Abstract

Professor Irwin Fridovich
Dr. Irwin Fridovich (Ph.D., 1955) is recognized here as a Redox Pioneer because as first/last author he has published at least 1 paper on antioxidant/redox biology that has been cited over 1000 times and has published at least 10 papers each cited over 100 times. In collaboration with his graduate student, Joe McCord, Dr. Fridovich discovered the activity of superoxide dismutase (SOD). Subsequently, he and his colleagues demonstrated that the enzyme is ubiquitous among aerobic biota and comprises a critical defense against oxidative stress. With coworkers, Dr. Fridovich identified the first physiological targets of superoxide, the iron–sulfur clusters of dehydratases. They also showed that SOD is just one of several strategies by which cells fend off oxidative stress. It is now clear that organisms are chronically exposed to endogenous superoxide; further, microbes, plants, and mammals all employ superoxide as a weapon to poison their competitors. Thus, the achievement of Fridovich's laboratory was not only the seminal discovery of SOD but also the painstaking work over the subsequent decades that illuminated its place in biology. Antioxid. Redox Signal. 14, 355–340.
When, by chance, you make an observation that cannot be explained in terms of current knowledge, do not hesitate to pursue it even though it may seem esoteric or unimportant. It may well lead you to discoveries of considerable importance.
—Professor Irwin Fridovich
Educational and Professional Training of Dr. Fridovich
Dr. Fridovich is a native of New York City and received his bachelor's degree at the City College of New York. He then earned his Ph.D. under the direction of Phil Handler in the Biochemistry Department at Duke University. He continued as a postdoc with Handler and ultimately joined the department as a faculty member.
Summary of Dr. Fridovich's Top Contributions
Dr. Fridovich and colleagues discovered superoxide dismutase (SOD). Much of his subsequent work tested the implication of this discovery: that superoxide is formed in aerobic cells and, unless scavenged, can damage cells. These predictions were affirmed, particularly through biochemical and physiological studies of Escherichia coli. Other workers subsequently demonstrated similar results in yeast, Caenorhabolitis elegans, Drosophila, mice, etc., such that superoxide and its partner, hydrogen peroxide, are now regarded as fundamental hazards to all organisms that dwell in the presence of oxygen.
Background, Development, and Training
Dr. Fridovich was a native of New York City, where as a high-school student he attended the Bronx School of Science. As an undergraduate he majored in chemistry at the City College of New York; upon graduating, he spent an extra year isolating a vasopressor from hog kidneys for his biochemistry professor, Abe Mazur. Mazur then encouraged him to go to graduate school at Duke, where Mazur's friend, Phil Handler, chaired the biochemistry department. It was a fateful suggestion: Irwin went to Duke, joining Handler's laboratory to work on the problem of sulfite oxidation, and ultimately spent the remainder of his career there.
Area of Interest in Redox Biology
The beginning: a biochemical problem
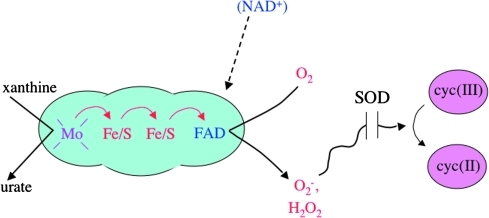
Xanthine oxidase catalyzes consecutive steps in the purine salvage pathway, delivering electrons from substrate via a molybdopterin cofactor, two iron–sulfur clusters, and a flavin to molecular oxygen. It effectively constitutes a small electron-transport chain, and so it was an intriguing enzyme for biochemists to consider as they worked out the rules of redox enzymes. Dr. Fridovich's interest in this enzyme arose when, working with Phil Handler, he found that it could initiate a free-radical chain of sulfite oxidation (10). He also noted that it could use cytochrome c as an artificial electron acceptor. In itself this observation was not surprising, because univalent redox enzymes are notoriously nonspecific in their use of substrates—but, curiously, cytochrome c could only oxidize xanthine oxidase in the presence of oxygen. This fact led him to propose that oxygen, bound to the enzyme, effectively comprised a bridge through which electrons traveled (11). That is, a bound molecule of superoxide might be an intermediate. Why did he not posit superoxide as a diffusible product? Radiation chemists had impressed upon biologists the instability of superoxide, which seemed to argue against the likelihood that it could be generated as a free species. More to the point, a quirk of the enzyme kinetics seemingly suggested that xanthine oxidase possessed two discrete binding sites for oxygen.
In 1967, Joe McCord, a graduate student in Fridovich's laboratory, noted that the apparent Michaelis constant of the enzyme for cytochrome c depended upon the oxygen concentration. The implication was, of course, that the Michaelis constant was determined by something other than a binding constant between the two proteins—and McCord and Fridovich realized that the proteins must not bind together at all and that superoxide must in fact move from one to the other as a free, diffusible species (Fig. 1). Further, although previous experiments had shown that preparations of myoglobin and carbonic anhydrase could inhibit the process, the initial presumption that these proteins did so by outcompeting cytochrome c for the putative binding site had to be revised (32). Instead, these protein preparations contained something that catalytically degraded the nascent superoxide before it could encounter and reduce cytochrome c. Suspecting carbonic anhydrase itself, McCord purified the inhibitor from bovine blood. He arrived instead at a bluish, copper-containing protein that had been described but for which no function was known—erythrocuprein.
FIG. 1.
Xanthine oxidase, the first well-studied enzymatic source of superoxide. Electrons originating on xanthine move from molybdopterin to flavin, the site of oxygen reduction. The enzyme was ultimately found to be a damaged form of xanthine dehydrogenase whose native NAD-binding site had been disrupted by proteolysis or sulfhydryl oxidation (7, 19). SOD, superoxide dismutase; NAD, nicotinamide adenine dinucleotide. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Their proof of its activity was quick and elegant. Erythrocuprein inhibited electron transfer reactions whether the superoxide was generated by an enzyme or an electrode, and whether the acceptor was a protein or a dye. Erythrocuprein did so catalytically, and it was shown to do so by dismutation:
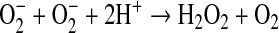 |
In November 1969, McCord and Fridovich published their results in the Journal of Biological Chemistry (33). That paper has been cited more than 7900 times.
Description of Key Finding 1
Circumstantial evidence that SOD protects cells from oxygen
Was the preceding work simply a masterful solution to a laboratory artifact? Or did it have biological meaning? The chemists knew that superoxide is so unstable that, in reagent quantities, it is explosive; therefore, it seemed unlikely that biological systems could create it, and if they somehow did, it seemed certain that spontaneous dismutation would be rapid enough that cells would not require a scavenging enzyme. Thus, it seemed plausible that the dismutation reaction of erythrocuprein was merely the adventitious activity of a protein that harbored copper for some other purpose. And so, at this point, what had been an enzymological puzzle became a physiological one.
McCord and Fridovich's first gambit was to test how widely the enzyme was distributed across the biota. They found that it was ubiquitous among animals and plants. Bacteria also contained SODs; interestingly, the first one that was isolated, from E. coli, used manganese rather than copper as its catalytic metal (22). The laboratory subsequently discovered a similar one in mitochondria, which would ultimately support the endosymbiotic theory of organellular evolution (38). But the more critical test was this: If the enzyme truly served to degrade a species derived from molecular oxygen, then one might expect it to be absent from organisms that live in the absence of oxygen, for they would have no need for it. Enlisting colleagues to provide samples, McCord and Fridovich found that SOD was substantial in aerobic microbes and only slightly less so in aerotolerant anaerobes. Most tellingly, the strict anaerobes that were sampled lacked detectable activity. This correlation, McCord and Fridovich concluded, supported the notion that SOD proteins were useful only in aerobic habitats, and it was consistent with its proposed role in fending off superoxide (34).
Physical studies also indicated that the enzyme was exquisitely evolved to scavenge superoxide. Duke colleagues discovered that the catalytic efficiency of the copper enzyme actually exceeded the presumptive limitation created by substrate diffusion (14, 36). The reason is that a cationic channel actively pulls substrate molecules from bulk solution toward the active-site metal. It was hard to square such perfection with adventitious chemistry. Moreover, because the enzymatic dismutation reaction consists of consecutive half reactions, it is kinetically first-order in substrate concentration, in contrast to the second-order spontaneous reaction. Thus, the enzyme-catalyzed reaction is far, far faster at degrading moderate levels of superoxide.
If superoxide warranted a scavenging system, then it must be generated somehow in diverse types of cells, and it must be capable of damaging at least some biomolecules. Both points gave pause. It was not immediately clear how cells might generate enough superoxide to require defenses, and it was even less obvious what kind of biological molecules superoxide might damage. Over the next few years, for some people this uncertainty hardened into strong skepticism.
Gene regulation is a useful indicator of protein function, and so Fridovich's laboratory attacked this problem by inspecting the pattern of SOD synthesis. Mick Gregory showed that E. coli did not synthesize its manganese-containing SOD (MnSOD) isozyme in anaerobic habitats, but the enzyme was made in moderate amounts in aerobic environments and was induced to very high levels when cultures were exposed to pressurized oxygen (15). Hosni Hassan then demonstrated that a similarly high level of MnSOD synthesis could be achieved under normoxic conditions if redox-cycling antibiotics, such as paraquat or streptonigrin, were added to the media (17). These compounds penetrate cells, where they abstract electrons from redox enzymes and transfer them to oxygen, forming superoxide. The apparent implication was that SOD was induced by its substrate. Even more strikingly, cells that had been preinduced to make much SOD were especially resistant to the toxicity of hyperoxia and of redox-cycling drugs (18). Collectively, the evidence supported the conclusion that SOD exists to dismute superoxide.
Description of Key Finding 2
Is superoxide formed in vivo? Is it harmful?
With the exceptions of aldehyde/xanthine oxidases and, perhaps, hemoglobin—proteins that certainly are not universal among cell types—prospective biological sources of superoxide were not immediately obvious. Lynch and Fridovich demonstrated that because superoxide is a charged species at physiological pH (pKa = 4.8), it requires a channel to penetrate lipid bilayers (29). Thus, environmental oxidation reactions should not create a need for intracellular SOD activity in bacteria, for example, which lack passive ion channels.
An alternative explanation was quickly found. Vince Massey and colleagues used the cytochrome c assay to show that reduced flavoproteins adventitiously transfer electrons to dissolved oxygen, releasing superoxide as a detectable product (30). The observation provided a foundation for the idea that superoxide is chronically formed inside aerobic cells. Boveris and Cadenas subsequently showed that the mitochondrial bc1 respiratory complex of eukaryotes leaks electrons to oxygen from both its Qo and Qi semiquinone intermediates (3). Superoxide is thereby released on each side of the membrane, and indeed SOD isozymes were discovered in both the mitochondrial matrix and the intermembrane space.
These advances reassured workers that they were on the right track, but the problem of damage mechanism was still daunting. Superoxide acts alternatively as a univalent reductant and oxidant in the dismutation reaction (midpoint reduction potential for 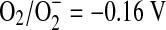 ; midpoint reduction potential for
; midpoint reduction potential for 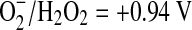 ); indeed, Fridovich's laboratory had documented its capacity to both reduce cytochrome c and oxidize dyes. However, the basic molecules of which organisms are comprised—amino acids, carbohydrates, nucleic acids, lipids—are neither reducible nor easily oxidized. Notably, the oxidizing capacity of superoxide is held in check because, as a physiological anion, it is not electrophilic and cannot abstract electrons at all unless it is first protonated. So how might superoxide harm a cell?
); indeed, Fridovich's laboratory had documented its capacity to both reduce cytochrome c and oxidize dyes. However, the basic molecules of which organisms are comprised—amino acids, carbohydrates, nucleic acids, lipids—are neither reducible nor easily oxidized. Notably, the oxidizing capacity of superoxide is held in check because, as a physiological anion, it is not electrophilic and cannot abstract electrons at all unless it is first protonated. So how might superoxide harm a cell?
Fridovich's group found that superoxide and hydrogen peroxide collaborate in vitro in the formation of hydroxyl radicals, which are potent oxidants of most organic molecules (1). McCord and Day subsequently showed that the process depended upon the reduction of trace ferric iron by superoxide; ferrous iron then transferred the electron to hydrogen peroxide, generating a hydroxyl radical (31). For a period this process stood as a plausible mechanism of superoxide toxicity in vivo. In retrospect, its greatest impact was to focus attention upon the ability of superoxide to react with iron.
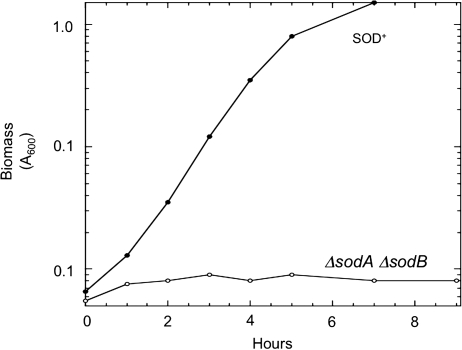
The genetic era was dawning. In the earliest days, McCord and Fridovich had attempted to isolate SOD-deficient mutants of E. coli, but they had failed; it was now apparent that their simple strategy had been thwarted by the existence of two SOD isozymes. However, in 1986, Daniele Touati used antibodies to identify the manganese- and iron-containing SOD (MnSOD and FeSOD) structural genes of E. coli; she then knocked out both (5). The outcome still stands as the best proof of the critical role of SOD: although double mutants grew well in glucose medium when they were cultured in the absence of oxygen, they could not grow at all if the culture was aerated (Fig. 2). Amino acid supplements restored growth, and Touati determined that superoxide somehow blocked the syntheses of branched-chain, aromatic, and sulfurous amino acids. Further work showed that the mutants were unable to catabolize standard E. coli carbon sources such as acetate and succinate. They also exhibited a high rate of mutagenesis (8).
FIG. 2.
Proof of SOD function. Escherichia coli mutants that lack cytoplasmic SOD cannot grow in aerobic minimal glucose medium (5). Anaerobic growth is unaffected (not shown). Figure courtesy of K.R.C. Imlay.
Still, as the molecular mechanisms of damage were yet to be revealed, everyone was not convinced. Some suggested that the biological purposes of MnSOD and FeSOD might be to traffic or store their cognate metals, functions that might be expendable under anaerobic conditions. But this possibility was answered by one final experiment: the phenotypic deficits of the double SOD mutant were fully complemented by a plasmid that expressed the human copper/zinc-containing SOD enzyme (35). The latter protein was structurally unrelated to the bacterial enzymes, and it was incapable of binding either iron or manganese; the only feature it had in common with them was that it rapidly dismuted superoxide. And with that, the proof was completed.
Description of Key Finding 3
So how does superoxide damage cells?
The aforementioned genetic result did not immediately reveal the mechanism of superoxide toxicity, but it provided a hint. Brown and colleagues had previously observed that hyperoxia blocked the branched-chain biosynthetic pathway of E. coli, and they had tracked the defect to inactivation of dihydroxyacid dehydratase, an enzyme in that pathway (2, 4). Now Touati had shown that the same growth phenotype could be generated by superoxide stress. Suspecting, then, that superoxide was the direct effector of hyperoxic toxicity, Kuo and Fridovich were able to poison the dehydratase by growing E. coli in the presence of redox-cycling antibiotics. And finally, they demonstrated that superoxide could directly inactivate the enzyme in vitro (24). For the first time, a physiological target of superoxide toxicity had been identified.
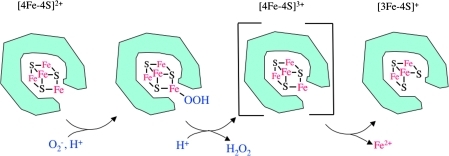
In Fridovich's laboratory, Paul Gardner and Stefan Liochev quickly found that dihydroxyacid dehydratase was not unique. Superoxide also inactivated related dehydratases, both in vivo and in vitro: aconitase, 6-phosphogluconate dehydratase, and fumarases A and B (12, 13, 25). The common feature of these enzymes is that they all employ solvent-exposed [4Fe-4S] clusters to bind and dehydrate their substrates. So how does superoxide inactivate them? Flint and colleagues showed that anionic superoxide ligands and oxidizes the exposed catalytic iron atom, shifting the cluster from a stable +2 valence to an unstable +3 one. The catalytic iron atom then dissociates, the enzyme loses activity, and the pathway fails (Fig. 3) (9).
FIG. 3.
Superoxide oxidizes and degrades the iron–sulfur clusters of dehydratases. Superoxide directly oxidizes the iron atom. Exposure of this iron atom to solutes is essential for enzyme function, because it coordinates and activates the natural substrates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
As aconitase and fumarase are essential for tricarboxylic acid cycle function, this discovery also explained the acetate and succinate growth defects. It additionally revealed how superoxide promotes mutagenesis: the iron that is released from damaged clusters accumulates in the cell, where it participates in Fenton chemistry and thereby catalyzes a high rate of hydroxyl-radical formation (23, 26). A complete exegesis of superoxide toxicity had finally been achieved.
Other Achievements: From Physiology to Ecology
Once its natural sources and targets had been identified, superoxide was gradually recognized as a weapon of choice in the interspecies warfare that occurs continuously in the biological world. Both plants and bacteria release redox-cycling antibiotics to poison their competitors. The induction of MnSOD in response to such agents, first noted by Hassan and Fridovich, was discovered to be just one part of a regulated response that also controls enzymes that modify or export these drugs (16, 37). Interestingly, Liochev and Fridovich found that this response additionally includes the induction of a cluster-free fumarase isozyme, which replaces the vulnerable isozymes when intracellular antibiotic levels are high (25).
Famously, the mammalian macrophage deliberately toxifies captured bacteria by spraying them with superoxide. The cellular target that this superoxide attacks is not yet known, but we do know that pathogenic bacteria rely on a periplasmic or cell-surface–associated copper/zinc-containing SOD to defray its effects (6). In addition, superoxide is an obligatory precursor to peroxynitrite, which exerts toxic effects of its own.
What about those anaerobic microbes? Interestingly, investigators ultimately found that many of these organisms, like their aerobic peers, have superoxide-scavenging enzymes, but they contain superoxide reductases rather than dismutases (21, 28). Work in Fridovich's laboratory also helped facilitate this discovery (27). Dismutation is simply the preferred device of aerobes, either because they can tolerate the oxygen that it produces, or because their redox poise, unlike that of anaerobes, is suitable for dismutation rather than reduction (20). It turns out that all organisms periodically confront oxygen, and so they all need to have defenses ready and waiting.
The big view has now come into focus. The geological record informs us that life originated and evolved on an Earth that was essentially anaerobic. During this long period, the familiar enzymatic mechanisms, biochemical pathways, and metabolic networks evolved. Only after 2 billion years did oxygenic photosynthetic microbes gradually create our contemporary aerobic environment and, with it, oxidative stress. We now see that the appearance of SOD was an important part of evolution's effort to protect metabolism—invented in an anaerobic world, but now employed in an aerobic one—from this epochal transformation of the environment.
Current Position
Dr. Fridovich is currently an emeritus professor in the Department of Biochemistry at Duke. The biochemical lesson of his work may be that mistakes happen, and in redox biology, they happen a lot. Superoxide is formed when molecular oxygen slips into the active sites of redox enzymes and oxidizes their flavins or quinones; the superoxide subsequently poisons cells by entering the active sites of dehydratases and oxidizing their iron–sulfur clusters. In that sense the promiscuity of redox enzymes has come home to roost. Dr. Fridovich suggests that the lesson for researchers might be to attend to the odd artifact—“When, by chance, you make an observation that cannot be explained in terms of current knowledge, do not hesitate to pursue it even though it may seem esoteric or unimportant. It may well lead you to discoveries of considerable importance.” To funding agencies, he adds, “Do not be so focused on practical results, currently called ‘translational research,’ that you fail to fund work that merely seeks explanations for puzzling phenomena. It is such ‘basic’ research that provides the facts that may then be translated into practical benefits.”
Supplementary Material
Abbreviations Used
- FeSOD
iron-containing SOD
- MnSOD
manganese-containing SOD
- SOD
superoxide dismutase
Footnotes
Reviewing Editors: Joseph S. Beckman, James Crapo, Thomas Kietzmann, Joe M. McCord, Daret St. Clair, and John A. Tainer
Author note: The author worked as a postdoctoral fellow with Dr. Fridovich from 1987 to 1992, and together they investigated the mechanisms of superoxide formation inside Escherichia coli. He is currently a professor in the Microbiology Department at the University of Illinois.
For a list of frequently cited articles published by Prof. Fridovich, see Supplemental Tables 1 and 2, available online at www.liebertonline.com/ars.
Acknowledgments
Professor Fridovich thanks the people with whom he worked over the years: “I should acknowledge the talented and hard-working students and postdocs whose efforts produced most of the edifice of knowledge that emerged from my lab. It has been a joy and a privilege to have known and labored with them.” The work in the lab of J.A.I. was supported by a grant from NIH (GM49640).
References
- 1.Beauchamp C. Fridovich I. A mechanism for the production of ethylene from methional. The generation of the hydroxyl radical by xanthine oxidase. J Biol Chem. 1970;245:5214–5222. [PubMed] [Google Scholar]
- 2.Boehme DE. Vincent K. Brown OR. Oxygen and toxicity: inhibition of amino acid biosynthesis. Nature. 1976;262:418–420. doi: 10.1038/262418a0. [DOI] [PubMed] [Google Scholar]
- 3.Boveris A. Cadenas E. Production of superoxide radicals and hydrogen peroxide in mitochondria. In: Oberley LW, editor. Superoxide Dismutase. Vol. 2. Boca Raton, FL: CRC Press; 1982. pp. 15–30. [Google Scholar]
- 4.Brown OR. Yein F. Dihydroxyacid dehydratase: the site of hyperbaric oxygen poisoning in branch-chain amino acid biosynthesis. Biochem Biophys Res Commun. 1978;85:1219–1224. doi: 10.1016/0006-291x(78)90672-1. [DOI] [PubMed] [Google Scholar]
- 5.Carlioz A. Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeGroote MA. Ochsner UA. Shiloh MU. Nathan C. McCord JM. Dinauer MC. Libby SJ. Vazquez-Torres A. Xu Y. Fang FC. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enroth C. Eger BT. Okamoto K. Nishino T. Nishino T. Pai EF. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc Natl Acad Sci USA. 2000;97:10723–10728. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farr SB. D'Ari R. Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint DH. Tuminello JF. Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 10.Fridovich I. Handler P. Xanthine oxidase. III. Sulfite oxidation as an ultrasensitive assay. J Biol Chem. 1958;233:1578–1580. [PubMed] [Google Scholar]
- 11.Fridovich I. Handler P. Xanthine oxidase. V. Differential inhibition of the reduction of various electron acceptors. J Biol Chem. 1962;237:916–921. [PubMed] [Google Scholar]
- 12.Gardner PR. Fridovich I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 13.Gardner PR. Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 14.Getzoff ED. Tainer JA. Weiner PK. Kollman PA. Richardson JS. Richardson DC. Electrostatic recognition between supeoxide and copper, zinc superoxide dismutase. Nature. 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 15.Gregory EM. Fridovich I. Induction of superoxide dismutase by molecular oxygen. J Bacteriol. 1973;114:543–548. doi: 10.1128/jb.114.2.543-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg JT. Monach P. Chou JH. Josephy PD. Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in E. coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassan HM. Fridovich I. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J Biol Chem. 1977;252:7667–7672. [PubMed] [Google Scholar]
- 18.Hassan HM. Fridovich I. Superoxide radical and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J Biol Chem. 1978;253:8143–8148. [PubMed] [Google Scholar]
- 19.Hille R. Nishino T. Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. FASEB J. 1995;9:995–1003. [PubMed] [Google Scholar]
- 20.Imlay JA. What biological purpose is sereved by superoxide reductase? J Biol Inorg Chem. 2002;7:659–663. doi: 10.1007/s00775-002-0361-3. [DOI] [PubMed] [Google Scholar]
- 21.Jenney FE., Jr. Verhagen MFJM. Cui X. Adams MWW. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 22.Keele BB., Jr. McCord JM. Fridovich I. Superoxide dismutase from Escherichia coli B. A new manganese-containing enzyme. J Biol Chem. 1970;245:6176–6181. [PubMed] [Google Scholar]
- 23.Keyer K. Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo CF. Mashino T. Fridovich I. α,β-dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 25.Liochev SI. Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liochev SI. Fridovich I. The role of O2- in the production of HO.: in vitro and in vivo. Free Rad Biol Med. 1994;16:29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 27.Liochev SI. Fridovich I. A mechanism for complementation of the sodA sodB defect in Escherichia coli by overproduction of the rbo gene product (desulfoferrodoxin) from Desulfoarculus baarsii. J Biol Chem. 1997;272:25573–25575. doi: 10.1074/jbc.272.41.25573. [DOI] [PubMed] [Google Scholar]
- 28.Lombard M. Fontecave M. Touati D. Niviere V. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J Biol Chem. 2000;275:115–121. doi: 10.1074/jbc.275.1.115. [DOI] [PubMed] [Google Scholar]
- 29.Lynch RE. Fridovich I. Permeation of the erythrocyte stroma by superoxide radical. J Biol Chem. 1978;253:4697–4699. [PubMed] [Google Scholar]
- 30.Massey V. Strickland S. Mayhew SG. Howell LG. Engel PC. Matthews RG. Schuman M. Sullivan PA. The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem Biophys Res Commun. 1969;36:891–897. doi: 10.1016/0006-291x(69)90287-3. [DOI] [PubMed] [Google Scholar]
- 31.McCord JM. Day ED. Superoxide-dependent production of hydroxyl radical catalyzed by iron-EDTA complex. FEBS Letts. 1978;86:139–142. doi: 10.1016/0014-5793(78)80116-1. [DOI] [PubMed] [Google Scholar]
- 32.McCord JM. Fridovich I. The reduction of cytochrome c by milk xanthine oxidase. J Biol Chem. 1968;243:5753–5760. [PubMed] [Google Scholar]
- 33.McCord JM. Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 34.McCord JM. Keele BB., Jr. Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Natvig DO. Imlay K. Touati D. Hallewell RA. Human copper-zinc superoxide dismutase complements superoxide dismutase-deficient Escherichia coli mutants. J Biol Chem. 1987;262:14697–14701. [PubMed] [Google Scholar]
- 36.Tainer JA. Getzoff ED. Richardson JS. Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983;306:284–286. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 37.Tsaneva IR. Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisiger R. Fridovich I. Superoxide dismutase. Organelle specificity. J Biol Chem. 1973;248:3582–3592. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.