Abstract
The tumor suppressor p53 is a central regulator of cell-cycle arrest and apoptosis by acting as a transcription factor to regulate numerous genes. We identified all human p53-regulated mRNAs by microarray analyses and searched for protein-coding genes which contain intronic miRNAs. Among others, this analysis yielded the panthothenate kinase 1 (PANK1) gene and its intronic miRNA-107. We showed that miRNA-107 and PANK1 are coregulated by p53 in different cell systems. The PANK1 protein, which catalyzes the rate-limiting step of coenzyme A biosynthesis, is also upregulated by p53. We observed that p53 directly activates PANK1 and miRNA-107 transcription through a binding site in the PANK1 promoter. Furthermore, p53 is recruited to the PANK1 promoter after DNA damage. In order to get more insight into miRNA-107 function we investigated its potential target genes. Cell-cycle regulators are significantly enriched among predicted miRNA-107 targets. We found miRNA-107-dependent regulation of two important regulators of G1/S progression, CDK6 and the RB-related 2 gene RBL2 (p130). CDK6 and p130 proteins are downregulated upon miRNA-107 expression. Our results uncover a novel miRNA-dependent signaling pathway which leads to downregulation of cell cycle proteins in the absence of transcriptional repression.
INTRODUCTION
miRNAs are short non-coding RNAs which have functions in numerous cellular processes like differentiation, cell survival and proliferation (1). Originally found in Caenorhabditis elegans by their ability to interfere the expression of the developmentally regulated lin-14 (2), it later became apparent that miRNAs are expressed in a wide variety of organisms including humans (3,4). They act as adaptors in a large protein complex known as RNA-induced silencing complex (RISC) (5) and direct sequence-specific target-mRNA recognition through imperfect base pairing to their untranslated regions (6,7). miRNAs can act on mRNA stability by recruitment of the CAF1-CCR4-NOT deadenylation complex and the decapping enzymes DCP-1 and DCP-2 (8,9). Furthermore, miRNAs can interfere with translation of target mRNAs (9–12). Elucidating the contribution of these mechanisms to the repression effect of a miRNA is controversial. It has been shown that in mouse Krebs-2 ascites cell extracts inhibition of target mRNA translation is initially induced and followed by degradation of the mRNA (13). Therefore, translational repression and initiation of RNA degradation may act synergistically on target mRNAs. Many miRNAs were predicted to have hundreds of target mRNAs in a cell due to the short seed sequences which are needed to guide miRNA/mRNA binding. Proteome screening studies after overexpression of different miRNAs have indeed shown that hundreds of proteins are efficiently repressed, albeit to a modest degree (14,15). Importantly, it has been shown that only a few target genes are sufficient to mimic a miRNA-dependent complex phenotype. As an example miRNA-31 is predicted to target hundreds of genes and is able to inhibit breast cancer cell metastasis (16). After re-expression of only three target genes, the cancer cells were able to form metastases again, showing that expression of only a small proportion of miRNA target genes are sufficient for a specific signaling pathway (17). This and other reports showed that miRNAs are specifically over- or underexpressed in certain tumor types (18–20). From such results it is obvious that elucidating transcriptional regulation of miRNA genes is important for gaining more insight into the function of miRNAs in tumorigenesis.
The transcription factor p53 is a well known tumor suppressor which is able to bind to specific palindromic sequences (21). p53 is able to regulate a plethora of target genes which function mostly in cell cycle control and apoptosis induction (22). Cell cycle arrest is achieved through induction of important CDK inhibitors like p21CIP1/WAF1 (23) and through transcriptional repression of central cell cycle genes like Cdc25C (24), Cdc25A (25), Cks2 (26) or Cyclin B (27). In addition to ‘classical’ functions, p53 also influences ‘non-classical’ pathways like controlling metabolism (28). One example of p53′s impact on metabolism is the enhancement of mitochondrial electron transport by inducing the cytochrome c regulator synthesis of cytochrome c oxidase 2 (SCO2) gene (29). Furthermore, glycolysis is shut down by p53 through transcriptional induction of the TP53-induced glycolysis and apoptosis regulator (TIGAR) gene (30) and through repression of the isomerase phosphoglycerate mutase (PGM) gene (31). The regulation of these genes prevents the shift of energy metabolism to glycolysis which is characteristic for nearly all cancers (32). The p53 target gene guanidinoacetate methyltransferase (GAMT) is a key downstream effector after nutrient deprival. GAMT induces fatty acid oxidation after glucose starvation which restores energy supply (33).
Recently, p53 has been shown independently by several groups to regulate miRNA transcription (34–38). The miRNA-34 family is transcriptionally upregulated by p53 and assists in initiating apoptosis. Also miRNA-192, -215 and -194 are induced by p53, which are mostly involved in the regulation of cell cycle arrest (39,40). Intronic miRNAs are processed by the RNase Drosha during splicing (41) and are frequently co-expressed with their host genes (42). However, there is only limited data on details of this coregulation. Often it has not been investigated whether miRNAs are regulated through the same promoters or the same transcription factors as their host genes. For some systems such studies have been carried out, e.g. E2F1 was shown to regulate miRNA-449 together with its host gene Cdc20B (43). The miRNA-25, -93, -106b cluster, which is intronic to the MCM7 gene, was shown to be repressed by p53. This repression is mediated by inhibition of E2F1 activity, which also controls MCM7 expression (44).
We studied intronic miRNAs of host genes regulated by the tumor suppressor p53. By DNA microarray analyses we observed that the host gene of miRNA-107, the panthothenate kinase 1 (PANK1) gene, is upregulated by p53. The coregulation of PANK1 and its intronic miRNA-107 were characterized. The PANK1 locus was observed to be activated through a p53-binding site in the PANK1 promoter. Finally, we identified CDK6 and the RB-related protein p130 as targets of miRNA-107 which have important functions at the G1/S transition of the cell cycle.
MATERIAL AND METHODS
Cell culture, transient transfections and FACS analyses
HCT116, SaOS-2, D53wt and D53mut cell lines were cultured as described (27). Human colon carcinoma HCT116 cells wild-type or with targeted deletions of p53 (HCT116 p53−/−) were treated with doxorubicin at a final concentration of 200 ng/ml and harvested after 24 and 48 h. Treatments with Mdm2-inhibiting nutlin-3 were performed at 5 µM for 24, 48 and 72 h. Derivatives of the colorectal carcinoma cell line DLD-1 were kindly provided by Bert Vogelstein (D53wt, D53mut) (45). These DLD-1 cells harboring an inactive 241F p53 mutant are stably transfected with a tetracycline-responsive p53 expression system. Inductions of p53wt and of the DNA-binding-deficient mutant p53R175H (p53mut) were performed by removal of tetracycline from the cell culture media for 6, 9 or 15 h.
Transfection of PANK1 expression plasmids into HCT116 cells were done with FuGENE 6 (Roche, Mannheim, Germany) according to the manufacturer’s instructions. Transfection of control siRNA (medium GC content, Invitrogen, Karlsruhe, Germany) and validated p53 siRNA (Invitrogen) into HCT116 cells were done with Dharmafect-1 (Dharmacon, Chicago, IL, USA) at a final concentration of 50 nM. Cells were treated with doxorubicin (800 ng/ml) 24 h after transfection. D53wt cells were transfected with synthetic pre-miRNA-107 (Ambion, Austin, TX, USA) at a final concentration of 100 and 150 nM, respectively, using the siPORTTM NeoFXTM transfection agent (Ambion). Cy3-labeled control pre-miRNA (Ambion) was transfected at 150 nM and cells were harvested after 48 h. miRNA-107-targeting antisense RNAs (AntagomiR-107, Ambion) were transfected into D53wt cells at a final concentration of 60 nM. Cy3-labeled control AntagomiR at 60 nM was used as a control. Additionally, we controlled the experiments by transfection of D53wt cells without RNA (mock). For luciferase reporter assays, SaOS-2 cells were seeded at 50 000 cells per well into 24-well plates. Cotransfections were carried out with FuGENE 6 as described above. The 400-ng PANK1 promoter constructs in pGL4.10 (Promega, Madison, MA, USA) were co-transfected together with 25-ng promoterless Renilla luciferase (pGL4.70, Promega) and 25-ng expression plasmid (p53wt, p53R175H or pcDNA3.1HisBlacZ). Luciferase activity was normalized to co-transfected Renilla luciferase activity in order to compensate for different transfection efficiencies. Dual Luciferase Assays (Promega) were performed after 24 h. All experiments were done in triplicate and repeated independently at least three times. For testing putative miRNA-107-binding sites, co-transfections of pGL4.10 UTR reporter constructs together with pre-miRNA-107 and control-pre-miRNA with a final concentration of 100 nM were performed with Dharmafect Duo (Dharmacon) as suggested by the manufacturer.
For FACS analyses, D53wt cells were trypsinized, centrifuged and resuspended in PBS (1 mM EDTA). Cells were fixed by addition of three volumes of absolute ethanol and incubation at 4°C overnight. Prior to FACS analyses, cells were centrifuged, resuspended in PBS (1 mM EDTA, with RNase A) and incubated with propidiumiodide at 10 µg/ml (SIGMA-Aldrich, Steinheim, Germany) for 5 min. All measurements were performed on a FACScan instrument (Becton Dickinson, NJ, USA). Data analysis was done with WinMDI 2.9 software.
Cloning and site-directed mutagenesis
PANK1β- and PANK1γ-isoform cDNAs were cloned from D53wt cDNA generated with Superscript III (Invitrogen). PCR was performed with High Fidelity Polymerase (Fermentas, St Leon-Rot, Germany) as suggested by the manufacturer and PCR fragments were cloned into pcDNA3.1(+) (Invitrogen). The PANK1α-isoform in pcDNA3.1(−) was generously provided by Dr Jackowski (46). The PANK1 promoter was cloned from human genomic DNA into pGL4.10. Shorter constructs were generated by PCR and subsequently cloned in pGL4.10. Site-directed mutagenesis was performed by quick-change-PCR with mutated primer pairs and Pfu Turbo polymerase (Stratagene). For investigation of putative miRNA-107 binding sites, UTR fragments of the CDK6 and p130 genes were cloned into the XbaI site of pGL4.10. All inserts were sequenced to control successful cloning. All primers used are outlined in Supplementary Table S5.
Microarray analyses, real-time RT–PCR and northern analysis
Total RNA preparations were done with TRIzol® as suggested by the manufacturer. Total RNA from D53wt cells (control or after 9 h induction) were labeled by using standard procedures (Affymetrix, Santa Clara, CA, USA). The RNA was hybridized on Affymetrix U133 Plus 2.0 GeneChips. Scanned images were evaluated using the Expression Console Software (Affymetrix). All probe sets with a detection P < 0.01 were assumed as being significantly expressed. Microarray data was deposited at the Gene Expression Omnibus database under accession ID GSE21105. Real-time RT–PCRs for mRNA measurements were performed as previously described with the QuantiTect SYBR Green RT–PCR kit (QIAGEN, Hilden, Germany) on a LightCycler 3 system (Roche, Mannheim, Germany). All measurements were normalized to expression of GAPDH mRNA employing the ΔΔCt-method. All RT–PCR primers used are described in the Supplementary Tables. Expression of mature miRNA-107 was measured by using TaqMan miRNA Assays (Applied Biosystems, Foster City, CA, USA). Total RNA was used for reverse transcription with the MicroRNA Reverse Transcription Kit (Applied Biosystems) according to the standard protocol of the supplier. PCR was done on cDNA with specific miRNA primer/probe set and Universal PCR Mastermix (Applied Biosystems). miRNA expression was normalized to expression of snoRNA U48 or snRNA U6B.
Northern analyses were performed essentially as described (47). Briefly, D53wt, D53mut or HCT116 total RNA was enriched for poly A RNA by using the Oligotex mRNA Mini Kit (QIAGEN). Two micrograms of poly A RNA were separated on a 2% agarose gel containing 1% formaldehyde. After transfer on northern-blot membranes (Bio-Rad, München, Germany) RNA was crosslinked by UV exposure. Probes were generated by T7 in vitro transcription of PANK1 cDNA sub-cloned in pSPT18 (Roche, Mannheim, Germany) with 50 µCi α-32P-CTP. Signal detection was carried out using a PhosphoImager system (Fuji). GAPDH mRNA was detected on the same blot as a loading control.
Western blot
For western-blot analyses protein was precipitated according to the TRIzol® standard protocol and dissolved in 8 M urea buffer (50 mM phosphate, 200 mM NaCl, pH 8.0). Western blot was done essentially as described previously (27,48). After SDS gel electrophoresis and transfer of proteins to PVDF membranes, 5% milk powder dissolved in TBS buffer containing 0.1% Tween-20 was used to block unspecific binding. The following primary antibodies were used: DO1 mouse anti-p53 (1:2500, DO1, Calbiochem, Darmstadt, Germany), mouse anti-PANK1 (1:1000, Abnova, Taipei, Taiwan), mouse anti-CDK6 (1:500, B-10, SantaCruz Biotech., Santa Cruz, CA, USA), rabbit anti-p130 (1:1000, C-20, Santa Cruz Biotech.), mouse anti-β-actin (1:5000, SIGMA-Aldrich) and rabbit-anti-GAPDH (1:5000, SIGMA-Aldrich). Secondary antibodies were rabbit anti-mouse (1:5000, Thermo Scientific, Waltham, MA, USA) and mouse anti-rabbit (1:5000, Thermo Scientific).
Chromatin immunoprecipitation
HCT116 p53+/+ cells were treated with doxorubicin for 24 h or left untreated as a control. Cellular proteins were fixed in 1% formaldehyde for 10 min. Chromatin immunoprecipitations were essentially done as described earlier (49–51). The monoclonal DO1 p53 antibody was used for precipitation of p53 and a non-targeting mouse antibody was used as a control for unspecific signals. We also included controls in which precipitation was carried out without antibody. For all precipitations Protein G Dynabeads (Invitrogen) were used. qPCR was carried out with the QuantiTect SYBR Green PCR kit (QIAGEN) and all primers were checked for equal amplification efficiency. All PCR results were normalized to input controls. ChIP primers are listed in Supplementary Tables.
Luciferase assays
Twenty-four hours after transfection, SaOS-2 cells were harvested with Passive Lysis Buffer (Promega). Firefly and Renilla luciferase activity were measured with the Dual Luciferase Assay system (Promega). Relative light units were calculated by normalizing firefly luciferase values with Renilla luciferase activities. All measurements were performed in triplicate and repeated independently at least three times. Regulation by p53 was calculated by dividing the relative light units of the p53wt-transfected wells by the p53mut (p53R175H)-transfected wells.
RESULTS
Identification of p53-regulated transcripts with intronic miRNAs
Intronic miRNAs have been shown to be frequently co-expressed with their host genes (42). These miRNAs could also have similar regulation patterns as their protein-coding host genes. We were interested in finding p53 target genes with intronic miRNAs. To this end, we carried out DNA microarray analyses with RNA from D53wt cells which carry an inducible system for wild-type p53 in a functionally p53-negative cellular background. Induction of transgenic p53 is detectable already 6 h after removal of tetracycline (Supplementary Figure S1A). Cell cycle arrest and apoptosis induced by p53 was monitored by FACS (Supplementary Figure S1B). In order to measure early effects of p53, we hybridized RNA from cells which were induced for 9 h and from non-induced control cells on DNA microarrays representing all human mRNAs. In this initial experiment, we identified 1425 transcripts which were detected significantly (P-value cutoff <0.01) and regulated more than 3-fold (Supplementary Table S1). By comparison of these potential direct targets with the chromosomal localization of all miRNAs in the human genome [miRBase release 14 (52)], 23 candidate genes were identified (Supplementary Table S2). Interestingly, we observed downregulation of MCM7 harboring the miRNA-25, -93, -106b cluster which was shown to be regulated by p53 (44). The PANK1/miRNA-107 gene locus was chosen for further studies since miRNA-107 had been reported to induce cell cycle arrest in G1 upon overexpression (53).
Regulation of PANK1 and miRNA-107 after induction of p53
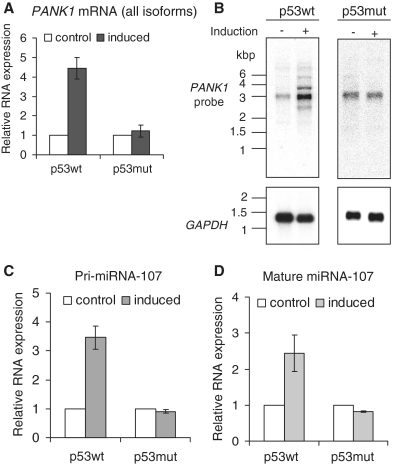
miRNA-107 is encoded in intron 5 of the PANK1 gene. PANK1 is transcribed into three different isoforms (PANK1α, β and γ). PANK1α is generated through an alternative transcription start upstream of the PANK1β and PANK1γ transcription start site (46). PANK1β and γ only differ in exon 4 which is spliced out in the PANK1γ isoform. In order to verify the results of the microarray experiment, regulation of all PANK1 isoforms in D53wt cells was tested. Induced expression of wild-type p53 led to a 4.5-fold increase of PANK1 mRNA (Figure 1A). As a control, the DNA-binding-deficient p53 mutant R175H was induced in the same cell system (D53mut) yielding no significant changes in expression of PANK1. Then we addressed the question whether p53 induces all isoforms or has only an impact on specific ones. Poly A-enriched RNA preparations of D53wt and D53mut cells before and after induction of the transgene were analyzed by northern-blot assays. The RNA probe directed against all PANK1 isoforms detected transcripts of four different lengths which show an increased expression after p53 induction (Figure 1B). The estimated lengths of the RNA molecules correspond well to the annotated PANK1 isoforms (PANK1γ, 2525 bp; PANK1β, 2702 bp; PANK1α, 3367 bp). The upper band at ∼5 kb does not correspond to a known isoform of PANK1. As a negative control, induction of p53 mutant protein does not change the expression of any detected RNA (Figure 1B).
Figure 1.
Regulation of PANK1 mRNA and miRNA-107 after induction of p53wt. D53wt or D53mut cells were induced for 9 h and total RNA was extracted. (A) PANK1 mRNA was measured by real-time RT–PCR using primers which detect all PANK1 isoforms. GAPDH expression was used for normalization. (B) Northern analyses of PANK1 isoforms after induction of p53wt and p53mut. An in vitro transcribed 32P-labeled RNA probe was employed for detection of PANK1 isoforms in poly A-enriched RNA preparations. GAPDH mRNA was detected on the same blots as a control for equal loading. (C) Pri-miRNA-107 was measured by RT–PCR with primers located in intron 5 of the PANK1 gene. Fold-changes in RNA expression relative to the non-induced cells are shown. (D) Real-time RT–PCR analyses of mature miRNA-107 expression in D53 cells. The small snoRNA U48 served as an endogenous control and was used for normalization. All RT–PCR results show averages and standard deviations of three independent experiments.
Expression of all PANK1 isoforms is expected to concomitantly lead to an increase of precursor miRNA-107 levels. Thus, we measured pri-miRNA-107 levels by real-time RT–PCR and detected an increase after induction of wild-type p53 but not after induction of p53 mutant protein (Figure 1C). Consistently, also the mature miRNA-107 is differentially expressed after p53 induction (Figure 1D). These analyses indicate that p53 upregulates miRNA-107 in tandem with PANK1 mRNAs. The regulation of the PANK1/miRNA-107 gene locus is dependent on a functional p53 DNA-binding domain.
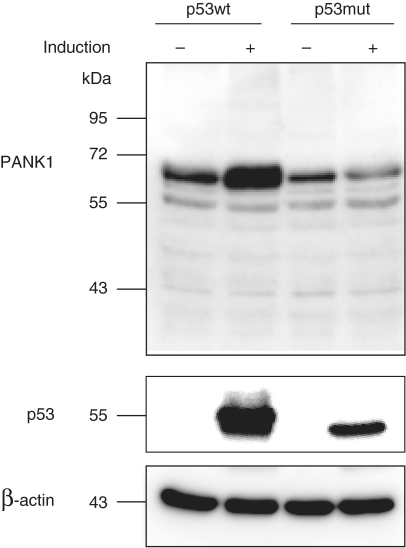
PANK1 protein is also upregulated by p53 (Figure 2). Interestingly, PANK1 protein whose molecular weight corresponds to the PANK1α isoform (∼64 kDa) appeared to be the only protein isoform induced by p53 and detected with a PANK1-specific antibody (Figure 2). In order to ascertain that the antibody employed recognizes all isoforms, we transfected expression plasmids coding for PANK1α, β and γ into HCT116 cells. Western analysis showed that all isoforms are detected by this antibody (Supplementary Figure S2). Thus, p53 upregulates the PANK1α protein. The other isoforms may be expressed and could also be regulated, but their expression is below the detection limit.
Figure 2.
PANK1 protein regulation after induction of p53. Protein extracts from D53wt and D53mut cells were used for western-blot analyses. PANK1 was detected before and after induction of p53wt and p53mut. As a control, the induction of the transgenes and the expression of β-actin were detected on the same blot.
Regulation of PANK1 and miRNA-107 is also observed with endogenous p53 levels
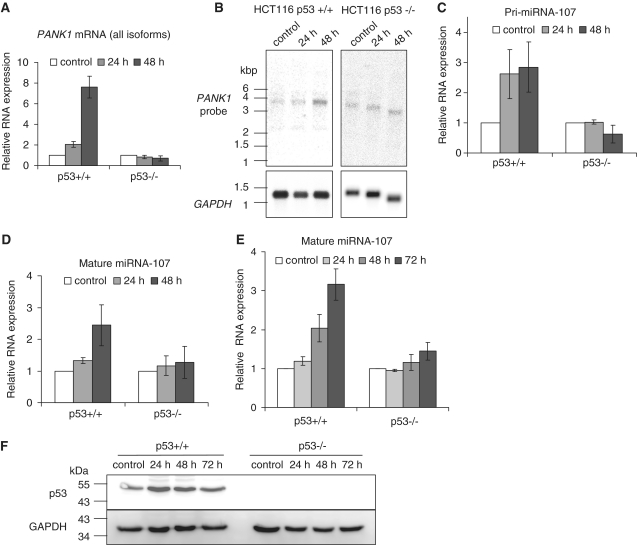
In order to investigate the regulation of PANK1 and miRNA-107 in another well-controlled cell system we used HCT116 p53+/+ and HCT116 p53−/− knockout cells. Induction of PANK1 and miRNA-107 was tested after doxorubicin-induced DNA damage. This treatment leads to stabilization of p53 expressed from the endogenous alleles. PANK1 mRNA is upregulated in HCT116 p53+/+ cells after 48 h of doxorubicin treatment, but not after treatment of HCT116 p53−/− cells (Figure 3A). Northern analyses detected only one isoform which is upregulated after DNA damage in these cells. The length of the RNA corresponds to PANK1α (Figure 3B). Expression of pri-miRNA-107 and mature miRNA-107 is upregulated in HCT116 p53+/+ after doxorubicin treatment, but remains essentially unchanged in HCT116 p53−/− cells (Figure 3C and D). In order to test whether p53 stabilization alone is sufficient to induce miRNA-107 expression, we treated HCT116 cells with the MDM2 inhibitor nutlin-3 (54). Also after nutlin-3-induced p53 stabilization mature miRNA-107 is upregulated in HCT116 p53+/+ cells but not in p53−/− knockout cells (Figure 3E). Thus, p53 is able to regulate mature miRNA-107 expression in the absence of other stress signals like DNA damage.
Figure 3.
PANK1 mRNA and miRNA-107 regulation in HCT116 cells after doxorubicin treatment. (A) HCT116 p53wt+/+ and p53wt−/− cells were treated with doxorubicin and harvested at different time points. PANK1 mRNA was measured and normalized to GAPDH expression. (B) HCT116 polyA-enriched RNA was subjected to northern-blot analyses. 32P-labeled RNA probes targeted against all three isoforms were used for hybridization. GAPDH mRNA was detected on the same blot to control for loading. (C) pri-miRNA-107 was measured after doxorubicin treatment of HCT116 cells. Untreated cells were used as a control. (D) Mature miRNA expression in HCT116 cells after treatment. Expression of snRNA U6 was used for normalization of small RNA expression. (E) miRNA-107 expression after nutlin-3 treatment of HCT116 cells harvested at different time points. All data represent the average and standard deviation of three independent experiments. (F) Western analyses were carried out to control for stabilization of p53 after nutlin-3 treatment of HCT116 cells.
The p53-dependent regulation of pri-miRNA-107 indicates a regulation on the transcriptional level. However, it has been shown that p53 is able to induce miRNA expression by enhancing Drosha/DGCR8-complex-dependent pre-miRNA processing of specific miRNAs (55). This seems not to be the case for miRNA-107 because pri-miRNA-107 is upregulated in D53wt and HCT116 p53+/+ cells. In order to control for non-specific effects caused by the doxorubicin treatment protocol, pri-miRNA-145 and pri-miRNA-34 expression were compared. It would be expected that pri-miRNA-145 levels remain unchanged after treatment because it is controlled through pre-miRNA processing, and pri-miRNA-34 was described to be upregulated by p53 on the transcriptional level. We observed the differential expression with an upregulation of pri-miRNA-34 and pri-miRNA-145 remaining essentially unchanged after doxorubicin treatment, which is consistent with earlier observations (55) (Supplementary Figure S3).
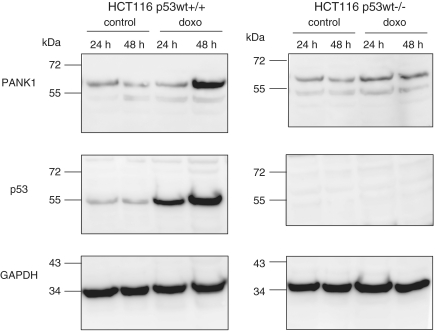
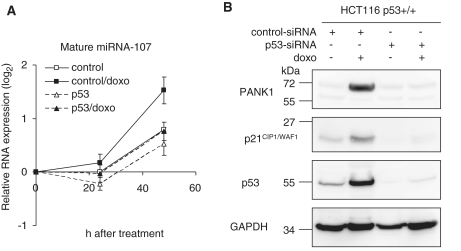
PANK1 protein levels before and after doxorubicin treatment of HCT116 cells were also tested. Consistent with the mRNA levels only PANK1α appeared differentially expressed (Figure 4). PANK1α increased after 48 h of doxorubicin treatment in HCT116 p53+/+ cells. In p53−/− knockout cells, PANK1α expression remained almost constant, indicating that its regulation is dependent on functional p53 (Figure 4). In another experimental approach we transfected a p53-targeting siRNA into HCT116 p53+/+ cells and treated them with doxorubicin. miRNA-107 induction in response to this treatment after 48 h was clearly impaired compared to the controls when p53 siRNA was transfected (Figure 5A). Western analyses show that PANK1α protein was upregulated after 48 h of doxorubicin treatment in cells transfected with the control siRNA (Figure 5B) but not after transfection with p53 siRNA. These observations confirm the p53-dependent regulation of the PANK1/miRNA-107 gene locus in an isogenic cellular context. Control experiments show that p53 expression was efficiently blocked by the p53 siRNA and regulation of the known target gene p21CIP1/WAF1 was abrogated after knockdown of p53 (Figure 5B). Taken together, these results show that expression of PANK1 and miRNA-107 is dependent on functional p53.
Figure 4.
Regulation of PANK1 protein expression in HCT116 cells. Cells were treated with doxorubicin for the indicated time or left untreated. Whole cell lysates were used for western analyses. The p53 was detected on the same blot to control for its successful stabilization after doxorubicin treatment. β-Actin served as a loading control.
Figure 5.
Regulation of miRNA-107 and PANK1 protein after knockdown of p53. (A) HCT116 p53+/+ cells were transfected with a p53-targeting siRNA (p53) or control siRNA (control). After 24 h, cells were treated with doxorubicin for different time periods. miRNA-107 was measured and normalized to the snRNA U6 control. RNA expression was calculated as the log2 transformed ratio relative to the 0 h time point. (B) Western-blot analyses of PANK1 after 48 h of doxorubicin treatment. As a control, p53, p21CIP1/WAF1 and GAPDH were detected on the same blot.
The PANK1/miRNA-107 gene locus is a direct target of p53
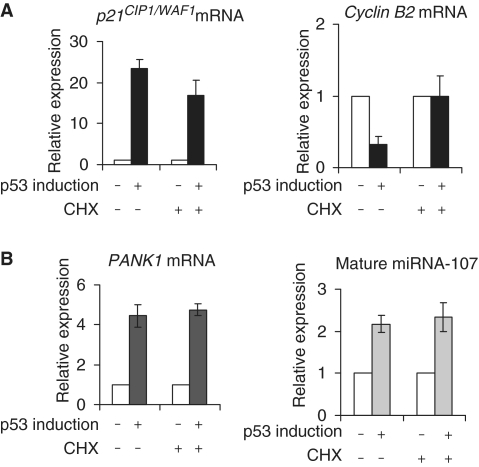
Our results indicated that PANK1 and miRNA-107 are regulated at the transcriptional level. Direct p53 targets are also expected to be regulated when translation is blocked. However, indirect target genes are expected to depend on regulation of intermediate proteins by p53. Inhibition of translation should alleviate regulation of these target genes. As positive and negative controls, we tested two p53 targets with or without treatment of the cells with the translation inhibitor cycloheximide (Figure 6A). The direct transcriptional target gene p21CIP1/WAF1 is upregulated after induction of wild-type p53 irrespective of functional translation. As an example for a gene indirectly controlled by p53, Cyclin B2 was tested. Cyclin B2 mRNA is downregulated in the control experiment but is not significantly changed when translation has been inhibited (Figure 6A). Interestingly, RNA levels of PANK1 and mature miRNA-107 are induced after p53 expression independent of functional translation (Figure 6B). This supports the notion that the PANK1/miRNA-107 gene locus is a direct target of p53.
Figure 6.
PANK1 and miRNA-107 are direct target genes of p53. D53wt cells were induced for 3 h, treated with cycloheximide (CHX) and incubated another 6 h. As a control, D53wt cells were induced but not treated with CHX. (A) mRNA induction of the direct target gene p21CIP1/WAF1 and downregulation of indirectly controlled Cyclin B2 mRNA. (B) PANK1 mRNA and mature miRNA induction were investigated by RT–PCR with or without prior CHX treatment. The fold regulations of the transcripts relative to the respective controls are shown.
Bioinformatic analyses of putative promoter regions in the PANK1 gene
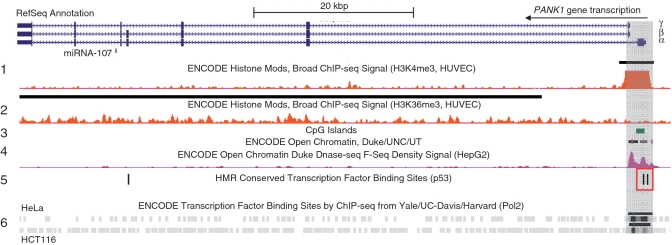
Promoter regions exhibit characteristic epigenetic modifications like histone H3K4 trimethylation (H3K4me3), a high sensitivity to DNase treatment and/or an accumulation of CpG islands. We searched several data sets from genome-wide screening experiments provided by the UCSC Genome Browser (56–58) for hints regarding potential promoters in the PANK1/miRNA-107 locus (Figure 7). Interestingly, only the region surrounding the transcriptional starts of PANK1α, β and γ exhibits classical promoter properties. H3K4me3 peaks at exon 1 of PANK1α and DNase sensitivity shows two peaks corresponding to the regions just upstream of the two different transcriptional start sites. PANK1α exon 1 displays a high-GC content with an annotated CpG island. However, these data do not show whether the CpG island is functional or not. Generally, transcribed regions are associated with histone H3K36 trimethylation. This is observed throughout the whole gene. Interestingly, there are two conserved p53-binding sites overlapping with the first exon of PANK1α (Figure 7, red box). Chromatin immunoprecipitation combined with high-throughput DNA sequencing in HCT116 and HeLa cells indicates recruitment of RNA polymerase II to the genomic region adjacent to the p53-binding sites and the transcriptional start sites. There is no evidence for intronic promoters in this gene. The bioinformatic analyses support the hypothesis that miRNA-107 expression is controlled by the promoter of the PANK1 host gene.
Figure 7.
Analyses of potential promoter regions in the PANK1 gene responsible for p53-dependent regulation. Gene structure of PANK1 from the RefSeq annotation is shown together with data from the UCSC Genome Browser. Signals from H3K4me3 (track 1) and H3K36me3 (track 2) ChIP-Seq analyses of HUVEC cells show putative promoter and transcribed regions. CpG islands are marked in green (track 3) and open chromatin in violet (track 4). Conserved p53-binding sites in the PANK1 gene are indicated as black bars (track 5) and two of them overlap with H3K4me3 (red box). RNA polymerase ChIP-Seq signals from HeLa and HCT116 cells are shown at the bottom (track 6). The genomic region harboring the functional PANK1 promoter is shaded in grey.
The p53 is recruited to the basal PANK1 promoter after DNA damage
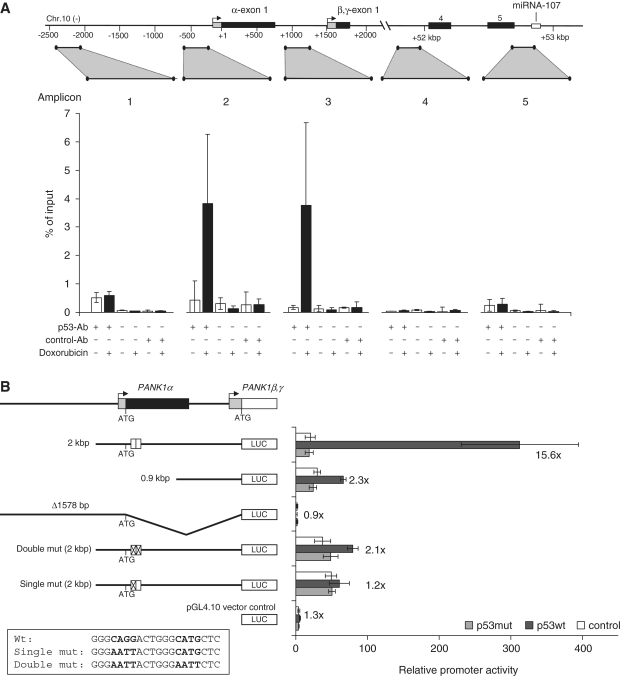
In order to test if p53 can bind to the potential promoter region of the PANK1/miRNA-107 locus, chromatin immunoprecipitations were carried out before and after induction of DNA damage in HCT116 cells. From the precipitates several regions were PCR-amplified from the PANK1 gene (Figure 8A). No differential p53 recruitment in the region 2200-bp upstream of the PANK1α start codon was measured (amplicon 1). Interestingly, p53 is recruited specifically to the region near the PANK1α, β and γ transcriptional start sites after induction of DNA damage as shown with detection of amplicons 2 and 3. We measured no differential recruitment near miRNA-107, neither to intron 3 nor intron 5 (Figure 8A). These results indicate that p53 is recruited to the PANK1 promoter region and not to intronic DNA segments close to miRNA-107.
Figure 8.
(A) Differential recruitment of endogenous p53 on the PANK1/miRNA-107 gene locus before and after DNA damage. HCT116 cells were treated with doxorubicin for 24 h. The p53 was precipitated from cross-linked chromatin (p53-Ab). PANK1 genomic regions were amplified by real-time qPCR. All signals are given relative to input. All primer pairs were tested for equal amplification efficiencies. A no-antibody control and a non-targeting isotype control (control-Ab) were used to determine nonspecific signals. Averages and standard deviations of two independent experiments are shown. Each genomic region was measured at least twice. (B) Regulation of the PANK1 promoter by p53. Promoter constructs were cotransfected with expression plasmids for p53wt, p53mut (R175H) or pcDNA3.1HisBlacZ as a control. The relative promoter activity normalized to Renilla luciferase expression is given. Fold-changes were calculated relative to the p53mut transfection. As a control, promoterless pGL4.10 was cotransfected with a p53-expressing plasmid to control for background luciferase activity. The p53-binding site mutants are indicated at the bottom.
The PANK1 promoter is inducible by p53
In order to test whether the PANK1 promoter is inducible by p53, a 2021-bp fragment starting upstream of the PANK1β and γ start codon was cloned into a reporter vector. Cotransfection experiments with p53wt and p53R175H-expressing plasmids (p53mut) in p53-negative SaOS-2 cells were carried out. The full-length 2-kbp construct was induced 15.6-fold when coexpressed with p53wt relative to the promoter activity obtained with p53mut (Figure 8B). A truncated promoter (0.9 kbp) showed almost no induction after p53wt expression. We also cloned 2026 bases upstream of the PANK1α start codon (Δ1578 bp) which also encompasses a putative p53-binding site 1874 bases upstream from the start codon as predicted by p53FamTaG (59). However, this promoter construct exhibited no activity and was not inducible by p53 (Figure 8B). Furthermore, we tested if a putative intronic promoter directly upstream of miRNA-107 drives its expression. However, neither activity nor inducibility of this construct indicated that miRNA-107 is regulated by an additional intronic promoter (data not shown), which further supports the notion that miRNA-107 expression is driven by the fragment starting upstream of the PANK1β and γ start codon.
Following these results, we searched for p53 binding sites in the PANK1 promoter construct upstream of the PANK1β, γ start codon. Two putative-binding sites were predicted based on comparison between human, mouse and rat sequences (Figure 7, red box). One of these two sites was not functional (data not shown). However, after mutation of the core regions of the other p53-binding motif, inducibility of the promoter construct was almost lost (Figure 8B). Even mutation of one of the two core regions diminishes the ability of p53wt to induce the promoter. Taken together, the results imply that the PANK1/miRNA-107 locus is directly regulated by p53 and that this regulation is mediated through p53 recruitment to the PANK1 promoter.
Bioinformatic prediction of miRNA-107 targets
After investigating the mechanism of PANK1/miRNA-107 locus regulation, we focused on possible miRNA-107 functions in the context of p53 signaling. Different algorithms for in silico prediction of miRNA target genes exist. One single algorithm mostly predicts hundreds of putative targets. In order to yield a low false positive rate, we employed the three prediction algorithms miRanda (60), PicTar (61) and Targetscan (62). We found 115 putative targets which were common to predictions by the three algorithms (Supplementary Table S3). These results were subjected to gene ontology analyses employing the FatiGO algorithm (63). We noticed 17 terms which were significantly enriched in the list of putative targets, among them were the terms ‘cell cycle’, ‘cell division’ and ‘regulation of cell cycle’ (Supplementary Table S4). Thirteen cell cycle genes were identified with these terms: CDK6, PNN, NEDD9, KIF23, FGF2, LATS2, SUFU, MTSS1, RASSF5, CDC37L1, NF1, MYB and YWHAH. One of these is the cyclin-dependent kinase 6 (CDK6). The protein product of the gene initiates G1/S progression when bound by cyclin D.
Important G1/S cell cycle phase proteins are regulated by miRNA-107 by interference with translation
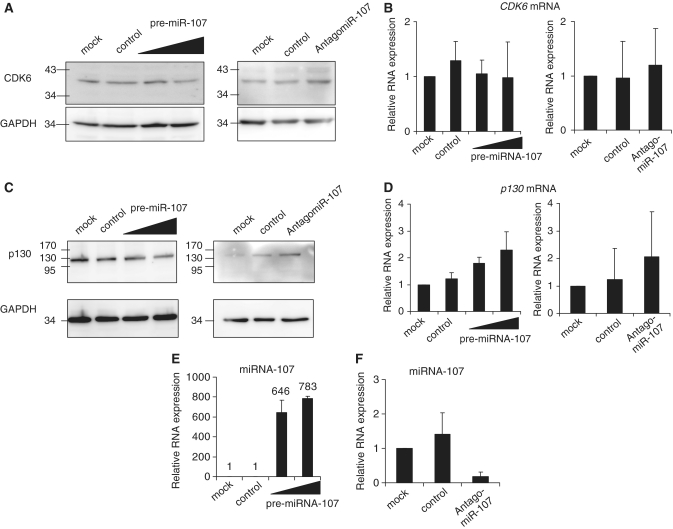
CDK6 had already been implicated as a miRNA-107 target (53). Expression of the kinase was assessed only after overexpressing miRNA-107, and it was left open whether CDK6 is regulated solely at the protein level or if CDK6 mRNA degradation is induced by miRNA-107. In order to characterize the regulation of CDK6 by miRNA-107, we transfected synthetic miRNA-precursors into D53wt cells and analyzed protein expression. CDK6 protein expression appeared slightly lower after transfection of pre-miRNA-107 compared to the control-pre-miRNA (Figure 9A, left panel) which is in agreement with earlier reports (53). The inverse experiment knocking down endogenous miRNA-107 with specific AntagomiR-107 yielded higher protein expression of CDK6 compared to the controls (Figure 9A, right panel). In order to ensure that changes in CDK6 protein levels are not due to a shift in cell-cycle distribution, we performed FACS analyses. Under the experimental conditions employed, the cell cycle-distribution of D53wt cells remained constant (Supplementary Figure S4). CDK6 mRNA was essentially unchanged under all experimental conditions (Figure 9B). Therefore, it is likely that CDK6 is regulated by interference with translation and not by initiation of RNA degradation.
Figure 9.
Regulation of CDK6 and p130 gene expression by miRNA-107. (A) D53wt cells were transfected with increasing amounts of pre-miRNA-107, with a control-pre-miRNA (control) or without RNA (mock). For knockdown experiments D53wt cells were transfected with AntagomiR-107 and a related control. CDK6 protein was detected by western analyses from whole cell lysates. GAPDH expression served as a loading control. (B) CDK6 mRNA was measured by real-time RT–PCR in transfected D53wt cells. Expression values from three independent experiments were normalized to GAPDH expression. (C) Western analysis of p130 protein expression in transfected D53wt cells and GAPDH control. (D) The p130 mRNA was measured in total RNA samples from transfected D53wt cells. (E) and (F) As a control, miRNA-107 levels in controls and pre-miRNA-107- or AntagomiR-107-transfected cells were assayed. Averages and standard deviations from three independent experiments are shown.
Furthermore, we tested whether other proteins which are important for progression from G1 to S phase are also under control of miRNA-107 like the RB-related pocket protein p130 (RBL2). Analysis of protein expression showed that p130 levels decrease after overexpression of miRNA-107. In contrast, AntagomiR-directed knockdown of endogenous miRNA-107 leads to an elevated p130 expression compared to the control (Figure 9C). Expression of p130 mRNA increased slightly after miRNA-107 overexpression and knockdown of miRNA-107 (Figure 9D). Thus, its expression is not correlated with protein expression. Control RT–PCR experiments showed that overexpression and knock-down of miRNA-107 was successful (Figure 9E and F). In order to investigate whether CDK6 and p130 are directly targeted by miRNA-107 we cloned putative miRNA-107-binding sites into the 3′-UTR of the luciferase gene in the pGL4.10 reporter vector. We used the RNAhybrid algorithm (64) to predict miRNA-107-binding sites in the p130 3′-UTR. One region was identified which could serve as such a binding site and we cloned it into the 3′-UTR of a luciferase gene. From the CDK6 3′-UTR one conserved element was chosen which had been investigated earlier (55). Overexpression of miRNA-107 led to a decrease of CDK6 reporter activity (Supplementary Figure S5). This supports the notion that CDK6 is directly controlled by miRNA-107. The p130 UTR construct, however, only slightly changed reporter activity after overexpression of miRNA-107. Knockdown of miRNA-107 had no effect on either reporter constructs. In summary, the results indicate that CDK6 and p130 protein levels are under control of miRNA-107 but p130 may not be a direct target of this miRNA.
DISCUSSION
The tumor suppressor p53 regulates transcription of numerous genes including miRNA genes. We showed that p53 is able to coregulate intronic miRNA-107 by transcriptional induction of its host gene PANK1. PANK1 mRNA and protein appear to be regulated in different cell systems. In HCT116 cells we could show that p53 stabilization even in the absence of stress signals is sufficient to induce miRNA-107. The PANK1/miRNA-107 gene is a direct target of p53 and is transcriptionally induced by specific recruitment of p53 to the PANK1 promoter after induction of DNA damage. We identified a conserved p53-binding site in the PANK1 promoter which overlaps with exon 1 of the PANK1α isoform. An intronic promoter which drives miRNA-107 expression under normal conditions, explicitly in a regular non-DNA damage situation, has been described very recently (65). However, here we observed that p53 is neither recruited to this genomic region after DNA damage nor is this DNA segment able to induce expression in a reporter assay. Taken together, the collected data is consistent with the idea that the p53-inducible promoter is located in the genomic region upstream of the transcription start site of the PANK1β and γ isoforms.
While this article was in preparation, a report was published which identified a p53-binding site 1811-bp away from the transcriptional start site of PANK1α as being responsible for p53-dependent induction of the PANK1 gene (66). This binding site is predicted by the p53FamTaG algorithm and is included in the Δ1578 construct which was investigated here in reporter assays. However, the region upstream of the PANK1α start codon has no significant promoter activity. Basal promoter activity is generated by the 0.9-kbp upstream of the PANK1β, γ start codon. Furthermore, in our assays this binding site neither contributes to induction of the PANK1 promoter activity nor is p53 recruited to this genomic position after DNA damage. Our data suggest that p53 regulates the PANK1 promoter through binding to a conserved binding site downstream of the PANK1α start codon.
PANK1 protein is involved in coenzyme A biosynthesis which is of great importance to the maintenance of metabolic homeostasis (46,67). It catalyses the rate-limiting step of coenzyme A biosynthesis. The interesting observation that the PANK1α protein is induced after DNA damage in a p53-dependent manner raises the exciting question of a possible influence of p53 on coenzyme A levels in the cell. PANK1α transcription is also regulated by PPARα which leads to higher PANK1 enzyme activity (46). Thus, a p53-dependent induction of PANK1α could lead to increased coenzyme A levels in the cell. Coenzyme A is needed for a plethora of metabolic and anabolic pathways. As an example, the tricarboxylic acid cycle drives the acetyl-coenzyme A-dependent generation of NADH which is oxidized in the mitochondrial electron transport chain. An increased PANK1α level could guarantee the supply with coenzyme A for this metabolic pathway under stress conditions. The p53 target gene guanidinoacetate methyltransferase (GAMT) initiates fatty acid β-oxidation (33), a metabolic pathway leading to fatty acid degradation by successive oxidation and transfer of acetyl groups to coenzyme A. PANK1 induction by p53 could be important for the maintenance of this alternative energy supply pathway. Interestingly, some tumors have a lower coenzyme A level as the corresponding normal tissue (68).
Finally, we identified two proteins regulated by miRNA-107, CDK6 and the RB-related 2 gene RBL2 (p130). After knockdown of miRNA-107 CDK6 and p130 protein levels slightly increase. Also a reduced expression of CDK6 and p130 on the protein level is observed upon miRNA-107 overexpression. The small extend of protein repression has been observed for numerous targets by proteome analyses. Quantification of protein expression by SILAC after miRNA overexpression or miRNA knockdown often yields numerous regulated targets. Only some of them were changed >2-fold (14,15). This indicates that miRNAs usually fine-tune protein expression. miRNAs mediate their effects through binding sites in the 3′-UTR of target mRNAs. We identified a binding site in the CDK6 3′-UTR which downregulates reporter activity upon overexpression of miRNA-107. A predicted miRNA-107-binding site in the p130 UTR, however, did not change reporter activity. This indicates that p130 may be indirectly targeted by miRNA-107, through the regulation of other proteins. We also performed knockdown of miRNA-107 together with cotransfection of UTR reporter constructs, but luciferase reporter activity did not change compared to controls (data not shown). The 5-fold downregulation of endogenous miRNA-107 by the AntagomiR employed appears not to be sufficient to induce a significant increase in luciferase protein which is transcribed from an exogenously introduced plasmid.
CDK6 and p130 exert important functions in the progression from G1 to S phase. p53 mediates a very fast cell cycle arrest. This function is mediated by numerous target genes, for example by induction of p21CIP1/WAF1 (23) or repression of cyclin B (27) and many other cell cycle genes. The regulation of miRNA-107 and the resulting decrease in CDK6 and p130 protein could therefore assist cell cycle arrest at the G1/S transition. In fact, overexpression of miRNA-107 has been shown to induce G1/S cell cycle arrest in human non-small cell lung cancer cell lines (53). Additionally, the data presented here show another way of p53-dependent repression of important cell-cycle regulators which is not mediated by transcriptional effects but by the interference of a regulated miRNA with translation. Although the effect of miRNA-107 on individual proteins is rather low, the synergistic regulation of several targets acting in the same pathway could lead to cell cycle deregulation. It should be noted, however, that also other p53-dependent regulatory pathways may exist leading to altered expression of CDK6 and p130 proteins, making it difficult to determine miRNA-107-specific contributions following p53 activation.
miRNA-107 was recently shown to inhibit tumor angiogenesis through regulation of the target gene HIF1β (66). Downregulation of HIF1β protein led to decreased VEGF production which significantly reduced angiogenesis in a tumor xenograft mouse model. These results show that miRNA-107 acts as a tumor suppressor. Together with data described here, this hypothesis is further supported since central cell-cycle proteins are repressed by miRNA-107.
The data presented here provide a novel p53-dependent pathway for cell cycle protein repression. It sheds light on the complex regulation patterns which have evolved in the human genome. As the transcriptome displays an amazing complexity, the emerging underlying regulatory pathways may be more interwoven than appreciated to date.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
formel.1 Junior research grant (to L.B.); Interdisciplinary Center for Clinical Research (IZKF) Leipzig and the TransGenomix project (EU, SMWK) (to K.E.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are indebted to Carola Koschke, Andrea Rothe and Jana Lorenz for expert technical assistance, Suzanne Jackowski for kindly sending PANK1α cDNA and Bert Vogelstein for generously providing inducible cell lines and plasmids. We would like to thank Knut Krohn and Andreas Lösche together with their teams at the IZKF Leipzig for performing microarray and FACS analyses.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 10.Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- 11.Kiriakidou M, Tan GS, Lamprinaki S, Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 13.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 16.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–2597. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 20.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.el Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 22.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 23.el Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 24.Krause K, Haugwitz U, Wasner M, Wiedmann M, Mossner J, Engeland K. Expression of the cell cycle phosphatase cdc25C is down-regulated by the tumor suppressor protein p53 but not by p73. Biochem. Biophys. Res. Commun. 2001;284:743–750. doi: 10.1006/bbrc.2001.5040. [DOI] [PubMed] [Google Scholar]
- 25.Rother K, Kirschner R, Sanger K, Bohlig L, Mossner J, Engeland K. p53 downregulates expression of the G1/S cell cycle phosphatase Cdc25A. Oncogene. 2007;26:1949–1953. doi: 10.1038/sj.onc.1209989. [DOI] [PubMed] [Google Scholar]
- 26.Rother K, Dengl M, Lorenz J, Tschop K, Kirschner R, Mossner J, Engeland K. Gene expression of cyclin-dependent kinase subunit Cks2 is repressed by the tumor suppressor p53 but not by the related proteins p63 or p73. FEBS Lett. 2007;581:1166–1172. doi: 10.1016/j.febslet.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Krause K, Wasner M, Reinhard W, Haugwitz U, Dohna CL, Mossner J, Engeland K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vousden KH, Ryan KM. p53 and metabolism. Nat. Rev. Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 29.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 30.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 31.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 32.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, el Deiry WS, Aaronson SA, Lee SW. GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol. Cell. 2009;36:379–392. doi: 10.1016/j.molcel.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 35.He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 37.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georges SA, Chau BN, Braun CJ, Zhang X, Dobbelstein M. Cell cycle arrest or apoptosis by p53: are microRNAs-192/215 and -34 making the decision? Cell Cycle. 2009;8:680–681. [PubMed] [Google Scholar]
- 41.Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with the spliceosome. Mol. Cell. Biol. 2009;29:3243–3254. doi: 10.1128/MCB.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death. Differ. 2010;17:452–458. doi: 10.1038/cdd.2009.188. [DOI] [PubMed] [Google Scholar]
- 44.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Zhang L, Hwang PM, Rago C, Kinzler KW, Vogelstein B. Identification and classification of p53-regulated genes. Proc. Natl Acad. Sci. USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramaswamy G, Karim MA, Murti KG, Jackowski S. PPARalpha controls the intracellular coenzyme A concentration via regulation of PANK1alpha gene expression. J. Lipid Res. 2004;45:17–31. doi: 10.1194/jlr.M300279-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 2009;4:37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- 48.Bohlig L, Metzger R, Rother K, Till H, Engeland K. The CCN3 gene coding for an extracellular adhesion-related protein is transcriptionally activated by the p53 tumor suppressor. Cell Cycle. 2008;7:1254–1261. doi: 10.4161/cc.7.9.5812. [DOI] [PubMed] [Google Scholar]
- 49.Friedrich M, Bohlig L, Kirschner RD, Engeland K, Hauschildt S. Identification of two regulatory binding sites which confer myotube specific expression of the mono-ADP-ribosyltransferase ART1 gene. BMC Mol. Biol. 2008;9:91. doi: 10.1186/1471-2199-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasner M, Tschop K, Spiesbach K, Haugwitz U, Johne C, Mossner J, Mantovani R, Engeland K. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 2003;536:66–70. doi: 10.1016/s0014-5793(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 51.Wasner M, Haugwitz U, Reinhard W, Tschop K, Spiesbach K, Lorenz J, Mossner J, Engeland K. Three CCAAT-boxes and a single cell cycle genes homology region (CHR) are the major regulating sites for transcription from the human cyclin B2 promoter. Gene. 2003;312:225–237. doi: 10.1016/s0378-1119(03)00618-8. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 56.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D'Erchia AM, et al. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl. 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 62.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 63.Al Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl Acad. Sci. USA. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rock CO, Karim MA, Zhang YM, Jackowski S. The murine pantothenate kinase (Pank1) gene encodes two differentially regulated pantothenate kinase isozymes. Gene. 2002;291:35–43. doi: 10.1016/s0378-1119(02)00564-4. [DOI] [PubMed] [Google Scholar]
- 68.Rapp GW. Some systemic effects of malignant tumors. I. Co-enzyme A levels. Cancer. 1973;31:357–360. doi: 10.1002/1097-0142(197302)31:2<357::aid-cncr2820310214>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.