – 22.4 | 8.6 | 0.0034 | 6.45; 2.4–29.4 |
| Grade | 0.01 | 0.94 | ND | 0.01 | 0.92 | ND |
| Lymph node | 4.53 | 0.033* | 3.71; 1.3 – 16.4 | 5.82 | 0.016 | 3.9; 1.5–13.9 |
| Gender | 0.1 | 0.77 | ND | 0.00 | 1.0 | ND |
| Age | 0.13 | 0.73 | ND | 1.23 | 0.27 | ND |
| CIS | 0.74 | 0.39 | ND | 0.15 | 0.7 | ND |
| HYAL-1 | 7.1 | 0.008* | 1.76; 1.27 – 3.0 | 5.71 | 0.017 | 1.63; 1.2–2.7 |
| HAS1 | 5.13 | 0.024* | 1.83; 1.21 – 3.5 | 3.8 | 0.049 | 1.31; 1.1–2.0 |
| HAS2 | 1.78 | 0.18 | ND | 0.02 | 0.9 | ND |
| HAS3 | 2.86 | 0.09 | ND | 3.5 | 0.06 | ND |
| RHAMM | 0.46 | 0.5 | ND | 0 | 0.97 | ND |
| CD44s | 2.43 | 0.1 | ND | 0.77 | 0.38 | ND |
| CD44v | 0.7 | 0.4 | ND | 0.11 | 0.74 | ND |
| CD44v/CD44s | 0.81 | 0.37 | ND | 0.13 | 0.72 | ND |
| HYAL-1 Staining* | 10.9 | 0.009* | 1.03; 1.02 – 1.06 | 4.13 | 0.04 | 1.36; 1.02 – 2.4 |
| Table 3B: Multivariate analysis | ||||||
| Parameter | Chi-Square | P-value | Risk ratio; 95% CI | Chi-Square | P-value | Risk ratio; 95% CI |
| Metastasis | Disease specific survival | |||||
| Stage | 5.66 | 0.0174* | 5.7; 1.4–59.4 | 5.47 | 0.0193* | 2.92; 1.2–8.3 |
| Lymph node | 7.26 | 0.007* | 10.2; 1.9–71.4 | 1.9 | 0.17 | ND |
| HYAL-1 | 5.35 | 0.019* | 1.76; 1.1–2.85 | 3.98 | 0.046* | 1.15; 1.05–1.28 |
| HAS1 | 4.97 | 0.027* | 1.37; 1.2–1.98 | 1.88 | 0.17 | ND |
| HYAL-1-Staining | 5.33 | 0.021* | 1.65; 1.05–2.93 | 0.3 | 0.58 | ND |
A: Univariate analysis: Logistic regression analysis was used to determine the association of pre-and post-operative parameters and biomarker levels with outcome. ND: Not determined for parameters that did not reach significance.
: significant parameter.
B: Multivariate analysis: Cox proportional hazards analysis was performed by including all of the pre- and post-operative parameters and either the transcript levels or the staining scores of the HA-family molecules.
Although, the number of specimens was limited, HYAL-1 mRNA (cut-off limit: 9.5) and HAS1 mRNA (cut-off 1.83) levels had 80%, 87.5% sensitivity; and 100%, 73.7% specificity to associate with metastasis, respectively. For HYAL-1 staining (cut-off 270), the sensitivity and specificity were 93.7% and 100%, respectively. However to predict disease-specific mortality, the sensitivity and specificity were modest for HYAL-1 mRNA and HYAL-1 staining: 70.2%, 80% sensitivity; and 80% and 65.7% specificity, respectively,.
HA-synthase, HYAL-1 expression is increased in exfoliated urothelial cells from BCa patients
We used the Q-PCR assay to measure the transcript levels of HA-family members in exfoliated urothelial cells present in urine specimens. HAS1, HAS2, HAS3 and HYAL-1 levels were elevated in urine specimens from BCa patients, when compared to the control categories (Figure 3). CD44s expression was low in exfoliated cells and it decreased in BCa patients by 3–7-fold. CD44v and RHAMM transcripts levels did not alter significantly among various categories. Univariate analyses showed that HAS1, HAS2, HAS3 and HYAL-1 mRNA levels were significantly associated with the presence of BCa (Table 4). For HAS1, HAS2 and HAS3, only the differences in mRNA levels among high-grade BCa patients and each control subgroup were statistically significant (P<0.001; data not shown). HYAL-1 mRNA levels were significantly elevated in both high-grade (P<0.0001) and low-grade (P=0.0011) patients’ exfoliated cells. Among HA-receptors, only CD44s mRNA levels were significantly different among BCa patients and the control category (Table 4).
Figure 3. Scatter diagrams of HA-synthase and HYAL-1 mRNA levels in exfoliated cells.
The distribution of the mRNA levels of each of the seven HA-family members among the study cohort is shown. The mean ± SD scores for each biomarker are indicated.
Table 4. Determination of the association between the presence of BCa and the levels of HA-synthase(s), HYAL-1 and HA-receptor transcripts in exfoliated urothelium.
Logistic regression single parameter analysis was used to determine the association between the presence of BCa and a biomarker.
| Biomarker | Chi-square | P-value | Odds Ratio | 95%-CI |
|---|---|---|---|---|
| HAS1 | 20.2 | < 0.0001 | 1.47 | 1.26 – 1.75 |
| HAS2 | 25.4 | < 0.0001 | 1.57 | 1.33 – 1.89 |
| HAS3 | 15.4 | < 0.0001 | 1.65 | 1.3 – 2.14 |
| HYAL-1 | 20.2 | < 0.0001 | 1.34 | 1.21 – 1.59 |
| HAS1+HYAL-1 | 21.09 | < 0.0001 | 2.85 | 1.95 – 4.76 |
| HAS2+HYAL-1 | 23.5 | < 0.0001 | 3.44 | 2.23 – 6.13 |
| HAS3+HYAL-1 | 22.3 | < 0.0001 | 1.94 | 1.5 – 2.63 |
| HAS1+HAS2 | 25.9 | < 0.0001 | 2.85 | 2.0 – 4.5 |
| HAS1+HAS3 | 20.9 | < 0.0001 | 2.44 | 1.7 – 3.76 |
| HAS2+HAS3 | 25.5 | < 0.001 | 1.77 | 1.44 – 2.26 |
| RHAMM | 0.96 | 0.34 | 0.99 | 0.98 – 1.00 |
| CD44s | 8.1 | 0.005 | 54.7 | 5.3 – 1140 |
| CD44v | 0.44 | 0.51 | 1.0 | 0.97 – 1.01 |
| CD44v/CD44s | 0.65 | 0.42 | 0.99 | 0.98 – 1.01 |
For normal individuals, the data shown in Figure 3 and Table 4 pertain to 18 volunteers categorized as Group 1 (as shown in Table 1). The mean and median age of these individuals were significantly lower than the other control categories of individuals (BGU and HxBCa) and BCa patients. The mean and median age of Group 2 individuals (Table 1) were not significantly different from the patients with BGU conditions, HxBCa or BCa. The mean and median levels of all seven markers in Group 2 normal individuals were as follows: HAS1: 1.3 ± 1.1, 1.1; HAS2: 3.5 ± 1.9, 2.8; HAS3: 0.9 ± 0.43, 0.93; HYAL-1: 2.3 ± 1.3, 2.3; CD44s: 0.22 ± 0.18, 0.12; CD44v: 3.8 ± 1.9, 3.7; RHAMM: 0.13 ± 0.24; 0.15. The differences in biomarker levels between Group 1 and the age-matched Group 2 were not statistically significant (P > 0.05), showing that the significant alterations observed among BCa and control categories of individuals were not related to age.
Efficacy of HA-family molecules to detect BCa
Based on the cut-off values generated by ROC curves all three HA-synthases and HYAL-1 markers had the same sensitivity (72.9%); however, the specificity was higher for HAS2 and HYAL-1 (81–83%; Table 5). The levels of CD44s, CD44v or RHAMM had either low sensitivity or specificity to detect BCa. HA-synthase markers had low sensitivity (≤50%) to detect low-grade BCa, but high-sensitivity (>80%) to detect high-grade BCa; contrarily, HYAL-1 had similar sensitivity to detect both low- (71.4%) and high-grade BCa (73.5%; Table 6).
Table 5. Determination of sensitivity, specificity and accuracy of HA-family molecules for detecting BCa.
Sensitivity, specificity and accuracy of the biomarkers were determined using the cut-off values determined from the ROC curves. AUC: area under the curve. For all markers except CD44s, transcript levels ≥ cut-off value indicated a positive inference. For CD44s, the levels ≤ 0.026 indicated a positive inference (either true positive (i.e., BCa) or false positive). Six patients with prostate cancer were not included in the non-BCa category for specificity calculations. The mean and 95% CI for sensitivity, specificity and accuracy of each biomarker were computed by bootstrap modeling.
| Biomarker | AUC | Cut-off | Sensitivity (Mean; 95% CI) | Specificity | Accuracy |
|---|---|---|---|---|---|
| HAS1 | 0.80 | 4.0 | 72.9% (35/48) (77.85; 74.5 – 81.2) |
71.1% (59/83 68.5; 65.2 – 72) |
71.8% (94/131) 73.3; 72.4 – 74.1 |
| HAS2 | 0.86 | 6.6 | 72.9% (35/48) (67.0; 64.5 – 69.5) |
83.1% (69/83) (86.4; 84 – 88.8) |
79.4% (104/131) 76.7; 75.8 – 77.6 |
| HAS3 | 0.77 | 2.4 | 72.9% (35/48) (80.1; 77.6 – 82.5) |
74.7% (62/83) (66.6; 64 – 69.1) |
74.1% (97/131) 73.3; 72.4 – 74.1 |
| HYAL-1 | 0.83 | 6.6 | 72.9% (35/48) (68.5; 65.7 – 71.4) |
81.9% (68/83) (85.8; 83.2 – 88.3) |
78.6% (103/131 ) 77.2; 76.2 – 78.2 |
| HAS1-HYAL-1 | 0.85 | 5.0 | 77.1% (37/48) 75; 72.9 – 77.1 |
78.3% (65/83) 84.1; 82.3 – 85.9 |
77.9% (102/131) 79.6; 78.9 – 80.3 |
| HAS2-HYAL-1 | 0.89 | 5.2 | 85.4% (41/48) 83.8; 82.1 – 85.5 |
79.5% (66/83) 83.1; 81.5 – 84.7 |
81.7% (107/131) 83.4; 82.8 – 84.1 |
| HAS3-HYAL-1 | 0.84 | 5.7 | 70.8% (34/48) 76.9; 74.7 – 79.3 |
79.5% (66/83) 80.6; 78.4 – 82.8 |
76.3% (100/131) 78.8; 78.1 – 79.5 |
| HAS1-HAS2 | 0.87 | 6.1 | 75% (36/48) 77.1; 74.8 – 79.5 |
80.7% (67/83) 84.4; 82.3 – 86.6 |
78.6% (103/131) 80.8; 80.2 – 81.4 |
| HAS2-HAS3 | 0.85 | 7.1 | 87.5% (42/48) 85.2; 83.5 – 86.9 |
69.9% (58/83) 74.1; 72.4 – 75.8 |
76.3% (100/131) 79.7; 79.0 – 80.4 |
| HAS1-HAS3 | 0.81 | 4.1 | 75% (36/48) 85.2; 83.3 – 87.2 |
69.9% (58/83) 67.9; 65.7 – 70.2 |
71.8% (94/131) 76.6; 75.9 – 77.2 |
| RHAMM | 0.52 | 0.17 | 39.6% (19/48) 45.1; 41 – 49.3 |
78.3% (65/83) 75.3; 71.4 – 79.3 |
64.1% (84/131) 60.3; 59.3 – 61.2 |
| CD44S | 0.74 | 0.026 | 62.5% (30/48) 66.3; 63.4 – 69.2 |
73.5% (61/83) 80.8; 77.6 – 80.04 |
69.5% (91/131) 73.6; 72.5 – 74.6 |
| CD44v | 0.45 | 4.4 | 81.3% (39/48) 78.5; 73.8 – 82.3 |
26.5% (22/83) 40.9; 36.3 – 45.8 |
46.7% (61/130) 59.7; 58.9 – 60.5 |
| CD44v/CD44s | 0.7 | 223 | 64.5% (31/48) 74.3; 70.3 – 78.3 |
72.3% (60/83) 58.5; 52.4 – 64.7 |
69.5% (91/131) 66.4; 64.6 – 68.2 |
Table 6. Analysis of sensitivity by tumor grade and of specificity by non-BCa conditions.
For each marker, the cut-off limit generated for by the ROC curve was used to determine the sensitivity and specificity of each marker for tumor grade and non-BCa conditions (normal, HXBCa and BGU), respectively.
| Marker | Sensitivity | Specificity | |||
|---|---|---|---|---|---|
| LG | HG | Normal | HXBCa | BGU | |
| HAS1 | 50% (7/14) | 82.4% (28/34) | 94.4% (17/18) | 64.5% (20/31) | 64.7% (22/34) |
| HAS2 | 42.9% (6/14) | 85.3% (29/34) | 100% (18/18) | 80.6% (25/31) | 76.5% (26/34) |
| HAS3 | 57.1% (8/14) | 79.4% (27/34) | 100% (18/18) | 67.7% (21/31) | 67.7% (23/34) |
| HYAL-1 | 71.4% (10/14) | 73.5% (25/34) | 100% (18/18) | 80.6% (25/31) | 73.5% (25/34) |
| HAS1-HYAL-1 | 1.4% (10/14) | 79.4% (27/34) | 100% (18/18) | 77.4% (24/31) | 67.7% (23/34) |
| HAS2-HYAL-1 | 78.6% (11/14) | 88.2% (30/34) | 100% (18/18) | 77.4% (24/31) | 70.6% (24/34) |
| HAS3-HYAL-1 | 64.3% (9/14) | 73.5% (25/34) | 100% (18/18) | 74.2% (24/31) | 70.6% (24/34) |
| HAS1-HAS2 | 42.9% (6/14) | 82.4% (28/34) | 94.4% (17/18) | 80.6% (25/31) | 73.5% (25/34) |
| HAS2-HAS3 | 78.6% (11/14) | 91.2% (31/34) | 100% (17/18) | 71% (22/31) | 55.9% (19/34) |
| HAS1-HAS3 | 57.1% (8/14) | 82.4% (28/34) | 94.4% (17/18) | 70.1% (20/31) | 64.7% (21/34) |
Specificity of all seven biomarkers among normal individuals was high (Table 6). However, only HYAL-1 and HAS2 had reasonable specificity for the BGU (76.5% and 73.5%) and HXBCa (80.6%) categories, respectively. Among the 31 patients with HXBCa, five recurred within 6 months. As shown in Table 7, HYAL-1 and HAS2 mRNA levels significantly associated with recurrence within 6-months.
Table 7. Mantael-Hanszel chi-square analysis to evaluate the predictive potential of HA-family markers for BCa recurrence.
Chi-square analyses was performed to determine the predictive value of each marker.
| Parameter | Chi-square | P-value | Risk Ratio | 95% CI |
|---|---|---|---|---|
| HAS1 | 2.9 | 0.088 | ND | ND |
| HAS2 | 19.5 | < 0.0001 | 20.8 | 3 – 147 |
| HAS3 | 4.0 | 0.045 | 4.2 | 0.92 – 19.2 |
| HYAL-1 | 10.7 | 0.001 | 8.3 | 2.0 – 35.4 |
| HAS1-HYAL-1 | 8.3 | 0.004 | 6.9 | 1.6 – 30 |
| HAS2-HYAL-1 | 8.3 | 0.004 | 6.7 | 1.7 – 30 |
| HAS3-HYAL-1 | 8.3 | 0.004 | 6.7 | 1.7 – 30 |
| HAS1-HAS2 | 10.7 | 0.001 | 8.3 | 1.9 – 35.3 |
| HAS1-HAS3 | 1.6 | 0.21 | ND | ND |
| HAS2-HAS3 | 5.1 | 0.024 | 4.9 | 1.1 – 22.1 |
Combination of HA-synthase and HYAL-1 biomarkers to detect BCa
As shown in Table 4, all six HA-synthase and/or HYAL-1 combinations significantly associated with BCa. The HAS2-HYAL-1 combination had the highest efficacy for detecting BCa (Table 5), with high sensitivity for low- and high-grade tumors and reasonable specificity for symptomatic controls. The HAS2-HYAL-1 combination also significantly associated with recurrence within 6 months (Table 7). These results show that the combination of HAS2 and HYAL-1 significantly increases the efficacy for detecting BCa, and to predict BCa recurrence before its clinical detection.
In this study, the biomarker expression was also evaluated in exfoliated urothelial cells present in urine specimens from six prostate cancer patients. HAS3 and the HAS2+HYAL-1 combination detected five patients (83.3%), while the other markers individually or in combination detected 50% – 66% of the patients. This suggests that if prostate tumor cells are shed in urine, they can be detected by HA family markers.
DISCUSSION
HA-family molecules have been shown to promote tumor growth, metastasis and angiogenesis, but some of them, such as HA-synthases and HA-receptors have overlapping functions. It is relatively unknown whether these proteins with overlapping functions are simultaneously expressed in normal and tumor tissues and what if any, correlation exists between their expression and the invasive potential of a tumor. In BCa, urinary HA levels are elevated (9–12), however, it is unknown which one or all three HA-synthases contribute to these elevated levels. Some qualitative PCR studies have reported increased HYAL-1 expression in urine sediments of BCa patients (26,31). However, HYAL-1 transcript levels have not been measured in tissues and exfoliated urothelial cells, to establish whether the increased urinary HAase levels in high-grade BCa patients are due to increased HYAL-1 levels. For the HA-receptors, different CD44 isoforms appear to have different clinical correlations. For example, the loss of CD44 and CD44v6 has been shown to associate with tumor stage and to poor outcome (19, 32, 33). However, CD44v8-10 expression may potentiate tumor progression (34) and increased CD44 mRNA expression in exfoliated cells has been reported as a marker for BCa (20,21,35). No study has reported RHAMM expression at the mRNA levels in BCa. Therefore, this is the first study that simultaneously evaluated the expression of all HA-family of molecules and at both the transcript and protein levels in any tissue.
The second objective of the study was to compare the diagnostic and prognostic potentials of all seven members of the HA-family in prospectively collected specimens. The transcript levels of HA-family molecules measured either in tissue or exfoliated cells had similar sensitivity and specificity to distinguish between normal and tumor specimens. The overlap observed among normal and BCa tissues with respect to the expression of various markers could be because of the genetic variability found in bladder tumors (36), since in this study we did not analyze multiple tumor foci from the same patient or use laser dissection to analyze different portions of the same tissue. In the case of exfoliated cells, such sorting of cells and their analysis, even by a sensitive technique such as the Q-PCR, will not be practical due to the detection limit and issues regarding RNA quality.
IHC was performed mainly to examine the pattern of expression of the HA-family molecules. Nevertheless, the sensitivity and specificity of the staining of HA-family molecules in bladder tissue specimens were albeit higher. This could be attributable to the fact that IHC is semi-quantitative and requires higher expression for detection than is required for the detection by Q-PCR. The semi-quantitative nature of IHC most likely contributed to increasing the differences in NBL and tumor tissues in terms of staining scores, and which in turn, resulted in higher specificity (or less overlap between NBL and BCa tissues). The use of quantitative fluorescence imaging technique could improve the quantification of the expression of HA-family molecules at the protein level (37–39). Nevertheless, unlike urine markers which have utility in the diagnostic arena, tissue markers have the most utility in providing prognostic information.
Our study demonstrates that while HAS1 and HYAL-1 expression in BCa tissues may correlate with metastasis (and disease-specific survival – HYAL-1), the combined HAS2-HYAL-1 mRNA levels in exfoliated urothelial cells display high sensitivity to detect BCa and may predict BCa recurrence. Although these conclusions are similar to those reported before regarding the increased HA and HAase levels (9–12), the present study reveals the molecular basis for this increase, i.e., increased HA-synthase and HYAL-1 transcript levels. Since HAS2 levels associate significantly with BCa diagnosis, but HAS1 levels associate with tumor metastasis, it suggests that there are functional differences among HA-synthases regarding tumor behavior. It remains to be determined why all three HA-synthases are elevated in BCa, despite all three genes are present in different chromosomes in the human genome (HAS1: 19q13.3-q13.4; HAS2: 8q24.12; HAS3: 16q22.1; 40).
Although in this study the number of patients with follow-up was small, it was sufficient to demonstrate that increased HYAL-1 expression was an independent prognostic indicator for BCa metastasis and survival and HAS1 expression associated with metastasis. However, one of the reasons why some study individuals did not develop metastasis might be the fact that the follow-up time for those patients was not sufficient. Only a study with large cohort of patients and sufficient follow-up time will definitively address whether transcript levels of the HA family members can identify patients that subsequently develop metastasis.
In regards to the diagnostic potential of seven HA family members, the combined inference of HAS2 and HYAL-1 mRNA levels increases the sensitivity for BCa detection. This is consistent with our previous observation regarding the HA-HAase test, which has higher accuracy to detect BCa than the individual HA and HAase tests, which measure urinary HA and HAase levels (11). The increased transcript levels of HAS2 and HYAL-1 in tumor cells confirm the molecular basis for increased urinary HA and HAase levels.
The major limitation of any Q-PCR assay is RNA stability. The ZR urine isolation kit™ allows on-site capturing of urothelial cells from urine and resuspending them in a lysis buffer. The lysis buffer stabilizes the RNA, which can be then shipped to a reference laboratory for Q-PCR assays. One limitation of our study is that this was a single institution study and the number of patients with high-grade BCa with variable clinical follow-up; the latter possibly could have skewed inferences regarding the prognostic capability of HA-family of markers. The second limitation may be that patients in BCa, BGU and HxBCa categories were not age-matched. However, this was not a case-control study and specimens were obtained from consecutive patients, to mimic the clinical scenario. Since the biomarker levels were not different among normal individuals (both groups 1 and 2), BGU patients and patients with HxBCa, it demonstrates that neither gender nor age influences the biomarker levels, however the presence of BCa does. Another perceived limitation of this study could be that the transcript levels were measured in mixed cell populations, where the ratio of cell populations (tumor versus normal) may change. This is a common limitation for any biomarker for any type of tumor, unless, the test itself distinguishes between normal and tumor cells (morphology or immunofluorescence-based tests). Although, theoretically it is possible to first isolate the tumor cells using a cell-based technique and then performing biomarker assays such those described here, such tests will not be feasible given their technical complexity, the associated cost and the inherent variability of such an approach. Our study shows that normalization of biomarker levels to actin provides a reliable method to distinguish between control and BCa categories of individuals with relatively high sensitivity and specificity.
Taken together, this study showed that HYAL-1 and HAS1 expression are likely independent predictors of BCa metastasis (and possibly disease-specific survival) and combined HAS2-HYAL-1 mRNA expression in exfoliated urothelial cells is a biomarker for BCa detection and plausibly for monitoring recurrence. These findings need to be confirmed in an independent set of samples using the cut-points established in this study for each biomarker.
Acknowledgments
We gratefully acknowledge the statistical advice provided by Dr. Robert C. Duncan, Department of Epidemiology and Public Health, University of Miami - Miller School of Medicine and of Ms. Sue Walsh from JMP Technical Support, SAS-Institute. We thank Ms. Anaid Benitez and Ms. Estrelle Crespo for their help.
Grant support: 2R01CA072821–11 (VBL); R01 CA 123063-03 (VBL); Florida Department of Health-James and Esther King Biomedical Research Program (10KT-01), CURED Department of Urology (Kristell Acosta), The Woman’s Cancer Association of the University Of Miami (MSS). Mario Kramer was supported by the International Academy of Life Sciences, Biomedical Science Exchange Program fellowship
Abbreviations used
- BCa
bladder cancer
- HA
hyaluronic acid
- HAase
hyaluronidase
- HAS
Hyaluronic acid synthase
- HG
high grade
- LG
low grade
- NBL
normal bladder
- NBL-O
NBL tissue obtained from organ donors
- NBL-B
NBL tissue obtained from BCa patients at the time of cystectomy
- RR
risk ratio
Contributor Information
Mario W. Kramer, Email: mariokramer@gmail.com, Department of Urology, Hannover Medical School (MHH) and Department of Urology, University of Miami, Miller School of Medicine, P.O. Box 016960, Miami, Florida, 33101 Phone: 49-(0)173-4525565; Fax: 305-243-6893.
Diogo O. Escudero, Email: descudero@med.miami.edu, Department of Cell Biology and Anatomy, University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893.
Soum D. Lokeshwar, Email: soumlokeshwar@yahoo.com, Department of Urology and Sylvester Comprehensive Cancer Center University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893
Roozbeh Golshani, Email: rgolshani@illumina.com, Department of Cell Biology and Anatomy, University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893
Obi O. Ekwenna, Email: oekwenna@med.miami.edu, Department of Urology, University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893
Kristell Acosta, Email: kacosta@med.miami.edu, Department of Urology, University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893
Axel S. Mersegurger, Email: Merseburger.Axel@mh-hannover.de, Department of Urology, Hannover Medical School (MHH) Carl-Neuberg-Str. 1, 30625 Hannover, Germany; Phone: +49-511-532-2925
Mark Soloway, Email: msoloway@med.miami.edu, Department of Urology, University of Miami School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6596; Fax: (305) 243-6893
Vinata B. Lokeshwar, Email: vlokeshw@med.miami.edu, Departments of Urology and Cell Biology and Anatomy, University of Miami Miller School of Medicine, P.O. Box 016960, Miami, Florida. Phone: (305) 243-6321; Fax: (305) 243-6893
References
- 1.Habuchi T, Marberger M, Droller MJ, et al. Prognostic markers for bladder cancer: International Consensus Panel on bladder tumor markers. Urology. 2005;66:64–74. doi: 10.1016/j.urology.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 2.Heldin P, Karousou E, Bernert B, Porsch H, Nishitsuka K, Skandalis SS. Importance of hyaluronan-CD44 interactions in inflammation and tumorigenesis. Connect Tissue Res. 2008;49:215–8. doi: 10.1080/03008200802143323. [DOI] [PubMed] [Google Scholar]
- 3.Simpson MA, Lokeshwar VB. Hyaluronan and hyaluronidase in genitourinary tumors. Front Biosci. 2008;13:5664–80. doi: 10.2741/3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tammi RH, Kultti A, Kosma VM, Pirinen R, Auvinen P, Tammi MI. Hyaluronan in human tumors: pathobiological and prognostic messages from cell-associated and stromal hyaluronan. Semin Cancer Biol. 2008;18:288–95. doi: 10.1016/j.semcancer.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Bharadwaj AG, Rector K, Simpson MA. Inducible hyaluronan production reveals differential effects on prostate tumor cell growth and tumor angiogenesis. J Biol Chem. 2007;282:20561–72. doi: 10.1074/jbc.M702964200. [DOI] [PubMed] [Google Scholar]
- 6.Misra S, Toole BP, Ghatak S. Hyaluronan constitutively regulates activation of multiple receptor tyrosine kinases in epithelial and carcinoma cells. J Biol Chem. 2006;281:34936–41. doi: 10.1074/jbc.C600138200. [DOI] [PubMed] [Google Scholar]
- 7.Passerotti CC, Bonfim A, Martins JR, et al. Urinary hyaluronan as a marker for the presence of residual transitional cell carcinoma of the urinary bladder. Eur Urol. 2006;49:71–5. doi: 10.1016/j.eururo.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Aboughalia AH. Elevation of hyaluronidase-1 and soluble intercellular adhesion molecule-1 helps select bladder cancer patients at risk of invasion. Arch Med Res. 2006;37:109–16. doi: 10.1016/j.arcmed.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Hautmann S, Toma M, Lorenzo Gomez MF, et al. Immunocyt and the HA-HAase urine tests for the detection of bladder cancer: a side-by-side comparison. Eur Urol. 2004;46:466–71. doi: 10.1016/j.eururo.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder GL, Lorenzo-Gomez MF, Hautmann SH, et al. A side by side comparison of cytology and biomarkers for bladder cancer detection. J Urol. 2004;172:1123–6. doi: 10.1097/01.ju.0000134347.14643.ab. [DOI] [PubMed] [Google Scholar]
- 11.Lokeshwar VB, Obek C, Pham HT, et al. Urinary hyaluronic acid and hyaluronidase: markers for bladder cancer detection and evaluation of grade. J Urol. 2000;163:348–56. doi: 10.1016/s0022-5347(05)68050-0. [DOI] [PubMed] [Google Scholar]
- 12.Lokeshwar VB, Schroeder GL, Selzer MG, et al. Bladder tumor markers for monitoring recurrence and screening comparison of hyaluronic acid-hyaluronidase and BTA-Stat tests. Cancer. 2002;95:61–72. doi: 10.1002/cncr.10652. [DOI] [PubMed] [Google Scholar]
- 13.Itano N, Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–9. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- 14.Golshani R, Hautmann SH, Estrella V, Cohen BL, Kyle CC, Manoharan M, Jorda M, Soloway MS, Lokeshwar VB. HAS1 expression in bladder cancer and its relation to urinary HA test. Int J Cancer. 2007;120:1712–20. doi: 10.1002/ijc.22222. [DOI] [PubMed] [Google Scholar]
- 15.Golshani R, Lopez L, Estrella V, Kramer M, Iida N, Lokeshwar VB. Hyaluronic acid synthase-1 expression regulates bladder cancer growth, invasion, and angiogenesis through CD44. Cancer Res. 2008;68:483–91. doi: 10.1158/0008-5472.CAN-07-2140. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–67. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 17.Bharadwaj AG, Kovar JL, Loughman E, Elowsky C, Oakley GG, Simpson MA. Spontaneous metastasis of prostate cancer is promoted by excess hyaluronan synthesis and processing. Am J Pathol. 2009;174:1027–36. doi: 10.2353/ajpath.2009.080501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong QY, Liu J, Chen XY, Wang XW, Sun Y, Li H. Differential expression patterns of hyaluronan receptors CD44 and RHAMM in transitional cell carcinomas of urinary bladder. Oncol Rep. 2003;10:51–5. [PubMed] [Google Scholar]
- 19.Stavropoulos NE, Filliadis I, Ioachim E, et al. CD44 standard form expression as a predictor of progression in high risk superficial bladder tumors. Int Urol Nephrol. 2001;33:479–83. doi: 10.1023/a:1019589923706. [DOI] [PubMed] [Google Scholar]
- 20.Miyake H, Eto H, Arakawa S, Kamidono S, Hara I. Over expression of CD44V8-10 in urinary exfoliated cells as an independent prognostic predictor in patients with urothelial cancer. J Urol. 2002;167:1282–7. [PubMed] [Google Scholar]
- 21.Miyake H, Hara I, Arakawa S, Kamidono S. Utility of Competitive Reverse Transcription-Polymerase Chain Reaction Analysis of Specific CD44 Variant RNA for Detecting Upper Urinary Tract Transitional-Cell Carcinoma. Mol Urol. 1999;3:365–370. [PubMed] [Google Scholar]
- 22.Maxwell CA, McCarthy J, Turley E. Cell-surface and mitotic-spindle RHAMM: moonlighting or dual oncogenic functions? J Cell Sci. 2008;121:925–32. doi: 10.1242/jcs.022038. [DOI] [PubMed] [Google Scholar]
- 23.Lokeshwar VB, Cerwinka WH, Lokeshwar BL. HYAL-1 hyaluronidase: a molecular determinant of bladder tumor growth and invasion. Cancer Res. 2005;65:2243–50. doi: 10.1158/0008-5472.CAN-04-2805. [DOI] [PubMed] [Google Scholar]
- 24.Ekici S, Cerwinka WH, Duncan R, et al. Comparison of the prognostic potential of hyaluronic acid, hyaluronidase (HYAL-1), CD44v6 and microvessel density for prostate cancer. Int J Cancer. 2004;112:121–9. doi: 10.1002/ijc.20368. [DOI] [PubMed] [Google Scholar]
- 25.Gomez C, Gomez P, Knapp J, Jorda M, Soloway MS, Lokeshwar VB. Hyaluronic acid and HYAL-1 expression in prostate biopsy specimens: predictors of biochemical recurrence. J Urol. 2009;182:1350–1356. doi: 10.1016/j.juro.2009.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eissa S, Swellam M, Shehata H, El-Khouly IM, El-Zayat T, El-Ahmady O. Expression of HYAL1 and survivin RNA as diagnostic molecular markers for bladder cancer. J Urol. 2010;183:493–8. doi: 10.1016/j.juro.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Kramer MW, Golshani R, Merseburger AS, et al. HYAL-1 Hyaluronidase: A potential prognostic indicator for progression to muscle invasion and recurrence in bladder cancer. Eur Urol. 2009;57:86–93. doi: 10.1016/j.eururo.2009.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lokeshwar VB, Lopez LE, Munoz D, et al. Anti-tumor activity of hyaluronic acid synthesis inhibitor 4-methylumbelliferone in prostate cancer cells. Cancer Res. 2010;70:2613–23. doi: 10.1158/0008-5472.CAN-09-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, Hanna J, et al. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner RB, Hurst RE, Rao J, Hemstreet G. Instrumentation, accuracy and quality control in development of quantitative fluorescence image analysis. In: Hanausek M, Walaszek Z, editors. Tumor Marker Protocols. Vol. 14. Totowa, NJ: Humana Press; 1998. pp. 181–205. chapt. 11. [Google Scholar]
- 31.Eissa S, Swellam M, Shehata H, El-Khouly IM, El-Zayat T, El-Ahmady O. Expression of HYAL1 and survivin RNA as diagnostic molecular markers for bladder cancer. J Urol. 2010;183:493–498. doi: 10.1016/j.juro.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 32.Klatte T, Seligson DB, Rao JY, Yu H, de Martino M, Garraway I, Wong SG, Belldegrun AS, Pantuck AJ. Absent CD44v6 expression is an independent predictor of poor urothelial bladder cancer outcome. J Urol. 2010 Jun;183(6):2403–2408. doi: 10.1016/j.juro.2010.01.064. [DOI] [PubMed] [Google Scholar]
- 33.Gadalla HA, Kamel NA, Badary FA, Elanany FG. Expression of CD44 protein in bilharzial and non-bilharzial bladder cancers. BJU Int. 2004;93:151–15. doi: 10.1111/j.1464-410x.2004.04575.x. [DOI] [PubMed] [Google Scholar]
- 34.Muramaki M, Miyake H, Kamidono S, Hara I. Over expression of CD44V8-10 in human bladder cancer cells decreases their interaction with hyaluronic acid and potentiates their malignant progression. J Urol. 2004;171:426–430. doi: 10.1097/01.ju.0000093446.54115.b6. [DOI] [PubMed] [Google Scholar]
- 35.Eissa S, Zohny SF, Swellam M, Mahmoud MH, El-Zayat TM, Salem AM. Comparison of CD44 and cytokeratin 20 mRNA in voided urine samples as diagnostic tools for bladder cancer. Clin Biochem. 2008;41:1335–341. doi: 10.1016/j.clinbiochem.2008.08.085. [DOI] [PubMed] [Google Scholar]
- 36.Hemstreet GP. Genetic instability and tumor cell variation. In: Mackiewicz A, Sehgal PB, editors. Molecular aspects of cancer and its therapy. Basel; Boston, MA: Birkhauser Verlag; 1998. pp. 179–234. [Google Scholar]
- 37.Rao J, Seligson D, Hemstreet GP. Protein expression analysis using quantitative fluorescence image analysis on tissue microarray slides. Biotechniques. 2002;32:924. doi: 10.2144/02324pt04. [DOI] [PubMed] [Google Scholar]
- 38.Huang D, Casale GP, Tian J, Wehbi NK, Abrahams NA, Kaleem Z, et al. Quantitative fluorescence imaging analysis for cancer biomarker discovery: application to beta-catenin in archived prostate specimens. Cancer Epidemiol Biomarkers Prev. 2007;16:1371–1381. doi: 10.1158/1055-9965.EPI-06-0718. [DOI] [PubMed] [Google Scholar]
- 39.Huang D, Casale GP, Tian J, Lele SM, Pisarev VM, Simpson MA, et al. Udp-glucose dehydrogenase as a novel field-specific candidate biomarker of prostate cancer. Int J Cancer. 2010;126:315–327. doi: 10.1002/ijc.24820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spicer AP, Seldin MF, Olsen AS, Brown N, Wells DE, Doggett NA, et al. Chromosomal localization of the human and mouse hyaluronan synthase genes. Genomics. 1997;41:493–497. doi: 10.1006/geno.1997.4696. [DOI] [PubMed] [Google Scholar]