Abstract
In the developing Drosophila optic lobe, eyeless, apterous and distal-less, three genes that encode transcription factors with important functions during development, are expressed in broad subsets of medulla neurons. Medulla cortex cells follow two patterns of cell movements to acquire their final position: first, neurons are arranged in columns below each neuroblast. Then, during pupation, they migrate laterally, intermingling with each other to reach their retinotopic position in the adult optic lobe. eyeless, which encodes a Pax6 transcription factor, is expressed early in progenitors and controls aspects of this cell migration. Its loss in medulla neurons leads to overgrowth and a failure of lateral migration during pupation. These defects in cell migration among medulla cortex cells can be rescued by removing DE-Cadherin. Thus, eyeless links neurogenesis and neuronal migration.
Keywords: Cell adhesion, Cell migration, Neuroblast, Optic lobe
INTRODUCTION
The adult optic lobe is one of the major structures in the Drosophila brain. It is composed of ∼60,000 cells (Hofbauer and Campos-Ortega, 1990) and can be divided into four neuropils: lamina, medulla, lobula and lobula plate (Meinertzhagen and Hanson, 1993). The optic lobe derives from an embryonic optic placode (Green et al., 1993). During larval development, this primordium contains two proliferation centers: the outer and the inner proliferation centers (OPC and IPC) (Fig. 1A,C) (Hofbauer and Campos-Ortega, 1990). Successive cell divisions of a limited number of neuroblasts, which last until early pupation (Hofbauer and Campos-Ortega, 1990; Ito and Hotta, 1992; White and Kankel, 1978), generate the correct number of cells present in the adult optic lobe: the IPC gives rise to neurons of the lobula complex and proximal medulla (presumably cells from the medulla rim), while the OPC generates most medulla neurons (presumably cells from the medulla cortex) as well as cells that will generate the lamina (Meinertzhagen and Hanson, 1993).
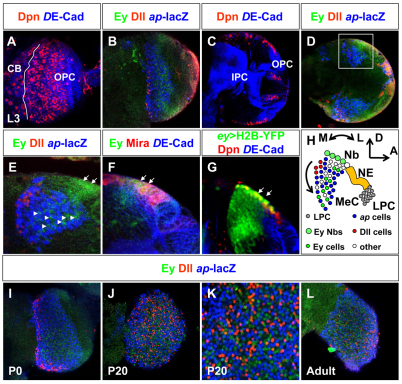
Fig. 1.
Medulla cortex organogenesis. (A,C) Anterior (A) and middle (C) sections of late L3 larval brains. Neuroepithelial cells were visualized with DE-Cadherin (blue) and neuroblasts with Deadpan (red). Solid white line in A separates optic lobe from central brain (CB). IPC, inner proliferation center; OPC, outer proliferation center. (B,D,E) Expression patterns of Eyeless (green), Distal-less (red) and ap-lacZ (blue) in cell populations in anterior (B) and middle (D,E) sections of late L3 larvae. Arrowheads in E indicate Ey expression in postmitotic cells and arrows indicate Ey-positive neuroblasts. (F,G) Expression pattern of Ey (F) and ey-Gal4 driving UAS-H2B-YFP (G) in OPC-derived neuroblasts (red). Neuroblasts were labeled with Miranda in F and Deadpan in G. Neuroepithelial cells were visualized with DE-Cadherin (blue). Arrows indicate neuroblasts. (H) Neurogenesis in the OPC. Lamina precursor cells (LPC) differentiate into lamina neurons (gray) in the most lateral part of the neuroepithelium (NE) (yellow). Medulla neurons (MeC) derive from eyeless neuroblasts (Nb) on the medial side of the NE. Postmitotic Ey-positive cells (green), ap-positive cells (blue), Dll-positive cells (red) and other cell types (white) are shown. A, anterior; D, dorsal; M, medial; L, lateral. (I-L) Expression patterns of Ey (green), Dll (red) and ap-lacZ (blue) in cell populations in anterior sections at P0 (I), P20 (J,K) and adult (L).
The medulla represents the largest structure in the adult optic lobe with an estimated 40,000 neurons (Hofbauer and Campos-Ortega, 1990), the cell bodies of which are located either in the medulla cortex, the region between the lamina and the medulla neuropil, or the medulla rim, the region between the medulla and the lobula plate. These two populations have been proposed to have different larval origins (Meinertzhagen and Hanson, 1993). We have identified three genes encoding transcription factors that are expressed in broad subsets of medulla cortex cells: eyeless (ey), apterous (ap) and distal-less (dll). These three transcription factors play important functions in the development of a large number of organs in Drosophila, as well as in vertebrates. In the adult medulla, their expression is mostly non-overlapping and covers over 90% of all medulla neurons (Morante and Desplan, 2008). In previous work, we had expanded on the work of Ramón y Cajal, Strausfeld and Fischbach (Ramón y Cajal and Sanchez, 1915; Fischbach and Dittrich, 1989; Strausfeld, 1976), and identified at least 63 distinct neuronal cell types that express these transcription factors in the medulla and might be involved in processing of visual information (Morante and Desplan, 2008). ey-positive cells are present only in the adult medulla cortex, whereas ap- and dll-expressing cells are both present in the adult medulla cortex and medulla rim (Morante and Desplan, 2008). Therefore, at least five different cell populations marked by ey, ap or Dll coexist in the adult medulla.
To investigate how each of these medulla cortex cell types is first determined in the OPC and comes to occupy its final position, we followed the early expression patterns of ey, ap and dll during larval and pupal development using reporter constructs or antibodies. We confirm that medulla cortex cells originate from the OPC, the main body of the crescent-shaped OPC.
During larval development, the progeny of each medulla cortex neuroblast forms columns where newly born neurons displace older neurons away from their neuroblast. We show that, later in pupation, cells marked with each transcription factor disperse laterally, mixing with other cell types to reach their final position. We also show that one of the genes, ey/Pax6, is expressed in OPC neuroblasts and controls migration of a subpopulation of cells in the optic lobe. Loss of ey function causes overproliferation of neuroblasts and prevents dispersion, leading to large clusters of ey-positive neurons. Formation of these large clusters can be rescued by removal of DE-Cadherin (Shotgun – FlyBase), which regulates cell adhesion and neuroblast proliferation (Dumstrei et al., 2003). Thus, eyeless links neurogenesis and migration in the optic lobe, revealing new mechanisms of brain formation in Drosophila that are similar to cortical development in mammals (Ge et al., 2006).
MATERIALS AND METHODS
Fly stocks
The following lines were used for the study: act-FRT-stop-FRT-Gal4; apmd544-Gal4, aprK568-lacZ, dllmd23-Gal4, eyOK107-Gal4, G-TRACE (Evans et al., 2009), shg-lacZ, UAS-p35, UAS-UEE3 and UAS-UEE7 (referred as eyDN) (Niimi et al., 2002), tub-Gal80ts, UAS-shg-GFP, UAS-CD8-GFP, UAS-H2B-YFP, UAS-nuGFP and FRTG13 shgR69.
G-TRACE analysis
G-TRACE analysis with ap- and dll-Gal4 caused larval lethality; thus, we made use of the tub-Gal80ts transgene (McGuire et al., 2004). Larvae containing ap- or dll-Gal4, G-TRACE and tub-Gal80ts were raised at 18°C until the second instar stage to allow Gal80 repression of Gal4 activity. They were then incubated at 30°C until third instar larval stages or P40 to relieve repression and allow induction of G-TRACE. G-TRACE analysis was performed using the following stocks (Evans et al., 2009): for ap-Gal4, yw hs-Flp122; ap-Gal4/tub-Gal80ts; UAS-flp, UAS-RFP, ubip63-FRT-stop-FRT-nuGFP/+; for dll-Gal4, yw hs-Flp122; dll-Gal4/tub-Gal80ts; UAS-flp, UAS-RFP, ubip63-FRT-stop-FRT-nuGFP/+; for ey-Gal4, yw hs-Flp122; tub-Gal80ts/+; UAS-flp, UAS-RFP, ubip63-FRT-stop-FRT-nuGFP/+; ey-Gal4.
MARCM clonal analysis
MARCM clones were induced in larvae 72 hours after egg laying (AEL) with a 20-minute heat-shock at 37°C using the following stocks: for ap-Gal, yw hs-Flp122 UAS-CD8-GFP; ap-Gal4/CyO; FRT82B tubP-Gal80/FRT82B; for dll-Gal, yw hs-Flp122 UAS-CD8-GFP; FRT40A tubP-Gal80 dll-Gal4/FRT40A; for ey-Gal, yw hs-Flp122 UAS-CD8-GFP; FRT82B tubP-Gal80/FRT82B; ey-Gal4/+, yw hs-Flp122 UAS-CD8-GFP; UAS-UEE3/+; FRT82B tubP-Gal80/FRT82B; ey-Gal4/+, yw hs-Flp122 UAS-CD8-GFP; FRT42D/FRT42D tubP-Gal80; UAS-UEE7/TM2; ey-Gal4/+ and yw hs-Flp122 UAS-CD8-GFP; FRTG13 shgR69/FRTG13 tubP-Gal80; UAS-UEE7/TM2; ey-Gal4/+; for tub-Gal, yw hs-Flp122 UAS-CD8-GFP;UAS-p35/+; tub-Gal4 FRT82B tubP-Gal80/FRT82B. Brains were dissected at L3, P5, P20, P44, P68 and adulthood.
To establish the cell migration pattern of clonally related cells, a 5-minute heat-shock (37°C) was induced in larvae 72 hours AEL using the following stock: yw hs-Flp122 UAS-CD8-GFP;ap-lacZ/+; tub-Gal4 FRT82B tubP-Gal80/FRT82B. Brains were dissected at L3, P10 and around P40.
Immunocytochemistry and confocal microscopy
Flies were raised on standard medium at 25°C. Larval, pupal and adult brains were dissected in cold PBS and fixed in PFA (4%) for 20 minutes (Morante and Desplan, 2010). Samples were incubated in a cocktail of primary antibodies diluted in PBST (0.3% Triton X-100) overnight at room temperature.
Primary antibodies used were as follows: guinea pig anti-Dll (dilution 1/3000, from R. Mann, Columbia University, New York, NY, USA), guinea pig anti-Dpn (1/3000, from J. Skeath, Washington University, St Louis, MO, USA), mouse anti-24B10 (1/50, DSHB), mouse anti-β-Gal (1/500, Promega), mouse anti-Elav (Embryonic lethal abnormal vision) (1/50, DSHB), mouse anti-Ey (1/10, from P. Callaerts, VIB, Leuven, Belgium), mouse anti-Mira (1/50, from F. Matsuzaki, RIKEN, Kobe, Japan), mouse anti-nc82 (1/50, DSHB), rabbit anti-GFP (1/1000, Molecular Probes), rabbit anti-β-Gal (1/20000, Cappel), rabbit anti-Mira (1/1000, from F. Matsuzaki), rabbit anti-pH3 (1/2000, Upstate), rat anti-DE-Cad (1/50, DSHB), rat anti-DN-Cad (1/50, DSHB) and rat anti-Elav (1/50, DSHB).
Brains were washed three times for 5 minutes in PBS and then incubated in secondary antibodies diluted in PBST for 3 hours.
Secondary antibodies were used as follows: donkey anti-rabbit 405 (1/400, Molecular Probes), donkey anti-guinea pig Alexa488 (1/400, Molecular Probes), donkey anti-mouse Alexa488 (1/1000, Molecular Probes), donkey anti-rabbit Alexa488 (1/1000, Molecular Probes), donkey anti-mouse Alexa555 (1/1000, Molecular Probes), donkey anti-rabbit Alexa555 (1/1000, Molecular Probes), donkey anti-rat Cy5 (1/400, Jackson ImmunoResearch), goat anti-rat Alexa555 (1/500, Molecular Probes), goat anti-guinea pig Alexa555 (1/500, Molecular Probes), goat anti-guinea pig Cy5 (1/500, Jackson ImmunoResearch) and donkey anti-mouse Alexa647 (1/200, Molecular Probes).
In Fig. S3 in the supplementary material, samples were incubated in a PBST solution containing DAPI (Sigma) for 30 minutes.
After washing overnight, brains were mounted in Vectashield (Vector Labs) keeping their 3D configuration.
Samples were imaged using a Leica TCS SP2 confocal using a 20× immersion lens. Images were assembled using Photoshop (Adobe).
Fig. S1A-C in the supplementary material was made with Volocity 4 (Improvision) using a 80 μm sample acquisition from a late L3 larval brain showing expression of ey (red), Distal-less (green), ap (white) and DE-Cadherin (blue).
Number of pH3 cells per clone in pupal optic lobes
MARCM clonal analysis of ey-expressing and ey-misexpressing eyDN flies was carried out after a 20-minutes heat-shock (37°C) in larvae 72 hours AEL and dissecting P5 pupal optic lobes. To determine the number of pH3-positive cells per clone, we counted the number of pH3-positive cells in each clone.
Number of cells per clone in adult optic lobes
MARCM clonal analysis of ey-expressing, ey-misexpressing eyDN and ey-expressing shg eyDN flies was carried out after a 20 minutes heat-shock (37°C) in larvae at 72 hours AEL and in dissecting adults. To determine the number of neurons per clone in adult optic lobes, we fully reconstructed the optic lobes (25 per genotype), collected stacks every 2 μm (between 90-100 μm) and counted the number of cells positive for GFP in each cluster.
RESULTS
Generation of the medulla cortex
We analyzed the simultaneous expression patterns of Ey, ap and Dll using an antibody against Eyeless (Clements et al., 2008) (Fig. 1), or eyOK107-Gal4 (Connolly et al., 1996) crossed with UAS-nuGFP (Figs S1, S2 in the supplementary material), a lacZ enhancer trap in apterous (ap-lacZ) (Cohen et al., 1992). The third cell population was labeled with an antibody against Distal-less (Estella et al., 2008). To follow migratory paths, we looked at these three markers in different sections of optic lobes (Betschinger et al., 2006) from larval stages until adulthood.
During the late third instar larval stage (L3), we observed three distinct cell populations corresponding to each marker in horizontal views of anterior (Fig. 1B) and middle sections (Fig. 1D,E) of the OPC. In anterior sections (Fig. 1A), Ey, ap- and Dll were expressed in three parallel stripes of cells that represent rows of neurons that emerge from the OPC (Fig. 1B). They correspond to progeny from the youngest to oldest neuroblasts (Fig. 1B). In middle sections (Fig. 1C), Dll-positive cells were generated in the progeny of the oldest neuroblasts, with ap- and Ey-positive cells often placed below Dll-positive cells, i.e. in cells that had emerged earlier from these neuroblasts (Fig. 1D,E). In the progeny of younger neuroblasts, we detected only ap- and Ey-positive cells, and no Dll-positive cells (Fig. 1D,E). Furthermore, Ey and ey-Gal4 were co-expressed in neuroblasts with Miranda (Mira) and Deadpan (Dpn) (Fig. 1F,G) (Betschinger et al., 2006; Egger et al., 2007; Ikeshima-Kataoka et al., 1997; Ohshiro et al., 2000; Yasugi et al., 2008). Therefore, three cell populations marked by Ey, ap- and Dll are already spatially and temporally segregated at this early stage (Fig. 1H; see Fig. S1 in the supplementary material).
We then followed how the expression patterns of the different cell populations in the OPC evolved during development until adulthood (Fig. 1I-L; see Fig. S2 in the supplementary material). By the beginning of pupation (P0), the number of cells originating from the OPC had increased (Fig. 1I). A major reorganization of the optic lobe structure occurred around P20: cells became intermingled with each other, making the three medial stripes observed earlier no longer distinguishable (compare Fig. 1B with Fig. 1J,K). Thus, the three Ey-, ap- and Dll-positive cell populations lost their spatial segregation and were interspersed within the adult medulla cortex (Fig. 1L).
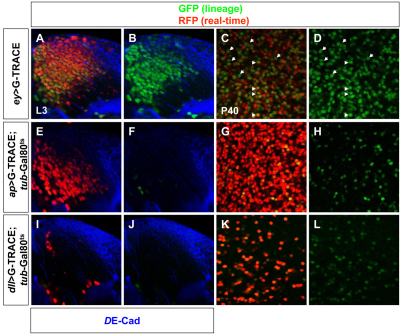
Next, we used the G-TRACE system to mark the cell lineages derived from cells expressing a given Gal4 driver (GFP) and, at the same time, to reveal real-time larval expression (RFP) of the given Gal4 driver that serves as a medulla cell marker (Fig. 2) (Evans et al., 2009). In G-trace, Gal4 mediates the expression of FLP recombinase that, in turn, removes an FRT-flanked transcriptional termination cassette inserted between Ubiquitin-p63E promoter fragment and the GFP open reading frame. Thereafter, the Ubi-p63E promoter maintains GFP expression perpetually in all subsequent daughter cells, independently of Gal4 activity.
Fig. 2.
G-TRACE analysis of OPC-derived neurons. (A-L) G-TRACE analysis with ey-Gal4 (A-D), ap-Gal4 (E-H) and dll-Gal4 (I-L) reporter lines. Images show OPC-derived cells at late L3 (A,B,E,F,I,J) and P40 (C,D,G,H,K,L). Neuroepithelial cells in A,B,E,F,I,J were labeled with DE-Cadherin (blue). Arrowheads in C,D indicate persisting ey expression in pupal cells (red and green) and arrows indicate cells with previous ey expression that no longer express the reporter (green).
With ey-Gal4, this allowed permanent expression of GFP in all the progeny of cells that expressed ey-Gal4 at some point (Evans et al., 2009). In L3, ey-Gal4 was expressed in neuroblasts (Fig. 1G) and in the vast majority of neurons derived from ey-positive neuroblasts that still maintained ey-Gal4 expression (most cells were both red and green; Fig. 2A-B), although this might be due to perdurance of Gal4 during L3 (see below).
Expression of G-TRACE with ap- and dll-Gal4 caused larval lethality and precluded the analysis in L3 larval stage. To avoid these early developmental defects, we made use of the tub-Gal80ts construct (McGuire et al., 2004). We raised at 18°C larvae containing ap- or dll-Gal4, G-TRACE and tub-Gal80ts until the second instar stage to allow Gal80 repression of Gal4 activity. Then, these larvae were incubated at 30°C for 2 days during third instar larval stages to relieve repression and to allow induction of G-TRACE when neuroblasts actively divide (Fig. 2E,F,I,J). ap-Gal4 was observed in neurons derived from ey-positive neuroblasts (Fig. 2E,F) but was not expressed in neuroblasts (see Fig. S3A,B in the supplementary material). dll-Gal4 (which precisely mimics Dll expression) showed expression in a small population of neurons located at the most proximal part of the OPC, i.e. in neurons generated by the oldest neuroblasts to have emerged from the neuroepithelium (Fig. 2I,J), but not in the neuroblasts themselves (see Fig. S3C,D in the supplementary material). Thus, ap and dll expression showed a delay before being turned on in maturing neurons.
Next, using the G-TRACE we analyzed the lineage followed by those larval cells in pupae (P40) to determine whether the expression of those Gal4 lines persisted during development. Indeed, ap and dll expression persisted during pupal development (Fig. 2G,H,K,L). By contrast, although ey-Gal4 expression was detectable in the vast majority of larval cells (Fig. 2A,B), its expression did not persist in pupae and a proportion of those cells were only green (Fig. 2C,D). Therefore, ey-Gal4 expression shows perdurance or is downregulated in most OPC-derived cells during pupal stages, with Ey being expressed in a subset of postmitotic cells that are negative for ap and Dll expression (Fig. 1).
Therefore, at least three different OPC-derived postmitotic cell populations marked by Ey, ap or Dll are spatially and temporally segregated early in larvae and pupae. These distinct cell populations appear to be pre-patterned early and subsequently become intermingled (Fig. 1, see Fig. S2 in the supplementary material).
Patterns of cell movements in optic lobe cells
The spatial and temporal segregation displayed by the different medulla cell populations coming from the OPC during larval and early pupal development is in sharp contrast with the extensive intermingling of cell types in the adult medulla cortex (compare Fig. 1B with 1L). This indicates that extensive cell movements take place in OPC-derived cells to achieve the final positioning of cell bodies and projections in the adult optic lobe. Although cell movement has been studied in detail in the Drosophila ovary and embryo (Kunwar et al., 2006; Rorth, 2007), its mechanisms have not been investigated in detail in the central nervous system.
The different cell populations that we observe might move similarly to each other, or each might use distinct mechanisms to achieve the final distribution of cells in the medulla cortex. We followed cell movements exhibited by ey-, ap- and dll-positive cells by performing mosaic analysis with a repressible cell marker (MARCM) (Lee and Luo, 1999) to generate neuroblast clones marked by the corresponding Gal4 drivers. The clones were generated in early L3 [72 hours after egg laying (AEL)] and brains were dissected at different stages throughout late larval and pupal development until adulthood (Fig. 3A-F and data not shown). This allowed us to visualize small subsets of cells.
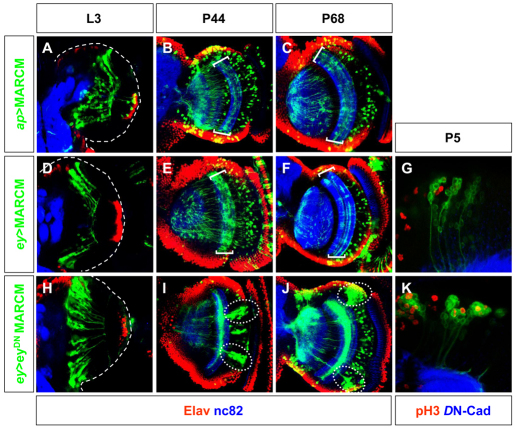
Fig. 3.
Cell migration pattern of eyeless and apterous cells. (A-F,H-J) Migration patterns of ap-(A-C), ey-(D-F) and ey-misexpressing eyDN (H-J) MARCM clones (green) generated 72 hours AEL and analyzed in late L3 (A,D,H), in P44 (B,E,I) and in P68 (C,F,J). Brackets in B,C,E,F indicate layers of medulla neuropils. Dotted circles in I,J highlight the accumulation of neurons in large clumps. Neurons were visualized with Elav (red) and the neuropil with nc82 (blue). (G,K) Proliferation analysis in ey-expressing (G) and ey-misexpressing eyDN (K) MARCM clones (green) generated 72 hours AEL and analyzed at P5. Mitosis was visualized with anti-phospho Histone 3 (red) and the neuropil with DN-Cad (blue).
In late L3 larvae, ey- and ap-marked OPC-derived clones presented a striking arrangement as discrete columns of cells (Fig. 3A,D), with the progeny of a neuroblast displacing cells away from the OPC (Egger et al., 2007). In the case of dll-positive cells coming from the OPC, the columnar organization was not evident in single confocal sections or through reconstructions (data not shown), probably owing to the very small number of dll-Gal4 neurons produced from each neuroblast.
By contrast, at mid-pupation (P44), profound reorganization occurred. Cells lost the columnar arrangement observed in larvae and became dispersed, while dendritic ramifications started to form hints of medulla layers (Fig. 3B,E). By late pupation (P68), medulla cells had reached their final position and were dispersed throughout the medulla cortex (Fig. 3C,F). Their dendritic arbors and axonal projections resembled those of the adult (Fischbach and Dittrich, 1989; Gao et al., 2008; Morante and Desplan, 2008), although we could still observe significant growth of branches during subsequent stages of development.
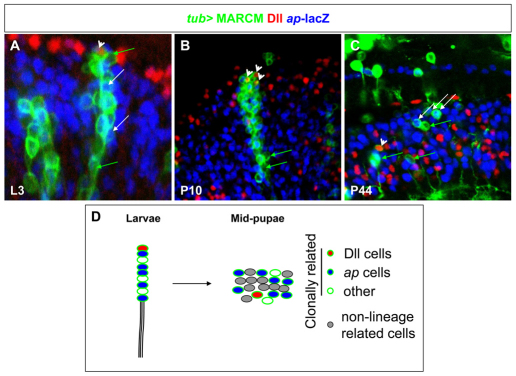
To visualize the patterns of cell movements followed by neurons derived from the OPC, we followed the fate of clonally marked cells derived from a single progenitor. We generated MARCM GFP clones using a tubulin-Gal4 driver and we studied the movement displayed by ap-positive and Dll-positive cells using fixed tissues. Clones were generated in early L3 (72 hours AEL) and brains were dissected at different stages throughout late larval and pupal development to follow their migratory paths (Fig. 4). In late L3 larval brains, tub-Gal4 MARCM clones (72 clones analyzed in 43 optic lobes) derived from the OPC showed a columnar organization (Fig. 4A). When we analyzed the progeny of the oldest neuroblasts that emerged from the neuroepithelium with ap-lacZ and Dll, we observed Dll in the latest born neurons, whereas ap-lacZ was expressed in older neurons (Fig. 4A). These cells were interspersed with unlabelled neurons that presumably expressed other markers (Fig. 4A). This confirms the columnar organization observed by ey- and ap-marked OPC-derived clones (Fig. 3A,D) and shows that a given neuroblast sequentially produces different cell types. The same columnar organization was maintained in OPC-derived clones analyzed during early pupal (P10) development (15 clones analyzed in 10 optic lobes) (Fig. 4B). By contrast, cells lost their columnar arrangement during mid-pupation and became dispersed in all axes throughout medulla cortex (70 clones analyzed in 38 optic lobes) (Fig. 4C). Despite this major dispersion, some medulla cells remained in small clusters of three or four cells.
Fig. 4.
Cell migration pattern of clonally related cells. (A-C) Migration patterns of tub-MARCM clones (green) generated 72 hours AEL and analyzed in late L3 (A), P10 (B) and P44 (C). Dll-positive cells visualized with anti-Dll (red) and ap-positive cells with ap-lacZ (blue). White arrows in A-C mark ap-positive cells, green arrows mark GFP-positive cells negative for ap and arrowheads label Dll-positive cells. (D) Schematic representation of a larval `column' in the OPC. During pupation, these OPC-derived cells lose their columnar organization and become dispersed throughout the medulla cortex with non-lineage-related cells (gray cells).
In order to address whether cell death plays a role in the dispersion of medulla cortex neurons, we generated MARCM GFP clones using a tub-Gal4 promoter mis-expressing UAS-p35, an apoptosis inhibitor. In adult brains (n=30 brains), medulla cortex cells did not show cell accumulation or alteration in the medulla neuropil lamination (data not shown).
Thus, optic lobe cells derived from the OPC follow two patterns of cell movement to achieve their final position in the adult optic lobe: first, neurons derived from the same progenitors are arranged in columns of cells, where neuroblasts sequentially produce cells that express different neuronal markers. These cells probably move away from the neuroblast because they are displaced by newly generated neurons, or they could undergo radial migration. Then, neurons lose this configuration in columns and disperse laterally to acquire their final position in the optic lobe and form the retinotopic map, with the different cell populations intermingling with each other (Fig. 4D).
Altered migration of optic lobe cells lacking eyeless function
As previously described, Ey is expressed in OPC-derived neuroblasts (Fig. 1). Thus, we addressed the function of ey in neuroblasts. For this purpose, we expressed a dominant-negative form of the Eyeless protein (hereafter referred as eyDN) in which the N-terminal domain is replaced by an Engrailed-repressor domain (Niimi et al., 2002), rendering the Ey activator a constitutive transcriptional repressor.
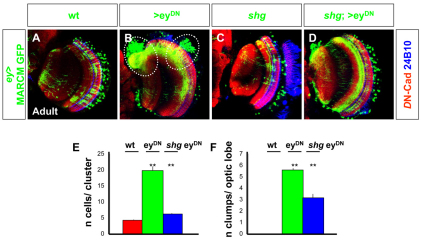
We generated MARCM clones expressing eyDN under the control of ey-Gal4. We compared the movement of these cells (Fig. 3H-J, Fig. 5B) with those of wild-type cells (Fig. 3D-F, Fig. 5A). Clones were generated in early L3 (72 hours AEL) and brains were dissected at different stages throughout larval and pupal development until adulthood. Wild-type clones forming columns of cells were observed in L3 and early pupation in wild type (Fig. 3D); by mid-pupation, these columns were lost, and dendritic ramifications were observed in the neuropil (Fig. 3E,F). The eyDN clones also formed columns of cells early (Fig. 3H). However, by mid-pupation, these clones formed clumps of cells (compare Fig. 3E,F with 3I,J). Clusters of cells significantly larger than in wild type could still be observed in adult brain sections [number of ey cells per cluster in wild-type adult brains (mean±s.e.m.), 4.08±1.47; in eyDN brains, 19.4±1.56) (Fig. 5E). This accumulation of clustered ey-cells disrupted the normal layering of the medulla as the cells were unable to disperse and reach their final retinotopic position.
Fig. 5.
Impaired optic lobe cell migration in eyDN MARCM clones rescued by DE-Cadherin. (A-D) Migration patterns of ey-positive cells in wild-type MARCM clones (A), in clones misexpressing eyDN (B), in clones lacking DE-Cadherin (C), and in clones misexpressing eyDN and lacking DE-Cadherin (D) (green) generated 72 hours AEL and analyzed in adult. Neuropil (red) was visualized with DN-Cadherin and photoreceptors with 24B10 (blue). Dotted circles in B highlight the accumulation of neurons in large clumps. (E) Number of cells per clone in ey-positive MARCM clones (mean±s.e.m.: 4.08±0.3), in clones mis-expressing eyDN (19.43±1.56), and in clones lacking DE-Cadherin and mis-expressing eyDN (6.04±0.31). (F) Number of large clumps per optic lobe in ey-positive MARCM clones (0), in clones mis-expressing eyDN (5.44±0.28), and in clones lacking DE-Cadherin and mis-expressing eyDN (3.11±0.35). **P<0.001.
To understand the genesis of clustering in eyDN MARCM clones, we analyzed the pattern of proliferation with phospho-Histone 3 in early pupal brains (P5). In wild-type MARCM clones (63 clones analyzed in 26 optic lobes), ey-positive cells formed columns of cells where some individuals cells started to move laterally (Fig. 3G). By contrast, eyDN MARCM clones (75 clones analyzed in 32 optic lobes) were rounder with many cells proliferating (mean±s.e.m.): 1.01±0.12 pH3-positive cells/clone in eyDN MARCM clones versus 0.2±0.04 pH3-positive cells/clone in control) and expressing neuroblast markers (Fig. 3K and data not shown), although other cells clearly showed signs of differentiation. In addition, these neuronal clusters exhibited thick axonal bundles (data not shown).
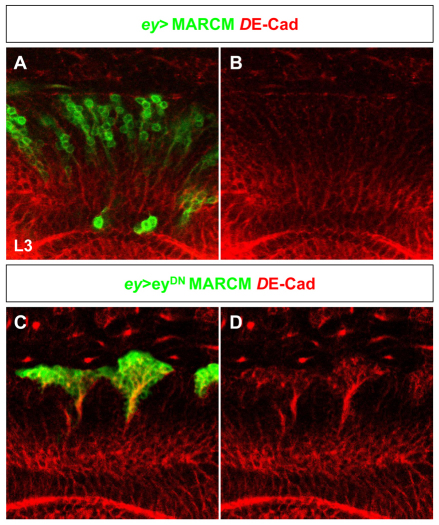
In larval eyDN MARCM clones, upregulation of DE-Cadherin was observed (Fig. 6C,D). In wild-type brains, DE-Cadherin is expressed by postembryonic neuroblasts (Fig. 1F,G) (Dumstrei et al., 2003). Dumstrei et al. (Dumstrei et al., 2003) showed that global expression of a dominant-negative form of DE-Cadherin driven by a heat pulse during early second instar results in a severe phenotype that includes deficits in neural proliferation: although neuroblasts appears in approximately normal numbers, they have highly reduced mitotic activity.
Fig. 6.
Increased expression of DE-Cadherin in eyDN MARCM clones. (A-D) DE-Cadherin (red) staining in ey-MARCM wild-type clones (A,B) and in ey-MARCM clones misexpressing eyDN (C,D) generated 72 hours AEL and analyzed in L3.
In order to test whether the clustering of cells in eyDN resulted from increased expression of DE-Cadherin, we removed DE-Cadherin using a shotgun (shg) mutation in MARCM clones that also expressed eyDN. These clones (Fig. 5D) formed clumps of cells (defined by the presence of more than six cells in a cluster and thick axonal bundles) at a much lower frequency [18/63 brains in eyDN shg exhibited clumps of cells (mean±s.e.m.): 3.11±0.35 clumps/optic lobe, when compared with 129/129 in eyDN brains with 5.44±0.28 clumps/optic lobe] (Fig. 5F), and most eyDN-expressing cells were found distributed throughout the medulla cortex (6.0±0.31 cells per cluster in eyDN shg compared with 19.4±1.56 in eyDN and 4.08±1.47 in wild-type clusters in adult brains) (Fig. 5E). Therefore, the loss of DE-cadherin appears to rescue the clumping phenotype of ey mutant cells, presumably by reducing the mitotic activity of neuroblasts and allowing neurons to migrate.
Thus, the lack of ey causes an increase in neuroblast proliferation and this impedes cells from reaching their final position in the medulla part of the optic lobe. This occurs at least in part by upregulation of DE-Cadherin. Therefore, eyeless links neurogenesis and migration in the optic lobe.
DISCUSSION
We show here that, as previously reported (Meinertzhagen and Hanson, 1993), medulla cortex cells indeed originate from the OPC (Fig. 1). In adults, there are ∼800 `columns' in the medulla, each formed by more than 60 different cell types that repetitively contact every R7-R8 fascicle (Morante and Desplan, 2008). The spatial segregation of ey-, ap- and dll-positive cells observed early in larval brain lobes is lost during pupal development through two modes of cell movements happening during larval (radial) and pupal life (mostly lateral) (Figs 3, 4). As a result, each adult `column' in wild-type medulla is composed of the precise complement of neurons necessary to process visual information (Gao et al., 2008; Morante and Desplan, 2008). Each R7/R8 termination pair is surrounded by at least 47 different cell types, while 13 other cell types do not appear to contact PRs (Morante and Desplan, 2008). Further experiments will determine whether the cell movements shown by medulla cortex cells during optic lobe organogenesis involve the dispersion of the entire neuron, or only the cell body.
ey-, dll- and ap-positive neurons originating from the OPC-derived neuroblasts show a radial organization, while the number of medulla neurons expands (Fig. 1H). This organization resembles the radial units present in the embryonic mammalian neocortex where cells reach their appropriate layer via radial migration along glial processes, expanding the size of the cerebral cortex (Noctor et al., 2001; Rakic, 1988; Rakic, 1992). However, it is unlikely that there is active radial migration in the medulla, as neurons appear to be simply displaced by newly formed neurons above them.
The lack of eyeless causes an increase in neuroblast proliferation and impedes cells from reaching their final position in the medulla. It should be noted that some eyDN cells still manage to disperse in the medulla cortex. They might represent cells that express ey-Gal4 but do not express or require ey for their migration, or cells that express lower levels of ey-Gal4 in which eyDN is not sufficient to affect function.
We also analyzed the effect of eyDN on eye imaginal discs and observed the same phenotype as that reported by Niimi et al. (Niimi et al., 2002); expression of eyDN resulted in underproliferation of eye imaginal disc cells and a partial or complete loss of eye structures (data not shown), which is similar to what is seen in eyeless mutants. Thus, the effects of eyDN in the eye disc are, surprisingly, the opposite of what we observe in medulla neuroblasts.
Furthermore, to address a possible role of eyeless in postmitotic cells, we mis-expressed eyDN using an act-FRT-stop-FRT-Gal4 construct both in larvae and in postmitotic cells (after P25). This mis-expression resulted in lethality, even with very short heat-shocks, probably owing to the strength of the act-Gal4 driver; this precludes analysis in adults. Thus, at this point, we cannot eliminate a possible role of eyeless in postmitotic cells.
Interestingly, Sey mice embryos, which are mutant for the mouse ortholog of ey (Pax6), exhibit an increase in the number of Cajal-Retzius cells, which migrate tangentially (Stoykova et al., 2003). Simultaneously, late-born cortical precursors show abnormalities in their patterns of radial migration, forming heterotopic clusters in the path towards their final position in the embryonic cerebral cortex (Caric et al., 1997). Although this defective neuronal radial migration has been proposed to be due to an altered radial glia morphology (Gotz et al., 1998), other studies have shown that these clusters exhibit an increase in adhesion molecules in neurons (Caric et al., 1997). Biochemical studies have demonstrated the presence of binding sites for the Pax6 transcription factor in the NCAM and L1 promoters (Holst et al., 1997; Meech et al., 1999). Thus, increased proliferation or adhesion among neurons in the clusters could prevent Sey mutant cells from detaching from each other to reach their final position in the cerebral cortex.
Therefore, development of medulla neurons of the optic lobe resembles the neurogenesis and migration mechanisms observed during the development of the embryonic mammalian brain.
Supplementary Material
Acknowledgments
We are very grateful to P. Callaerts, R. Carthew, C. Estella, T. Hayashi, R. Mann, F. Matsuzaki, J. Skeath, J. Treisman, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank at the University of Iowa for reagents, and to I. Tan for help with the Volocity software. We thank Justin Blau, Ching-Man Choi, Volker Hartenstein, Gord Fishell, Juan Luque, Daniel Vasiliauskas and members of the Desplan laboratory for very useful comments on the manuscript and for support and discussions. This work was supported by Grant NIH R01 EY017916 to C.D. We especially thank Maria Dominguez with whom this study was finished with the support of the Ministerio de Ciencia e Innovación (BFU2009-09074 and MEC-Consolider CSD2007-00023), Generalitat Valenciana (PROMETEO 2008/134), Fundacion Marcelino Botin and a European Union research grant UE-HEALTH-F2-2008-201666. J.M. was supported by a JAE-Doc fellowship funded by the Ministerio de Ciencia e Innovacion. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.056069/-/DC1
References
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241-1253 [DOI] [PubMed] [Google Scholar]
- Caric D., Gooday D., Hill R. E., McConnell S. K., Price D. J. (1997). Determination of the migratory capacity of embryonic cortical cells lacking the transcription factor Pax-6. Development 124, 5087-5096 [DOI] [PubMed] [Google Scholar]
- Clements J., Hens K., Francis C., Schellens A., Callaerts P. (2008). Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc. Natl. Acad. Sci. USA 105, 16183-16188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B., McGuffin M. E., Pfeifle C., Segal D., Cohen S. M. (1992). apterous, a gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes Dev. 6, 715-729 [DOI] [PubMed] [Google Scholar]
- Connolly J. B., Roberts I. J., Armstrong J. D., Kaiser K., Forte M., Tully T., O'Kane C. J. (1996). Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science 274, 2104-2107 [DOI] [PubMed] [Google Scholar]
- Dumstrei K., Wang F., Hartenstein V. (2003). Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J. Neurosci. 23, 3325-3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Boone J. Q., Stevens N. R., Brand A. H., Doe C. Q. (2007). Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C., McKay D. J., Mann R. S. (2008). Molecular integration of Wingless, Decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev. Cell 14, 86-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. J., Olson J. M., Ngo K. T., Kim E., Lee N. E., Kuoy E., Patananan A. N., Sitz D., Tran P., Do M. T., et al. (2009). G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach K. F., Dittrich A. P. M. (1989). The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441-475 [Google Scholar]
- Gao S., Takemura S. Y., Ting C. Y., Huang S., Lu Z., Luan H., Rister J., Thum A. S., Yang M., Hong S. T., et al. (2008). The neural substrate of spectral preference in Drosophila. Neuron 60, 328-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W., He F., Kim K. J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J. I., Martinowich K., et al. (2006). Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. USA 103, 1319-1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M., Stoykova A., Gruss P. (1998). Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031-1044 [DOI] [PubMed] [Google Scholar]
- Green P., Hartenstein A. Y., Hartenstein V. (1993). The embryonic development of the Drosophila visual system. Cell Tissue Res. 273, 583-598 [DOI] [PubMed] [Google Scholar]
- Hofbauer A., Campos-Ortega J. A. (1990). Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux's Arch. Dev. Biol. 198, 264-274 [DOI] [PubMed] [Google Scholar]
- Holst B. D., Wang Y., Jones F. S., Edelman G. M. (1997). A binding site for Pax proteins regulates expression of the gene for the neural cell adhesion molecule in the embryonic spinal cord. Proc. Natl. Acad. Sci. USA 94, 1465-1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J. B., Nabeshima Y., Doe C. Q., Matsuzaki F. (1997). Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625-629 [DOI] [PubMed] [Google Scholar]
- Ito K., Hotta Y. (1992). Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 149, 134-148 [DOI] [PubMed] [Google Scholar]
- Kunwar P. S., Siekhaus D. E., Lehmann R. (2006). In vivo migration: a germ cell perspective. Annu. Rev. Cell Dev. Biol. 22, 237-265 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461 [DOI] [PubMed] [Google Scholar]
- McGuire S. E., Mao Z., Davis R. L. (2004). Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004, pl6 [DOI] [PubMed] [Google Scholar]
- Meech R., Kallunki P., Edelman G. M., Jones F. S. (1999). A binding site for homeodomain and Pax proteins is necessary for L1 cell adhesion molecule gene expression by Pax-6 and bone morphogenetic proteins. Proc. Natl. Acad. Sci. USA 96, 2420-2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen I. A., Hanson T. E. (1993). The Development of the Optic Lobe. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Morante J., Desplan C. (2008). The color-vision circuit in the medulla of Drosophila. Curr. Biol. 18, 553-565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J., Desplan C. (2010). Dissecting and staining Drosophila optic lobes. In Drosophila Neurobiology: A Laboratory Manual (ed. Zhang B., Freeman M. R., Waddell S.), pp. 157-166 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Niimi T., Clements J., Gehring W. J., Callaerts P. (2002). Dominant-negative form of the Pax6 homolog eyeless for tissue-specific loss-of-function studies in the developing eye and brain in drosophila. Genesis 34, 74-75 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Flint A. C., Weissman T. A., Dammerman R. S., Kriegstein A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714-720 [DOI] [PubMed] [Google Scholar]
- Ohshiro T., Yagami T., Zhang C., Matsuzaki F. (2000). Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408, 593-596 [DOI] [PubMed] [Google Scholar]
- Rakic P. (1988). Specification of cerebral cortical areas. Science 241, 170-176 [DOI] [PubMed] [Google Scholar]
- Rakic P. (1992). Dividing up the neocortex. Science 258, 1421-1422 [PubMed] [Google Scholar]
- Ramón y Cajal S., Sanchez D. (1915). Contribucion al conocimiento de los centros nerviosos de los insectos. Trab. Lab. Invest. Biol. XIII, 1-167 [Google Scholar]
- Rorth P. (2007). Collective guidance of collective cell migration. Trends Cell Biol. 17, 575-579 [DOI] [PubMed] [Google Scholar]
- Stoykova A., Hatano O., Gruss P., Gotz M. (2003). Increase in reelin-positive cells in the marginal zone of Pax6 mutant mouse cortex. Cereb. Cortex 13, 560-571 [DOI] [PubMed] [Google Scholar]
- Strausfeld N. J. (1976). Atlas of an Insect Brain. Berlin, Heidelberg, New York: Springer-Verlag; [Google Scholar]
- White K., Kankel D. R. (1978). Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev. Biol. 65, 296-321 [DOI] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M., Tabata T. (2008). Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development 135, 1471-1480 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.