Abstract
Homing endonucleases recognize long target DNA sequences generating an accurate double-strand break that promotes gene targeting through homologous recombination. We have modified the homodimeric I-CreI endonuclease through protein engineering to target a specific DNA sequence within the human RAG1 gene. Mutations in RAG1 produce severe combined immunodeficiency (SCID), a monogenic disease leading to defective immune response in the individuals, leaving them vulnerable to infectious diseases. The structures of two engineered heterodimeric variants and one single-chain variant of I-CreI, in complex with a 24-bp oligonucleotide of the human RAG1 gene sequence, show how the DNA binding is achieved through interactions in the major groove. In addition, the introduction of the G19S mutation in the neighborhood of the catalytic site lowers the reaction energy barrier for DNA cleavage without compromising DNA recognition. Gene-targeting experiments in human cell lines show that the designed single-chain molecule preserves its in vivo activity with higher specificity, further enhanced by the G19S mutation. This is the first time that an engineered meganuclease variant targets the human RAG1 locus by stimulating homologous recombination in human cell lines up to 265 bp away from the cleavage site. Our analysis illustrates the key features for à la carte procedure in protein–DNA recognition design, opening new possibilities for SCID patients whose illness can be treated ex vivo.
INTRODUCTION
The generation of DNA double-strand breaks (DSBs) results in the activation of the DNA damage response. DSBs can be repaired by two major mechanisms: non-homologous end joining (NHEJ), an error-prone pathway that may generate short insertions or deletions at the cleavage site upon rejoining the ends of the DSBs, or by homologous recombination (HR). In contrast to NHEJ, HR uses a homologous DNA sequence as template and therefore is essentially an error-free mechanism for DSB repair. However, if the template includes extra sequences flanked by the homologous regions, HR repair results in sequence modification, and can be used for exchanging genetic information between endogenous and/or exogenous sequences (1). This characteristic of HR can be exploited in gene therapy to promote the exchange between an endogenous mutated chromosomal sequence and an exogenous repair DNA construct for gene-correction purposes. However, to trigger this mechanism, a very specific ‘tool’ that would provide a specific DSB close to or in the area of interest is essential. In the search to generate such an efficient and specific ‘tool’, homing endonucleases (HE) have emerged as perfect candidates due to their high specificity (≥12-bp target sequences), their low cleavage frequency in eukaryotic genomes and the fact that their action actively induces HR (2). Although several hundreds of HEs have been identified, the repertoire of cleavable sequences is limited. Consequently, HE engineering aimed at increasing the repertoire of recognized DNA sequences is necessary to obtain variants capable of accurately cleaving chosen sequences. Chimeric endonucleases resulting from the fusion of engineered zinc-finger domains with the catalytic domain of the FokI restriction enzyme have been used to induce targeted recombination in endogenous genes in human cells, Drosophila, plants, fish and other organisms (3–6). In a totally different approach, triplex-forming oligonucleotides (TFOs) have been conjugated with restriction enzymes (7) to target their cleavage activity to specific long sequences. The recent development of adapted protein-engineering strategies (8) has paved the way for the development of tailored endonucleases derived from natural HEs.
I-CreI is a well-characterized homodimeric member of the LAGLIDADG family of HE (9). This enzyme cleaves a 22-bp pseudo-palindromic target and, due to its large recognition sequence, minimizes the probability of generating nonspecific DSB in a mammalian genome, opening up the possibility to customize the enzyme for therapeutic purposes. The analysis of I-CreI bound to its natural target depicted the residues involved in direct interactions with seven nucleotides (10). The mutations of these DNA-interacting residues have been shown to induce the cleavage of novel targets in conjunction with the abolishment of cleavage at the natural site (8,11–14). An extensive study of the protein–DNA interactions of the I-CreI homodimer was used to generate libraries resulting from the randomization of two, three or even four amino acids that contact DNA bases, which were screened against all the potential cognate DNA sequences. Millions of combinations were screened, resulting in several hundreds of mutants with locally altered specificities that were later on used in a combinatorial strategy, and expanded to include homodimeric and heterodimeric variants of I-CreI (15). Following this approach, we previously achieved a redesigned meganuclease that targets the human XPC gene (13) in vivo. Energy calculations further helped to improve the engineering of meganuclease variants by highlighting key residues involved in target recognition and specificity (12,16,17).
In a similar way, we have generated several artificial meganucleases derived from the I-CreI that are capable of cleaving a DNA sequence from the human Recombination Activating Gene 1 (RAG1 gene) (8,14). The RAG1 gene product has been shown to form a complex with RAG2 that is responsible for the initiation of V(D)J recombination, an essential step in the maturation of immunoglobulins and T-lymphocyte receptors (18,19). Their inactivation causes severe combined immunodeficiency (SCID) due to the absence of T and B lymphocytes (20–22). SCID represents a model for monogenic diseases amenable to ex vivo treatment and certain types of SCID have already been treated using gene therapy (23). In this work, we have determined the crystal structure of the different variants of I-CreI that display high efficiency and specificity for RAG1 target cleavage, and thus analyzed the molecular basis for the new target DNA recognition by the engineered meganucleases. In vivo experiments demonstrate that these engineered enzymes are able to induce repair in the targeted sequences in human cells.
MATERIALS AND METHODS
Protein expression and purification
The I-CreI homodimeric mutants were cloned and expressed as in refs (13,24). The co-expression, purification and storage of the heterodimeric I-CreI derivatives were carried out as described (13). All the proteins were folded and possessed biophysical properties similar to those of the wild type (verified by circular dichroism and NMR; data not shown), and their oligomeric states in solution were identified by analytical ultracentrifugation.
Mass spectrometry
Mass determinations of intact proteins were performed (13) and the intact protein predominantly gave multiply protonated molecules corresponding to molecular masses of M = 19 634/19 906 Da (V2/V3), obtained by deconvoluting the multiply charged ions (data not shown) using MagTran software v.1.02 kindly provided by Dr Zhongqi Zhang (25). The mass determination of dissolved crystals was performed as described (13).
Biochemical and biophysical characterization
Circular dichroism (CD) measurements were performed as before (24). Analytical gel-filtration chromatography was done at room temperature with an ÄKTA FPLC system (GE) using a Superdex™ 200 10/300 GL column in 20 mM sodium phosphate buffer at pH 6.0 and 1 M NaCl 100 μl of the scV3V2(G19S) protein was injected at 0.3 mg/ml concentration and eluted from the column at a flow rate of 0.2 ml/min.
Heterodimer stability
The heterodimer dissociation and formation of homodimers were checked over a period of 3 days as described (13). Briefly, 4.5 mg of I-CreI V2V3 or V2(K7E-G19S)V3(E8K) heterodimers and I-CreI V2 His6-tagged homodimer as control were incubated at 37°C for 0, 1, 2, 4 and 72 h (lanes 1–5) (0 h for the control His6-tagged homodimer, lane 6). After incubation, the solutions were mixed with Strep-Tactin Superflow resin (IBA, Gmbh) or Talon Superflow resin (Clontech) loaded with Co2+ for 25 min at room temperature, then centrifuged and the supernatants loaded on 18% acrylamide-SDS gel to monitor the heterodimer dissociation. If homodimer formation takes place during that time, the homodimer containing the Strep-tag should not bind to the resin and must stay in the supernatant thus appearing as a band in the gel. The absence of this band demonstrates the stability of the selected heterodimer over this period of time. Lanes 7 and 8 correspond to the I-CreI V2 His6-tagged homodimer control and the I-CreI V2V3 heterodimers incubated without resin (Figure 1b). For all the heterodimers: V2 variants contain a His6 tail and V3 variants contain a Strep-tag tail. The quantification of the remaining heterodimers after incubation was performed by measuring the intensity of the bands in the scanned gels using ImageJ software (rsb.info.nih.gov/ij/).
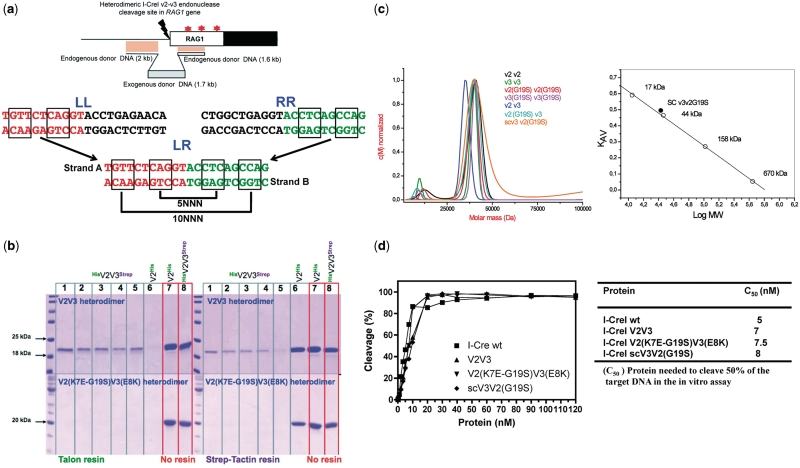
Figure 1.
Cleavage by I-CreI variants. (a) The upper panel displays the human RAG1 gene showing the V2V3 DSB site and the donor DNA construct used to verify the frequency of exogenous DNA integration, as a marker. In the genomic locus, the star shows the location of the mutation in the active core of the RAG protein that causes T-B-severe combined immunodeficiency or Omenn’s syndrome. In the lower panel—the LL: palindromic sequence cleaved by homodimer V2V2, RR: palindromic sequence cleaved by homodimer V3V3, LR: non-palindromic sequence cleaved by heterodimers V2V3 and V2(K7E-G19S)V3(E8K) and single-chain scV3V2(G19S). The LR sequence corresponds to a region of the human RAG1 gene (entry NM_000448.1 in NCBI nucleotide database) located 11 bp upstream of the coding exon (14). (b) Heterodimer stability. The possible formation of homodimers as a consequence of heterodimer dissociation was examined over a period of 3 days. 4.5 mg of V2V3 and V2(K7E-G19S)V3(E8K) (V2 His6-tagged and V3 Strep-tagged in both heterodimers) heterodimers (and the His6-tail V2V2 homodimer as control) were incubated at 37°C during 0 (lane 1), 1 (lane 2), 2 (lane 3), 4 (lane 4) and 72 (lane 5) h (zero hours for the control homodimer, lane 6). Lanes 7 and 8 correspond to the loading control of the V2V2 His6-tagged homodimer and the V2V3 His6- and Strep-tag heterodimer. After the mixing reaction, the samples were exposed to Talon or Strep-Tactin resin for 25 min, centrifuged and the supernatants loaded in an 18% acrylamide-SDS gel to monitor the heterodimer dissociation. If heterodimer dissociation occurs, the homodimer containing the Strep-tag (I-CreI V3 versions), should not bind to the Talon resin and must stay in the supernatant appearing as a band in the gel (left upper and lower panels). The same procedure is applied to test the formation of homodimers containing the His6-tag (I-CreI V2 versions) that do not bind to the Strep-Tactin resin and must stay in the supernatant, appearing as a band in the gel (right upper and lower panels). The absence of this band demonstrates the stability of the heterodimers over this period of time. (c) Left panel. Sedimentation velocity distribution measured by analytical ultracentrifugation of the I-CreI proteins. Right panel, calibration plot of the partition coefficient (KAV) versus the logarithm of the molecular mass using four protein standards (open circles) for an analytical Superdex 200 10/300 GL column. The values of the single-chain meganuclease are indicated with a filled circle (measured molecular mass = 34 kDa, theoretical molecular mass = 41 kDa). (d) Representation of the in vitro cleavage activity and C50 (concentration of enzyme required to cleave 50% of 2 nM of target DNA).
Analytical ultracentrifugation
The sedimentation velocity experiment was carried out as described (24).
In vitro cleavage assay conditions
The in vitro cleavage assay conditions were the same as those previously described (14).
Fluorescence polarization-binding assays
Solutions containing 25 nM 6-FAM-DNA and various concentrations (0–400 nM) of I-CreI proteins were prepared in a total volume of 50 μl binding buffer (10 mM Tris–HCl pH 8, 300 mM NaCl and 10 mM CaCl2). Following incubation at 37°C for 10 min, the fluorescence polarization was measured with a Wallac Victor2V 1420 Multilabel HTS counter (PerkinElmer). The dissociation constant (KD) was determined by non-linear least squares fitting (Prism, GraphPad).
Crystallization
Dynamic light scattering indicated essentially monodisperse solutions. The 24-bp-long RAG target DNA was purchased from Proligo and consisted of two strands of the following sequence: 5′-TTGTTCTCAGGTACCTCAGCCAGA-3′ and 5′-TCTGGCTGAGGTACCTGAGAACAA-3′, which form a 24-bp blunt-end duplex on mixing, heating and cooling. The protein–DNA complex was obtained in the presence of either 2 mM CaCl2 [for V2(K7E-G19S)/V3(E8K) and scV3V2(G19S)] or MgCl2 (for V2/V3) and MnCl2 [for scV3V2(G19S)] to obtain the bound and cleaved states of the target DNA, respectively, by pre-warming the meganuclease and the oligonucleotide samples at 37°C and mixing them in a 1.5:1 molar ratio (DNA:protein). The mixture was incubated for 50 min at this temperature, and then spun down for 5 min to remove insoluble material. The final concentration of protein in the DNA–protein complex solution was 4 mg/ml. Crystals were grown using the hanging-drop method at 290 K, in 2 μl droplets formed by 1 μl of the DNA–protein complex and 1 µl of precipitant solution consisting of 0.2 M ammonium acetate and 15% (w/v) PEG-4000 in 0.1 M sodium citrate pH 5.6 for V2(K7E-G19S)/V3(E8K)–DNA–Ca2+ and 40% (v/v) 1,2-propanediol, 0.05 M calcium acetate in 0.1 M calcium acetate pH 4.5, in the case of V2V3–DNA–Mg2+. The crystallization conditions for the scV3V2(G19S) were the same in the presence of Ca2+ or Mn2+ (20% (v/v) PEG 400, 0.1 M MES pH 6.5, 5% (v/v) ethanol and 10% (v/v) ethylene glycol). V2(K7E-G19S)/V3(E8K)–DNA–Ca2+ crystals were cryo-protected by adding 35% (v/v) methanol and 20% (v/v) glycerol to the mother liquor, while V2V3–DNA–Mg2+ and scV3V2(G19S) crystals were directly cooled in liquid nitrogen.
Data collection, structure solution, model building, refinement and structural analysis
All data were collected at 100 K, using synchrotron radiation at the PX beam line (SLS, Villigen, Switzerland). The diffraction pattern was recorded on a Mar225 CCD or Pilatus detectors. Data processing and scaling were accomplished with XDS (26) or MOSFLM (27) (see Supplementary Table SI). The structures were solved by molecular replacement as implemented in the programs MOLREP (28) or PHASER (29). The search model was based on a poly-alanine backbone derived from the PDB entry 1G9Z (I-CreI–DNA–Mg2+). The structures were then subjected to iterative cycles of model building and refinement with O (30), Coot (31) and PHENIX (32). The identification and analysis of the protein–DNA hydrogen Bonds and van der Waals contacts was done with the Protein Interfaces, Surfaces and Assemblies service PISA at the European Bioinformatics Institute (http://www.ebi.ac.uk/msdsrv/ prot_int/pistart.html).
Quantum chemical calculations
Taking cue from the mechanism proposed by Ivanov et al. for endonuclease IV (33), we built a reaction path connecting the crystallographic structures containing (i) the cleaved DNA and (ii) the non-cleaved DNA. The reaction path was obtained by formation of the phosphorus bond with the oxygen of the deoxyribose and cleaving the hydroxyl-phosphorus bond (Figure 4). At each step, all the atoms not directly involved in the reaction were fully relaxed. The system was divided into two parts using ONIOM (34). The quantum-mechanical part, formed by the phosphate group, the manganese ion and its ligands, was modeled via density functional theory using the B3LYP functional and a 6-311G(d,p) basis set. The non-quantum mechanical part, including all the atoms within a distance of 8 Å from the reacting phosphate, interacts with the QM subsystem by electronic embedding. After full relaxation, the reaction path (Figure 4) remained close to that of reference (33) confirming the validity of the initial guess. The potential energy profiles of the WT as well as the single- and double-G19S meganuclease mutants are reported in Figure 4. The energy differences between the penta-coordinate phosphorus and the non-cleaved DNA are 16 kcal/mol and 14 kcal/mol for the WT and G19S meganucleases, respectively. Interestingly, the double mutation, which experimentally hampers the catalysis, shows the highest energy barrier (17.5 kcal/mol).
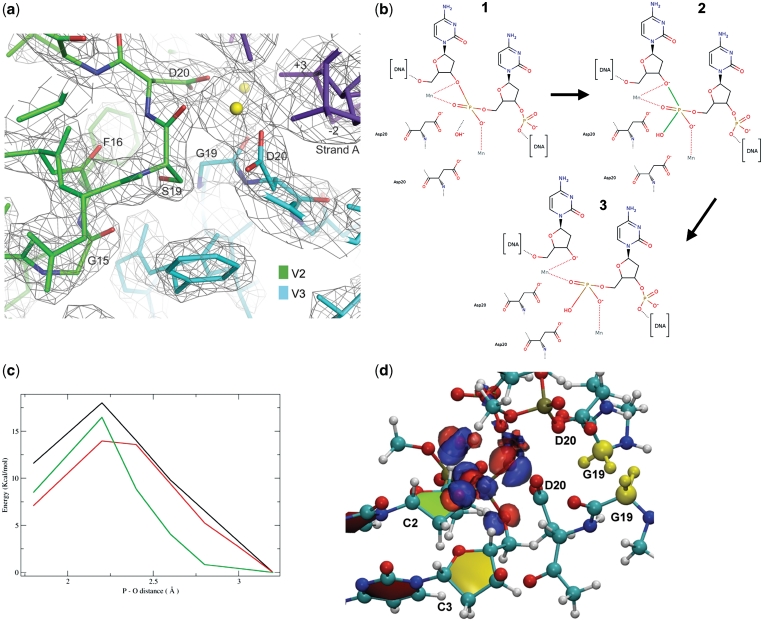
Figure 4.
The G19S mutation. (a) Detailed view of the G19S mutation in the V2(K7E-G19S)V3(E8K) heterodimeric protein bound to the RAG1 DNA duplex. A 3Fo−2Fc map calculated at 2.3 Å and contoured at 1σ, showing both the catalytic D20 residues, the G19S mutation in the V2 chain as well as the position of G15 and F16. The calcium atoms are colored in yellow and the DNA chain in purple. (b) Scheme of the reaction mechanism for DNA cleavage. (1) uncleaved DNA, (2) intermediate state with the formation of the pentacoordinated phosphorus atom—the two bonds involved in the reaction are colored in green and (3) cleaved DNA. (c) Energy profile of the P-O bond break between the phosphorus atom and the hydroxyl group. Black line: double mutant G19S-G19S modelled in the scV3V2(G19S) structure. Green line: I-CreI, G19-G19. Red line: scV3V2(G19S). The points at 3.2 Å correspond to the uncleaved DNA. (d) Enlargement of the catalytic centre of the wild type heterodimeric protein with a representation of the electronic density difference between wild type and single mutant (G19S). The isodensity surfaces in blue (red) correspond to an increase (decrease) in the electronic density due to the G19S mutation. The two glycines mutated in the single and double G19S variants are colored yellow.
The difference of the electronic densities between the wild-type and the single mutant is shown in Figure 4. From the figure, it is apparent that the mutation significantly polarizes the reacting hydroxyl group, effectively lowering the reaction barrier. These results, consistent with the experimental data, clearly support the role of the single mutation in the transition state of the DNA cleavage, which was reported to be the rate-limiting step in the DNA cleavage by endonucleases (33).
In silico specificity and DNA variability analysis
To predict ‘in silico’ the target specificity of the three structures with non-cleaved DNA, that is, the wild-type I-CreI (1G9Y), and the RAG1 binding proteins V2(K7E, G19S)V3(E8K) and scV3V2(G19S), we used the protein-design tool FoldX (35,36). First, the positions of the amino acid side chains and bases in the crystal structure were energetically optimized using the FoldX RepairPDB function. Then, each DNA base was mutated to the other three bases five times to increase the conformational space analyzed (FoldX does not guarantee convergence of the solution, since depending on the rotamers that are randomly selected from the library and the order of movement of the neighboring residues, it can offer different results due to occasional energy traps). Using the average value, the difference in protein–DNA interaction energy with respect to the wild-type (ΔΔGint) was calculated. To this difference, we added the difference in intramolecular clashes if they were 0.6 kcal/mol higher than that for the crystal structure (a residue can have good binding energy to DNA but be in a conformationally strained position with respect to the molecule to which it belongs). This function is graphically displayed as information content by means of the R package seqLogo, where the height of a given nucleotide is proportional to exp(−ΔΔGint/RT). In all the cases, the same physical conditions were assumed for the calculations: temperature of 298 K, pH of 7.0 and ionic strength of 150 mM.
To analyze the variability allowed at each nucleotide position that each DNA structure can tolerate on its own, we first mutated each triplet base pair in the DNA to all 64 possible combinations. Then, we evaluated the base-pair energies for the central base of all triplets. We averaged these energies, removing those resulting in an increase in base–base intramolecular or torsional clashes, above the FoldX standard deviation (0.8 kcal/mol) (35). The resulting averages for the two different DNA base pairs are ΔGAT = −1.8 ± 0.9 and ΔGCG = −2.8 ± 1.1 kcal/mol. We then considered as a valid context (out of the 16 possible combinations), for a nucleotide in a given position and structure, those that had a base-pair energy within 1 kcal/mol of the above averages.
Expression of meganucleases in mammalian cells
To avoid construct-stability issues, the two coding sequences fused in single-chain molecules contain silent mutations, and share only 72% nucleic acid sequence identity. Like I-CreI, the N-terminal mutant has 163 residues while the C-terminal mutant sequence begins at residue Asn6. Details of constructs are available upon request. The open reading frames of different meganucleases were amplified and the expression done as before (14).
Gene-targeting experiments
The human 293H and MRC5 cell lines were cultured in DMEM or MEM medium (Invitrogen Life Science), respectively, supplemented with 2 mM l-glutamine, penicillin (100 IU/ml), streptomycin (100 µg/ml), amphotericin B (Fongizone) (0.25 µg/ml) (Invitrogen Life Science) and 10% FBS (PAA Invitrogen Life Science). Gene-targeting experiments were carried out in 293H and/or MRC5 cell lines. The donor plasmid for gene-targeting experiments contained left and right homology arms generated by polymerase chain reaction (PCR) amplification of the human RAG1 locus. An exogenous DNA fragment was inserted between these two arms. These sequences consisted of a 1.7 kb DNA fragment derived from a neomycin expression plasmid. A donor plasmid carrying one nucleotide mutation in its homologous right arm was also designed. The mutation was located 265 bp downstream of the double-strand break. To prevent cleavage of the repair matrix by the meganuclease targeting the human RAG1 locus, a 9 bp deletion (comprising the nucleotides +4 to +12 from the meganuclease DNA target sequence) was also introduced. A day prior to transfection, cells were seeded at 1 × 106 (293H) and 5 × 105 (MRC5) cells per 10-cm dish. Cells were transfected with 3 µg of the meganuclease expression vector (14) and 2 µg of matrix vector, in the presence of lipofectamine 2000 (293H cells) or polyfect (MRC5 cells) transfection reagent in accordance with the manufacturer’s protocol (Invitrogen). After 48 h of incubation at 37°C, cells were treated with trypsin and dispensed at a density of 10 or 100 cells per well in 96-well plates. DNA was extracted with the ZR-96 genomic DNA kit (Zymo research) according to the manufacturer’s protocol. PCR amplification reactions performed with specific primers enabled us to selectively amplify gene-targeting events. Gene-insertion events were determined by specific PCR amplification carried out using a primer specific to the exogenous sequence (5′-AGGATCTCCTGTCATCTCAC-3′) and a primer specific to the RAG1 locus (5′-CTTTCACAGTCCTGTACATCTTGT-3′). For polymorphism analysis, PCR amplifications were performed with a primer specific to the RAG1 locus (5′-TTCACTGCTCTTCATGGCCTCCC-3′) and a primer overlapping with the 9 bp deletion of the DNA repair matrix (5′-GTTCTCAGGTACCATGGCAGC-3′). Meganuclease-induced mutagenesis was performed in 293H cells by transfecting them with 3 µg of the meganuclease expression vector and 2 µg of repair-matrix vector. As a negative control, empty vector alone was also used for transfection. After 48 h of incubation at 37°C, cells were treated with trypsin, re-plated and individual clones were picked and subsequently amplified. After 21 days, DNA was extracted with the ZR-96 genomic DNA kit and PCR amplification on RAG1 locus was performed with the primers
F9: 5′- GGCAAAGATGAATCAAAGATTCTGTCCT-3′ and
R9: 5′-GATCTCACCCGGAACAGCTTAAATTTC-3′. Mutagenesis events were determined by sequencing the PCR products.
RESULTS
Strategy for generation of the RAG1 variants
In a previous study (14), we scanned the sequence of the human RAG1 gene (NM_000448.1, NCBI nucleotide data base) for 22-bp sequences that could be potential targets of an I-CreI variant resulting from our combinatorial strategy. Eighteen DNA hits were identified, and the most promising one was located before the ATG codon of the human RAG1 gene (Figure 1a). This hit was then chosen to screen combinatorial libraries of I-CreI variants to repair mutations reported in the human RAG1 gene (20,21). Two combinatorial libraries of I-CreI variants cleaving the left- (LL) or right-hand (RR) palindromic sequences were generated, and a set of active heterodimeric variants cleaving the RAG1 sequence was identified. Target cleavage by this heterodimeric protein was further improved by random mutagenesis (12,13), resulting in the design of a new heterodimeric protein called V2V3, which displays high cleavage levels in yeast and CHO cells (14). However, the formation of the V2V3 heterodimer inside the cell requires co-expression of both V2 and V3, which results in a certain proportion of homodimers as by-products of the heterodimer formation (Figure 1b, upper panel). The presence of these homodimers decreases the overall efficiency and specificity of the engineered meganuclease, promoting deleterious effects in cells. To avoid these potentially toxic side effects, we redesigned this enzyme based on the I-CreI scaffold, first by modification of the dimerization interface to ensure heterodimerization (Figure 1b, lower panel), and second by the creation of a single-chain molecule joining the two monomers with a linker loop (Supplementary Figure SF1) (14). After checking the homodimeric conversion of the heterodimers (Figure 1b, ‘Material and Methods’ section), we noticed that the recovery of the V2 homodimer (tagged with the His6 sequence) was 5–15% (Figure 1b, right upper panel) and that of the V3 homodimer (tagged with the Strep sequence) ranged from 30 to 40% (Figure 1b, left upper panel). However, no homodimer formation could be observed when the same procedure was applied to the V2(K7E-G19S)V3(E8K) heterodimer (with the corresponding protomers tagged with the same Strep and the His6 sequences as above; Figure 1b, lower panels), indicating an increased stability of this variant due to the new inter-protomer interface). The mutation G19A was found in a number of active I-CreI variants (12,13) optimized by random mutagenesis, and the G19S mutation was serendipitously discovered to increase the specificity of the V2V3 heterodimer toward its non-palindromic DNA sequence (14).
These heterodimeric variants and some of the precursor homodimeric ones were subjected to biochemical and biophysical characterization (Figure 1c and d). Analytical ultracentrifugation sedimentation velocity analysis indicated that all the variants behave as dimers in solution (with a molecular mass that is double that of the I-CreI monomer of 20 kDa) including the monomeric single-chain variant scV3V2(G19S). This fact was also confirmed by size-exclusion chromatography. The analysis of the in vitro cleavage activity showed that all the variants behaved similar to the wild-type I-CreI with C50 between 5 and 8 nM (Figure 1d).
The crystal structures of V2V3, the dimerization-interface-modified-variant V2(K7E-G19S)V3(E8K) and the single-chain scV3V2(G19S) variant in complex with the RAG1 DNA were solved by molecular replacement (Figure 2, Supplementary Figure SF1 and Table SI). Overall, the structures of the protein moieties were similar to that of wild-type I-CreI (0.48–0.65 Å Cα r.m.s.d.), but the structure of the DNA duplex shows distortions with respect to the I-CreI wild-type DNA. Specific protein loops change their local conformation to fit DNA changes.
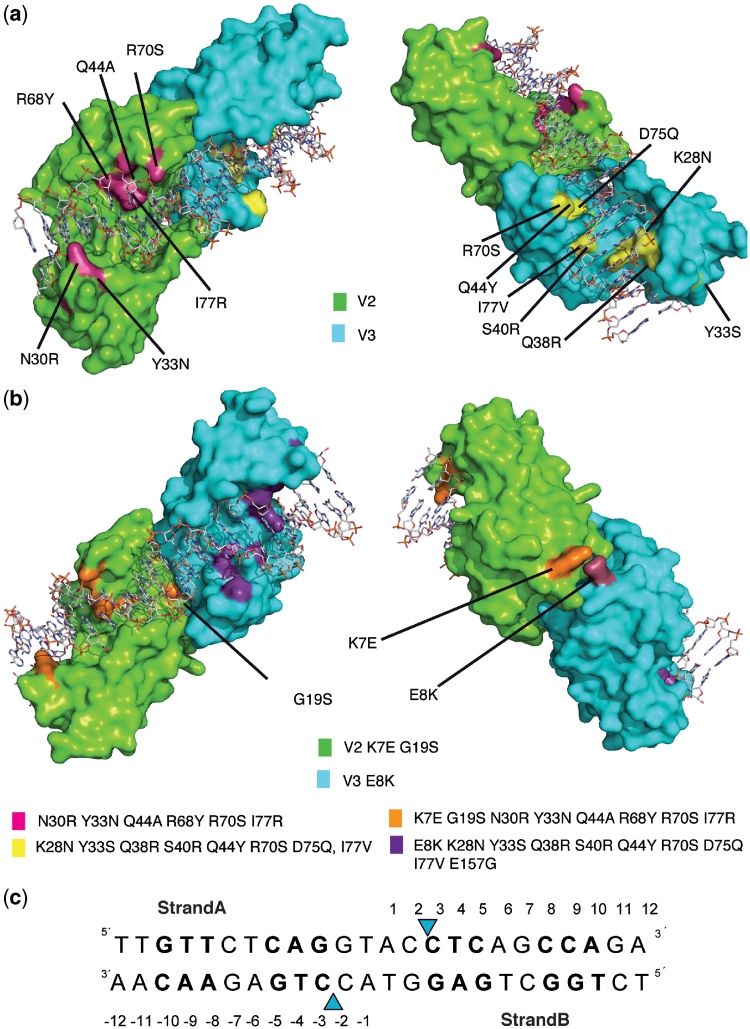
Figure 2.
Structures of the RAG variants. Crystal structures of the V2V3 (a) and V2(K7E-G19S)V3(E8K) (b) heterodimeric variants in complex with the RAG DNA target. (c) Mutations in each monomer are depicted in magenta for V2, yellow for V3, orange for V2(K7E-G19S) and violet for V3(E8K). The V2(K7E-G19S)V3(E8K) variant contains the same mutations as V2V3 plus K7E, G19S and E8K mutations. The regions of DNA with the new amino acid contacts are shown in bold. Phosphodiester bonds cleaved by the enzyme are indicated by blue triangles.
Analysis of the protein–DNA complexes
The I-CreI RAG1 variants display mutations clustered in two different DNA-binding subdomains, and cleave a RAG1 target sequence, which is different from the palindromic I-CreI C1221 target site of 22 bp (15). The residues that contact the bases at positions ±8, ±9 and ±10 or ±3, ±4 and ±5 were those mutated and screened for cleavage against an array of DNA sequences differing at these same positions (Figures 1a and 2). Consequently, the main structural changes in the protein moiety of the complex are located in the loops among K28-H37 (Figure 3a and b), the β–strand following it, P113-D120 (Figure 3a and c), and the loop between positions R68 and S79 (Figure 3a and d). For a detailed description of the protein–DNA interactions, see Supplementary Data and Supplementary Figures SF2 and SF3.
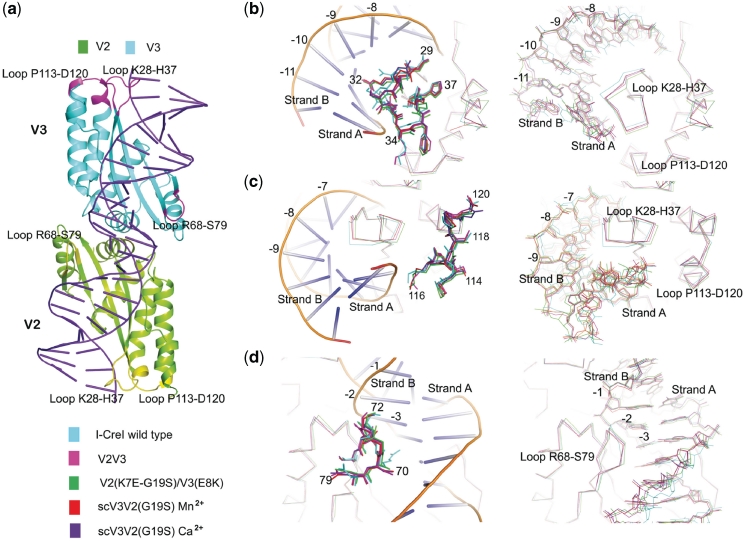
Figure 3.
Structural analysis of I-CreI wild type, V2V3, V2(K7E-G19S)V3(E8K), scV3V2(G19S)-Mn2+ and scV3V2(G19S)-Ca2+ with DNA. (a) Cartoon representation of I-CreI wild type (1G9Y). Mutations in magenta for V3 and yellow for V2 in the three loops where the major differences with respect the wild type are present, and which might affect some aspect of the binding to the DNA chain. Detailed views in panels (b), (c) and (d), highlight the structural changes in the protein and DNA molecules. Left: the proteins displaying amino acid side chains defining the loops, including the wild type DNA in cartoon representation; Right: Cα superposition of all the proteins and their corresponding DNA molecules. Bases from the Strand B are labeled in all the panels, and boxes with the color code used for each molecule are added for clarity. (b) The K28-H37 loop, and 8–11 bases of the bound DNA. (c) The P113-D120 loop and bases 7–9. (d) The R68-S79 loop and bases 1–3.
The selected mutations efficiently recognize and cleave the new DNA target. The symmetric recognition pattern of the wild-type protein is exchanged for the asymmetric pattern of the V2V3, the heterodimer stability improved in V2(K7E-G19S)V3(E8K) and the specificity further enhanced in scV3V2(G19S). This transformation induced by the engineering approach combines a reduction of the total number of contacts between the amino acids and the nucleic acid, with an increase in the proportion of hydrogen bonds along the new target-DNA major groove (Supplementary Figure SF3). It is noteworthy that the conformation of the RAG DNA differs from that of the wild type, with the largest differences localized in the major groove. The width of the major groove is 1–4 Å larger in the RAG target (depending on the region) than in the wild-type DNA, exposing a larger area to the protein. The conformational changes in the minor groove are less prominent, and as in the wild type, the RAG variants do not use this region of the DNA to effect recognition. Interestingly, the four central base pairs of the wild-type target sequence, where catalysis takes place, are found in a distorted minor groove with the bases at position ±1 and ±2 in both strands having van der Waals interactions with the protein. This pattern is altered in the RAG variants. The heterodimer V2V3 does not display any interaction with the ±1 and ±2 bases. However, the variants that contain the G19S mutation do show interactions in this region.
The linker introduced in the scV3V2(G19S) meganuclease variant contains 32 residues that cannot be observed in the electron-density map (Supplementary Figure SF1), probably due to the loop’s high flexibility. A similar situation was observed in the crystal structure of a single-chain variant based on I-MsoI (37), another member of the LAGLIDADG family, where only a few residues could be modeled. In our case, only one of the initial alanines was modeled into the electron density, indicating where the linker begins. The presence of the linker did not introduce significant changes in the I-CreI scaffold.
Cleavage activity and the G19S mutation
The DNA cleavage reaction in I-CreI has been extensively studied (38). However, there are open questions regarding the relevance of DNA bases located at the central positions ±2, ±1 in cleavage and specificity of the enzyme-DNA recognition (Figures 1a and 2c). These bases show van der Waals interactions with the protein, plus the interaction of the phosphate group between nucleotides −2 and −1 with the side chain of K139. We have found that the inclusion of the G19S mutation in the V2 or V3 monomers resulted in a large decrease in homodimer cleavage activity against their corresponding LL and RR DNA targets, displaying a correlation between the in vitro cleavage and the in vivo measurements (14). In contrast, its presence-enhanced cleavage activity against the LR target was observed when the G19S mutation was included in one of the protomers of the heterodimers. An additional increase in specificity is achieved for the LR target with the monomeric scV3V2(G19S) over the heterodimeric V2(G19S)V3.
The variant V2(K7E-G19S)V3(E8K), which displays maximal cleavage of the LR with high specificity, was chosen for further characterization. As previously mentioned, there are no interactions with the bases of the central four base pairs. G19 is close to the catalytic D20 and the metal centers involved in the cleavage of the phosphodiester bonds (Figure 4). In the V2V3 heterodimer, G19 interacts through its backbone carbonyl with one of the catalytic Mg2+ and is in the neighborhood of the carboxyl of D20 in the other monomer and a phosphate from the digested DNA. The G19S [V2(G19S)V3 and V2(K7E-G19S)V3(E8K)] mutants maintain these interactions, and through the side-chain hydroxyl, form a hydrogen bond with the backbone carbonyl of G15 in the other monomer. A close look at the structures indicates that the presence of S19 in both monomers would promote a steric clash that could hamper catalysis due to the distortion of the catalytic centre including the position of D20 in both monomers. A single G19S mutation in the heterodimer does not cause substantial conformational changes in this region. The enhancer effect of the G19S mutation was observed in several in vivo tests, with different proteins (14).
To decipher this effect, we performed quantum chemistry and quantum mechanical/molecular mechanical (QM/MM) calculations on the role of the mutation on the activation energy barrier of the DNA cleavage reaction (Figure 4b). Using a similar approach as the one adopted to study the mechanism of the endonuclease IV (33), we built a reaction path connecting the crystallographic structures containing the cleaved DNA and the non-cleaved DNA. The results are in agreement with the experiments in showing that the single G19S mutation lowers the activation energy of the reaction (Figure 4c). The energy difference between the transition state and the non-cleaved DNA is 16 and 14 kcal/mol for the V2V3 and the V2(G19S)V3 heterodimers, respectively (Figure 4 and Supplementary Data). These results agree with the experimental observation. Thus, the introduction of the G19S mutation only in one monomer increases the activity of the heterodimer (14). The calculations on a model structure of the V2(G19S)V3(G19S) heterodimer yielded a barrier of 17.5 kcal/mol, larger than that for V2V3, in agreement with the undetectable activity found for the heterodimers containing the G19S mutation in both monomers. As the breaking of the bond between the oxygen O3′ and the phosphorus was found to be the rate-limiting step in Endonuclease IV (33), these calculations provide computational and theoretical support to the biochemical findings.
Computational analysis of protein–DNA recognition
In silico analysis of the sequence–structure relationship of the DNA from I-CreI, and the RAG1 variants’ structures using FoldX (35,36) reveals how the conformations of residues in the previously mentioned loops (K28-H37 and A113-D120) influence DNA sequence recognition by the I-CreI scaffold. Some of these conformations may not promote the recognition of certain DNA sequences. This could help to explain the high energies observed for the RAG1 variants bound to the wild-type DNA (Supplementary Table SII). For example, the combination of the N30R mutation in V2 and the substitution of −2CstrandA of the wild-type target by a −2TstrandA promotes a movement of the loop away from the DNA (Figure 3). The thymine enforces a small shift of the DNA and the protein cannot fit an arginine in this region without moving the whole loop at once (same in the case of R70S in the 68–75 loop) (Figure 3). In addition, we observed that the R38 and R40 residues (Figure 3) force the DNA structure to move away from the protein, with concomitant changes in the tilting angles of the two new contacting bases. This movement therefore reveals that both the protein moiety and the nucleic acid change their conformation upon complex formation. Large losses of stability or a significant loss of interaction energy were observed while taking into account the DNA structure alone (except in the 2VBJ structure, Supplementary Table SIII).
Influence of the DNA conformation in recognition
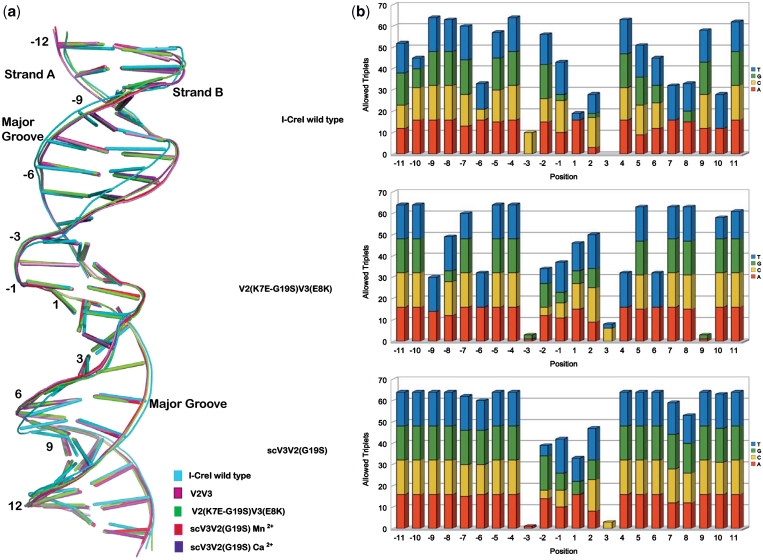
The superposition of the protein–DNA complexes using the protein Cα backbone reveals differences between the wild-type DNA and the RAG1 target conformation in the complexes (Figure 5a). Main differences can be observed in the DNA around positions −10 to −3 and 3 to 10 in strand A and their corresponding base pairs in strand B. These regions contain the ±10 to ±7 and ±3 to ±5 positions that were scanned during the combinatorial approach to search for new variants. We have used FoldX (35) to evaluate the tolerance to nucleotide variability at each position of the non-cleaved DNAs in the structures of the wild-type, V2(K7E-G19S)V3(E8K) and the scV3V2(G19S) complexes. The energies were calculated taking into account the 16 possible combinations of the surrounding base pairs. This analysis ranks the 64 possible triplets centered on a given position by their energy. The graph in Figure 5b shows the number of energetically favorable allowed triplets classified by the identity of the base at that position. We observe that position ±3 is always very restrictive and that the central four bases are more restrictive than the rest of the sequence. The comparison of the three Ca2+ bound structures shows that the DNA duplex bound to V2(K7E-G19S)V3(E8K) variant and the scV3V2(G19S) displays a higher degree of tolerance to base-pair variability while the wild-type DNA structure allows a minor degree of variability, with several positions only permitting an AT base pair (Supplementary Figure SF4). This is consistent with the higher DNA-binding affinity of the wild type when compared with scV3V2(G19S) (Supplementary Figure SF5).
Figure 5.
Overlay of the DNA structures. (a) Comparison of the RAG DNA conformations with those of the wild type DNA target. (b) In silico analysis of the non-cleaved DNA structures. For every nucleotide in each position of the DNA, the number of energetically possible combinations of surrounding nucleotides (out of 16) is shown. Less specific positions allow for different bases one position upstream or downstream, while highly specific positions are restricted to just a few combinations.
Gene-targeting efficiency with the use of single-chain meganucleases
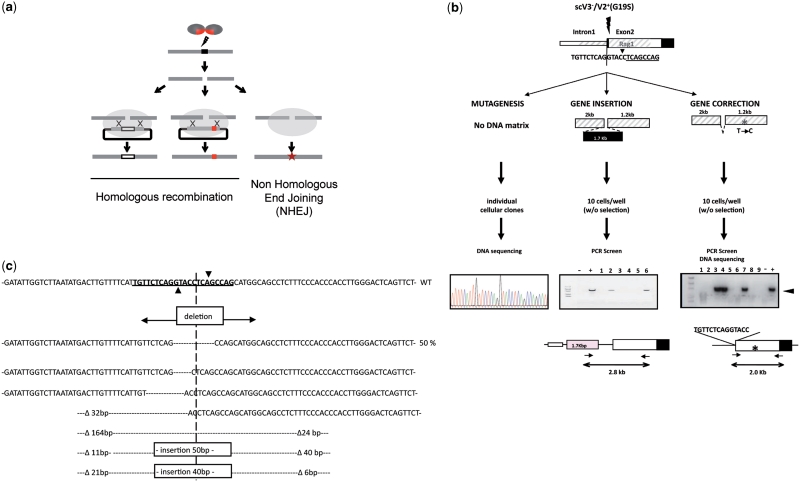
A mutation in the human RAG1 gene has been shown to be present in several patients with Omenn’s syndrome, an autosomal recessive form of SCID (39). This mutation is characterized by a two base-pair deletion Δ368A/369A (21) and introduces a frame shift at K86 that leads to the addition of 32 amino acids before a premature stop codon is encountered. Thus, we designed a DNA-repair matrix for gene correction, carrying one nucleotide mutation in its homologous region 265-bp downstream of the DSB site as a marker (Figure 6a and b). The repair matrix also contains a 9 bp deletion in the RAG1 recognition site. This deletion prevents cleavage of the repair matrix by the meganuclease and permits the specific detection of homologous recombination events by using a PCR primer that covers the deletion. After co-transfection of the repair matrix and the scV3(K7E-K96E)V2(E8K-E61R-G19S)-expressing vector (14), cells were seeded and cultured, and a 2 kb DNA fragment including the marker was amplified by specific PCR in order to detect gene-correction events. The estimated gene-correction events occurred in 5.3% of the transfected cells (Table 1). The meganuclease variant used in this analysis combined the redesigned interface and the linker of the single-chain variant that have been structurally characterized. The PCR-positive wells were analyzed by DNA sequencing and the frequency of the marker conversion was determined. In contrast to previous data (40), the single nucleotide polymorphism was observed in 66% of the analyzed pools, showing that a conversion tract of at least 265 bp in length occurred in up to two-third of the targeted cells (Table 1).
Figure 6.
Endonuclease-based strategies for genome modification. (a) Scheme of the main strategies to detect the three possible DSB-induced genome modifications. The meganuclease generates a DNA double-strand break at a specific locus. In the absence of DNA repair matrix, the DSB would be repaired by NHEJ. This error prone mechanism would eventually lead to small insertions and/or deletions. When targeting exonic sequences, this process can be useful for gene inactivation. In the presence of DNA repair matrix, homologous recombination mechanism will lead to targeted gene modification. Exogenous DNA sequence as well as mutation can be introduced at the cleavage site. Upon repair of the DSB, adjacent sequence can be converted, allowing for the correction of a mutation distant from the DSB. (b) In vivo activity of the meganuclease targeting the human RAG1 gene was monitored using three assays (see Materials and Methods for details). Meganuclease-induced mutagenesis activity was quantified by sequencing the RAG1 locus in cells transfected with a plasmid coding for the meganuclease. Targeted gene insertion and gene correction efficiencies were monitored in cells transfected with the meganuclease and a repair matrix, respectively, containing an exogenous sequence (1700 bp) or carrying 1 mutation located 265-bp downstream of the DSB. After transfection, cells were seeded at the density of 10 cells/well in 96-well plates and cultured without selection for 21 days prior to DNA extraction. Specific PCR was used to detect targeted recombination events. The presence of the polymorphism was detected by enzyme digestion and sequencing. To prevent cleavage by the meganuclease, the sequence underlined was deleted from the repair matrix used for gene correction. The asterisk indicates the mutation introduced in the left homologous arm. (c) Molecular analysis of the mutagenesis events induced by meganucleases in vivo. Top: DNA sequence of the coding strand of the human RAG1 gene surrounding the meganuclease cleavage site. The DNA sequence recognized by the meganuclease is underlined. The inverted filled triangle indicates the cleavage site. Bottom: Insertion and deletion events induced by the meganuclease. Sizes of the nucleotide deletion (open triangle) are numbered from the cleavage site.
Table 1.
Frequency of targeted polymorphism induced by meganuclease
| Total number cells analyzed | PCR positive (%) | Insertion polymorphism (%) | |
|---|---|---|---|
| Repair plasmid | 2256 | 0 | NA |
| RAG1 meganuclease and repair plasmid | 2632 | 139 (5.3%) | 27/41 (65.8%) |
The 293H cells were transfected with plasmid coding for the meganuclease targeting the human RAG1 gene and a repair matrix carrying a polymorphism located 265 bp downstream of the DSB.
The generation of site-specific DNA double-strand breaks is a powerful tool for genome engineering. When a meganuclease and a repair matrix are artificially introduced into the cell, they will trigger homologous recombination events at the meganuclease cleavage site and induce gene insertion or gene correction (Figure 6a). Alternatively, in the absence of a repair matrix, non-homologous end joining (NHEJ), an error-prone process that restores the initial sequence or results in small insertions or deletions of various sizes, will repair the DSB. These three endonuclease-based strategies (Figure 6a) are at the origin of the potential applications of this technology in the field of gene therapy. Therefore, we investigated the efficiency of meganucleases to promote different targeted approaches (Figure 6b).
In the present work, the ability of meganucleases to stimulate gene targeting (GT) in human cell lines was evaluated. As the chromatin accessibility might modulate the cleavage efficiency in a locus-dependent manner and thus affect the GT frequency, we quantified GT efficiency in two different cell types: the 293H (ATCC CRL-1573) cells and the normal diploid human cell line MRC5 (ATCC CCL-171) immortalized with SV40 large T antigen. Moreover, we also determined in these cell lines the recombination frequency induced by the heterodimeric variant V2(G19S)V3, the scV3V2(G19S) and the combined single-chain meganuclease scV3(K7E-K96E) V2(E8K-E61R-G19S), bearing the two engineered electrostatic interactions previously described (14). GT events were analyzed by PCR screen of the transfected cells. Our screen was first validated for its ability to detect GT events under these conditions (data not shown). The results of targeted recombination could be achieved in both cell lines at frequencies up to 5.6% and 4.4% of the transfected 293H and MRC5 cell pools, respectively (Table 2). In contrast, no GT event could be detected when a DNA-repair matrix was introduced in the absence of meganuclease. Interestingly, the combined single-chain molecule scV3(K7E-K96E)V2(E8K-E61R-G19S) was revealed to be the most effective meganuclease design in both cell lines while the heterodimer V2(G19S)V3 and its corresponding single-chain version scV3V2(G19S) display similar abilities to induce targeted recombination.
Table 2.
Meganuclease-induced gene targeting
| Cells/wells | Cells/wells | Total cells analyzeda | PCR positive | Estimated recombination frequencyb (%) | |
|---|---|---|---|---|---|
| 293H | V2(G19S)V3 | 10 | 1012 | 7 | 0.8 |
| scV3V2(G19S) | 10 | 2924 | 26 | 1 | |
| scV3(K7E-K96E)V2(E8K-E61R-G19S) | 10 | 3284 | 166 | 5.6 | |
| Repair matrix alone | 100 | 13 920 | 0 | NA | |
| MRC5 | V2(G19S)V3 | 10 | 960 | 8 | 1 |
| scV3V2(G19S) | 10 | 960 | 13 | 1.7 | |
| scV3(K7E-K96E)V2(E8K-E61R-G19S) | 10 | 1236 | 44 | 4.4 | |
| Repair matrix alone | 100 | 2630 | 0 | NA |
Gene-targeting efficiency was estimated by PCR screening. The 293H and MRC5 cells were transfected with plasmids coding for various designs of the meganuclease targeting the human RAG1 gene and a repair matrix carrying 1700 bp of exogenous sequence. Cells were then seeded at low density (10 cells/well) in 96 well plates. Targeted events were detected by specific PCR.
aCorrected for plating efficiency.
bCorrected for transfection efficiency.
NHEJ-dependent mutagenesis represents a powerful strategy to inactivate genes. Alternatively, it is also a valuable tool to detect in vivo cleavage activity at the endogenous site (Figure 6a). Indeed, repair of DSB by NHEJ can lead to mutations, usually small deletions or insertions at the cleavage site. Hence we investigated the impact of cleavage by scV3(K7E-K96E)V2(E8K-E61R-G19S) on NHEJ-dependent mutagenesis. The 293H cells were transfected with the vector expressing this protein without repair matrix and seeded at low density. After 3 weeks of culture without selection, individual cellular clones were analyzed. A 400-bp fragment surrounding the cleavage site was amplified by PCR and sequenced. The 284 DNA sequences were analyzed and 17 of them (6%) were mutated at the cleavage site (Figure 6c). In contrast, no mutation could be found within the recognition site when cells were transfected with a plasmid that did not express the meganuclease gene. It is noteworthy that the major part of the detected mutations consists of small deletions of a few nucleotides, with the deletion of nine nucleotides making up to 50% of the mutagenesis events (Figure 6c).
DISCUSSION
The generation of heterodimeric species inside the cell could introduce a new variable affecting specificity in meganuclease variants, with the potential to induce off-target cleavage, because of the undesired homodimers generated from the co-expression of monomers. Hence, these by-products could decrease the overall specificity of the meganucleases by cleaving additional targets. Although this effect was not observed in other engineered variants (13), we noticed a correlation between an increase in the dissociation of the V2V3 heterodimer in vitro (Figure 1b) and an increase in its toxicity (14). To overcome this limitation, we have suppressed homodimer formation by exploiting the dimerization interface and single-chain designs (14).
We combined the single-chain design with the dimerization interface variant, to produce the scV3(K7E-K96E)V2(E8K-E61R) and scV3(K7E-K96E) V2(E8K-E61R-G19S) molecules. The addition of the G19S mutation to V2 or V3 resulted in the largest specific decrease in homodimer activity with no decrease in heterodimer activity (14). We combined various sets of mutations and assessed the effects of these combinations. Even though this mutation does not introduce any distortion in the position of the protein main chain, or in the DNA chain, its presence increases the hydrogen bond network around the active site and our quantum-chemistry calculations show an increased polarization of the water-reacting hydroxyl group during catalysis that decreases the reaction energy barrier by >2 kcal/mol (Figure 4). Homodimer formation is prevented because although there is space for one serine at position 19, two serine residues will result in large van der Waals clashes.
Our study indicates that there are concerted conformational changes that occur between the protein loops and the DNA target molecule, emphasizing the need for as many different protein–DNA templates as possible to computationally design new variants that would target specific sequences. This idea also arose from the analysis of the XPC targeting heterodimers (13). Interestingly, the DNA structure of the XPC and wild-type DNA targets show more similarities between them than when compared with the RAG1 target DNA. The structure of RAG1 DNA is more divergent than that of XPC from the wild type, even though the change in the sequence with respect to the wild type is similar in both, 16 and 18 bp, respectively.
Our data show that the use of the LAGLIDADG scaffold can be generalized to target a good range of different DNA sequences (12,13,41). This work is a step toward the generation of à la carte meganuclease variants based on the I-CreI scaffold targeting DNA sequences of interest. However, the analysis and the comparison of these variants in complex with their targets suggests that this scaffold could not cover the conformational landscape of the multiple different DNA target sequences, indicating that new scaffolds must be explored to overcome this hurdle.
The repair mechanism of these newly designed variants on the targeted gene locus has also been examined in vivo. Recent data indicate that only one sequence of 22 bp for every 300 bp in a genome can be targeted by a specific I-CreI-based meganuclease (42). This limitation could make the meganuclease-mediated gene-correction strategy not available for all mutations. Pioneering work using gene-reporter systems has shown that short gene-conversion tracts of few dozens of nucleotides predominate (40) after chromosomal DSB induced gene conversion. In contrast, it has been recently shown in plants that targeted mutations could be introduced at a distant location from the DSB (43). In this work, we have demonstrated that, in human cells, a mutation located 265 nucleotides downstream of the DSB could be efficiently corrected, suggesting that mutations far away from the induced DSB also have the potential to be repaired using this approach.
Altogether, these data show that redesigned endonucleases represent one of the most promising tools for targeted approaches. The opening of a clinical trial for AIDS patients has recently shown the maturity of these strategies (http://www.cirm.ca.gov/content/zinc-finger-nuclease-based-stem-cell-therapy-aids-dr1-01490). However, there is still a ‘quest’ for the best reagents, in other words, the endonucleases providing the best efficacy–toxicity ratio. New advances in protein design have allowed the engineering of new scaffolds, such as meganucleases, and the landscape of existing methods is likely to change over the next few years, providing an exciting and effervescent area of research which could deliver new genome-editing ‘tools’ that could have a big impact in areas ranging from biotechnology to biomedicine.
ACCESSION NUMBERS
The coordinates and structure factors have been deposited in the PDB (3XE0, 3MXA, 3MX9, 3MXB).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online
FUNDING
European Union MEGATOOLS project (LSHG-CT-2006-037226); J.C. is funded by a fellowship from the Ministerio de Ciencia e Innovación (BFU2005-02403) to G.M. S.S. is funded by the Ministerio de Ciencia e Innovación (CSD2006-00023) to G.M. Funding for open access charge: European Union Project and Spanish Ministry of Science.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the beamline staff at the ESRF and SLS for helpful advice during data collection.
REFERENCES
- 1.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 3.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 4.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai CQ, Doyon Y, Ainley WM, Miller JC, Dekelver RC, Moehle EA, Rock JM, Lee YL, Garrison R, Schulenberg L, et al. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol. Biol. 2009;69:699–709. doi: 10.1007/s11103-008-9449-7. [DOI] [PubMed] [Google Scholar]
- 7.Eisenschmidt K, Lanio T, Jeltsch A, Pingoud A. A fluorimetric assay for on-line detection of DNA cleavage by restriction endonucleases. J. Biotechnol. 2002;96:185–191. doi: 10.1016/s0168-1656(02)00029-9. [DOI] [PubMed] [Google Scholar]
- 8.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Prieto PC, Redondo P, Blanco FJ, Bravo J, Montoya G, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Kim HH, Yuan X, Herrin DL. Purification, biochemical characterization and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 1997;25:3767–3776. doi: 10.1093/nar/25.19.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 11.Seligman LM, Stephens KM, Savage JH, Monnat RJ., Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 13.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 14.Grizot S, Smith J, Prieto J, Daboussi F, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 16.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005;16:378–384. doi: 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 19.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 20.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 21.Santagata S, Gomez CA, Sobacchi C, Bozzi F, Abinun M, Pasic S, Cortes P, Vezzoni P, Villa A. N-terminal RAG1 frameshift mutations in Omenn's syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc. Natl Acad. Sci. USA. 2000;97:14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karaca NE, Aksu G, Genel F, Gulez N, Can S, Aydinok Y, Aksoylar S, Karaca E, Altuglu I, Kutukculer N. Diverse phenotypic and genotypic presentation of RAG1 mutations in two cases with SCID. Clin. Exp. Med. 2009;9:339–342. doi: 10.1007/s10238-009-0053-1. [DOI] [PubMed] [Google Scholar]
- 23.Fischer A, Le Deist F, Hacein-Bey-Abina S, Andre-Schmutz I, Basile Gde S, de Villartay JP, Cavazzana-Calvo M. Severe combined immunodeficiency. A model disease for molecular immunology and therapy. Immunol. Rev. 2005;203:98–109. doi: 10.1111/j.0105-2896.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 24.Prieto J, Redondo P, Padro D, Arnould S, Epinat JC, Paques F, Blanco FJ, Montoya G. The C-terminal loop of the homing endonuclease I-CreI is essential for site recognition, DNA binding and cleavage. Nucleic Acids Res. 2007;35:3262–3271. doi: 10.1093/nar/gkm183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Marshall AG. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. XDS. Acta. Crystallogr. D. Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB. Newslett. Protein Crystallogr. 1992;26 [Google Scholar]
- 28.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta. Crystallogr. D. Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47 (Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov I, Tainer JA, McCammon JA. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc. Natl. Acad. Sci. USA. 2007;104:1465–1470. doi: 10.1073/pnas.0603468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ. A new ONIOM implementation in Gaussian 98. 1. The calculation of energies, gradients and vibrational frequencies and electric field derivatives. J. Mol. Struct. 1999;462:1–21. [Google Scholar]
- 35.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 36.Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl Acad. Sci. USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ., Jr Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, Lemieux C, Stephens K, Monnat RJ, Jr, Stoddard BL. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 39.Marcaida MJ, Munoz IG, Blanco FJ, Prieto J, Montoya G. Homing endonucleases: from basics to therapeutic applications. Cell Mol. Life. Sci. 67:727–748. doi: 10.1007/s00018-009-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galetto R, Duchateau P, Paques F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin. Biol. Ther. 2009;9:1289–1303. doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- 43.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.