Abstract
Recognition of genomic binding sites by transcription factors can occur through base-specific recognition, or by recognition of variations within the structure of the DNA macromolecule. In this article, we investigate what information can be retrieved from local DNA structural properties that is relevant to transcription factor binding and that cannot be captured by the nucleotide sequence alone. More specifically, we explore the benefit of employing the structural characteristics of DNA to create binding-site models that encompass indirect recognition for the Escherichia coli model organism. We developed a novel methodology [Conditional Random fields of Smoothed Structural Data (CRoSSeD)], based on structural scales and conditional random fields to model and predict regulator binding sites. The value of relying on local structural-DNA properties is demonstrated by improved classifier performance on a large number of biological datasets, and by the detection of novel binding sites which could be validated by independent data sources, and which could not be identified using sequence data alone. We further show that the CRoSSeD-binding-site models can be related to the actual molecular mechanisms of the transcription factor DNA binding, and thus cannot only be used for prediction of novel sites, but might also give valuable insights into unknown binding mechanisms of transcription factors.
INTRODUCTION
Transcriptional regulation allows cells to adapt their gene expression in response to changing conditions. Essential in the process of transcriptional regulation is the interaction between the transcription factor (TF) and its associated binding site or motif, upon which the TF will exert its inhibitory or activating effect. Considerable effort has been done to model these DNA motifs based on known binding sites in order to predict novel functional sites.
Position-specific weight (PWM) matrices or consensus representations are the most frequently used motif models: they describe the nucleotides which are shown to be common over a significant fraction of known binding sites (1). As PWMs or consensus models only describe the nucleotide sequence, they do not exploit the information contained within the DNA structure. It has indeed been shown that TF's can also recognize binding sites by their local DNA structure, a type of recognition that is less dependent on sequence conservation and that is commonly referred to as indirect binding or intramolecular read-out (2). Using the information contained within the DNA structure could therefore result in better classifiers for regulatory binding sites. For example, several prior studies have successfully used molecular modeling to predict target sequences for regulatory proteins, however these methods are restricted in their use as they require the native structure of the involved protein–DNA complexes to have been characterized (3–5). Other approaches circumvent the use of entire structures of the DNA–protein complex by focusing their model on the DNA site of the interaction. More specifically, they use specific structural properties of the DNA that are known to vary between different DNA regions within the genome and that might play a role in the protein–DNA recognition, e.g. the directional bendability of the DNA molecule (6). These methods usually rely on different structural profiles, where each profile represents per position in the genome the values of a specific DNA structural property. Based on the structural profiles characteristics of known binding sites, a classifier can distinguish true from false positive binding sites, as was first demonstrated by Karas et al. (7). Furthermore, the combination of these structural profiles with a higher order machine-learning classifier has demonstrated improved classification performance of true and false positive binding sites (8). Although structure-based methods have been shown to be successful in a cross validation setting on known binding sites (9–12), their ability to also improve upon the prediction of novel binding sites in a genome-wide setting has been largely understudied. In addition because most previous studies were very limited in scope (e.g. focusing on few TF’s only) it is still unknown to what extent local DNA-structural properties provide information to predict novel binding sites that cannot be captured by the nucleotide sequence alone i.e. to what extent a screening method that exploits local structural properties is redundant or complementary to a standard screening method based on PWM.
To study this, we developed a generic framework for screening any sequence for novel regulatory binding sites by using the local structural DNA properties of known binding sites, which we call Conditional Random fields of Smoothed Structural Data (CRoSSeD). This method shows an improved overall, performance on a synthetic dataset and on a large set of Escherichia coli regulons when compared to previous structure- and sequence-based methods. We further show that a set of novel predictions can be made using the proposed method that could be validated by using independent data sources and that could not be made using a traditional sequence-based model.
MATERIALS AND METHODS
Structural properties
The structural profiles can be obtained from the DNA sequence by using di- or trinucleotide structural scales (13). These scales rely on the principle that the structure of a DNA molecule depends largely on its sequence of nucleotides and that the overall structural properties, such as flexibility or stability of the helix (14,15), are caused by the interactions between neighboring base pairs (13,16). The origin of the values contained within the scale are either derived from experimental data, such as X-ray crystallography, or from molecular modeling of a DNA helix or a DNA–protein complex. Thus each scale contains complementary information and provides a unique insight into the structure of the DNA molecule (13,17,18).
For this implementation, we selected a number of scales which have been frequently used (14,17,19) and which capture structural properties that might be of importance for binding site recognition in prokaryotes (listed in Table 1). A single scale contains the values for a single structural property for each possible di- or trinucleotide. For more details about each of these structural scales, we refer to their respective references (6,20–29).
Table 1.
Overview of the utilized structural scales
| Scale name | Structural property | Order |
|---|---|---|
| A-DNA philicity24 | A-DNA conformation | Dinucleotide |
| DNase-I cutting frequency25 | Flexibility | Trinucleotide |
| B-DNA twist26 | Flexibility | Dinucleotide |
| Protein-induced deformability6 | Flexibility | Dinucleotide |
| Denaturation temperature23 | Stability | Dinucleotide |
| Disruption energy21 | Stability | Dinucleotide |
| Propeller twist28 | Flexibility | Dinucleotide |
| Protein-induced B-DNA twist6 | Flexibility | Dinucleotide |
| Stabilization energy29 | Stability | Dinucleotide |
| Base stacking energy20 | Stability | Dinucleotide |
| Persistence length27 | Flexibility | Dinucleotide |
| Z-DNA free energy22 | Z-DNA conformation | Dinucleotide |
Conditional random fields
For the purposes of this article, the CRF model is trained using the open-source software tool CRF++ (version 0.51) which was designed to label sequential data (available at http://crfpp.sourceforge.net/). Among other options, it allows defining which features the model should use. The features used in our binding-site model correspond to the value at each position of the profile, complemented with higher order features which link each position in the profile with its neighbors and the position mirrored on the opposite side of the motif for palindrome modeling. For more details, see Supplementary Data 1. The input of the training algorithm consists of the 12 structural profiles described above expanded with two additional vectors, one representing the GC content (a dinucleotide scale assigning a value identical to the number of guanines or cytosines the dinucleotide contains) and the other containing the plain nucleotide sequence information. Two extensions (the scale optimization extension and correction extension) were designed in order to allow the model training algorithm to deal with some of the specific aspects of the sample data and the structural property modeling problem (Supplementary Data 1). Both extensions are applied to the training data before the training of the final CRoSSeD model. These extensions were evaluated on the presented datasets and consistently improved performance of the model (data not shown).
Position-weight matrix
As a reference, the performance of the trained CRoSSeD models was compared to a PWM model with the same training set. Each entry in a PWM matrix represents the frequency of a certain nucleotide at a certain position over all known binding sites. A pseudo-count of 0.01 was added to any nucleotides with no instances in the entire training set at any of the positions. Test sequences were scored with the sum of the logarithms of the frequencies in the PWM. The corresponding motif logos for the PWMs of the 27 studied TF s can be found in Supplementary Data 2.
BioBayesNet
BioBayesNet (30) is a web application based on Bayesian Networks that allows inclusion of structural profiles. This methodology demonstrated a higher specificity than a simple PWM in predicting binding sites of four modeled motifs in a cross validation setting (11). We used this application for comparison as it is one of the most recent and the only publicly available structure-based methodology. The model was trained using default settings: all structural properties were used and no prior information on significant regions or known motifs were included.
CRFseq
To estimate the increased classification performance of the CRoSSeD method due contribution of the structural properties and not due to the implicit higher order nucleotide relationships, a comparison is made with a ‘CRFseq’ method during the cross-validation analyses. The CRFseq method uses the same training and testing algorithm as CRoSSeD, except that only sequence data is provided as input data and, as a consequence, neither of the structure-specific extensions could be used. Higher order relationships are also included between adjacent nucleotide positions up until trinucleotides (the highest order of the structural scales used for CRoSSeD) as these relationships are implicitly present in a structure-based model and inclusion of this type of positional interdependency has been shown to improve TF binding site classification (31,32). Higher order dinucleotide relationships that are explicitly defined in CRoSSeD, i.e. between positions symmetrically around the TF-binding site center, have also been included in CRFseq. All in all, CRFseq employs the exact same higher order nucleotide relations as those implicitly or explicitly present in CRoSSeD, but incorporates none of the actual values describing the structural DNA properties.
Datasets
We used both synthetic and biological datasets to evaluate our model. Each dataset consists of a positive and a negative set. Each sample in the dataset is a sequence of 41 nt with the center of the binding site corresponding to the 21st position, which is sufficient to encase the span of most currently known binding sites (9). From these nucleotide sequences fourteen different input vectors are generated, which include the 12 structural profiles, the GC-content vector and the nucleotide vector.
The synthetic positive dataset samples were created so that they resemble fit a flexibility profile with one region of high flexibility and one region of high rigidity. This was achieved by giving the positions in these regions a higher probability of containing a dinucleotide which corresponded to these structural properties. The negative dataset consists of 1000 samples with a random nucleotide sequence.
Real datasets were derived from experimentally confirmed binding sites of E. coli [as obtained from RegulonDB (TF binding sites table, version 6.2) (33)]. We constructed datasets for all TF with more than ten known binding sites. Positive samples consist of the nucleotide sequences of known binding sites. The negative samples consist of 1000 non-overlapping sequences that were randomly sampled from the remainder of the E. coli intergenic region. All sequence data was derived from the E. coli K12 genome MG1655 (NCBI release; NC_000913).
Cross validation
The methods’ performances were evaluated by a 10-fold cross validation, where a reduced training set was constructed by randomly leaving out 1/10 of both the positive and negative sequences from the original training set. The model trained on the reduced set is subsequently used to score the left out samples for their similarity to the positive set. This procedure is repeated ten times, each time leaving out a different set of samples so that at the end each sample was left-out exactly once. To remove training/test set bias from the cross validation, the entire procedure was iterated five times and the results reported here are averages of these repeats.
Validation with gene expression
In order to validate novel binding sites predicted by our CRF model, we used a gene expression compendium consisting of 870 publicly available microarrays (34). In a first step we retrieved, for each given TF, a set of genes which are co-expressed with the set of known target genes of that TF (the seed gene set) across a subset of conditions in the compendium (i.e. a bicluster). Target genes are defined here as the genes containing the known TF-binding sites. Assuming that co-expression with known TF targets might infer co-regulation, we considered the bicluster genes that were not part of the original seed gene set as novel potential targets for the corresponding TF. Using this potentially co-regulated gene set, we can validate the predicted binding sites by evaluating whether this gene set is enriched in high-scoring binding sites predicted by our model. Biclusters were built with the Iterative Signature Algorithm (ISA) (35) with default parameters and using the known target genes for each TF as the bicluster seed. Any final bicluster that had lost all of its original seed genes was removed from further analysis. To allow for a fair enrichment calculation, seed genes were not considered as members of the biclusters. Gene functions assigned to the E. coli genome were retrieved from EcoCyc (36). The functional enrichment of the biclusters were calculated through a cumulative hypergeometric function. As the significance cut-off, we used a Bonferroni corrected value which equals the cut-off 0.05 divided by the number of gene functions found in the bicluster. The significantly enriched gene functions were then compared to a list of gene functions related to the function of the TF or its known regulon. The statistical test used for evaluating enrichment of the biclusters with high-scoring predicted binding sites is a running sum as described by Keller et al. (37), applied per TF, where the novel binding site predictions are used to sort a ranked list of genes not known to be regulated by the TF and then compared to the first genes upstream in the same operon as the genes found to be co-expressed in the corresponding bicluster. The significance of the enrichment is tested by re-iterating the test several times with a gene set of identical size randomly selected from the sorted list and recalculating the running sum. The P-value is then defined as the fraction of random sampled gene sets that achieve a higher enrichment score.
Webtool
The structure-based CRoSSeD models used to predict novel binding sites for the TF’s in this article have been incorporated into a webtool. This application allows the input of any sequence to be screened for the binding sites of a discussed TF. It is currently available at: http://ibiza.biw.kuleuven.be/crossed/webtool.html. The scripts utilized in this article to create and use the CRoSSeD models, can also be downloaded from this location.
RESULTS
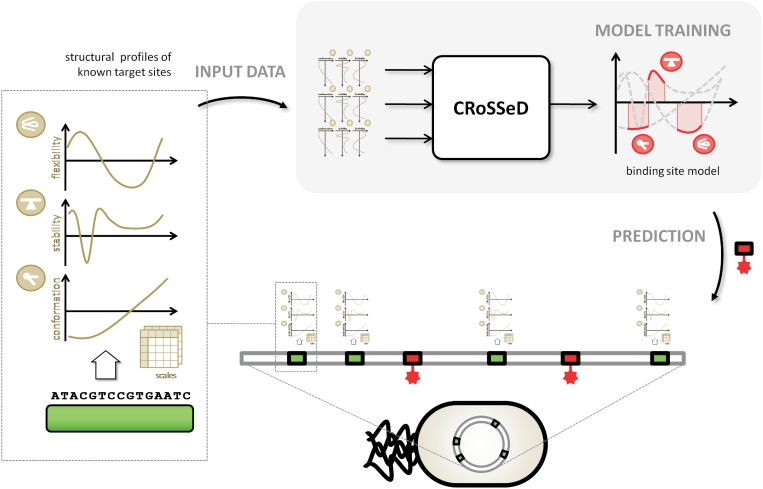
CRoSSeD is a supervised classifier based on conditional random fields’ (CRF) theory (38) that uses structural properties to model and predict novel binding sites. The local structural DNA profiles upon which CRoSSeD relies, are derived from the DNA sequence using di- or trinucleotide structural scales (39). See Figure 1 for an overview.
Figure 1.
Overview of the CRoSSeD methodology. The sequence of known TF-binding sites (green) are collected and used to create different structural profiles by applying structural scales. These structural scales are then used as input for the CRoSSeD model which will create a binding site model featuring strongly conserved structural profile characteristics in specific regions at the binding sites. These binding site models can then be used to predict other binding sites (red) for the given TF in the genome.
CRoSSeD performance
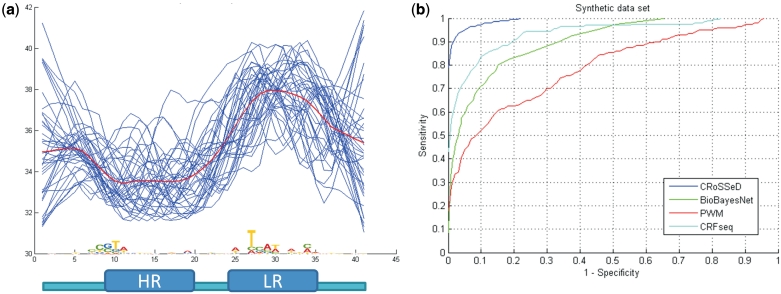
To evaluate the performance of CRoSSeD and to demonstrate the difference between a sequence and a structure-based method, we first created a synthetic dataset with a positive set of 40 samples that contain a region of both low and high rigidity (Figure 2a). This synthetic dataset was used to compare the predictive power of the CRoSSeD structure-based model to that of a standard PWM model, BioBayesNet (BBN) and CRFseq using a 10-fold cross validation. The resulting Receiver Operator Characteristic (ROC) curve is displayed in Figure 2b. The CRoSSeD model uncovers the common flexibility profile present in all positive samples of the synthetic dataset by assigning high weights to several structural properties that measure the rigidity of the molecule, such as propeller twist, intrinsic B-DNA twist and the propensity for the DNA molecule to exist in the more rigid A-form (40). The BBN model, which also makes use of structural properties, has a higher false positive rate for all sensitivities than the CRoSSeD model. The BBN model treats the structural properties as a global feature by using the mean value for the entire sequence, whereas the CRoSSeD model defines the structural properties as a local feature at each position in the sequence. Because the data set contains local regions of respectively high and low rigidity rather than global ones, these are difficult for the BBN model to capture. Adding prior knowledge on significant regions increases the performance of the BBN model (data not shown). However, when attempting to model TF-binding sites as is the goal here, the information given as prior is exactly the unknown information what we want to infer by applying the model to our training data. The predictive power of the PWM model for this dataset is rather poor and underperforms compared to the models that exploit the structural properties. This is to be expected as the dataset contains structural properties rather than sequence conservation: the different positive samples show a low-sequence conservation, but share a similar flexibility profile which can only indirectly be modeled by the PWMs if the structural conservation results in sufficient sequence similarities. The CRFseq model is also limited to sequence information but is able to capture di- and trinucleotide relationships, which can be related to the local DNA structure as discussed above, and therefore shows a better classification performance than the PWM which assumes independence between positions. The CRoSSeD method, which can look directly at structural conservation, still outperforms the CRFseq.
Figure 2.
(a) Flexibility profiles of all 40 positive synthetic samples (blue lines) as measured by the B-DNA twist scale, (lower values correspond to more flexible regions). The red line is the average profile. For comparison, the sequence conservation logo is also given for each position. At the bottom of the figure is the structural characteristic that was simulated (HR: high rigidity, LR: low rigidity). (b) ROC curve displaying the average result of five 10-fold cross validations for the CRoSSeD (blue line), BioBayesNet (green line), PWM (red line) and CRFseq (cyan line) model when applied to the synthetic data set.
While the synthetic dataset shows the superiority of the CRoSSeD model over its sequence-based counterpart CRFseq, a classical PWM and the structure-based BioBayesNet in capturing shared local structural properties, the question remains as to whether the CRoSSeD model will still outperform the other methodologies on real datasets, where such features might be less pronounced. Therefore, we evaluated the same three methods on datasets with known TF-binding sites. So as to base our conclusions on a wide variety of examples, we derived data sets for every E. coli TF with more than ten experimentally validated binding sites.
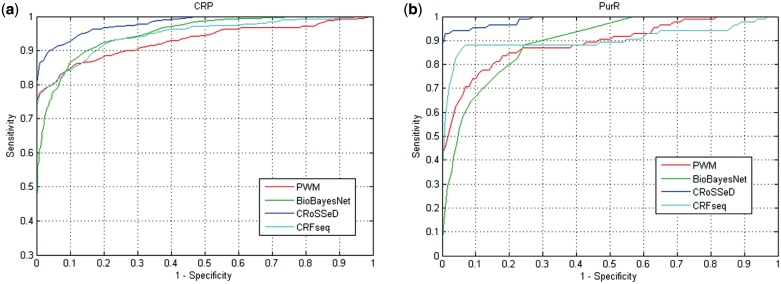
The datasets contain for each TF all its known binding sites as positive samples and as a negative set 1000 random intergenic samples from the E. coli genome. The performance of each of the compared methods was again evaluated using a 10-fold cross validation. The predictive power could then be summarized in the area under the curve (AUC) statistic (listed in Supplementary Data 3) for each of the resulting ROC curves. For nineteen out of the 27 TF datasets the CRoSSeD model outperformed both the BBN and PWM model. In only two of these 19 cases does the CRFseq model achieve a similar level of performance as the CRoSSeD model, with the former being outperformed by the latter in the remainder, implying that the structural properties provide a boost in performance that cannot be attributed to the implicit higher order nucleotide relationships. For these particular set of TF’s, the representation of the binding site by means of structural properties as is done in our CRoSSeD model clearly improves the prediction of binding sites. Our method thus captures structural properties seemingly important for TF-binding site recognition that could not be found or modeled by either the PWM or the BBN models. For instance, for both the cAMP receptor protein (CRP) and the purine repressor (PurR) of E. coli, the CRoSSeD model consistently shows for each specificity level the highest sensitivity of all methods (results shown in Figure 3). While the CRoSSeD models are outperformed by the other methods in a few cases (see Supplementary Data 4 for details), the CRoSSeD model does demonstrate a higher sensitivity at very high specificities for almost all of these cases (Supplementary Figure 2). High-specificities levels are the most interesting region on the ROC curve in a prediction scheme, as significance thresholds will be chosen to limit the number of false positives to reasonable levels. Thus while the PWM or BBN models present a better AUC score in some cases, the CRoSSeD model still displays greater predictive power at high specificities, where it is intended to be applied.
Figure 3.
Performance results of the different methods on the CRP (a) and PurR (b) data sets. The ROC curves display the trade-off between the sensitivity (the fraction of positive samples correctly identified as binding sites) and specificity (the fraction of incorrectly identified negative samples) of the results on the left out samples obtained at different probability thresholds for five 10-fold cross validations for the CRoSSeD (blue), CRFseq (cyan line), the PWM (red) and BioBayesNet model (green).
Screening for novel binding sites
After showing the ability of CRoSSeD to model binding sites in a cross validation setting, we used it to screen the genome for novel binding sites of the studied TFs. Models were trained on the complete training set and used as input for a genome-wide screening. The same was also done for the PWM models from the previous paragraph so as to be able to compare results from the CRoSSeD screening with those from a traditional screening. The available web-based implementation of BioBayesNet does not allow for a genome-wide analysis. Genes containing predicted binding sites, were considered as novel targets of the modeled TFs and they were evaluated using gene expression data and an extensive literature and database survey (see Supplementary Data 5).
To avoid having to place an arbitrary cut-off on the screening scores for any of the methods, as well as to minimize the possible effects of combinatorial regulation on observed co-expression patterns in the expression data, we applied the following strategy (see Materials and methods section for further details): we started by retrieving a set of genes which were co-expressed with the known target genes of a given TF under a subset of conditions in a large compendium of E. coli microarray data. We considered each co-expressed gene set as potential novel targets of the TF, provided that the gene set was enriched in a functional class to which the TF or its know target genes belong. All but three gene sets were significantly enriched with a function that could be directly linked to that of the TF. No enriched gene function was found for both the IHF and the H-NS gene set because these TF’s have no clearly defined function enriched in their target genes. We then tested to what extent each of these co-expressed gene sets that represent ‘novel targets of a TF’ were enriched with binding site predictions obtained by the genome wide motif screening.
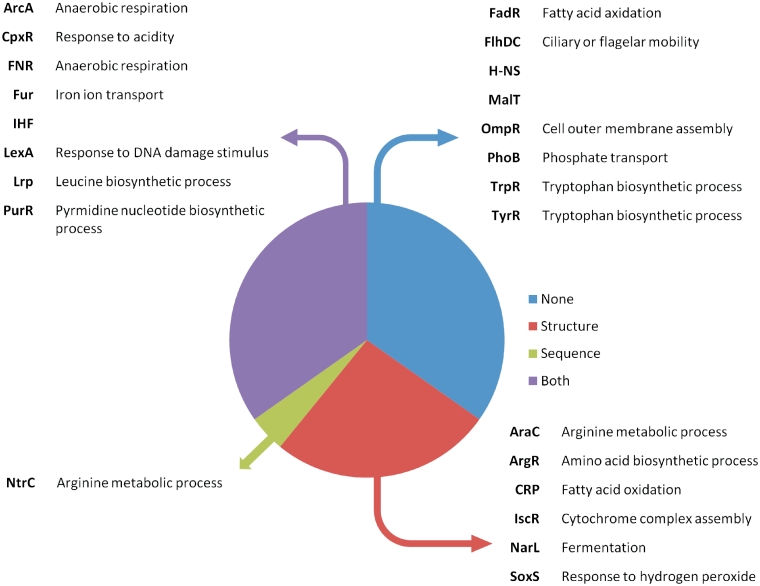
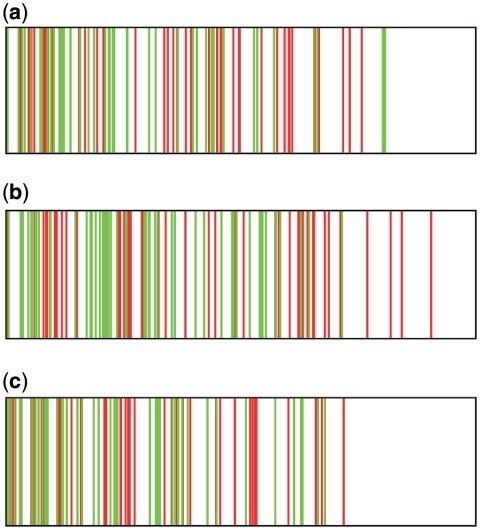
The analysis shows that 14 out of the 23 gene sets were enriched in high-ranking predicted targets of the structure-based model with a P-value of 0.05 or less (Figure 4). The probability of this enrichment occurring without any relation between the co-expression and the model screening is 3.2e – 13. Nine co-expressed gene sets were significantly enriched at the same cut-off with high-scoring PWM targets, at a random-occurrence probability of 8.4e – 7. There was substantial overlap between the sequence- and structure-based predictions as all but one of the gene sets significantly enriched for PWM-based predictions were also enriched for the CRoSSeD-based predictions. This indicates that most novel predictions made using a PWM-based method can also be made with a structure-based model, while the reverse is not always true. A list with the highest ranked predictions that were validated by the gene expression analysis can be found in Supplementary Data 5. Almost a third of the co-expressed gene sets were enriched for highly ranked sites predicted by the CRoSSeD model but not for highly ranked predictions obtained by the PWM model (red pie in Figures 4 and 5a and b). To verify whether the targets predicted only by the structure-based method might indeed correspond to true targets, we carefully checked the literature for additional experimental evidence. For example, the structure-based highest ranked novel predicted ArgR target that was co-expressed with known ArgR genes, aroP, had previously been confirmed as true ArgR target (41). Moreover, the location described by CRoSSeD as the most likely binding site corresponds to the region where the previously confirmed site was located. In contrast, aroP was only located in the fortieth percentile of the PWM ArgR ranked list. Another prediction of interest is sdhC, potentially regulated by SoxS, a transcriptional TF involved in oxidative-stress response. shdC is an integral part of the TCA cycle that produces NADPH, which plays a significant role to reduce oxidative stress (42). Furthermore, it was found that the expression levels of sdhC drop in a soxS-knock-out mutant (43). This corresponds well to the fact that sdhC was found to be co-expressed with known SoxS target genes and strongly supports our prediction of sdhC as a direct SoxS target. The structure-based screening ranks the sdhC gene as one of the most likely SoxS targets at rank 6 while with the PWM screening this gene only at rank 471, explaining why it has not been proposed as a SoxS target gene yet. A detailed list with other literature predictions is available in Supplementary Data 4.
Figure 4.
Overview of the co-expressed gene sets and their enrichment with high-scoring predicted binding sites obtained from respectively the structure- or sequence-based models. The pie chart represents all found co-expressed gene sets, divided into segments with no significant high-ranking predictions enrichment (blue) and gene sets that were found to be enriched and if this was for the binding sites predicted by the CRoSSeD (red), PWM (green) or both models (purple). A table is provided per segment, listing the related TF (in bold) and the most relevant significantly enriched gene function in the gene sets.
Figure 5.
Representation of high-scoring binding site predictions enrichment in co-expressed gene sets for respectively ArgR (a), SoxS (b) and PurR (c). Each plot corresponds the entire ranked gene list as obtained from the screening using the PWM (red) and CRoSSeD (green) motif models with decreasing confidence from left to right. Marked are the positions of the genes that were found co-expressed with the known target genes of the respective TFs.
The gene sets which were found to be significantly enriched for both the CRoSSeD and PWM predictions (purple pie in Figures 4 and 5c), were mostly those for global TF's, such as ArcA, FNR, Fis and IHF, and other well-characterized TFs, such as Fur, PurR and LexA. These TF’s have been the subject of many studies due to their importance in the general functioning of the organism and therefore their binding characteristics have been well documented. As a result, the sequence-based model has access to a large amount of high-confidence training data and is therefore very reliable. We succesfully predicted three target genes [polB and dinI as targets for LexA (44) and mntH as a target for Fur (45)] which were not reported in RegulonDB at the time of the analysis, but have been confirmed in the meantime and several others that have already been predicted in previous work. Other than rediscovering several previously predicted targets for PurR, a nucleotide biosynthesis TF, such as the purT gene, we also found several new targets that seem functionally related to the PurR regulon. Among these were the genes serA and serC, that code for enzymes necessary for the biosynthesis of serine (46,47), an important contributor of one-carbon units for the de novo biosynthesis of purines (48). Both serA and serC were assigned a high rank by the CRoSSeD model but very poor rankings by the PWM. Closer inspection of the predicted binding sites in serA and serC reveal that they indeed exhibit a lower sequence homology than other predicted binding sites but presented several structural profiles that were highly similar to those of the known PurR-binding sites and were therefore still assigned high scores by the CRoSSeD model.
Biological relevance of the CRoSSeD models
We have shown that CRoSSeD, as a structure-based methodology, can predict valid novel TF-binding sites which could not be made using sequence-based methods. In these cases, the structure-based models might have uncovered certain structural properties that play a biological role in the recognition of the binding sites by the regulator protein that remain unseen by sequence-based methods such as PWMs. This possibility is illustrated by comparing the used structure-based models to current knowledge of protein–DNA interaction for two well-studied TF’s, namely CRP and PurR.
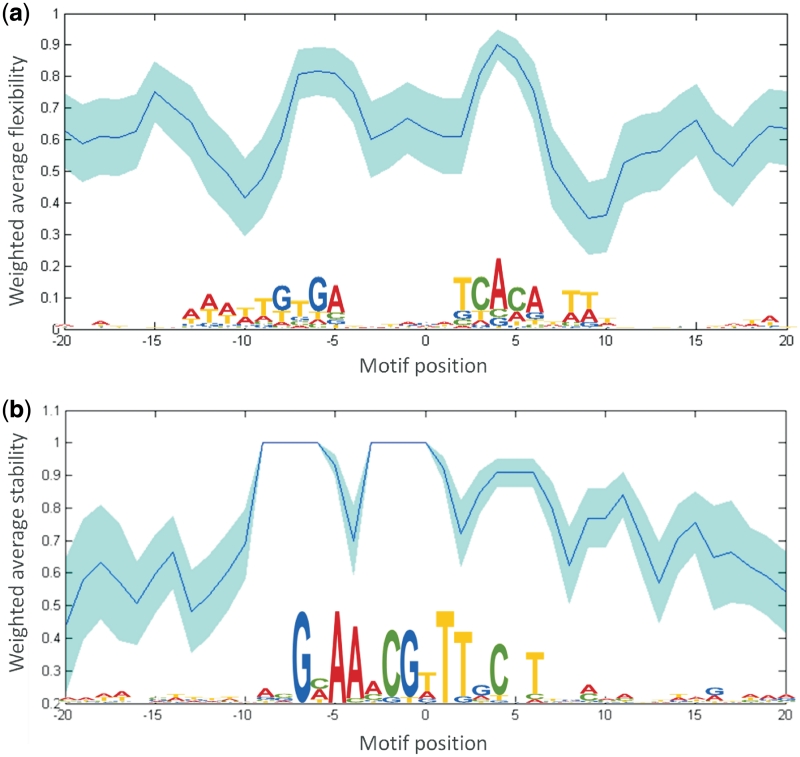
For CRP, it is known that it binds as a dimer and commonly introduces two kinks in the DNA molecule near its binding site, a ‘primary kink’ approximately at position +5/–5 in the motif and a ‘secondary kink’ around position +11/–11 which is located at about half a turn of the DNA helix from the primary kink (49). Both kinks result in the DNA-helix being bent towards the CRP protein complex. This is represented in the CRoSSeD-binding site model for CRP by the fact that the largest weights trained by the structure-based methodology were assigned to the flexibility property. Figure 6a shows a weighted average profile for this property. Because the values in this flexibility scale are derived from the cutting frequencies of DNase I, which is known to cut preferentially DNA that is bent towards the major groove, the sharp rise in the profile around position 5 will likely correspond to positions where the DNA is either intrinsically curved or is bendable towards the major groove. In the case of CRP, this will be the primary kink which is a bend in this direction, as shown by crystallography data (50). Similarly, the dip in the profile corresponds to the position where CRP introduces the secondary kink, now a bend towards the minor groove due to the intrinsic twist of the DNA molecule. Also striking is the symmetry in the flexibility profile, corresponding to the dimer nature of CRP, even in the areas which appear to have poor sequence conservation. The trained model therefore matches well with what is currently known about the mechanism of CRP binding.
Figure 6.
Important features contributing to the CRoSSeD model for, respectively, CRP (a) and PurR (b). In panel (a), the profile corresponds to the DNase-I cutting frequency (flexibility) profile based on the weights assigned to the CRP model. Plotted in the dark blue line is the weighted average of the property at each position in the motif and surrounding it in the light-blue area is the standard deviation on this average for each position. Panel (b) contains the disruption energy profile (stability) based on the PurR model.
It was elucidated from crystal structure data that the PurR dimer also induces a bend in its binding site upon binding, though through a mechanism that is different from that of CRP. PurR is known to induce a single kink at the central position of the DNA motif by intercalating a pair of leucine residues into the minor groove of the CG dinucleotide, resulting in a local disruption of the DNA structure (51). Despite this severe local deformation, the structure of the surrounding DNA sequence displays little to no alteration and it was shown to remain quite stable (51). This mechanism is also reflected in the profile. For PurR, the highest weight was not assigned to a structural profile related to DNA flexibility as was the case with CRP, but to a structural property related to DNA stability, the disruption energy (21) (Figure 6b). The large weights assigned to the disruption energy of the DNA sequence might correspond to the necessity of the binding site to remain stable, with as few deformations or deviations from the standard B-DNA form as possible, in close proximity to the disruption resulting from the leucine intercalation.
DISCUSSION
In this article, we investigated what information can be retrieved from local DNA structural properties, relevant to TF binding that cannot be captured by the nucleotide sequence alone, and to what extent this information can be used to model and predict TF-binding sites. We evaluated TF target predictions made by structure-based models compared to those of more traditional sequence-based methods. To this end, we developed a novel structure-based method which could screen for TF-binding sites on a large scale and still had a predictive power comparable, if not greater than, other methodologies which incorporate sequence information or structural properties (PWMs, CRFseq and BBN) as was shown by a cross validation analysis on known binding sites. PWMs represent the traditional sequence conservation models and BBN is a Bayesian network methodology that can include additional information, such as structural DNA characteristics, apart from the nucleotide sequence. The improved classification performance could not be replicated using a sequence-only higher order CRF method, namely CRFseq, thus demonstrating that the structural profiles contain additional information about TF-binding sites, which cannot be directly derived from the respective di- or trinucleotides. It is important to note that there might exist a bias in favor of sequence-based methods for several known binding site data sets (i.e. the model training/test data). These binding sites were often first identified using a sequence-based method and subsequently confirmed in an experimental setup. This will create a propensity for the known and confirmed sites to have very similar DNA sequences and thus might not be an optimal representation of all true target sites, considering the importance of indirect read-out for some TF’s (2). This means that the difference between the PWM model and the CRF model might be more pronounced than we were able to show here. Based on the local structural characteristics of the DNA at the known binding sites, the proposed method was able to make more confident predictions about the presence of binding sites in promoters of co-expressed genes than the sequence-based methodology. These results support previous theories that structural DNA information can improve classifier performance by providing a higher level data source that is explicitly different from the nucleotide sequence itself (7,8). We could also show that some of the novel, CRoSSeD predicted binding sites (e.g. serC) have strong structural similarities while exhibiting relatively low-sequence similarity. The independent validation analysis based on co-expression and a literature and database survey, suggests that some of these sites with low-sequence conservation are indeed true TF-binding sites. It seems possible that binding sites can compensate for a poor conservation of the sequence motif with strong conservation of particular structural signals. Whether this plays out in the form of a trade-off, a way to tune binding efficiency, or whether sequence conservation in these instances is only a by-product of the necessary presence of certain structural properties or vice versa, remains to be elucidated. Furthermore, we were also able to show that the CRoSSeD models and their inherent structural profiles are not simply another way to represent the nucleotide sequence, but that they can be related to the actual molecular mechanisms of the TF to DNA binding, as was illustrated for CRP and PurR. Structure-based models such as the ones created here by CRoSSeD can thus not only be used for the prediction of novel sites, but they might be able to give valuable insight into the binding mechanisms of TF’s for which currently little detailed information is known.
We limited the analysis in this article to prokaryotes to allow easier isolation of the contribution of structural properties to TF binding, as current understanding seems to indicate that less factors influence the gene regulation of TFs in these organisms. However there is no reason that the presented method could not be applied to eukaryotes, as the contribution of indirect readout to binding-site recognition is known to also occur in these organisms (2). CRoSSeD is a general framework and can therefore be applied to any TF for which sufficient reliable binding site information exists. Furthermore many more structural scales than those used to train CRoSSeD are available and in the future the performance of the models could possibly be further increased by the incorporation of additional structural properties, which could be specific to a target organism. Additionally, as this method focuses on structural properties, which is just one of the aspects used in TF recognition of binding sites, it will most likely be able to provide complementary information if combined with methods in the same scheme which use data from different sources, such as nucleosome binding information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
KULeuven Research Council [GOA/08/011, CoE EF/05/007—SymBioSys, CREA/08/023, OT 05-33, OT09/022]; Agency for Innovation by Science and Technology [SBO-BioFrame, SB-81297]; Interuniversity Attraction Poles [P6/25—BioMaGNet]; Research Foundation—Flanders [IOK-B9725-G.0329.09]; Human Frontier Science Program [RGY0079/2007C]. Funding for open access charge: Agency for Innovation by Science and Technology (IWT Flanders, grant no. SB-81297).
Conflict of interest statement. None declared.
REFERENCES
- 1.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 2.Gromiha MM, Siebers JG, Selvaraj S, Kono H, Sarai A. Role of inter and intramolecular interactions in protein-DNA recognition. Gene. 2005;364:108–113. doi: 10.1016/j.gene.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Kono H, Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 4.Morozov AV, Havranek JJ, Baker D, Siggia ED. Protein-DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005;33:5781–5798. doi: 10.1093/nar/gki875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angarica VE, Perez AG, Vasconcelos AT, Collado-Vides J, Contreras-Moreira B. Prediction of TF target sites based on atomistic models of protein-DNA complexes. BMC Bioinformatics. 2008;9:436. doi: 10.1186/1471-2105-9-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karas H, Knuppel R, Schulz W, Sklenar H, Wingender E. Combining structural analysis of DNA with search routines for the detection of transcription regulatory elements. Comput. Appl. Biosci. 1996;12:441–446. doi: 10.1093/bioinformatics/12.5.441. [DOI] [PubMed] [Google Scholar]
- 8.Thayer KM, Beveridge DL. Hidden Markov models from molecular dynamics simulations on DNA. Proc. Natl Acad. Sci. USA. 2002;99:8642–8647. doi: 10.1073/pnas.132148699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponomarenko JV, Ponomarenko MP, Frolov AS, Vorobyev DG, Overton GC, Kolchanov NA. Conformational and physicochemical DNA features specific for transcription factor binding sites. Bioinformatics. 1999;15:654–668. doi: 10.1093/bioinformatics/15.7.654. [DOI] [PubMed] [Google Scholar]
- 10.Liu R, Blackwell TW, States DJ. Conformational model for binding site recognition by the E.coli MetJ transcription factor. Bioinformatics. 2001;17:622–633. doi: 10.1093/bioinformatics/17.7.622. [DOI] [PubMed] [Google Scholar]
- 11.Pudimat R, Schukat-Talamazzini EG, Backofen R. A multiple-feature framework for modelling and predicting transcription factor binding sites. Bioinformatics. 2005;21:3082–3088. doi: 10.1093/bioinformatics/bti477. [DOI] [PubMed] [Google Scholar]
- 12.Gunewardena S, Zhang Z. Accounting for structural properties and nucleotide co-variations in the quantitative prediction of binding affinities of protein-DNA interactions. Pac. Symp. Biocomput. 2006:379–390. [PubMed] [Google Scholar]
- 13.Baldi P, Baisnee PF. Sequence analysis by additive scales: DNA structure for sequences and repeats of all lengths. Bioinformatics. 2000;16:865–889. doi: 10.1093/bioinformatics/16.10.865. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen AG, Jensen LJ, Brunak S, Staerfeldt HH, Ussery DW. A DNA structural atlas for Escherichia coli. J. Mol. Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- 15.Abeel T, Saeys Y, Bonnet E, Rouze P, Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18:310–323. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodsell DS, Dickerson RE. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2000;97:3347–3351. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii S, Kono H, Takenaka S, Go N, Sarai A. Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 2007;35:6063–6074. doi: 10.1093/nar/gkm627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Florquin K, Saeys Y, Degroeve S, Rouze P, Van de Peer Y. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res. 2005;33:4255–4264. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ornstein RL, Rein R, Breen DL, Macelroy RD. Optimized potential function for calculation of nucleic-acid interaction energies. 1. Base stacking. Biopolymers. 1978;17:2341–2360. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]
- 21.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986;5:2737–2744. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delcourt SG, Blake RD. Stacking energies in DNA. J. Biol. Chem. 1991;266:15160–15169. [PubMed] [Google Scholar]
- 24.Ivanov VI, Minchenkova LE. [The A-form of DNA: in search of the biological role] Mol. Biol. (Mosk) 1994;28:1258–1271. [PubMed] [Google Scholar]
- 25.Brukner I, Sanchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 27.Sivolob AV, Khrapunov SN. Translational positioning of nucleosomes on DNA - the role of sequence-dependent isotropic DNA bending stiffness. J. Mol. Biol. 1995;247:918–931. doi: 10.1006/jmbi.1994.0190. [DOI] [PubMed] [Google Scholar]
- 28.El Hassan MA, Calladine CR. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J. Mol. Biol. 1996;259:95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- 29.Sugimoto N, Nakano S, Yoneyama M, Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolajewa S, Pudimat R, Hiller M, Platzer M, Backofen R. BioBayesNet: a web server for feature extraction and Bayesian network modeling of biological sequence data. Nucleic Acids Res. 2007;35:W688–W693. doi: 10.1093/nar/gkm292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulyk ML, Johnson PL, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemmens K, De BT, Dhollander T, De Keersmaecker SC, Thijs IM, Schoofs G, De WA, De MB, Vanderleyden J, Collado-Vides J, et al. DISTILLER: a data integration framework to reveal condition dependency of complex regulons in Escherichia coli. Genome Biol. 2009;10:R27. doi: 10.1186/gb-2009-10-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergmann S, Ihmels J, Barkai N. Iterative signature algorithm for the analysis of large-scale gene expression data. Phys. Rev. E. Stat. Nonlin. Soft. Matter Phys. 2003;67:031902. doi: 10.1103/PhysRevE.67.031902. [DOI] [PubMed] [Google Scholar]
- 36.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller A, Backes C, Lenhof HP. Computation of significance scores of unweighted Gene Set Enrichment Analyses. BMC Bioinformatics. 2007;8:290. doi: 10.1186/1471-2105-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lafferty J, McCallum A, Pereira F. Proceedings of the 18th International Conference on Machine Learning, 2001. San Francisco, CA: MA Morgan Kaufmann; Conditional random fields: probabilistic models for segmenting and labeling sequence data; pp. 282–289. [Google Scholar]
- 39.Baldi P, Chauvin Y, Brunak S, Gorodkin J, Pedersen AG. Computational applications of DNA structural scales. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:35–42. [PubMed] [Google Scholar]
- 40.Charney E, Chen HH, Rau DC. The flexibility of A-form DNA. J. Biomol. Struct. Dyn. 1991;9:353–362. doi: 10.1080/07391102.1991.10507917. [DOI] [PubMed] [Google Scholar]
- 41.Bulyk ML, McGuire AM, Masuda N, Church GM. A motif co-occurrence approach for genome-wide prediction of transcription-factor-binding sites in Escherichia coli. Genome Res. 2004;14:201–208. doi: 10.1101/gr.1448004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl Acad. Sci. USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabir M, Shimizu K. Investigation into the effect of soxR and soxS genes deletion on the central metabolism of Escherichia coli based on gene expressions and enzyme activities. Biochem. Engineer. J. 2006;30:39–47. [Google Scholar]
- 44.Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- 45.Patzer SI, Hantke K. Dual repression by Fe(2+)-Fur and Mn(2+)-MntR of the mntH gene, encoding an NRAMP-like Mn(2+) transporter in Escherichia coli. J. Bacteriol. 2001;183:4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobey KL, Grant GA. The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase. J. Biol. Chem. 1986;261:12179–12183. [PubMed] [Google Scholar]
- 47.Duncan K, Coggins JR. The serC-aro A operon of Escherichia coli. A mixed function operon encoding enzymes from two different amino acid biosynthetic pathways. Biochem. J. 1986;234:49–57. doi: 10.1042/bj2340049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dev IK, Harvey RJ. Regulation of synthesis of serine hydroxymethyltransferase in chemostat cultures of Escherichia coli. J. Biol. Chem. 1984;259:8394–8401. [PubMed] [Google Scholar]
- 49.Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J. Mol. Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 50.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arvidson DN, Lu F, Faber C, Zalkin H, Brennan RG. The structure of PurR mutant L54M shows an alternative route to DNA kinking. Nat. Struct. Biol. 1998;5:436–441. doi: 10.1038/nsb0698-436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.