Abstract
Genetic diversity in RNA viruses is shaped by a variety of evolutionary processes, including the bottlenecks that may occur at inter-host transmission. However, how these processes structure genetic variation at the scale of individual hosts is only partly understood. We obtained intra-host sequence data for the coat protein (CP) gene of Zucchini yellow mosaic virus (ZYMV) from two horizontally transmitted populations – one via aphid, the other without – and with multiple samples from individual plants. We show that although mutations are generated relatively frequently within infected plants, attaining similar levels of genetic diversity to that seen in some animal RNA viruses (mean intra-sample diversity of 0.02%), most mutations are likely to be transient, deleterious, and purged rapidly. We also observed more population structure in the aphid transmitted viral population, including the same mutations in multiple clones, the presence of a sub-lineage, and evidence for the short-term complementation of defective genomes.
Keywords: evolution, intra-host diversity, Zucchini yellow mosaic virus, plant RNA virus, potyvirus, complementation
1. Introduction
Determining the extent and structure of genetic variation in RNA viruses is central to understanding the mechanisms that shape their evolution. The high levels of genetic diversity that characterize many RNA viruses have been linked to their ability to adapt rapidly to changing environments including new host species (Holmes, 2009; Jerzak et al., 2008; Woolhouse et al., 2001), and to evade mechanisms of host resistance (Feuer et al., 1999; Lech et al., 1996). Most estimates of the rate of molecular evolution in animal RNA viruses fall within approximately one order of magnitude of a mean rate of 1 × 10−3 nucleotide substitutions per site, per year (subs/site/year; Duffy et al., 2008). In contrast, it has previously been suggested that plant RNA viruses are characterized by lower rates of evolutionary change, in some cases by several orders of magnitude (Blok et al., 1987; Fraile et al., 1997; Kim et al., 2005; Marco and Aranda, 2005; Rodriguez-Cerezo et al., 1991). This major difference in evolutionary dynamics has been attributed to intrinsically lower mutation rates, weaker immune-mediated positive selection, and the frequent occurrence of population bottlenecks (Garcia-Arenal et al., 2001). However, more recent analyses using longitudinally sampled gene sequence data have resulted in substitution rate estimates in accord with those previously observed in animal RNA viruses, at least in the short term (Fargette et al., 2008; Gibbs et al., 2008; Gibbs et al., 2010; Pagán and Holmes, 2010). As a case in point, we previously reported a mean evolutionary rate of 5 × 10−4 subs/site/year for the coat protein (CP) of Zucchini yellow mosaic virus (ZYMV) (Simmons et al., 2008).
Most studies of genetic diversity in plant viruses have been conducted at the inter-host level. However, if plant RNA viruses do evolve as rapidly as suggested by the analysis of epidemiological scale sequence data then we would also expect them to exhibit measurable genetic diversity at the intra-host scale. Those studies undertaken to date have found varying levels of intra-host variation. Turturo et al. (2005) observed limited (<0.1%) intra-host genetic diversity in Grapevine leafroll-associated virus, while Jridi et al. (2006) noted that the nucleotide diversity of Plum pox virus measured over 13 years in a prunus tree ranged from 0 to 2.4%. Rather higher levels of intra-host diversity were observed in Banana mild mosaic virus, with divergence levels of more than 15% in a third of the sequences obtained (Teycheney et al. 2005).
Determining the extent and patterns of intra-host genetic diversity in plant RNA viruses is central to revealing the fundamental processes of viral evolution. Large-scale population bottlenecks are thought to result in effective population sizes for RNA viruses that are several orders of magnitude lower than consensus population numbers (García-Arenal et al., 2001). Indeed, population bottlenecks have been documented during aphid transmission in Cucumber mosaic virus (Ali et al., 2006; Betancourt et al., 2008) and Potato virus Y (Moury et al., 2007). Systemic bottlenecks (that occur as the virus moves from cell-to-cell and tissue-to-tissue) may reduce effective population sizes even further (French and Stenger, 2003; Sacristan et al., 2003; Li and Roossinck, 2004; Miyashita and Kishino, 2010). In these circumstances genetic drift is predicted to play a major role in the substitution dynamics of mutant alleles. However, little is known about the frequency and impact of population bottlenecks in natural virus populations (Li and Roossinck, 2004). As an exception, the extent of genetic diversity in Citrus tristeza virus transmitted via aphids was reduced by an order of magnitude compared to that found in the sweet orange (Citrus sinensis) host (Nolasco et al., 2008).
ZYMV was first isolated in 1973 in Italy, and since this time the virus has been found in more than 50 countries as a naturally occurring infection of the Cucurbitaceae (Desbiez and Lecoq, 1997; Desbiez et al., 2002). Viral symptoms include a distinctive yellow mottling in the leaves, stunting of the plant, and severe deformities in the fruits and leaves (Desbiez and Lecoq, 1997; Gal-on, 2007). Production of cucurbits in the United States is valued at approximately $1.5 billion per annum (Cantliffe et al., 2007), and as ZYMV infection can reduce agricultural yields by up to 94% (Blua and Perring, 1989), it is one of the most economically significant agricultural pathogens in cultivated cucurbits (squash, melon and cucumber). ZYMV is a member of the Potyviridae family of positive-sense, single-stranded encapsidated RNA viruses. The ~9.5 kb viral genome encodes a single polyprotein precursor that is cleaved into ten putative proteins (Gal-on, 2007). Transmission occurs primarily via aphids in a nonpersistent manner (Lisa et al., 1981) and, to date, 26 aphid species have been shown to transmit ZYMV (Katis et al., 2006). The viral coat protein (CP) is multifunctional and involved in cell-to-cell and systemic movement, the regulation of viral RNA amplification (Urcuqui-Inchima et al., 2001), encapsidation of the RNA, vector transmission (Urcuqui-Inchima et al., 2001; Shukla et al., 1991), and perhaps host specificity (Shukla et al. 1991). ZYMV transmission is the result of an interaction between the stylet of the aphid, the helper component protein (HC-Pro), and the conserved DAG (Asp-Ala-Gly) region of the CP (Pirone and Blanc, 1996). The highly variable N-terminus region of the CP is exposed on the surface of the coat protein and is thought to contain virus-specific epitopes. The core region and C-terminus are more conserved, although the last ten amino acids of the C-terminus may be exposed on the viral surface (Gal-On, 2007).
To obtain a better understanding of the patterns and processes of plant virus evolution at the scale of individual hosts, we analyzed the intra-host genetic diversity of ZYMV in Cucurbita pepo ssp texana (a wild gourd) under two distinct modes of transmission: aphid-vectored and mechanically-inoculated (i.e. without aphids). The aphid-vectored experiment was conducted in an experimental field and resulted in two types of data; a time series as the virus evolves within the host over the course of the infection, and epidemiological-scale data following the spread of the virus as it was transmitted by aphids between hosts during the growing season. Because the number of transmission events is not controlled, these data recapitulate the natural spread of the virus. Using data of the first type the extent of the bottleneck imposed by the aphid during transmission can be estimated. The second type of data allowed us to determine if mutations are transmitted between individuals or are generated anew within each individual.
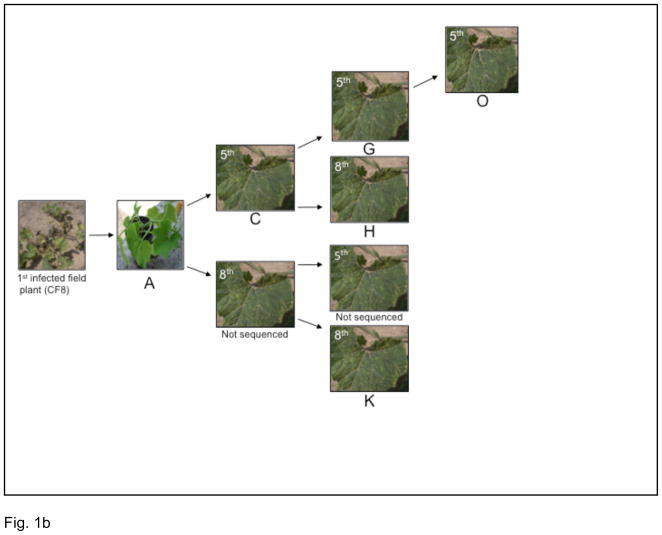
In the mechanical inoculation experiment, carried out in a greenhouse, ZYMV was serially passaged across four generations by mechanical inoculation. By comparing these data to those from the field study we were able to compare viral genetic diversity with and without the aphid-imposed bottleneck. To assess the effect of intra-host systemic bottlenecks, half of the fifth and eighth leaves from each mechanically-inoculated individual were used separately to inoculate another individual. This follows the design of two earlier studies which showed that the number of mutant clones present in a leaf decreased as a function of distance from the original inoculum source, presumably as a result of systemic bottlenecks (Li and Roossinck, 2004; Ali and Roossinck, 2010).
2. Methods
2.1. Field experiment
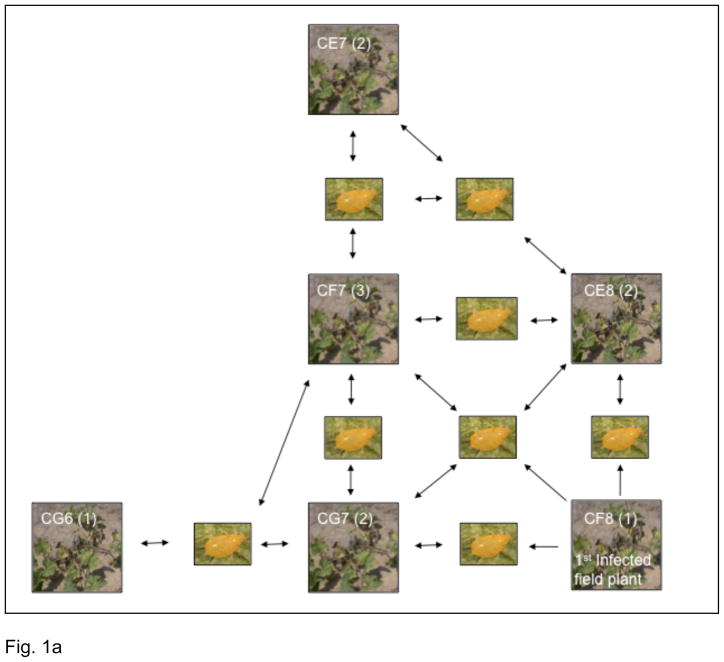
The field experiment was conducted at The Pennsylvania State University Agriculture Experiment Station at Rock Springs, Pennsylvania, USA, using Cucurbita pepo ssp. texana (a wild gourd). One 0.4-hectare field was laid out as a grid labeled A-L and 1–15, with approximately six meters between plants and 180 plants per field (Fig. 1a). In 2007 individual F-8 (located in the middle of one of the fields) was mechanically inoculated with ZYMV, the consensus sequence of which has been deposited in GenBank (accession number EU371649). When the inoculated plant, CF8, exhibited viral symptoms a leaf was collected. Plant labels are as follows: The first digit C designates that the sample was collected from the field, the next digit and number in this case F8, designate the plant coordinates within the field grid, and the number in parenthesis denotes the order in which samples where collected from an individual plant. As neighboring plants became infected, leaf samples were collected so that a leaf sample was gathered every two weeks from each individual that displayed disease symptoms from the onset of visible symptoms until the host plant died (approximately 9 weeks in total). Presence of ZYMV was detected immunologically using DAS-ELISA (Agdia, IN) and confirmed by polymerase chain reaction (PCR) and sequencing of the viral CP. The DAS-ELISA results not only confirmed the presence of ZYMV in the field plants but also revealed that all but one of the plants (CE7) was co-infected with another potyvirus. Leaf samples from confirmed ZYMV-infected plants were stored at −80°C. Although samples were collected from all of the infected plants in the field, eleven of these, which represents six individual plants, were selected for sequencing. One plant (CF7) was sampled at three time points (August 4th, August 28th, September 13th); three plants were sampled at two time points (CE7 on September 13th and September 20th, CE8 on August 8th and September 13th, and CG7 on August 30th and September 20th); and clonal sequences were sampled only once from two plants (CF8 and CG6; Table 1).
Fig. 1.
Experimental design of the current study. (a) Field experiments. The schematic shows the position of the field plants relative to each other. Plant labels are as follows: The first digit C designates that the sample was collected from the field, the next two digits designate the plant coordinates within the field grid, and the number in parenthesis denotes the number of samples collected from an individual plant. The boxed images that occur between the sampled field plants are of Aphis gossypii (cotton aphid), which serves to indicate that the spread of infection in the field occurred naturally (i.e. was aphid vectored). (b) Greenhouse experiments. The first field infected plant was used to infect plant A, the fifth leaf of which was used to infect C. The fifth leaf of C was used to infect G and the eighth leaf to infect H. The fifth leaf of G was used to infect O, and K was infected from the third generation from the eighth leaf of A.
Table 1.
Summary of the ZYMV CP sequences from each infected plant under aphid-vectored (field) and mechanically-inoculated (greenhouse) transmission
| Sample | Number of sequences | Total number variant sites | Mean pairwise divergence (%) | Mean dN/dS (CI 95%) | Stop codon mutations | Sequences with more than one mutation from consensus | Number of sequences with the same mutation |
|---|---|---|---|---|---|---|---|
| CE7 (1) | 40 | 4 | 0.03 | 0.41 (0.07–1.28) | 0 | 0 | 2 |
| CE7 (2) | 36 | 0 | 0 | NA (NA) | 0 | 0 | 0 |
| CE8 (1) | 39 | 1 | 0.02 | 7.97 (0.44–34.08) | 0 | 1 | 0 |
| CE8 (2) | 30 | 2 | 0.02 | 0.48 (0.028–2.13) | 0 | 1 | 0 |
| CF7 (1) | 38 | 3 | 0.02 | 0.18 (0.01–0.78) | 0 | 0 | 0 |
| CF7 (2) | 32 | 2 | 0.01 | 2.65 (0.15–11.73) | 0 | 0 | 0 |
| CF7 (3) | 37 | 3 | 0.02 | 0.50 (0.084–1.56) | 0 | 1 | 0 |
| CF8 | 38 | 0 | 0 | NA (NA) | 0 | 0 | 0 |
| CG6 | 36 | 1 | 0.01 | 1.52 (0.086–6.67) | 0 | 0 | 0 |
| CG7 (1) | 36 | 6 | 0.11 | 1.29 (0.32–3.35) | 1 | 21 | 2/7/7/13a |
| CG7 (2) | 16 | 1 | 0.04 | 0 (0–0.56) | 0 | 0 | 0 |
| Field total | 376 | 23 | 0.02 | 0.54 (0.23–0.84) | 1 | 24 | NA |
| GA5 | 35 | 1 | 0.01 | 1.37 (0.078–6.06) | 0 | 0 | 0 |
| GA8 | 39 | 1 | 0.01 | 0 (0–1.09) | 0 | 0 | 0 |
| GC5 | 40 | 1 | 0.01 | 2.46 (0.14–10.90) | 0 | 0 | 0 |
| GC8 | 36 | 5 | 0.03 | 1.31 (0.41–3.10) | 0 | 0 | 0 |
| GG5 | 37 | 0 | 0 | NA (NA) | 0 | 0 | 0 |
| GH5 | 38 | 2 | 0.01 | 1.85 (0.10–8.25) | 0 | 0 | 0 |
| GH8 | 35 | 4 | 0.03 | 0.29 (0.049–0.90) | 0 | 0 | 0 |
| GK5 | 36 | 2 | 0.01 | 5.25 (0.87–16.27) | 0 | 0 | 0 |
| GO5 | 32 | 2 | 0.05 | 1.61 (0.092–7.10) | 1 | 0 | 7 |
| Greenhouse Total | 325 | 18 | 0.02 | 0.66 (0.34–1.13) | 1 | 0 | NA |
Refers to four sites with four different mutations
2.2. Greenhouse experiment
Two individual plants were mechanically inoculated in January of 2008 with a ZYMV sample taken from the first diseased individual from the 2007 season (CF8). The mechanical inoculations performed in the greenhouse using carborundum powder (500gm). The infectious tissue was prepared from infected plant tissue diluted in a phosphate buffer (0.1 M Na2H/KH2PO4 buffer) in a 1:3 ratio. The carborundum powder was dusted on the surface of the leaf, and the inoculum was then applied with a pestle to the leaf surface. When the plants displayed disease symptoms and exhibited at least an additional eight leaves of growth from the inoculation site (typically 4 to 5 weeks), half of the fifth and eighth leaves (distance from the first inoculated leaf) each were each used separately to inoculate another individual and so on through four generations (Fig. 1b). The infection rate of the mechanical inoculations was 100%. The other half of each leaf was stored at −80°C. We generated clonal sequence data from nine samples representing one transmission chain. In summary, clones were generated from the fifth and eighth leaves of individual A, the fifth and eighth leaves of individual C (which was infected from the fifth leaf of A), the fifth leaf of individual G (which was infected from the fifth leaf of C), the fifth and eighth leaves of individual H (which was infected from the eighth leaf of C), and the fifth leaf of individual O (which was infected from the fifth leaf of G). In addition, we sequenced one sample (fifth leaf of K) from the third generation from the eighth leaf of A.
2.3. RNA isolation, PCR analysis, cloning and sequencing
RNA was isolated from frozen leaf samples using the RNeasy® Plant Mini Kit (Qiagen, CA). First-strand cDNA was synthesized from the extracted RNA following the protocol provided by the supplier using the Superscript™ III First-Strand kit (Invitrogen, CA). The target cDNA was then amplified directly via PCR using Phusion® High-Fidelity PCR Master Mix (Finnzyme, MA). Although we used a high fidelity Taq polymerase to reduce the number of ‘mutations’ introduced during the experimental procedure, it is impossible to fully eliminate RT-introduced errors from occurring (see Results). Prior to cloning with the TOPO® TA Cloning® Kit (Invitrogen, CA), each sample was purified using the QIAquick PCR Purification Kit (Qiagen, CA) and an A overhang was added to each sample. Before submitting samples for sequencing at The Pennsylvania State University Nucleic Acid Facility, each sample was purified with the QIAprep Spin Miniprep Kit (Qiagen, CA). The CP-specific primers used for the cDNA, PCR and steps were: forward: AAGTGAATTGGCACGCTA; reverse: CGGTAAATATTAGAATTACGTCG. To ensure that mutations were valid each clone was sequenced in forward and reverse and manually aligned. Any mutations occurring in one direction only were discarded. T7 forward and M13 reverse primers were used for clone sequencing. All sequences generated here have been submitted to GenBank and assigned accession numbers HM768168-HM768204.
2.4. Sequence analysis
All ZYMV sequences were manually aligned using Se-Al (2.0a11; kindly provided by Andrew Rambaut, University of Edinburgh) and trimmed to cover the coat protein region: from the CP start codon until the stop codon, for a total of 849 nucleotides (nt). Counts of the number of mutations in each sample were undertaken manually, while pairwise genetic distances were estimated using MEGA (version 3) (Kumar et al., 2004). Because of the very small number of mutations observed we utilized uncorrected genetic (p) distances. As the number of cloned sequences varies across individual plants or time points we performed a chi-squared goodness of fit test (using R 2.10.1; 2008) to correct for the number of mutations compared to the number of sequences. To estimate the number of nonsynonymous (dN) and synonymous substitutions (dS) per site (ratio dN/dS), itself a measure of selection pressure, we used the Single Likelihood Ancestor Counting (SLAC) algorithm employing the MG94 × HY85_3×4 substitution model in HyPhy (Kosakovsky Pond et al., 2005). Finally, minimum spanning trees for the field and greenhouse populations were estimated separately using the statistical parsimony approach available in the TCS 1.21 program (Clement et al., 2000).
3. Results
To determine the extent and structure of intra-host viral genetic diversity in ZYMV we sequenced clones from 20 viral samples representing both the greenhouse and field populations. In total, we obtained 706 clonal sequences, with an average of 35 sequences per leaf sample. Approximately 90% of the clones sequenced were identical to the consensus sequence. Pairwise genetic distances ranged from 0–0.11%, with an overall mean of 0.02% for the field and greenhouse populations combined (Table 1).
3.1. Mutational spectrum in field plants
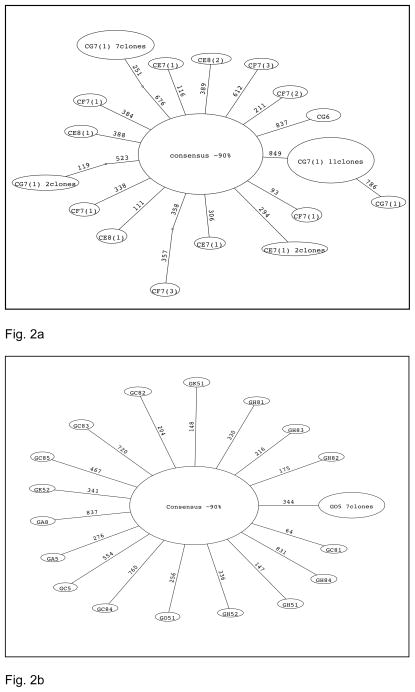
We generated a total of 378 clones from 11 field samples. Of these, 329 had no mutations and therefore matched the consensus sequence generated from the first-infected field plant. Clones from two of the individual plants, including the first inoculated plant – CF8 and CE7(2) – exhibited no mutations. Overall, there were total of 47 mutated sequences and 23 different mutations, 18 of which were singletons (occurred in one sequence only). This represents a mutational frequency of 1.47 × 10−4 mutations per nucleotide site. Ten of the mutations were synonymous; two sequences exhibited the same silent mutation, and 13 sequences from individual CG7 at time point 1 showed a change from a TAG stop codon to a TAA stop codon. There were 13 nonsynonymous mutations, three of which were found in multiple clones. Notably, one of these nonsynonymous mutations (TTG to TAG) resulted in a premature stop codon and was found in seven (19.4%) of the clones from plant CG7(1). A minimum spanning tree showing the structure of this genetic diversity is shown in Fig. 2a. Although most mutations are only one step away from the consensus, clear population structure was present in the form of three clones being two mutational steps away from the consensus, a number of mutations present in multiple clones, and in one case a mutant clone (at position 849) itself possessing a descendent mutation (at position 786). The latter is indicative of a distinct sub-lineage, although one that is only found at a single time-point in a single plant. Although we cannot exclude the possibility of a ZYMV infection other than our primary inoculant, given the low level of genetic diversity and the fact that ~90% of the sequenced clones match the consensus this seems extremely unlikely. DAS-ELISA tests undertaken by Agdia revealed that only one of the samples, CE7, was co-infected with another virus, in this case Watermelon mosaic virus-2 (WMV-2). There appears to be no significant difference in mean pairwise genetic divergence, or mean dN/dS, between this sample and the other field samples (Table 1).
Fig. 2.
Minimum spanning trees of the sequences collected here. (a) Field experiments. (b) Greenhouse experiments. The numbers along the branches represent the nucleotide position at which each mutation occurred. The number of clones with a particular mutation is one unless otherwise noted within the oval. Plants labeled as in Fig. 1.
Previous work has suggested 36 of the 42 amino acids of the N-terminus of the CP can be altered with no apparent effect on the viral life-cycle and hence are highly variable (Gal-on, 2007). In our study, only five of the total of 23 mutations occurred in this region, three of which were nonsynonymous. However, when correcting for sequence length we observed no significant difference in the number of mutations between the N-terminus and the rest of the CP (p= 0.2618). We also observed no mutations in the conserved DAG region known to be involved in aphid transmission.
Finally, the number of unique mutations did not differ significantly over time within individuals (CF7: p=0.944; CE7: p=0.0578; CG7: p=0.345; CE8: p=0.418). However, the total number of mutated sequences within an individual over time was significantly different for two individuals (CE7: p=0.0339 & CG7: p=0.0077 applying the same correction).
3.2. Mutational spectrum in the greenhouse plants
A total of 328 clones were generated from the nine greenhouse plants, 301 of which had no mutations and so matched the consensus sequence of the first-infected field plant. Only one individual plant, from the third generation, exhibited no mutations. There were a total of 24 mutated sequences and 18 different mutations, 17 of which were singletons, representing an error frequency of 8.7 × 10−5 mutations/site. Seven of the mutations were synonymous, and 11 were nonsynonymous, one of the latter being found in seven clones. One stop codon mutation was found in one sequence. Notably, none of the mutations were the same between transmission events. Three of the 18 mutations were found in the highly variable N-terminus region of the CP, although we again observed no mutations in the conserved DAG region. Finally, comparing the fifth and eighth leaves within a plant, we found that the number of mutations was the same between them in plant A, increased from one to five in plant C, and increased from two to four in plant H. Crucially, however, we identified no shared mutations between sequenced clones from the fifth and the eighth leaves, indicative of a rapid population turnover. Indeed, the minimum spanning tree of these data is striking in its marked lack of population structure, such that all the mutations are only one step away from the consensus (although one is present in seven clones; Fig. 2b).
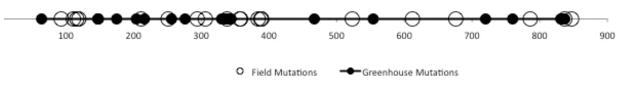
As the aphid vector was removed in the greenhouse experiment we might expect the extent of purifying selection to be stronger in the field than the greenhouse. However, we observed no marked difference in mean dN/dS ratios among these populations; a value of 0.54 (CI 95%: 0.23–0.84) was observed in the field compared to 0.66 (CI 95%: 0.34–1.13) in the greenhouse. The high dN/dS values (>1) observed in some individual samples likely reflect a large sampling error on the small number of mutations observed. Finally, it is notable that we observed no clear difference in the spatial distribution of mutations along the CP between the two experimental conditions (Fig. 3).
Fig. 3.
Spatial distribution of mutations in the CP gene from both the field and greenhouse experiments. The numbers below the horizontal line represent nucleotide positions.
3.3 Mutations introduced during the experimental procedure
The error rate for the reverse transcriptase (RT) enzyme used here is reported as 2.9 × 10−5 mutations/site/replication (personal communication, Invitrogen). Given our sequenced target region of 849 nt, the expected number of mutations per cDNA copy of the CP gene is therefore 0.0246 (2.9 × 10−5 mut/site/replication × 849 sites × single round of replication). We cloned 706 of these cDNA copies, leading to an overall expectation of 17.37 mutations among our 706 clones. The overall error rate including both the Phusion taq error rate and RT error rate is 0.0377 (calculated using the Phusion Taq error rate provided by Finnzymes and the RT error rate given above). Accounting for both the RT enzyme and Taq polymerase error rate we would expect the total number of artefactual mutations to be ~27. The actual number of mutations observed in our data was 71. Although is it clear that our data contains a number of artefactual mutations, as is likely to be true of any study of intra-host genetic variation in RNA viruses, many of the mutations observed here will be bona fide, especially as the reported error rate for RNA-dependent RNA polymerase is greater than of RT (Drake et al. 1998). In addition, we used great caution when calling mutations and only counted those that were present in both the forward and reverse alignments, and in some cases sequenced both directions twice. Hence, our reported introduced RT error rate is likely to be conservative. As such, it is highly unlikely that mutations at a frequency >1 are artefactual, including the stop codon mutation in plant CG7(1).
4. Discussion
Although the level of intra-host diversity we report for ZYMV (mean = 0.02%) is on average less than that recently observed in intra-host studies of animal influenza viruses using similar methodologies, there was considerable overlap among estimates and fewer clones were analyzed in this case (Hoelzer et al., 2010; Iqbal et al., 2009; Murcia et al., 2010). For example, a study of 2366 sequences of equine influenza virus resulted in a mean intra-host diversity of 0.04% (range 0.01 – 0.12% among samples) (Murcia et al. 2010). Hence, ZYMV appears to exhibit mutational dynamics broadly similar to those observed in some rapidly evolving animal RNA viruses, and as expected given the intrinsically error-prone nature of replication with RNA-dependent RNA polymerase. The possibility of artificially induced mutations should therefore be explored for those plant RNA viruses in which far higher levels of intra-host genetic diversity are observed.
It is also striking that most mutations in ZYMV are transient in nature, only being observed at a single sampling point. Indeed, we observed no mutations that were shared between time points from individual plants. Although a certain proportion of the mutations observed are clearly artefactual and an inherent outcome of the experimental procedures employed, particularly singleton mutations which should be treated with caution, our results are compatible with the notion that the majority of intra-host mutations in ZYMV are deleterious and removed by purifying selection between sampling times. The relatively high number of stop codon mutations observed supports this hypothesis, as does the marked difference in mean dN/dS values within (~0.6; herein) and between (0.108; Simmons et al., 2008) hosts. A similar turnover of apparently transient deleterious mutations has been observed in a number of animal RNA viruses (Holmes, 2003; Holmes, 2009; Hoelzer et al., 2010; Murcia et al., 2010), is supported by experimental studies of fitness distributions in RNA viruses (Sanjuán et al., 2004), and may therefore be a common component of intra-host viral genetic diversity. Despite this, it is notable that some short-lived population structure was present in the field samples – manifest as clones that differed in multiple mutations from the consensus, the same mutations present in multiple clones, and at least one distinct viral sub-lineage – yet not so in the greenhouse experiment. It is therefore possible that transmission mode impacts the structure of viral genetic diversity, even at the scale of individual plants, although this is evidently an issue that needs to be reassessed with a far larger number of clones than generated here.
Importantly, the discontinuity of mutations within individuals over time extends to transmission: no lineages were shared between individuals during aphid transmission. This suggests that the bottleneck imposed by the aphid is substantial, although it is also possible that our sample size is insufficient to sample minor lineages. As the aphid-imposed bottleneck is absent from the greenhouse experiment we might have expected to see more lineages transferred between hosts in this case. That this does not appear to the case from the data generated here suggests that the intra- and inter-plant population bottlenecks are generally severe enough to remove most genetic variation. In addition, that the number of unique mutations did not increase during serial passaging in the greenhouse indicates that the aphid-imposed bottleneck is not the only factor restricting genetic diversity, although this will clearly need to be explored further using a larger number of serial passages. Irrespective of sample size, the existence of strong population bottlenecks means that genetic drift will play a major role in substitution dynamics.
One of the most striking observations of our study was that seven clones sampled from one leaf at one time point from one field plant contained the same stop codon mutation. Such a high frequency of what is likely to be a deleterious mutation is suggestive of the action of transient complementation, although this will require future experimental verification. Indeed, that the stop codon mutation was not found at later time-points in this individual argues against both recurrent mutation and polymerase read-through as both would be expected to have longer-term effects.
Complementation has previously been reported in experimental infections of plant viruses (Fraile et al. 2008; Osbourn et al., 1990). For example, a mutant Tobacco mosaic virus with a frameshift and premature stop codon mutation in the CP was fully complemented in transgenic plants that expressed the wild-type CP gene (Holt and Beachy, 1991). Complementation has also been documented during viral co-infections, including truncated CP mutants of Pepper huasteco virus that were complemented by coinfection with Taino tomato mottle virus (Guevara-Gonzales et al. 1999). Not only is viral co-infection a frequent occurrence in nature, but the use of transgenic squash is now commonplace in agricultural settings. Complementation in these circumstances could theoretically lead to the inhibition of gene silencing (Qu et al., 2003, Thomas et al., 2003), the correction of defects in movement (Callaway et al., 2004), and perhaps even the expansion of host range (Latham and Wilson, 2008; Spitsin et al., 1999). Given the threat that RNA viruses such as ZYMV pose to staple crop production worldwide, the frequency and consequences of complementation in natural populations of plant viruses clearly needs to be investigated in greater detail.
Acknowledgments
This work was supported by Biotechnology Risk Assessment Program Grant no. 2009-33120-20093 from the USDA National Institute of Food and Agriculture. ECH is supported in part by grant R01 GM080533 from the National Institutes of Health. We are grateful to Joseph Dunham for his invaluable advice and assistance, and thank Melinda Bothe, Sarah Scanlon and James Parker for their help performing mini-preps.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali A, Li H, Schneider ML, Sherman DJ, Grey S, Smith D, Roossnick MJ. Analysis of genetic bottlenecks during horizontal transmission of Cucumber Mosaic Virus. J Virol. 2006;80:8345–8350. doi: 10.1128/JVI.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Roossnick MJ. Genetic bottlenecks during systemic movement of Cucumber Mosaic virus may vary in different host plants. Virology. 2010;404:279–283. doi: 10.1016/j.virol.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Betancourt M, Fereres A, Fraile A, Garcia-Arenal F. Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J Virol. 2008;82:12416–12421. doi: 10.1128/JVI.01542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok J, Mackenzie A, Guy P, Gibbs AJ. Nucleotide sequence comparisons of turnip yellow mosaic virus isolates from Australia and Europe. Arch Virol. 1987;97:283–295. doi: 10.1007/BF01314427. [DOI] [PubMed] [Google Scholar]
- Blua M, Perring T. Effect of Zucchini Yellow Mosaic Virus on development and yield of cantaloupe (cucumis melo) Plant Dis. 1989;73:317–320. [Google Scholar]
- Callaway AS, George CG, Lommel SA. A Sobemovirus coat protein gene complements long-distance movement of a coat protein-null Dianthovirus. Virology. 2004;330:186–195. doi: 10.1016/j.virol.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Cantliffe DJ, Shaw NL, Stoffella PJ. Current trends in cucurbit production in the U.S. Acta Hort. 2007;731:473–478. [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Debiez C, Lecoq H. Zucchini yellow mosaic virus. Plant Path. 1997;46:809–829. [Google Scholar]
- Desbiez C, Wipf-Scheibel C, Lecoq H. Biological and serological variability, evolution and molecular epidemiology of Zucchini yellow mosaic virus Island of Martinique. Plant Dis. 2002;80:203–207. doi: 10.1016/s0168-1702(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Fargette D, Pinel A, Rakotomalala M, Sangu E, Traoré O, Sérémé D, Sorho F, Issaka S, Hébrard E, Séré Y, Kanyeka Z, Konaté G. Rice Yellow Mottle Virus, an RNA plant virus, evolves as rapidly as most RNA animal viruses. J Virol. 2008;82:3584–3589. doi: 10.1128/JVI.02506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuer R, Boone JD, Netski D, Morzunov SP, St Jeor SC. Temporal and spatial analysis of Sin Nombre virus quasispecies in naturally infected rodents. J Virol. 1999;73:9544–9554. doi: 10.1128/jvi.73.11.9544-9554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile A, Sacristán S, García-Arenal F. A quantitative analysis of complementation of deleterious mutants in plant virus populations. Spanish J Ag Res. 2008;6:195–200. [Google Scholar]
- Fraile A, Escriu F, Aranda MA, Malpica JM, Gibbs AJ, García-Arenal F. A century of tobamovirus evolution in an Australian population of Nicotiana glauca. J Virol. 1997;71:8316–8320. doi: 10.1128/jvi.71.11.8316-8320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R, Stenger DC. Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu Rev Phytopathol. 2003;41:199–214. doi: 10.1146/annurev.phyto.41.052002.095559. [DOI] [PubMed] [Google Scholar]
- Gal-On A. Zucchini yellow mosaic virus: insect transmission and pathogenicity – the tails of two proteins. Mol Plant Pathol. 2007;8:139–150. doi: 10.1111/j.1364-3703.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- García-Arenal F, Fraile A, Malpica JM. Variability and genetic structure of plant virus populations. Annu Rev Phytopathol. 2001;39:157–186. doi: 10.1146/annurev.phyto.39.1.157. [DOI] [PubMed] [Google Scholar]
- García-Arenal F, Fraile A, Malpica JM. Variation and evolution of plant virus populations. Int Microbiol. 2003;6:225–232. doi: 10.1007/s10123-003-0142-z. [DOI] [PubMed] [Google Scholar]
- Gibbs AJ, Fargette D, García-Arenal F, Gibbs MJ. Time – the emerging dimension of plant virus studies. J Gen Virol. 2010;91:13–22. doi: 10.1099/vir.0.015925-0. [DOI] [PubMed] [Google Scholar]
- Gibbs AJ, Ohshima K, Phillips MJ, Gibbs MJ. The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS ONE. 2008;3:e2523. doi: 10.1371/journal.pone.0002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-González RG, Ramos PL, Rivera-Bustamante RF. Complementation of coat protein mutants of pepper huasteco geminivirus in transgenic tobacco plants. Phytopathol. 1999;89:540–545. doi: 10.1094/PHYTO.1999.89.7.540. [DOI] [PubMed] [Google Scholar]
- Hoelzer K, Murcia P, Baillie GJ, Wood JLN, Metzger S, Osterrieder K, Dubovi EJ, Holmes EC, Parrish CR. Intra-host evolutionary dynamics of canine influenza virus in naïve and partially immune dogs. J Virol. 2010;84:5329–5335. doi: 10.1128/JVI.02469-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. Patterns of intra- and inter-host nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. The Evolution and Emergence of RNA Viruses. In: Harvey PH, May RM, editors. Oxford Series in Ecology and Evolution. Oxford University Press; Oxford: 2009. [Google Scholar]
- Holt CA, Beachy RN. In vivo complementation of infectious transcripts from mutant tobacco mosaic virus cDNAs in transgenic plants. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- Iqbal M, Xiao H, Baillie G, Warry A, Essen SC, Londt B, Brookes SM, Brown IH, McCauley JW. Within-host variation of avian influenza viruses. Phil Trans R Soc Lond B. 2009;364:2739–2747. doi: 10.1098/rstb.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jridi C, Martin JF, Marie-Jeanne V, Labonne G, Blanc S. Distinct viral populations differentiate and evolve independently in a single perennial host plant. J Virol. 2006;80:2349–2357. doi: 10.1128/JVI.80.5.2349-2357.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak GVS, Brown I, Shi P, Kramer LD, Ebel GD. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology. 2008;374:256–260. doi: 10.1016/j.virol.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis NI, Tsitsipsi JA, Lykouressis DP, Papapanayotou A, Kokinis GM, Perdikis DC, Manoussopoulos IN. Transmission of Zucchini yellow mosaic virus by colonizing and non-colonizing aphids in Greece and new aphid vectors of the virus. J Phytopathol. 2006;154:293–302. [Google Scholar]
- Kim T, Youn MY, Min BE, Choi SH, Kim M, Ryu KH. Molecular analysis of quasispecies of Kyuri green mottle mosaic virus. Virus Res. 2005;110:161–167. doi: 10.1016/j.virusres.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Kumar S, Koichiro T, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Latham JR, Wilson AK. Transcomplementation and synergism in plants: implications for viral transgenes? Mol Plant Path. 2008;9:85–103. doi: 10.1111/j.1364-3703.2007.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech WJ, Wang G, Yang YL, Chee Y, Dorman K, McCrae D, Lazzeroni LCJ, Erickson W, Sinsheimer JS, Kaplan AH. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Roossinck MJ. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J Virol. 2004;78:10582–10587. doi: 10.1128/JVI.78.19.10582-10587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisa V, Boccardo G, D’Agostino G, Dellavalle G, D’Aquilio M. Characterization of a potyvirus that causes Zucchini yellow mosaic. Phytopathol. 1981;71:667–672. [Google Scholar]
- Marco CF, Aranda MA. Genetic diversity of a natural population of Cucurbit yellow stunting disorder virus. J Gen Virol. 2005;86:815–822. doi: 10.1099/vir.0.80584-0. [DOI] [PubMed] [Google Scholar]
- Miyashita S, Kishino H. Estimation of the size of genetic bottlenecks in cell-to-cell movement of Soil-borne wheat mosaic virus and the possible role of the bottlencks in speeding up selection of variations in trans acting genes or elements. J Virol. 2010;84:1828–1837. doi: 10.1128/JVI.01890-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Fabre F, Senoussi R. Estimation of the number of virus particles transmitted by an insect vector. Proc Natl Acad Sci USA. 2007;104:17891–17896. doi: 10.1073/pnas.0702739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia P, Baillie GJ, Daley J, Elton D, Jervis C, Mumford JA, Newton R, Parrish CR, Hoelzer K, Dougan G, Parkhill J, Lennard N, Ormond D, Moule S, Whitwham A, McKinley TJ, McCauley JW, Holmes EC, Grenfell BT, Wood JLN. The intra-and inter-host evolutionary dynamics of equine influenza virus. J Virol. 2010;84:6943–6954. doi: 10.1128/JVI.00112-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolasco G, Fonseca F, Silva G. Occurrence of genetic bottlenecks during citrus tristeza virus acquistion by Toxoptera citricida under field conditions. Arch Virol. 2008;153:259–271. doi: 10.1007/s00705-007-1089-8. [DOI] [PubMed] [Google Scholar]
- Osbourn JK, Sarkar S, Wilson MA. Complementation of coat protein-defective TMV mutants in transgenic tobacco plants expressing TMV coat protein. Virology. 1990;179:921–925. doi: 10.1016/0042-6822(90)90169-r. [DOI] [PubMed] [Google Scholar]
- Pagán I, Holmes EC. Long-term evolution of the Luteoviridae: time-scale and mode of virus speciation. J Virol. 2010;84:6177–6187. doi: 10.1128/JVI.02160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirone TP, Blanc S. Helper-dependent vector transmission of plant viruses. Annu Rev Phytopathol. 1996;34:227–247. doi: 10.1146/annurev.phyto.34.1.227. [DOI] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. ( http://www.R-project.org) [Google Scholar]
- Rodríguez-Cerezo E, Elena SF, Moya A, García-Arenal F. High genetic stability in natural populations of the plant RNA virus tobacco mild green mosaic virus. J Mol Evol. 1991;32:328–332. [Google Scholar]
- Sacristán S, Malpica JM, Fraile A, García-Arenal F. Estimation of population bottlenecks during systemic movement of Tobacco mosaic virus in tobacco plants. J Virol. 2003;77:9906–9911. doi: 10.1128/JVI.77.18.9906-9911.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci USA. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla DD, Frenkel MJ, Ward CW. Structure and function of the potyvirus genome with special reference to the coat protein coding region. Canadian J Plant Path. 1991;13:178–191. [Google Scholar]
- Simmons HE, Holmes EC, Stephenson AG. Rapid evolutionary dynamics of zucchini yellow mosaic virus. J Gen Virol. 2008;89:1081–1085. doi: 10.1099/vir.0.83543-0. [DOI] [PubMed] [Google Scholar]
- Spitsin S, Steplewski K, Fleysh N, Belanger H, Mikheeva T, Shivprasad S, Dawson W, Koprowski H, Yusibov V. Expression of alfalfa mosaic virus coat protein in tobacco mosaic virus (TMV) deficient in the production of its native coat protein supports long-distance movement of a chimeric TMV. Proc Natl Acad Sci USA. 1999;96:2549–2553. doi: 10.1073/pnas.96.5.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teycheney PY, Laboureau N, Iskra-Caruana ML, Candresse T. High genetic variability and evidence for plant-to-plant transfer of Banana mild mosaic virus. J Gen Virol. 2005;86:3179–3187. doi: 10.1099/vir.0.81197-0. [DOI] [PubMed] [Google Scholar]
- Thomas CL, Leh V, Lederer C, Maule AJ. Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology. 2003;306:33–41. doi: 10.1016/s0042-6822(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Turturo C, Saldarelli P, Yafeng D, Digiaro M, Minafra A, Savino V, Martelli GP. Genetic variability and population structure of Grapevine leafroll-associated virus 3 isolates. J Gen Virol. 2005;86:217–224. doi: 10.1099/vir.0.80395-0. [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima S, Haenni A, Bernardi F. Potyvirus proteins: a wealth of functions. Virus Res. 2001;74:157–175. doi: 10.1016/s0168-1702(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]