Abstract
Although cell fate specification is tightly controlled to yield highly reproducible results and avoid extreme variation, developmental programs often incorporate stochastic mechanisms to diversify cell types. Stochastic specification phenomena are observed in a wide range of species and an assorted set of developmental contexts. In bacteria, stochastic mechanisms are utilized to generate transient subpopulations capable of surviving adverse environmental conditions. In vertebrate, insect, and worm nervous systems, stochastic fate choices are used to increase the repertoire of sensory and motor neuron subtypes. Random fate choices are also integrated into developmental programs controlling organogenesis. Although stochastic decisions can be maintained to produce a mosaic of fates within a population of cells, they can also be compensated for or directed to yield robust and reproducible outcomes.
Keywords: bet-hedging, color vision, olfaction, spinal cord, lateral inhibition
INTRODUCTION
The development of an organism requires the integration of lineage and signaling cues to yield reproducible outcomes. Specific mechanisms provide robustness to cell fate decisions to generate evolutionarily favored body plans and thus prevent extreme variation. Although the majority of cell fate determination mechanisms induce reproducible outcomes, some cell fate decisions are made at random. Stochastic mechanisms of cell fate specification are observed in species ranging from bacteria to humans and are utilized to generate cell fate diversity. Understanding of the mechanisms controlling these phenomena in different species is at vastly different levels, ranging from the initial identification of molecular players to the deep characterization of the source and regulation of noise within a genetic circuit. The comparison of these seemingly disparate systems should allow for the generation of new, testable hypotheses and the identification of unifying features. The first goal of this review is to describe the current understanding of the mechanisms involved in stochastic fate choices using examples in bacteria, worms, flies, and vertebrates (Table 1).
Table 1.
Stochastic cell fate phenomena that yield random or robust final outcomes
| Organism | Phenomenon | Stochastic mechanism | Robustness mechanism |
Final outcome |
|---|---|---|---|---|
| Escherichia coli | Bacterial persistence | Unknown mechanism | NA | Stochastic subset of bacteria can tolerate harsh conditions |
| Bacillus subtilis | Competence to take up DNA | Noise in gene regulation | NA | Transient stochastic windows of competence |
| Primates | Color vision | 1. Unknown locus selection 2. LCR promoter selection |
NA | Stochastic expression of photopigments |
| Mus musculus | Olfaction | 1. Unknown locus selection 2. LCR promoter selection |
NA | Stochastic expression of olfactory receptors |
| Drosophila melanogaster | Color vision | Unknown mechanism regulating the spineless TF | NA | Stochastic expression of photopigments |
| Vertebrates | Motor neuron specification | Unknown regulation of Hox genes | Cell positioning | Reproducible motor pools in positioning and constituency |
| Caenorhabditis elegans | Uterus development | Lateral inhibition via Notch/LIN-12 | Cell positioning and division | Reproducible uterus structure |
| Drosophila melanogaster | R3/R4 photoreceptor specification | Lateral inhibition via Notch/LIN-12 | Frizzled activity gradient directs Notch activity | R3/R4 patterning across the retina is uniform |
| Drosophila melanogaster | R7 photoreceptor specification | Lateral inhibition via Notch/LIN-12 | Cff-inhibition of Notch activity in R1/R6 cells | One R7 is specified per ommatidium |
| Caenorhabditis elegans | AWC olfactory neuron lateral asymmetry | Lateral inhibition via differential signaling through gap junctions | NA | Stochastic expression of olfactory receptors |
NA, not applicable; LCR, locus control region; TF, transcription factor.
Stochastic mechanisms of specification result in diversified cell fates arranged randomly within a tissue or population of cells. In geno-typically identical bacteria, a percentage of the population is maintained in a transient, differential cell state to allow for survival in adverse environmental conditions. In vertebrate, fly, and worm nervous systems, sensory neuronal subtypes can be stochastically specified to increase the spectrum of responses to environmental stimuli. These phenomena have random final outcomes in that a transient subpopulation or stochastic pattern of cell fates is maintained.
In contrast, stochastic fate decisions can be intermediate steps in otherwise robust developmental programs. In other words, mechanisms acting after stochastic choices can compensate to yield reproducible outcomes. Furthermore, developmental programs can direct inherently stochastic specification mechanisms to ensure robustness. The second goal of this review is to outline how biological systems compensate for or direct stochastic choices to yield robust final outcomes.
We conclude this review with a general discussion of the preponderance of stochastic cell fate mechanisms in biological systems and how the study of variable mutant phenotypes may reveal new paradigms.
STOCHASTIC CHOICES LEADING TO RANDOM OUTCOMES
Bacteria Hedge Their Bets
Bacteria of identical genotype can exist in highly variable conditions. Biochemical pathways respond to changes in the environment to ensure survival (also called response switching). Nevertheless, molecular machinery within similar cells can only compensate for environmental variation to a limited degree. In severely harsh conditions (e.g., high antibiotic levels), bacteria would surely perish. However, bacterial populations are prepared for such potentially disastrous situations by bet-hedging strategies. To hedge a bet means to bet on several possible outcomes to minimize losses. Bacterial populations hedge their bets by maintaining a variety of cell fates within a genetically identical population, thus anticipating ever-changing environmental conditions. Typically, these diversified cell fates are transient. Therefore, the rate of entry into and exit from a given state must be carefully controlled to maintain an ideal percentage of the population in that particular state. The random entry into and exit from a state is also called stochastic switching (for a review, see Veening et al. 2008).
Thus, bacteria have two means of dealing with environmental changes: response switching and stochastic switching. Kussel & Leibler (2005) developed a mathematical model describing these two mechanisms and found that response switching would be favored in constantly changing environments whereas stochastic switching would be favored when changes are less frequent. Below, we discuss two examples in which bet-hedging strategies stochastically generate diversified subpopulations of bacteria.
Bet-hedging bacterial persisters
Bigger (1944a,b) found that penicillin kills staphylococci rather than simply being bacteriostatic. During his treatment experiments, he identified a tiny population of bacteria that survived and named them persisters. In the years since this discovery, the phenomenon of persistence has been observed in numerous bacterial species (for a review, see Gefen & Balaban 2009).
Persistence is essentially a bet-hedging strategy in which bacterial populations maintain small, stochastic subpopulations in either slow-growing or dormant states. Upon application of antibiotics, the normally growing cells die, but the persisters survive. Upon removal of the antibiotic, these persister cells start dividing again, which yields a population of normally growing cells and a small subpopulation of persister cells (Figure 1a). Hence, the generation of the minute population of persister cells is not due to genotypic differences but rather is a case of stochastic cell-state induction (for a review see Gefen & Balaban 2009).
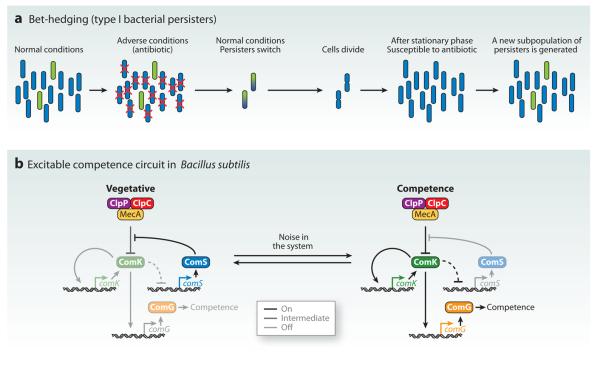
Figure 1.
Stochastic bet-hedging in bacteria generates population diversity. (a) Type I bacterial persisters. A small, stochastic subpopulation of dormant persisters survive adverse conditions. Upon return to normal growth conditions, these cells divide and reestablish the population. Finally, a new persister subpopulation is determined. (b) The DNA uptake competence cycle in Bacillus subtilis. Noise within the system drives the transition from the vegetative state to the competent state.
The phenomenon of bacterial persistence is challenging to study because the stochastic incidence of persister bacteria is extremely low [i.e., estimated at 1 in 106 in Escherichia coli as first measured by Bigger (1944a,b)]. The identification and characterization of E. coli mutants that have a higher rate of persistence have provided initial insights into this phenomenon. In particular, Balaban et al. (2004) conducted a thorough analysis of the cell growth dynamics of two high-persister mutants, hipA7 and hipQ, which allowed them to identify two types of persister cells.
hipA7 mutants were originally isolated in screens for increased persistence by Moyed & Bertrand (1983) and later cloned by Moyed & Broderick (1986). The hipA gene encodes a toxin and is part of a toxin-antitoxin (TA) module with its antitoxin hipB (Black et al. 1991). In TA modules in general, the antitoxin functions as a repressor of the operon and prevents expression of the toxin molecule in normal conditions. However, it is believed that under specific conditions, the antitoxin is rendered nonfunctional, which allows the toxin to affect the cell (for a review, see Gerdes et al. 2005).
The hipA7 mutants contain two point mutations in the hipA gene (Korch et al. 2003). Both mutations are required to induce hipA7 phenotypes, which suggests that hipA7 is a gain-of-function allele and that the HipA toxin functions to promote persistence. Growth arrest is also observed upon overexpression of wild-type hipA, further supporting its role in persistence induction (Correia et al. 2006, Falla & Chopra 1998, Korch & Hill 2006). However, deletion of the hipA gene does not affect persistence rate. This may be due to redundancy in other TA modules (see below). The molecular mechanism by which hipA regulates persistence is unknown (for a review, see Gefen & Balaban 2009).
Balaban and colleagues (2004) discovered that cultures of hipA7 mutants contain a population of persisters at stationary phase that is directly proportional to the overall number of stationary phase cells. When placed in fresh media, these persisters switch back to growing cells, which leads to repopulation. The persisters observed in hipA7 mutants are termed Type I persisters (Balaban et al. 2004).
When they enter persistence, Type I persisters arrest protein production. Gefen and colleagues (2008) found that protein synthesis resumes during a short window when dormant type I persister cells exit stationary phase following inoculation into fresh media. This new protein translation suggested that these bacteria may be susceptible to antibiotic treatment. Indeed, during this short window, the bacterial population is more vulnerable to antibiotics before it generates a new subpopulation of dormant persisters (Gefen et al. 2008; Figure 1a).
The hipQ mutants were identified in screens for persistence to quinolones (synthetic broad-spectrum antibiotics) and have not yet been molecularly characterized (Wolfson et al. 1990). Cultures of hipQ mutants contain a population of persisters whose mechanism of determination is independent of stationary phase. These “Type II persisters” are a slow-growing population in normal growth culture conditions. Whereas type I persisters cease growth during antibiotic treatment, Type II persisters maintain growth but at a much slower rate than nonpersisters. The slow growth rate is believed to be critical for Type II persister survival in the presence of antibiotics (Balaban et al. 2004).
When assessing wild-type E. coli, Balaban et al. (2004) found that the bacterial population consisted of both Type I and Type II persisters. Their mathematical modeling of stochastic switching into persistence agreed well with observations of these two types of persisters, but they could not rule out the possibility of additional persister types (Balaban et al. 2004).
Although these studies suggest at least two mechanisms of persister determination, little is known about the actual molecular mechanisms controlling this stochastic phenomenon. Isolation of large numbers of wild-type persister populations has only recently become possible. Assuming that all persister cells (dormant type I and slow-growing type II) have a low level of translation, Shah et al. (2006) sorted bacteria with a lower level of green fluorescent protein (GFP) expression from a population. They found an upregulation in the gene expression of TA modules (Shah et al. 2006), which suggests a role for these genes in persister determination (as also suggested by the role of hipA). However, the changes in TA module expression were uncovered from isolated persister cells and thus may represent an end result of persister specification rather than its inductive mechanism. Nevertheless, isolation of persister cells will enable the identification of key molecular players and eventually the elucidation of the mechanisms controlling persister formation.
Once the mechanisms that control persister formation are elucidated, the next step will be to study how these mechanisms function stochastically to maintain miniscule, random subpopulations of persisters. Studies of other bet-hedging phenomena in bacteria may provide some clues to the mechanisms controlling the stochastic generation of persister cells. In the next section, we discuss the genetic circuit controlling another example of bet-hedging in which uptake of foreign DNA in Bacillus subtilis allows a small population of cells to adapt to novel conditions. This circuit is so well understood that some groups have begun to study how and why noise in the system creates stochastic, transient cell states.
Bet-hedging competence in Bacillus subtilis
Diversification of bacterial populations is also achieved by maintaining a subpopulation that is competent for DNA uptake. This competence can allow for rapid adaptation to a changing environment but is also a potentially dangerous intake of foreign DNA. The mechanisms controlling natural competence for DNA up-take have been well studied, particularly in the gram-positive bacteria Bacillus subtilis and Streptococcus pneumoniae and the gram-negative bacteria Neisseria gonorrhoeae and Haemophilus influenzae (Dubnau 1999). Recently, new, highly quantitative methodologies have been applied to the study of competence in B. subtilis to derive fundamental principles about the nature of the excitable genetic circuit controlling this phenomenon.
Approximately 10% of the B. subtilis population is competent for DNA uptake at a given time (Nester & Stocker 1963). Competence is transient, as cells remain in this state for a limited period of time, and therefore different members of the population gain and lose this property. In this system, the comK gene appears to play a critical role, as its expression controls several downstream targets that induce competence (van Sinderen et al. 1995). In the majority vegetative state, comK is transcribed at low levels, but the ComK protein is rapidly degraded by the activity of the MecA/ClpP/ClpC complex of proteases (Turgay et al. 1997, 1998). The ComS peptide competes for the activity of the MecA/ClpP/ClpC complex, which allows some buildup of ComK levels (Solomon et al. 1995). When ComK surpasses a critical threshold, it starts a loop of autoregulation that induces its own expression to reach high levels (Hamoen et al. 1998, van Sinderen & Venema 1994). When this switch is thrown, high ComK levels activate downstream genes including comG to induce competence (Hahn et al. 1994, 1996). ComK is believed to indirectly negatively regulate ComS. During competence, the decreasing levels of ComS can no longer compete with ComK for MecA/ClpP/ClpC complex activity. Thus, over time, ComK levels are reduced by MecA/ClpP/ClpC-mediated degradation, and the cell transitions back from competence to the vegetative state (Suel et al. 2006; Figure 1b).
With such a well-understood genetic circuit, recent work has focused on the dynamic nature of the B. subtilis competence cycle. In other words, researchers have started to get at the nature of the stochasticity within the system and how it is controlled and utilized. First, Suel et al. (2006) developed a mathematical model suggesting the excitable nature of the competence circuit, that is, the circuit is susceptible to activation by noisy perturbations above a threshold. They also found that competence induction is stochastic and not based on the cell's history (i.e., the rate of competence is nearly the same in naive cells and cells that have reverted from competence). They went on to show an inverse relationship between ComK and ComS and that maintained expression of ComS can lock the system in the competent state (Suel et al. 2006).
These authors followed up this work by evaluating the dynamics of the system when promoter strengths are modified (Suel et al. 2007). They placed comK or comS under the control of an inducible promoter to precisely vary their basal expression rates. They found that increasing ComK levels could drive the system into an oscillatory state, as predicted mathematically. At extremely high levels of ComK, all cells enter competence. Although increasing levels of ComK have a marked effect on the probability of competence initiation, little effect on this parameter was observed when ComS levels were modified. Rather, increasing ComS increases the duration of competence. They modeled these effects and were able to identify a parameter set that allowed for both maintenance of excitability in high ComS conditions and independent tunability by ComS and ComK (Suel et al. 2007).
To test the role of noise in the system, Suel et al. (2007) generated cells that lacked septation (i.e., cells replicate DNA but do not divide). These cells share diffusible contents, which reduces noise within the system without affecting average concentration. As these cells undergo cell division without septation and elongate, a reduction in the initiation of competence is observed, suggesting that noise within the system is necessary for stochastic induction of competence (Suel et al. 2007). Maamar et al. (2007) corroborated the noisy nature of the competence cycle by observing individual mRNA molecules. They found a high variation in mRNA levels between the endogenous comK gene and comK reporter transgenes, suggesting that transcription of this gene is intrinsically noisy. Furthermore, they tested the role of noisy comK transcription by increasing the transcription of comK while reducing its translation, thus presumably decreasing overall variability. They found a reduced incidence of competence events, which supports a role for noisy gene regulation in this excitable system (Maamar et al. 2007).
Although noise has been implicated in the induction of competence, the source of this noise is still not clear. Both Suel et al. (2007) and Maamar et al. (2007) suggested that transcription of comK is critical because comK is a key parameter controlling induction and because comK transcription is inherently noisy. However, the experiments of both groups involve gross, fairly nonspecific manipulations of cells (i.e., inhibiting septation, reducing translation). It will be interesting to determine the relative importance of noisy regulation of each component of the circuit. The Suel group (Cagatay et al. 2009) has provided some insight into this question by showing that the architecture of the circuit is critical for noise regulation. When they artificially reversed the order of interactions in the ComK ⊣ ComS → ComK cycle (Figure 1b) using ComK → MecA ⊣ ComK, they reduced the variability in competence duration. However, this reorganized network also causes a reduction in overall competence duration and DNA uptake efficiency. The authors concluded that different circuit architectures may be selected for optimal fitness in specific (and possibly variable) environmental conditions (Cagatay et al. 2009). These types of approaches will allow for the deduction of the importance of circuit organization in regulating the noise that controls stochastic systems.
Cell Autonomous Mechanisms of Fate Specification in Multicellular Organisms
Bacteria utilize stochastic cell state induction to maintain subpopulations and hedge their bets against ever-changing environmental conditions. Such global variation would be detrimental to multicellular organisms that need to tightly control cell fate specification to avoid wild variation in body form. However, multicellular organisms do utilize stochastic mechanisms to generate cellular diversity in specific developmental programs. Below, we discuss how stochastic mechanisms are used to generate random patterns of sensory receptor selection in mammals and flies.
Locus control regions regulate stochastic expression of local sensory receptor genes
Mammalian systems utilize locus control regions (LCRs) to control multiple genes from one genomic locus. The β-globin gene locus is a well-studied case in which one LCR is required during development for the sequential transcriptional activation of the five types of globin genes within a cluster (for a review, see Levings & Bungert 2002). LCRs are also critical for the control of sensory receptor expression, and we discuss the cases of primate trichromacy (i.e., three-color vision) and mouse olfaction below.
Color vision photopigment selection in primates
The color vision system in primates is an interesting example of stochastic gene expression. In trichromatic primates, a red-, green-, or blue-detecting photopigment (also called L, M, or S) is selected at random for expression in a cone cell to the exclusion of the other two (Nathans et al. 1986a,b, 1989; Roorda & Williams 1999). The result is a mosaic of cell fates across the retina (Figure 2c). Trichromacy is observed in both New World (Central and South American origins) and Old World (African and Asian origins including humans) primates utilizing mechanisms of specification that have distinct similarities and differences. In both cases, there is a poorly understood, stochastic selection step between the blue opsin on one hand and the red or green opsins on the other hand. If the red/green opsin locus is chosen, different mechanisms are employed in New and Old World primates to control a second selection step between the red and green opsins (Jacobs & Nathans 2009).
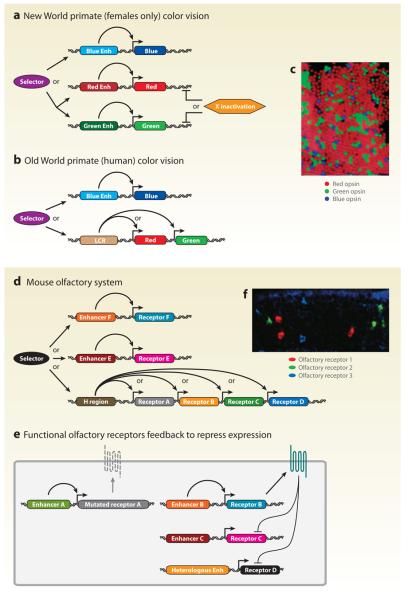
Figure 2.
The primate color vision and mouse olfactory systems utilize stochastic locus selection and locus control region (LCR)-mediated mechanisms. (a) Stochastic photopigment selection in New World primates requires locus selection and X inactivation. (b) Stochastic photopigment selection in Old World primates requires locus selection and LCR-mediated exclusive activation. (c) Stochastic expression of blue, red, and green photopigments in humans (adapted from Roorda & Williams 1999). (d) Stochastic olfactory receptor selection in mice requires locus selection and H region/LCR-mediated activation. (e) Functional olfactory receptors feed back to repress expression of other olfactory receptor gene alleles. (f) Stochastic expression of olfactory receptors in mice (courtesy of T. Ishii and P. Mombaerts, The Rockefeller University). Enh, enhancer.
In New World primates, the red and green opsin genes are actually two variant alleles of a single locus on the X chromosome. Thus, in females that are heterozygous for the red and green opsin genes, stochastic X chromosome inactivation leads to repression of one allele in each cell and allows for the activation of the other (Figure 2a). With such an X chromosome–based system, heterozygous females are trichromats whereas homozygous females and hemizygous males are dichromats (Hunt et al. 1998; Jacobs 1993; Jacobs et al. 1981, 1996; Kainz et al. 1998).
In contrast, Old World primates have one copy of each of the red and green opsin genes in close proximity on the X chromosome, an arrangement most likely established through an ancestral unequal crossing-over event that led to local gene duplication (Jacobs & Nathans 2009, Jacobs et al. 2007, Nathans 1999, Vollrath et al. 1988). In this case, if the red/green locus is chosen, the secondary decision between these opsins involves stochastic promoter selection. Both genes require a common upstream regulatory element for expression, an LCR located ~3.5 kb upstream of the red opsin locus. However, this LCR can activate only one of the two neighboring genes present in the cluster (Figure 2b). Distance from the LCR element, opsin-specific minimal promoter differences, and LCR copy number are critical in determining the final red-to-green ratio, which varies widely from individual to individual in human populations (Ibbotson et al. 1992; Smallwood et al. 2002; Wang et al. 1992, 1999).
In contrast to New World primate color vision, which allows for trichromacy in only a subpopulation of females, the regulatory system controlling Old World primate color vision enables trichromacy for all individuals. Both males and females carry both red and green alleles whose expression is chosen at random via an LCR-mediated mechanism. Coexpression of opsins from the two alleles in females is prevented via X inactivation. This elegant system has enabled better perception of red/green colors in the whole population.
Olfactory receptor selection in mice
The mouse olfactory system exhibits a similar case of stochastic receptor expression, although to a much higher level of complexity. Each olfactory sensory neuron (OSN) expresses one allele of a single olfactory receptor (OR) gene out of a set of approximately 1,300 OR genes, i.e., nearly 4% of the mouse repertoire of genes (Buck & Axel 1991, Chess et al. 1994, Godfrey et al. 2004, Zhang & Firestein 2002; Figure 2f). This selection appears to be random (though regionally biased; see Miyamichi et al. 2005, Ressler et al. 1993, Vassar et al. 1993) and cell autonomous. Initially, a recombination-based mechanism similar to those used to produce immunoglobulins was posited to account for the decision mechanism. Such a mechanism would lead to permanent genomic changes. Two groups ruled out such a mechanism by cloning mice from a single OSN and observing that the OR gene choice was still reset in the cloned mice (Eggan et al. 2004, Li et al. 2004).
Major insights into the mechanism of OR selection came from Serizawa and colleagues (2003). These authors identified a ~2 kb region called the H region for its high homology to humans. This H region is found upstream of a cassette of four OR genes and is required for their exclusive expression in transgenic reporters (Figure 2d). Similar to the LCR in primate trichromacy, modulating the distance from the H region of each gene changes its expression frequency (Serizawa et al. 2003).
More recently, Lomvardas et al. (2006) proposed that the H region controls not only the cis cassette of OR genes but also all OR genes in trans. This hypothesis was based on the colocalization of the H region with actively expressed OR genes and the biochemical isolation of the H region in proximity with trans OR genes (Lomvardas et al. 2006). However, this hypothesis was called into question when the Mombaerts and Sakano groups (Fuss et al. 2007, Nishizumi et al. 2007) deleted the H region and found that only cis OR gene expression was lost. Thus, the H region likely acts like an LCR to control selection of local OR genes. The mechanism for choosing an LCR/gene cluster or individual enhancer/gene (i.e., locus selection) remains unknown (Figure 2d).
To prevent OR coexpression, transcriptional activation of an OR gene must be coupled with repression of all other OR genes. Serizawa and colleagues (2003) showed that prevention of coexpression is mediated by the OR protein itself. OR transgenic reporters that include the receptor coding regions were never coexpressed with other OR genes. However, deletion of the coding region allowed for coexpression. Corroborating this idea, a frameshift mutation in an OR gene resulted in coexpression with other OR genes. This feedback role for a functional membrane receptor is further supported by the fact that OR pseudogenes are coexpressed with other ORs (Lewcock & Reed 2004, Serizawa et al. 2003, Shykind et al. 2004). Recently, Tian & Ma (2008) showed that OR signaling activity is required for these exclusionary mechanisms. Nguyen and coworkers (2007) showed that this negative feedback must act on DNA repressor elements that are located within the OR protein-coding region because OR transgenes that only contain the OR coding sequence under the control of a heterologous promoter can still be repressed (Figure 2e).
Similarities in sensory receptor selection
Selection of OR gene expression from a local cluster appears to have shared features with photopigment selection in Old World primates. First, both phenomena require mechanisms that select an enhancer/LCR from different chromosomes, e.g., blue versus red/green (Figure 2a,b,d). In both systems, this step is still poorly understood. If an LCR (or H-like region) is selected, a second, better-understood selection step occurs in which the promoter of a local receptor gene is chosen for activation to the exclusion of other local genes (Figure 2a,b,d). Such intra- and interchromosomal regulatory interactions have been observed in other biological phenomena including the controlled regulation of cytokine genes in the immune system (for example, Spilianakis et al. 2005). In the future, it will be exciting to see if the locus selection mechanisms and the LCR-mediated promoter selection machinery are similar in these systems.
Stochastic expression of a transcription factor controls the color vision mosaic in flies
Although they are morphologically divergent and evolutionarily independent, the human and Drosophila color vision systems share specific features including stochastic expression of genes encoding light-sensing opsin proteins. The fly eye is composed of approximately 800 ommatidia (i.e., unit eyes). Each ommatidium contains eight photoreceptors (PRs) called R1 through R8 (Wolff & Ready 1991). The R7 and R8 PRs are specialized for color vision. Specific rhodopsin (rh) expression in the R7 and R8 PRs defines two main subtypes of ommatidia. In the pale (p) subtype, Rh3 is expressed in R7 coupled with Rh5 expression in R8. In the yellow (y) subtype, Rh4 is expressed in R7 coupled with Rh6 expression in R8. The retina contains ~30% p ommatidia and 70% y ommatidia arranged at random (Figure 3a; Bell et al. 2007; Chou et al. 1996; Fortini & Rubin 1990; Franceschini et al. 1981; Montell et al. 1987; Papatsenko et al. 1997; Zuker et al. 1985, 1987).
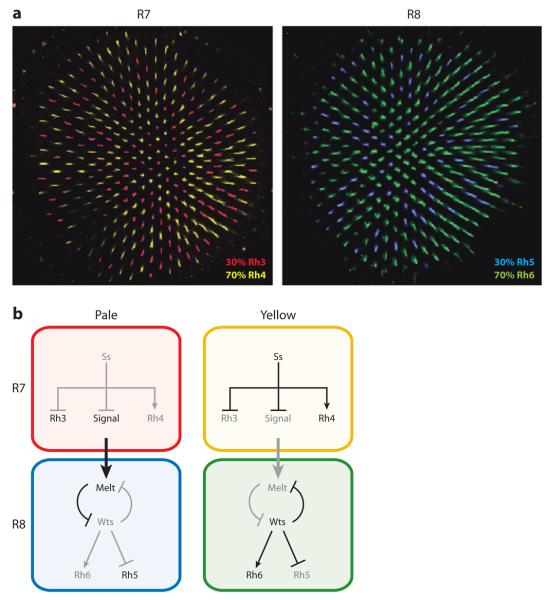
Figure 3.
The fly eye is a stochastic mosaic of two ommatidial subtypes. (a) Rhodopsin (Rh)3 and Rh4 are stochastically distributed in R7 neurons. Rh5 and Rh6 are stochastically and exclusively distributed in R8 neurons, and their expression is coupled to R7 subtype specification. (b) The yellow subtype is determined by the stochastic expression of the Spineless (Ss) transcription factor in R7s. Pale subtype fate is signaled from R7 to R8. Wts, Warts; Melt, Melted.
The stochastic ommatidial subtype decision is made in R7 and conferred upon R8. In sevenless mutants that lack R7 cells, nearly all R8s take on the y subtype fate, which suggests that pR7s must signal to R8s to induce pR8 fate (Chou et al. 1999). R8 subtype fate is controlled by a bistable feedback loop of two critical regulators, warts (wts)and melted (melt). In R8s that receive the p signal from R7, melt is upregulated, which represses wts and yields activation of rh5 and repression of rh6. In R8s that do not receive the R7 signal, melt remains repressed, allowing for wts expression and activation of rh6 and repression of rh5 (Figure 3b; Mikeladze-Dvali et al. 2005).
Because the stochastic ommatidial subtype decision is made in R7 and conferred upon R8, the underlying mechanism must not only control Rh expression but also signaling. Wernet and colleagues (2006) identified the PAS-bHLH transcription factor, Spineless (Ss), as a critical player regulating these features. ss is expressed in approximately 70% of R7s, corresponding to the ~70% of R7s that take on the yR7 subtype fate (Figure 3b). In ss mutants, all R7s take on the pR7 fate, displaying expression of Rh3 and complete loss of Rh4. Because R7 controls R8 subtype fate, nearly all R8 cells attain pR8 fate in ss mutants including expression of Rh5 and absence of Rh6. Ectopic expression of ss is sufficient to induce yR7 fate (i.e., expression of Rh4 and loss of Rh3) and indirectly to promote y fate in R8 (Wernet et al. 2006).
Although ss has been identified as an important player in the ommatidial subtype decision, little is known about how its stochastic expression is regulated or how it controls its downstream target gene network. Although this stochastic phenomenon is found in all analyzed higher dipteran lineages, mosquitoes (in a distantly related lower dipteran branch) display a regionalized, nonstochastic array of Rh expression (Hu et al. 2009). As our understanding of the mechanisms controlling stochastic ss expression/ommatidial specification increases, it will be interesting to test whether the key mechanistic difference with mosquitoes reflects a cis-regulatory difference in the ss gene locus, a change in the activity of an upstream transcription factor, or the invention of a novel mechanism unique to higher dipterans.
Stochastic opsin expression occurs in both flies and primates, but it is unclear if the mechanisms controlling these phenomena share similar features. Mechanisms for LCR promoter selection found in Old World monkeys do not appear to be utilized in flies, as evidenced by the use of binary fate switches controlled by regulatory factors and the great genomic distances between Rh genes. However, selection between the blue and red/green opsins may occur via a binary switch specification step similar to the one controlled by ss. As the mechanisms of stochastic opsin expression are elucidated, it will be interesting to see which features are shared between these systems.
STOCHASTIC CHOICES LEADING TO ROBUST OUTCOMES
The examples of stochastic fate specification discussed above lead to random outcomes as they produce diverse cell fates that are randomly distributed within a population of cells. However, other developmental programs compensate for, or direct, stochastic fate specification to yield reproducible and robust outcomes. We discuss examples of these types of biological phenomena below.
Motor Neuron Subtype Specification in the Vertebrate Spinal Cord
The vertebrate spinal cord is made up of pools of interneurons and motor neurons that establish specific axonal connections to other neurons or muscles. The motor neurons within the spinal cord are organized into discrete columns innervating different muscles. These columns are thought to be determined along the rostrocaudal axis by gradients of the signaling molecules retinoic acid, fibroblast growth factor (FGF), and Gdf11 (a TGFβ family member).
One specific motor column, the lateral motor column (LMC), is further subdivided into groups of motor neurons called motor pools. The 50 motor pools are marked by the expression of distinct transcription factor profiles. All neurons from a pool project to a single muscle target in the limb, and all receive monosynaptic input from proprioceptive sensory neurons from the same muscle target. Additionally, the neurons from a given pool are electrically coupled (for a review, see Dasen & Jessell 2009).
The diversification of neuronal subtype fate in motor pools is a complex process involving directed and stochastic processes. Dasen and colleagues (2003, 2005) have implicated the Hox transcription factors in the specification of a subset of motor pools. In this context, expression of the Hox transcription factors is generally controlled by selective, cross-repressive interactions that occur both rostrocaudally and within segments of the spinal cord. Within the brachial LMC, spatial information determines expression of the Hoxa5 and Hoxc5 proteins rostrally and Hoxc8 caudally. Loss of Hoxc8 function results in an expansion of Hox5 expression caudally, which suggests that Hoxc8 is required to determine caudal motor pool fate (Figure 4a). This directed specification step generates rostral Hox5+ motor pools and caudal Hoxc8+ motor pools (Figure 4b, purple and black, respectively).
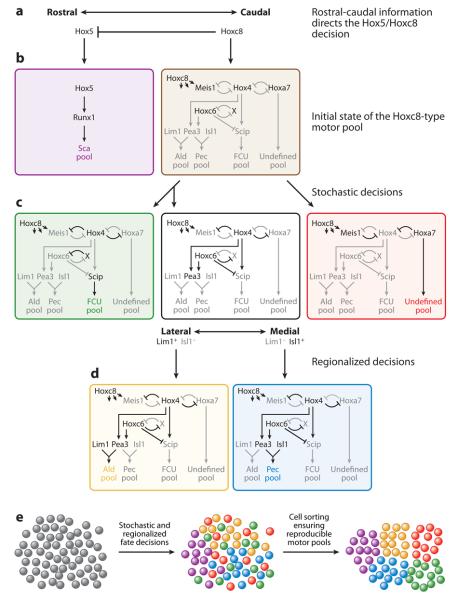
Figure 4.
Neuronal migration compensates for stochastic cell fate specification mechanisms to yield robust motor pools. (a) Rostral/caudal gradients of retinoic acid, fibroblast growth factor (FGF), and Gdf11 (a TGFβ family member) determine expression of Hox5 rostrally and Hoxc8 caudally. (b) Hox5 determines the scapulohumeralis posterior (Sca) muscle motor pool (purple). Hoxc8-expressing cells coexpress several Hox genes before undergoing additional stochastic and directional specification steps (brown). (c) The Hoxc8-expressing cells stochastically choose between expression of Hox4 or Meis1/Hoxa7. Hox4-expressing cells undergo a second stochastic decision to express Hoxc6 or not. Hox4+ Hoxc6− cells are specified as the flexor carpi ulnaris (FCU) pool (green). Hox4+ Hoxc6+ cells undergo an additional directed specification step (black). Meis1/Hoxa7-expressing cells determine an undefined motor pool (red). (d) Directed regionalization mechanisms determine Lim1 or Isl1 expression in Hox4+ Hoxc6+ cells. Lim1-expressing cells are specified as the anterior latissimus dorsi (Ald) motor pool (yellow). Isl1-expressing cells are specified as the pectoralis muscle (Pec) motor pool (blue). (e) Stochastic cell fate specification is compensated for via neuronal migration into coherent motor pools.
The Hox5 proteins specify approximately 25 of the 50 motor pools in the brachial LMC. However, little is known about the gene expression batteries or specification mechanisms that constitute these motor pools. The motor pool that projects to the scapulohumeralis posterior (Sca) muscle is specified by the combination of the Hox5 proteins and the Runx1 transcription factor (Runt family) (Theriault et al. 2004) (Figure 4b, purple).
Similarly, Hoxc8 controls specification of approximately 25 motor pools within the brachial LMC. A subset of the Hoxc8+ motor pools has been characterized to proceed through several additional specification events. Initially, the Hoxc8+ motor pool is characterized by the expression of a set of transcription factors including Meis1, Hox4, Hoxa7, and Hoxc6 (Figure 4b, black). As specification occurs, expression of these factors is refined, which leads to distinct expression profiles for each motor pool.
Several stochastic and directed cell fate decisions generate the diversity of Hoxc8+ motor pools. The first decision is made stochastically to maintain expression of Hox4 (Figure 4c, green and black) or Meis1/Hoxa7 (Figure 4c, red) coupled with repression of the alternate option. This decision involves two double negative feedback loops centered around Hox4. These bistable loops allow the cells to robustly choose one fate or the other in response to unknown variations in the expression of these genes.
If Meis1/Hoxa7 expression is maintained, a motor pool of unknown type is specified (Figure 4c, red). If Hox4 expression is maintained, additional specification steps ensue. A second stochastic decision is made within the Hox4+ pools regarding whether or not to maintain expression of Hoxc6. This choice appears to occur through yet another bistable double negative regulatory loop between Hoxc6 and an unknown factor X. This decision further subdivides the Hox4+ pool into Hoxc6+ (Figure 4c, black) and Hoxc6− pools (Figure 4c, green).
In the Hox4+/Hoxc6− pool (Figure 4c, green), Hox4 activates the Scip transcription factor that controls flexor carpi ulnaris (FCU) pool identity (Helmbacher et al. 2003). In the Hox4+/Hoxc6+ pool (Figure 4c, black), Hoxc6 activates expression of the ETS transcription factor Pea3 and represses Scip to prevent adoption of FCU (green) pool characteristics (Figure 4c,d). The Hox4+/Hoxc6+ pool undergoes a final regionally directed fate decision between expression of the Lim1 or Isl1 transcription factors (determined by medial or lateral position of the pool in the spinal cord). Lateral Lim1 expression drives anterior latissimus dorsi (Ald) pool fate (Figure 4d, yellow) whereas medial Isl1 expression induces pectoralis muscle (Pec) pool fate (Lin et al. 1998) (Figure 4d, blue).
The stochastic and spatial mechanisms controlling motor pool identity initially leave a mosaic of cell fates within the spinal cord. However, these neurons must be clustered to become electrically coupled, innervate their targets, and ensure generation of reproducible coherent motor pools (Figure 4e). Motor neuron pool sorting appears to involve cadherins. Price et al. (2002) showed that motor pools have unique combinatorial cadherin expression profiles. They demonstrated that altering expression of a specific cadherin gene (i.e., MN-cadherin) impaired pooling (Price et al. 2002). Furthermore, Livet and colleagues (2002) showed that Pea3 mutant mice displayed a loss of Cadherin-8 expression and a failure to properly cluster motor pools. Together, these data suggest that motor pool–specific cadherin activity is critical for pool sorting.
For the vertebrate spinal cord, repositioning neurons after stochastic cell fate specification yields robust motor pools. It is not yet clear how these stochastic choices are made, but the end result of these specification mechanisms is clear: distinct and exclusive cell fate. These fates are not only defined by discrete transcription factor constituency but also by unique expression patterns of specific cadherin genes. These cadherins putatively drive neuronal migration to produce the final robust outcome. Therefore, it appears that stochastic choices lead to the specification of different neuronal cell types distributed randomly within a field of cells. The repositioning of these cells compensates for random fate determination, which leads to the clustering of functionally similar but independently specified neurons that need to work together.
Lateral Inhibition Coordinates Stochastic Cell Fate Choices
The stochastic decisions described above appear to be cell autonomous. However, other aspects of stochastic choices are not cell autonomous and instead involve a turf war between two or more cells. In many of the best-studied cases, the Notch signaling pathway is used to achieve lateral inhibition, i.e., a competition for exclusive fates through cross-signaling between two or more cells. Schematically, activation of the Notch receptor induces its own upregulation and repression of its ligand Delta. This leads to increased sensitivity in one cell to the Delta signal from a neighboring cell. This neighboring cell receives less signal, leading to the downregulation of Notch expression and upregulation of Delta expression. In this way, a minimal initial difference in signaling between two cells can lead to dramatically different fates between these two cells, one being Notch+ and the other Notch−. This type of feedback loop forms a bistable system that ensures unambiguous stochastic decisions (for reviews, see Bray 2006, Greenwald 1998).
Notch signaling is used in numerous developmental programs. However, if the basis of Notch lateral inhibition is stochastic, how do biological systems compensate to generate robust outcomes? Below, we discuss the ways in which Notch signaling is directed or compensated for to yield reproducible outcomes. Furthermore, we discuss a special type of Notch-independent lateral inhibition that relies on random biasing of signaling through gap junctions to produce stochastic cell fate specification.
Coping with stochastic Notch signaling: the worm anchor cell/ventral uterine precursor decision
The worm Caenorhabditis elegans is well known for its mostly lineage-based mechanism of cell fate specification. The worm is composed of 959 somatic cells whose lineages have been precisely defined. Despite this predominantly determinate cell fate strategy, C. elegans development incorporates stochastic mechanisms to further diversify cellular subtypes.
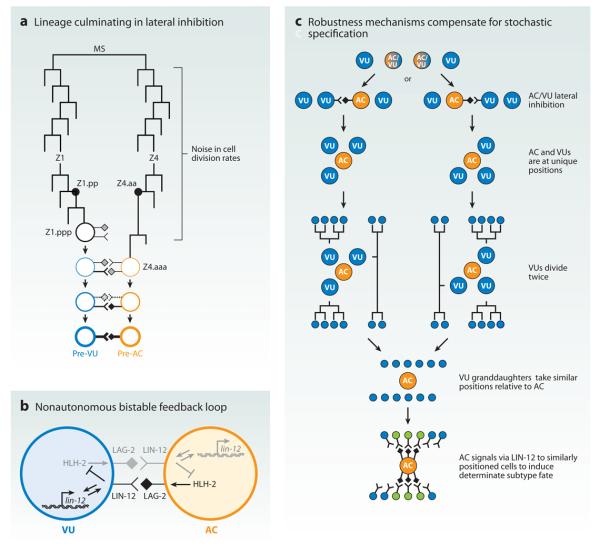
One of the best-described examples of nonautonomous stochastic cell fate specification occurs in gonadogenesis. Lateral inhibition mediated by Notch (LIN-12 in worms) signaling is critical to coordinate two distinct cell fates that are stochastically determined. The gonad precursor cells Z1.ppp and Z4.aaa are equipotent for the anchor cell (AC) and ventral uterine precursor (VU) fates. Both of these cells express the LIN-12/Notch receptor and the LAG-2/Delta ligand. A small difference in LIN-12 activity leads to upregulation of lin-12 and repression of lag-2 in the presumptive VU and the opposite effect in the presumptive AC (Figure 5b). In the absence of LIN-12 activity (as in lin-12 null mutants), both cells take on the AC fate, whereas high, ectopic LIN-12 activity in both cells is sufficient to induce VU fate (Greenwald 1985, 1987, 1998; Greenwald & Seydoux 1990; Greenwald et al. 1983, 1987; Kimble 1981; Kimble & Hirsh 1979; Seydoux & Greenwald 1989; Wilkinson et al. 1994; Yochem et al. 1988).
Figure 5.
Cell positioning and signaling compensate for stochastic lateral inhibition in the worm vulval developmental pathway. (a) The precursor cells of the anchor cell (AC, orange) and the ventral uterine precursor (VU, blue) have similar lineage origins. Variability (i.e., noise) in cell division rates results in stochastic precursor birth order. The first-born precursor cell has higher LIN-12/Notch activity that biases lateral inhibition. The cell with high LIN-12/Notch takes on the VU fate, whereas the cell with low LIN-12/Notch activity takes on the AC fate. The first division at the MS cell stage has been inverted for illustration purposes (adapted from Karp & Greenwald 2003). (b) LIN-12 lateral inhibition generates two exclusive fates via a nonautonomous bistable feedback loop (adapted from Karp & Greenwald 2003). The bHLH transcription factor, HLH-2, mediates this feedback loop. (c) Robustness mechanisms, including reproducible cell positioning and directional LIN-12 signaling, compensate for the stochastic AC/VU decision.
What is the source of the stochastic difference between the two cells? Karp & Greenwald (2003) demonstrated that the cell born first is more frequently destined to take on the VU fate (i.e., high LIN-12/Notch activity). They posited that the first-born cell either expresses more Notch receptor or is at a cell cycle stage that is more receptive when signaling occurs. Alternatively, it might be able to receive some signaling from other surrounding cells and therefore be biased to higher LIN-12 activity than the naive cell that is born later. It appears that the ultimate cause of this random decision is the temporal noise inherent in the cell divisions preceding the final division leading to Z1.ppp and Z4.aaa (Figure 5a; Karp & Greenwald 2003).
Interestingly, it ultimately does not matter which cell becomes the AC or VU. The three VU cells (one dependent on the AC/VU decision and two that are independently determined) divide twice and take up similar positions in relation to the AC irrespective of the stochastic AC/VU decision. The AC then uses LIN-12 signaling unidirectionally to induce Pi cell fate (a particular ventral uterine intermediate precursor fate) in a specifically positioned subset of six VU descendants, thus ensuring robustness and reproducibility of uterine formation (Figure 5c; Newman et al. 1995). Thus, one cannot distinguish the AC/VU cell fate decision in adults who have completed development. However, the important feature of the system is that it ensures the determination of one AC and one VU from these equipotent precursors because deviation leads to gross uterine and vulval abnormalities. In this way, the developmental program uses stochastic lateral inhibition to generate cell diversity (i.e., AC/VU cell fate specification), but then incorporates other mechanisms (i.e., cell positioning and signaling) to ensure robustness and reproducibility.
The robustness of the vulval/uterine developmental program has been rigorously examined in different species and genetic backgrounds and in response to environmental variation (Braendle & Felix 2008, Felix 2007, Kiontke et al. 2007, Milloz et al. 2008). In the future, it will be interesting to see whether there are subtle differences in the robustness of vulval/uterine development and overall fitness that are dependent on the stochastic AC/VU decision.
Directing Notch signaling in the fly eye
As illustrated for the AC/VU decision in worms, Notch signaling can result in stochastic cell fate specification. In that case, downstream cell positioning and signaling mechanisms ensure reproducible organogenesis. In this way, the system compensates for the randomness of Notch signaling after the stochastic cell fate specification event. However, other systems use distinct mechanisms to predetermine Notch signaling to yield robust outcomes. Below, we describe two such mechanisms incorporated in the fly eye.
Using a gradient to bias Notch signaling: R3/R4 specification
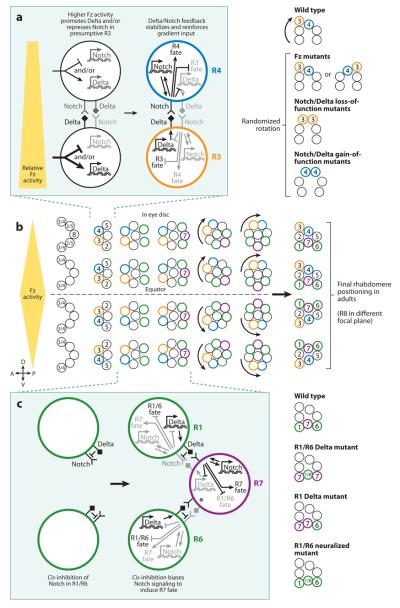
As mentioned above, the fly eye is made up of ~800 ommatidia composed of eight PRs. The six outer PRs (R1-R6) are arranged in a distinct trapezoidal shape around the inner PRs (R7 and R8) (Wolff & Ready 1991). The determination of these eight PRs incorporates distinct mechanisms involving signaling between specific cells. The R8 cell is the master regulator that recruits the other PRs through epidermal growth factor receptor (EGFR) signaling. R2/R5 are recruited first, followed by R3/R4, then R1/R6, and finally R7. Of the pairs of photoreceptor cells (i.e., R2/R5, R3/R4, and R1/R6), R3 and R4 appear to be the only ones that undergo further determinate specification (Figure 6b; Fanto et al. 1998).
Figure 6.
A gradient of Frizzled (Fz) activity and cis-inhibition directs stochastic Notch signaling in the fly eye. (a) A gradient of Fz activity biases Notch activity such that R3 has high Notch activity and R4 has low Notch activity. As in the AC/VU example, feedback loops reinforce the fate decisions. (b) Development of the fly retina. A 5-cell precluster is determined followed by specification of R3 or R4 fate via Fz-directed Notch signaling. R1 and R6 are recruited to the cluster and then signal via Notch to induce R7 fate (completing photoreceptor recruitment). The clusters of cells rotate to their final position. (c) Cis-inhibition of Notch activity by Delta in R1/R6 cells induces directional Notch signaling to specify R7 fate. As in the AC/VU example, feedback loops reinforce the fate decisions. For simplicity, * indicates the Notch-mediated inputs via signaling from R6.
Upon recruitment, the presumptive photoreceptors R3 and R4 in each ommatidium have the capacity to take on either cell fate. Lateral inhibition via Notch signaling ensures that only one cell becomes R3 whereas the other becomes R4. This distinction between R3 and R4 allows proper positioning of the six outer photoreceptor cells to form a chiral trapezoid that is necessary for the rotation of the photoreceptor cluster and the formation of the regular array of ommatidia. This mechanism works exactly as in the worm gonad, where the cell with high Notch activity upregulates Notch receptor transcription and downregulates Delta ligand expression and the cell with low Notch activity acts in the opposite way. Here, the cell with high Notch becomes the R4 cell whereas the cell with low Notch becomes the R3 cell, the same way that the cell with high LIN-12 activity becomes the VU whereas the cell with low LIN-12 becomes the AC (Figure 6a; Cooper & Bray 1999, Fanto & Mlodzik 1999, Tomlinson & Struhl 1999).
Excluding the R7/R8 subtype specification discussed above, the fly eye relies on highly robust cell fate specification mechanisms to generate its highly ordered crystalline structure. So, how is stochastic R3/R4 specification directed to yield reproducible outcomes? The fly eye uses a gradient system to bias the lateral inhibition system. There is a gradient of frizzled (fz) activity that is strongest at the dorsal/ventral equator of the eye and dissipates toward the polar regions (Boutros et al. 1998, Strutt et al. 1997, Tomlinson et al. 1997, Zheng et al. 1995). Fz is a canonical Wingless/Wnt receptor. However, in this context, Fz must be activated via a different mechanism because wingless/wnt mutants have no detectable defects. Rather, opposing expression gradients of the transmembrane/secreted protein Four-jointed (strongest equatorially/weakest distally) and the cadherin Dachsous (weakest equatorially/strongest distally) regulate the function of another cadherin, Fat, to control graded Fz activity (Yang et al. 2002).
Because, in each ommatidium, R3 is always positioned closer than R4 to the equator in the dorsal/ventral axis, the presumptive R3 is subject to a slightly higher level of Frizzled activity. Higher Frizzled activity in R3 leads to upregulation of Delta and/or downregulation of Notch that causes slightly lower Notch activity in R3 relative to R4. This bias is reinforced by the classical Notch feedback loops, which lead to low Notch activity and R3 fate in the cell positioned closer to the equator and high Notch activity and R4 fate in the distal cell (Figure 6a). Thus, in fz mutants, the bias is absent and the lateral inhibition determining R3 and R4 is now stochastic, which leads to randomized trapezoid polarity and random rotation of the cluster with disruption of the ommatidial array. In Notch mutants, both cells take on R3 fate, leading to a characteristic dual R3 symmetric cluster and randomized rotation. Notch gain of function leads to both cells assuming symmetric R4 fates with their distinct cell positions and randomized rotation (Figure 6a; Cooper & Bray 1999, Fanto & Mlodzik 1999, Tomlinson & Struhl 1999).
Cis-inhibition biases Notch signaling: R7 specification
The last step in photoreceptor specification is the recruitment and determination of the R7 photoreceptor. This process involves a combination of EGFR and Sevenless/RAS signaling from R8 and Notch signaling from R1/R6 to specify the presumptive R7 cell. Loss of any of these signaling events prevents proper R7 specification. Particularly, loss of Notch signaling causes the presumptive R7 to take on the R1/R6 fate. In contrast, ectopic Notch activation in R1/R6 induces R7 fate, which suggests that R1, R6, and R7 are all competent for the R1/R6 or R7 fate, but directing Notch signaling ensures that the cells take on the proper fates in a highly reproducible way (Cooper & Bray 2000, Tomlinson & Struhl 2001, Voas & Rebay 2004).
The directional nature of Notch signaling in R7 specification must involve biasing that would prevent Notch activity in R1/R6 while inducing high activity in R7. Miller and coworkers (2009) showed that cis-inhibition of Notch activity plays a critical role in this bias. Cis-inhibition occurs when the Delta ligand inhibits Notch activity when expressed in the same cell (Cordle et al. 2008, Jacobsen et al. 1998, Klein et al. 1997, Ladi et al. 2005, Micchelli et al. 1997, Sakamoto et al. 2002). Miller and coworkers (2009) found that, as expected, when R1 and R6 precursors are mutant for Delta, they both assume R7 fate whereas the R7 precursor takes on the R1/R6 fate. This result shows that the R1/R6 signaling interaction with R7 must be a case of directed lateral inhibition. They next tested cases in which a single R1 or R6 cell was mutant for Delta. In the classical feedback model of Notch-mediated lateral inhibition, the fates should be reversed because the bias of Notch signaling from the mutant R1 or R6 to R7 has been flipped. However, they found that when a single R1 or R6 precursor is mutant for the Delta ligand, that cell and the R7 precursor both take on R7 fate, whereas the wild-type R1/R6 precursor retains R1/R6 fate. This observation suggested that Delta might play a cell-autonomous role in R1/R6 to prevent high Notch activity and R7 fate. The simplest idea is that Delta acts via cis-inhibition in R1/R6 to repress Notch activity to control the bias in this directional lateral inhibition phenomenon (Figure 6c).
Miller and coworkers (2009) corroborated this cis-inhibition model by generating R1/R6 precursors mutant for the E3 ubiquitin ligase Neuralized. Neuralized is required in the Delta signaling cell to efficiently activate Notch in the neighboring cell (Lai et al. 2001, Pavlopoulos et al. 2001). In retinas in which both R1 and R6 are mutant for neuralized, Delta is present in these cells but is unable to induce high Notch activity in R7 (Li & Baker 2004). In the classical feedback model, this should result in a flip-ping of the lateral inhibition interaction such that the R1/R6 precursors take on R7 fate and the R7 precursor takes on R1/R6 fate. In this mutant condition, the R7 precursor does indeed assume the R1/R6 fate, but the R1/R6 precursors retain R1/R6 fate. The most parsimonious explanation is that although Delta in R1/R6 is not able to signal high Notch activity in R7 in neuralized mutants, it is able to suppress Notch activity autonomously via cis-inhibition. The end result is that all three cells take on the R1/R6 fate because R7 lacks Notch activation via Delta signaling and R1/R6 autonomously represses Notch activity (Figure 6c).
Cis-inhibition of Notch activity in R1/R6 is critical for directed lateral inhibition, but how is this directionality set up? It appears that the timing of Delta expression is important. Delta is expressed in R1/R6 hours before it is expressed in R7 (Parks et al. 1995). When Delta is ectopically expressed early in the R7 precursor, this cell takes on the R1/R6 fate, suggesting that the R7 precursor is susceptible to Notch cis-inhibition and that the timing of Delta expression is key to setting up the directionality. Presumably, the R1/R6 precursors remain susceptible to cis-inhibition even when Delta is ectopically expressed in R7, which completely prevents lateral inhibition and results in R1, R6, and R7 specifying R1/R6 fate. Thus, these data suggest that Delta is expressed early in R1/R6, which sets up cis-inhibition in these cells. This low Notch activity in R1/R6 biases the signaling interaction with R7 such that R7 now receives high Notch activation.
These observations raise the question of the general role of cis-inhibition in regulating Notch-mediated lateral inhibition. Because the study of the R7 versus R1/R6 decision is the first to directly implicate the role of cis-inhibition in directing lateral inhibition, the answer to this question is unclear. A thorough investigation of this phenomenon will elucidate whether cis-inhibition is a sparsely used adaptation of the Notch pathway or a common feature utilized in all cases of lateral inhibition.
Temporal input into Notch-mediated lateral inhibition
As in the AC/VU decision, timing is critical in determining the outcome of Notch-mediated lateral inhibition between R1/R6 and R7. However, it appears that the Notch activity outcome is opposite for these two cases. In the R1/R6 interaction with R7, low Notch activity occurs in the first-recruited cells (i.e., R1 and R6), whereas high Notch activity is activated in the later-recruited R7 cell (Figure 6c). For the AC/VU decision, the first-born cell has high Notch activity and becomes the VU cell, whereas the last-born cell has low activity and becomes the AC cell (Figure 5a). Cis-inhibition can explain the temporal ordering for the R1/R6 and R7 decision. For the AC/VU decision, the mechanism is less clear and perhaps more intriguing. How would the early cell attain higher Notch activity? It seems that cis-inhibition can likely be ruled out because the opposite phenotype would be predicted in such a case (i.e., the firstborn cell would be inhibited for Notch activity and thus take on the AC fate). It will be interesting to see how higher Notch activity is established in the firstborn cell, leading to the bias in signaling between the AC/VU precursors.
Lateral Inhibition through Gap Junctions Controls Stochastic Neuronal Asymmetry
The examples mentioned above illustrate how Notch-mediated lateral inhibition can be directed or compensated for to yield robust outcomes. In this section, we describe a similar phenomenon of lateral inhibition that utilizes signaling through gap junctions as a completely novel specification mechanism. As opposed to the other examples of lateral inhibition, this case generates a random outcome incorporating additional dedicated mechanisms to maintain the stable fate choice.
As described above, C. elegans development is characterized by an almost completely determinate cell lineage. Among its ~1,000 somatic cells, 302 are specified as neurons, 198 of which occur in bilateral pairs (Hobert et al. 2002, Wes & Bargmann 2001). Thus, the worm seems to have a limited repertoire of neuronal types and, hence, functions. The complement of neuronal functions is increased by diversification of gene expression and functions laterally, resulting in left/right asymmetry (Pierce-Shimomura et al. 2001, Wes & Bargmann 2001). One well-studied case involves the gustatory neurons, ASE left (ASEL) and ASE right (ASER). This example is stereotypical of much of worm development in that it is determinate and relies on cell fate decisions made at early stages (Poole & Hobert 2006). By an unknown mechanism, early inductive events are translated into a binary fate decision mediated in postmitotic neurons by a bistable feedback loop of microRNAs and transcription factors (Chang et al. 2003, 2004; Johnston & Hobert 2003, 2005; Johnston et al. 2005, 2006; Sarin et al. 2007).
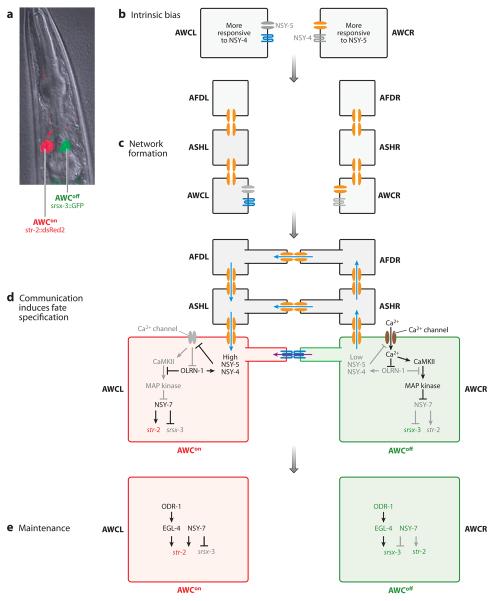
In contrast, the AWC pair of olfactory neurons uses stochastic cell fate specification to diversify gene expression and function. In wild-type animals, one AWC neuron takes on the AWCon fate whereas the other establishes the AWCoff fate. In 50% of animals, the AWC on the left side takes on AWCon fate with the AWC on the right side specified as the AWCoff fate. The other 50% establish the reverse combination of fate specification. AWCon fate is defined by the expression of str-2 and absence of srsx-3, whereas AWCoff fate is marked by the expression of srsx-3 and absence of str-2 (Figure 7a,d,e; Troemel et al. 1999).
Figure 7.
A gap junction–mediated mechanism of lateral inhibition controls stochastic lateral olfactory neuronal fate in worms. (a) The AWC on the left side (AWCL) and the AWC on the right side (AWCR) stochastically and exclusively choose the AWCon or AWCoff fate. AWCon is marked by expression of str-2::dsRed2 whereas AWCoff is marked by expression of srsx-3::green fluorescent protein (GFP). Image from Lesch et al. (2009). (b) AWCL is intrinsically biased to be more responsive to NSY-4 activity whereas AWCR is intrinsically biased to be more responsive to NSY-5 activity. Both cells pursue AWCon fate but via different mechanisms. (c) A gap junction network mediated by NSY-5 forms between ipsilateral AFD, ASH, and AWC neurons in embryonic stages. (d) The network is completed when bilateral neuronal pairs create gap junctions. The AWC pair generates gap junctions via the NSY-5 innexin protein. Signaling through gap junctions is stochastically directed to an AWC cell (e.g., AWCL). Directionality of signaling is reinforced by upregulation of NSY-4 and NSY-5, which represses Ca2+ influx and prevents activation of the MAP kinase (MAPK) cascade. NSY-7 is active in the absence of MAPK activity, which leads to AWCon fate (red). The cell with lower NSY-4 and NSY-5 activity allows Ca2+ influx. Ca2+ activates CAMKII and the MAPK cascade. The MAPK pathway represses NSY-7, which leads to AWCoff fate (green). (e)AWCon fate is maintained by NSY-7. ODR-1 and EGL-4 are generally required for maintenance of both AWCon and AWCoff fates.
The first indication that a lateral inhibition mechanism controls AWC asymmetry was the observation that laser ablation of an AWC precursor at the L1 larval stage results in the other cell taking the default AWCoff fate 100% of the time. Ablation after the L1/L2 transition did not affect fate specification, which suggests that the fates are already established and maintained at this point. Furthermore, mutations affecting axon guidance also cause both AWCs to take on the AWCoff state, which suggests that cell-cell contact is required for this mechanism (Troemel et al. 1999).
This case of lateral inhibition does not involve Notch signaling (Troemel et al. 1999). Rather, the Bargmann lab (Vanhoven et al. 2006, Chuang et al. 2007) has identified several components of a new type of lateral inhibition mechanism. The key molecular players appear to be the NSY-4 claudin-like transmembrane protein (Vanhoven et al. 2006) and the NSY-5 innexin (Chuang et al. 2007), which mediate gap junction formation. High activity of either NSY-4 or NSY-5 can drive AWCon fate. These proteins are expressed in both AWC neurons. However, each neuron appears to be more responsive to a particular regulator. The AWC on the left side is biased to high NSY-4 activity, whereas the AWC on the right side is more responsive to NSY-5 activity (Figure 7b). Thus, both AWC cells pursue AWCon fate, but in different ways.
The mechanism controlling the determination of AWC asymmetry is complex. First, an embryonic neural network of NSY-5-mediated gap functions is initiated (Figure 7c). Later, NSY-5 and NSY-4 participate in the completion of this neural network. Once this network is established, it is hypothesized that a difference in membrane potential between the left and right AWC cells is required for their specification as AWCon or AWCoff (Chuang et al. 2007) (Figure 7d).
Once this bias is established, a signaling cascade reinforces the cell fate decision. In the AWCon cell, high signaling through gap junctions is associated with active NSY-4 and NSY-5. High NSY-4 and NSY-5 activity prevents calcium influx, which leads to the repression of CaMKII and MAP kinase function. Repression of the CaMKII/MAPK pathway and activation of NSY-4 and NSY-5 is reinforced by the RAW-repeat protein OLRN-1. In the absence of MAP kinase activity, the transcription factor NSY-7 upregulates str-2 and represses srsx-3. In the presumptive AWCoff cell, low signaling through gap junctions induces the influx of calcium, which leads to the upregulation of CaMKII and MAP kinase activity. MAP kinase activity represses NSY-7 expression, which leads to the upregulation of srsx-3 and repression of str-2 (Figure 7d; Bauer Huang et al. 2007, Chuang & Bargmann 2005, Chuang et al. 2007, Lesch et al. 2009, Sagasti et al. 2001, Tanaka-Hino et al. 2002, Vanhoven et al. 2006).
Cellular communication between AWCL and AWCR is required during the embryonic and L1 larval stages. During the L1/L2 transition, the fates have been set, and disruption of this cell-cell interaction (or ablation of one of the cells) causes no effect (Troemel et al. 1999). Thus, AWC fate must be maintained independent of the initial signaling spec-ification events. The NSY-7 transcription factor is induced in response to the initiation of AWCon fate and persists to maintain expression of str-2 and repression of srsx-3. Additionally, the ODR-1 guanylate cyclase and EGL-4 cGMP-dependent kinase are required to maintain expression of str-2 in AWCon and srsx-3 in AWCoff. Because these genes are required for normal olfaction and olfactory plasticity in AWC, neuronal activity appears to be critical to maintain receptor expression in this system (Figure 7e; Lesch et al. 2009).
CONCLUSION: ARE STOCHASTIC CELL FATE SPECIFICATION MECHANISMS COMMON IN DEVELOPMENTAL PATHWAYS?
In this review, we discuss stochastic cell fate specification mechanisms that are either utilized to generate random outcomes or compensated for/directed to yield robust, reproducible outcomes. In both cases the function of stochastic cell fate specification is to diversify cell fates, whether it is to generate subpopulations of different bacterial states or to produce a larger variety of neuronal subtypes.
Are other stochastic mechanisms utilized in robust developmental phenomena? Although Notch-mediated lateral inhibition is used often throughout development, it is not clear whether other stochastic mechanisms are common. The answer may lie in the evaluation of genetic mutations. Although some mutations cause complete and reproducible phenotypes, others result in variable or stochastic outcomes. Examples of incomplete penetrance (the percentage of individuals that exhibit a mutant phenotype) or expressivity (variable phenotypes between individuals) are common to many mutant phenotypes. Of course, in many of these cases, a directed mechanism may simply stochastically surpass required molecular thresholds for function. However, in other cases, the variable outcome may be the result of an underlying stochastic mechanism that is only revealed when a robustness mechanism is ablated. An example of the identification of such a mechanism based on mutant analysis is the stochastic R3/R4 chirality observed in frizzled mutants (Figure 6c). As the mechanisms controlling robustness are elucidated, it will be interesting to see the commonality of underlying stochastic mechanisms.
SUMMARY POINTS.
Stochastic cell fate specification mechanisms are used to generate cellular diversity.
Stochastic fate determination can lead to random outcomes (as is often seen in the nervous system). However, it can be compensated for or directed by using robustness mechanisms.
Stochastic bet-hedging in bacteria diversifies a genetically clonal population, which allows some cells to survive adverse conditions.
Different stochastic fate specification processes may utilize similar mechanisms (e.g., primate color vision and mouse olfaction).
General strategies of stochastic mechanisms can be utilized by very different systems using quite different molecular players to generate cellular diversity (i.e., lateral inhibition in worm gonadal development and AWC lateral asymmetry).
FUTURE ISSUES.
For most of these examples of stochastic cell fate specification, the identification of new molecular players will be necessary before true insight into the mechanisms controlling these phenomena can be achieved. This is particularly true of the apparently cell-autonomous cases in vertebrates and flies. The difficulties in identifying critical factors controlling these mechanisms suggest that they may involve genes that are critical for basic functions of the cell/organism (e.g., factors that mediate intra- or interchromosomal interactions).
Once the molecular players have been identified, precise genetic and biochemical pathways must be established to elucidate the distinct cell fate specification mechanisms.
The phenomena that have well-characterized mechanisms (e.g., Notch-based lateral inhibition) can then be analyzed to identify the sources of noise and/or bias in the system (e.g., cell birth order). Of course, this may lead to reevaluation of the mechanism (e.g., how does cell birth order control lateral inhibition in the worm uterus?).
Careful characterization of each step in a given mechanism will reveal the importance of the logical organization of a gene network/cellular process (e.g., the order of gene interactions in the ComK negative feedback loop).
Development is often thought of as a directed process leading to a final robust outcome. However, many of the examples here illustrate that stochastic cell fate mechanisms can generate cell diversity, but these processes can be directed or compensated for by other means to ensure robust outcomes. The identification of new cases of stochastic cell fate specification may lead to the characterization of new stochastic fate determination and robustness mechanisms.
ACKNOWLEDGMENTS
We would like to thank Cori Bargmann, Jeremy Dasen, Ted Erclik, Xantha Karp, Sasha Levy, Dan Pollard, Jens Rister, Mark Siegal, Gurol Suel, and Daniel Vasiliauskas for helpful discussions and critical reading of the manuscript. C.D. is supported by NIH R01 EY013010. R.J.J. Jr. was supported by a Jane Coffin Childs Memorial Fund fellowship.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–25. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, et al. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural. Dev. 2007;2:24. doi: 10.1186/1749-8104-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ML, Earl JB, Britt SG. Two types of Drosophila R7 photoreceptor cells are arranged randomly: a model for stochastic cell-fate determination. J. Comp. Neurol. 2007;502:75–85. doi: 10.1002/cne.21298. [DOI] [PubMed] [Google Scholar]
- Bigger JW. The bactericidal action of penicillin on Staphylococcus pyogenes. Irish J. Med. Sci. 1944a;227:553–68. [Google Scholar]
- Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944b:497–500. [Google Scholar]
- Black DS, Kelly AJ, Mardis MJ, Moyed HS. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1991;173:5732–39. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–18. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- Braendle C, Felix MA. Plasticity and errors of a robust developmental system in different environments. Dev. Cell. 2008;15:714–24. doi: 10.1016/j.devcel.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signaling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–87. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Suel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–22. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–89. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–37. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–34. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, et al. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–15. doi: 10.1016/s0896-6273(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Chou WH, Huber A, Bentrop J, Schulz S, Schwab K, et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–16. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–81. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, Bargmann CI. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell. 2007;129:787–99. doi: 10.1016/j.cell.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. Frizzled regulation of Notch signaling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–30. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr. Biol. 2000;10:1507–10. doi: 10.1016/s0960-9822(00)00826-5. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 2008;15:849–57. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, et al. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 2006;188:8360–67. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–33. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–91. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53:217–44. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, et al. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Falla TJ, Chopra I. Joint tolerance to β-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 1998;42:3282–84. doi: 10.1128/aac.42.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M, Mayes CA, Mlodzik M. Linking cell-fate specification to planar polarity: determination of the R3/R4 photoreceptors is a prerequisite for the interpretation of the Frizzled mediated polarity signal. Mech. Dev. 1998;74:51–58. doi: 10.1016/s0925-4773(98)00063-x. [DOI] [PubMed] [Google Scholar]
- Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–26. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- Felix MA. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 2007;17:103–14. doi: 10.1016/j.cub.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Rubin GM. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 1990;4:444–63. doi: 10.1101/gad.4.3.444. [DOI] [PubMed] [Google Scholar]
- Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–67. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–84. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Gefen O, Balaban NQ. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 2009;33:704–17. doi: 10.1111/j.1574-6976.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:6145–49. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–82. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA. 2004;101:2156–61. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor. Cell. 1985;43:583–90. doi: 10.1016/0092-8674(85)90230-2. [DOI] [PubMed] [Google Scholar]
- Greenwald I. The lin-12 locus of Caenorhabditis elegans. Bioessays. 1987;6:70–73. doi: 10.1002/bies.950060207. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–62. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Greenwald I, Coulson A, Sulston J, Priess J. Correlation of the physical and genetic maps in the lin-12 region of Caenorhabditis elegans. Nucleic Acids Res. 1987;15:2295–307. doi: 10.1093/nar/15.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I, Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990;346:197–99. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–44. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hahn J, Kong L, Dubnau D. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J. Bacteriol. 1994;176:5753–61. doi: 10.1128/jb.176.18.5753-5761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Luttinger A, Dubnau D. Regulatory inputs for the synthesis of ComK, the competence transcription factor of Bacillus subtilis. Mol. Microbiol. 1996;21:763–75. doi: 10.1046/j.1365-2958.1996.371407.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Van Werkhoven AF, Bijlsma JJ, Dubnau D, Venema G. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 1998;12:1539–50. doi: 10.1101/gad.12.10.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmbacher F, Dessaud E, Arber S, deLapeyrière O, Henderson CE, et al. Met signaling is required for recruitment of motor neurons to PEA3-positive motor pools. Neuron. 2003;39:767–77. doi: 10.1016/s0896-6273(03)00493-8. [DOI] [PubMed] [Google Scholar]
- Hobert O, Johnston RJ, Jr, Chang S. Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat. Rev. Neurosci. 2002;3:629–40. doi: 10.1038/nrn897. [DOI] [PubMed] [Google Scholar]
- Hu X, England JH, Lani AC, Tung JJ, Ward NJ, et al. Patterned rhodopsin expression in R7 photoreceptors of mosquito retina: implications for species-specific behavior. J. Comp. Neurol. 2009;516:334–42. doi: 10.1002/cne.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DM, Dulai KS, Cowing JA, Julliot C, Mollon JD, et al. Molecular evolution of trichromacy in primates. Vision. Res. 1998;38:3299–306. doi: 10.1016/s0042-6989(97)00443-4. [DOI] [PubMed] [Google Scholar]
- Ibbotson RE, Hunt DM, Bowmaker JK, Mollon JD. Sequence divergence and copy number of the middle- and long-wave photopigment genes in Old World monkeys. Proc. Biol. Sci. 1992;247:145–54. doi: 10.1098/rspb.1992.0021. [DOI] [PubMed] [Google Scholar]
- Jacobs GH. The distribution and nature of color vision among the mammals. Biol. Rev. Camb. Philos. Soc. 1993;68:413–71. doi: 10.1111/j.1469-185x.1993.tb00738.x. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Bowmaker JK, Mollon JD. Behavioural and microspectro-photometric measurements of color vision in monkeys. Nature. 1981;292:541–43. doi: 10.1038/292541a0. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Nathans J. The evolution of primate color vision. Sci. Am. 2009;300:56–63. doi: 10.1038/scientificamerican0409-56. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Neitz M, Deegan JF, Neitz J. Trichromatic color vision in New World monkeys. Nature. 1996;382:156–58. doi: 10.1038/382156a0. [DOI] [PubMed] [Google Scholar]
- Jacobs GH, Williams GA, Cahill H, Nathans J. Emergence of novel color vision in mice engineered to express a human cone photopigment. Science. 2007;315:1723–25. doi: 10.1126/science.1138838. [DOI] [PubMed] [Google Scholar]
- Jacobsen TL, Brennan K, Arias AM, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signaling in Drosophila. Development. 1998;125:4531–40. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–49. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA. 2005;102:12449–54. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Copeland JW, Fasnacht M, Etchberger JF, Liu J, et al. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development. 2006;133:3317–28. doi: 10.1242/dev.02494. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Hobert O. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development. 2005;132:5451–60. doi: 10.1242/dev.02163. [DOI] [PubMed] [Google Scholar]
- Kainz PM, Neitz J, Neitz M. Recent evolution of uniform trichromacy in a New World monkey. Vision Res. 1998;38:3315–20. doi: 10.1016/s0042-6989(98)00078-9. [DOI] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 2003;17:3100–11. doi: 10.1101/gad.1160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kiontke K, Barriere A, Kolotuev I, Podbilewicz B, Sommer R, et al. Trends, stasis, and drift in the evolution of nematode vulva development. Curr. Biol. 2007;17:1925–37. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Klein T, Brennan K, Arias AM. An intrinsic dominant negative activity of Serrate that is modulated during wing development in Drosophila. Dev. Biol. 1997;189:123–34. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 2003;50:1199–213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Korch SB, Hill TM. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 2006;188:3826–36. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–78. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, et al. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 2005;170:983–92. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell. 2001;1:783–94. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Gehrke AR, Bulyk ML, Bargmann CI. Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev. 2009;23:345–58. doi: 10.1101/gad.1763509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings PP, Bungert J. The human β-globin locus control region. Eur. J. Biochem. 2002;269:1589–99. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl. Acad. Sci. USA. 2004;101:1069–74. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428:393–99. doi: 10.1038/nature02433. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev. Biol. 2004;4:5. doi: 10.1186/1471-213X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–92. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–29. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–95. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, et al. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–87. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr. Biol. 2009;19:1378–83. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milloz J, Duveau F, Nuez I, Felix MA. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008;22:3064–75. doi: 10.1101/gad.495308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J. Neurosci. 2005;25:3586–92. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Jones K, Zuker C, Rubin G. A second opsin gene expressed in the UV-sensitive R7 photoreceptor cells of Drosophila melanogaster. J. Neurosci. 1987;7:1558–66. doi: 10.1523/JNEUROSCI.07-05-01558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983;155:768–75. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed HS, Broderick SH. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J. The evolution and physiology of human color vision: insights from molecular genetic studies of visual pigments. Neuron. 1999;24:299–312. doi: 10.1016/s0896-6273(00)80845-4. [DOI] [PubMed] [Google Scholar]
- Nathans J, Davenport CM, Maumenee IH, Lewis RA, Hejtmancik JF, et al. Molecular genetics of human blue cone monochromacy. Science. 1989;245:831–38. doi: 10.1126/science.2788922. [DOI] [PubMed] [Google Scholar]
- Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986a;232:203–10. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986b;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- Nester EW, Stocker BA. Biosynthetic latency in early stages of deoxyribonucleic acid transformation in Bacillus subtilis. J. Bacteriol. 1963;86:785–96. doi: 10.1128/jb.86.4.785-796.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, White JG, Sternberg PW. The Caenorhabditis elegans lin-12 gene mediates induction of ventral uterine specialization by the anchor cell. Development. 1995;121:263–71. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–17. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizumi H, Kumasaka K, Inoue N, Nakashima A, Sakano H. Deletion of the core-H region in mice abolishes the expression of three proximal odorant receptor genes in cis. Proc. Natl. Acad. Sci. USA. 2007;104:20067–72. doi: 10.1073/pnas.0706544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–73. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech. Dev. 1995;50:201–16. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell. 2001;1:807–16. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, Lockery SR. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature. 2001;410:694–98. doi: 10.1038/35070575. [DOI] [PubMed] [Google Scholar]
- Poole RJ, Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr. Biol. 2006;16:2279–92. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell. 2002;109:205–16. doi: 10.1016/s0092-8674(02)00695-5. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Roorda A, Williams DR. The arrangement of the three cone classes in the living human eye. Nature. 1999;397:520–22. doi: 10.1038/17383. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–32. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Ohara O, Takagi M, Takeda S, Katsube K. Intracellular cell-autonomous association of Notch and its ligands: a novel mechanism of Notch signal modification. Dev. Biol. 2002;241:313–26. doi: 10.1006/dbio.2001.0517. [DOI] [PubMed] [Google Scholar]
- Sarin S, O'Meara MM, Flowers EB, Antonio C, Poole RJ, et al. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics. 2007;176:2109–30. doi: 10.1534/genetics.107.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–94. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57:1237–45. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O'Donnell S, Nemes A, Mendelsohn M, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–15. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Smallwood PM, Wang Y, Nathans J. Role of a locus control region in the mutually exclusive expression of human red and green cone pigment genes. Proc. Natl. Acad. Sci. USA. 2002;99:1008–11. doi: 10.1073/pnas.022629799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Magnuson R, Srivastava A, Grossman AD. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995;9:547–58. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signaling. Nature. 1997;387:292–95. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–50. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–19. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, et al. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault FM, Roy P, Stifani S. AML1/Runx1 is important for the development of hindbrain cholinergic branchiovisceral motor neurons and selected cranial sensory neurons. Proc. Natl. Acad. Sci. USA. 2004;101:10343–48. doi: 10.1073/pnas.0400768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol. Cell Neurosci. 2008;38:484–88. doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Strapps WR, Heemskerk J. Linking Frizzled and Wnt signaling in Drosophila development. Development. 1997;124:4515–21. doi: 10.1242/dev.124.22.4515. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development. 1999;126:5725–38. doi: 10.1242/dev.126.24.5725. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol. Cell. 2001;7:487–95. doi: 10.1016/s1097-2765(01)00196-4. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–98. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–38. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K, Hamoen LW, Venema G, Dubnau D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 1997;11:119–28. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 1995;15:455–62. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- van Sinderen D, Venema G. comK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J. Bacteriol. 1994;176:5762–70. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoven MK, Bauer Huang SL, Albin SD, Bargmann CI. The claudin superfamily protein nsy-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron. 2006;51:291–302. doi: 10.1016/j.neuron.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–18. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 2004;229:162–75. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Vollrath D, Nathans J, Davis RW. Tandem array of human visual pigment genes at Xq28. Science. 1988;240:1669–72. doi: 10.1126/science.2837827. [DOI] [PubMed] [Google Scholar]
- Wang Y, Macke JP, Merbs SL, Zack DJ, Klaunberg B, et al. A locus control region adjacent to the human red and green visual pigment genes. Neuron. 1992;9:429–40. doi: 10.1016/0896-6273(92)90181-c. [DOI] [PubMed] [Google Scholar]
- Wang Y, Smallwood PM, Cowan M, Blesh D, Lawler A, Nathans J. Mutually exclusive expression of human red and green visual pigment-reporter transgenes occurs at high frequency in murine cone photoreceptors. Proc. Natl. Acad. Sci. USA. 1999;96:5251–56. doi: 10.1073/pnas.96.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for color vision. Nature. 2006;440:174–80. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- Wilkinson HA, Fitzgerald K, Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994;79:1187–98. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morpho-genetic furrow and the second mitotic wave. Development. 1991;113:841–50. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and β-lactam antimicrobial agents. Antimicrob. Agents Chemother. 1990;34:1938–43. doi: 10.1128/aac.34.10.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–88. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Yochem J, Weston K, Greenwald I. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. 1988;335:547–50. doi: 10.1038/335547a0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5:124–33. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhang J, Carthew RW. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–55. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]
- Zuker CS, Cowman AF, Rubin GM. Isolation and structure of a rhodopsin gene from D. melanogaster. Cell. 1985;40:851–58. doi: 10.1016/0092-8674(85)90344-7. [DOI] [PubMed] [Google Scholar]
- Zuker CS, Montell C, Jones K, Laverty T, Rubin GM. A rhodopsin gene expressed in photoreceptor cell R7 of the Drosophila eye: homologies with other signal-transducing molecules. J. Neurosci. 1987;7:1550–57. doi: 10.1523/JNEUROSCI.07-05-01550.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]