Abstract
This live cell study of chromatin dynamics in four dimensions (space and time) in cycling human cells provides direct evidence for three hypotheses first proposed by Theodor Boveri in seminal studies of fixed blastomeres from Parascaris equorum embryos: (I) Chromosome territory (CT) arrangements are stably maintained during interphase. (II) Chromosome proximity patterns change profoundly during prometaphase. (III) Similar CT proximity patterns in pairs of daughter nuclei reflect symmetrical chromosomal movements during anaphase and telophase, but differ substantially from the arrangement in mother cell nucleus. Hypothesis I could be confirmed for the majority of interphase cells. A minority, however, showed complex, rotational movements of CT assemblies with large-scale changes of CT proximity patterns, while radial nuclear arrangements were maintained. A new model of chromatin dynamics is proposed. It suggests that long-range DNA-DNA interactions in cell nuclei may depend on a combination of rotational CT movements and locally constrained chromatin movements.
Key words: chromatin dynamics, 4D live-cell microscopy, Theodor Boveri, mitosis, nuclear rotation, long range chromatin movements
Introduction
Chromosome territories (CTs) have become accepted as a basic feature of nuclear architecture in animal and plant species (reviewed in refs. 1–5). Recently, the question of chromatin dynamics in nuclei of living cells6 and cell-type specific CT rearrangements, which bring genes together in specific nuclear domains either for silencing or expression has gained prominence.7–9 A rapidly increasing body of evidence has strongly supported the concept that “gene kissing” events can occur both in cis, i.e., genes harbored in the same chromosome, and in trans, i.e., genes located on different chromosomes10–13 (see also Discussion).
In order to elucidate the mechanisms involved in “gene kissing,” it is important to know whether global chromosome order or at least the order of specific chromatin assemblies in trans can be faithfully propagated through mitosis or whether a cell type specific nuclear assembly of CTs and/or chromatin loops is generated during interphase. Several groups have studied this problem in living cells taking advantage of cell lines, which express core histones tagged with fluorescent proteins.14–16 Laser-microirradiation was used to produce fluorescent chromatin patterns, which were then followed through interphase and mitosis. Gerlich et al.15 concluded that global chromosome positions are maintained in cycling mammalian cells and proposed a mechanism acting at anaphase to restore the loss of order. Further evidence in favor of a global inheritance of chromosome order throughout mitosis was published by Essers et al.17 Evidence reported by other groups, however, including ours,16 argued for pronounced changes of CT neighborhood arrangements from one cell cycle to the next. Walter et al.16 detected increased chromatin mobility during the first 2 hrs of G1, which arguably provides a time window for the restoration of positional order. Thomson et al.18 tracked specific human loci after exit from mitosis and concluded that the organization of chromatin in the nucleus is not passed down precisely from one cell to its descendants but is more plastic and becomes refined during early G1. Cvacková et al.14 demonstrated major changes of chromatin arrangements between mother and daughter nuclei, but noted that the CT order present in the mother nucleus was not entirely randomized despite a significant stochastic component associated with reassortment of chromosome territories/chromatin. A partial maintenance of order could happen by chance and would not require a special mitotic mechanism. In the present study we wished to test the claim for an anaphase mechanism, which restores the loss of order during prometaphase.
Back to the Future: Theodor Boveri's Hypotheses on Chromosome Order and Mobility in Cycling Cells
The term “chromosome territory” (CT) was coined in 1909 by Theodor Boveri (1862–1915),19 although Carl Rabl (1853–1917) was the first to propose this seminal concept in a study of two amphibian species, Proteus anguineus and Salamandra maculosa.20 Boveri studied early cleavage stages of fertilized Parascaris equorum eggs or Ascaris megalocephala as the horse roundworm was called in Boveri's days.19,21 Two varieties of this species exist: Ascaris megalocephala univalens and Ascaris megalocephala bivalens, which contain one respectively two pairs of chromosomes in blastomeres, but undergo chromosome diminuation during later development. A.m. bivalens was first introduced as a model system by Edouard van Beneden (1846–1910) to study the material nature of heredity (Fig. 1A and B). In his 1909 publication Boveri proposed three bold hypotheses about higher order chromatin arrangements and their dynamics during the cell cycle (reviewed in refs. 22 and 23):
Chromosome territory (CT) arrangements are stably maintained during interphase.
Chromosome proximity patterns change profoundly during prometaphase.
Similar CT arrangements in pairs of daughter nuclei reflect symmetrical chromosomal movements during anaphase and telophase. Whereas CT proximitypatterns differ substantially from the mother cell nucleus, radial chromatin arrangements are maintained.
Figure 1.
Parascaris equorum, an early model to study cytological features of the chromosome theory of heredity and higher order chromatin arrangements. (A) Edouard van Beneden's drawing (1883) of the female and male pronuclus in the fertilized egg of Parascaris equorum, formerly termed Ascaris megalocephala, variety bivalens.84 (B) Edouard van Beneden observed that the female and male pronucleus delivers two chromosomes of equal size and shape. He argued that the hereditary substance of the two pronuclei does not blend as expected in case of two hereditary fluids, but that chromatin threads (baptized as chromosomes by Wilhelm Waldeyer in 1888) split into two halves (now called sister chromatids) and that each daughter cell receives one chromatid. His findings argued for an equal contribution of chromosomes provided by each parent and became an early cornerstone of all theories implicating chromosomes as the hereditary material. (C and D) Sister nuclei from 2-cell embryos of Ascaris megalocephala, variety univalens, drawn by Theodor Boveri at prophase19 show a single pair of chromosomes. Chromosomal ends stick out into protuberances of the nuclear envelope (thin arrows). Note that the positions of the protuberances and the number of chromosome ends located in them is similar in both nuclei of a given embryo, while the positions can vary largely from embryo to embryo (compare C with D). (E) This 4-cell embryo drawn by Boveri presents each cell in interphase.19 Arguably, pairs of nuclei with similar arrangements of their nuclear protrusions represent daughter cells derived from the same mitotic event, whereas nuclei with striking differences represent 1st grade cousins derived from different mitotic events. Boveri made use of these protrusions as markers for the interphase positions of the hypothetical chromosome territories.
Boveri supported these hypotheses with observations he made in blastomeres of fixed 2- and 4-cell embryos of Parascaris equorum (Fig. 1C–E; for further explanation of Boveri's arguments for chromosome territories and their arrangements see supporting online material). Despite Boveri's ingenuity all his conclusions about higher order chromatin dynamics in cycling cells were hampered by the fact that he could only study fixed cells at his time, since he lacked the means to visualize individual chromosomes directly in the cell nucleus. Compelling evidence in favor of CTs was only obtained in the 1970s and 1980s (reviewed in refs. 22 and 23).
The following 4D (space and time) analysis of chromatin dynamics in somatic human cell nuclei provides direct evidence for Boveri's hypotheses that chromosome arrangements present in prophase nuclei change strongly during prometaphase. The claim for an anaphase mechanism, which counteracts the loss of order during prometaphase,24 could not be confirmed in the present investigation. Moreover, our study has revealed that in his claim for stability of interphase CT arrangements Boveri missed the possibility that CT proximity patterns can change during complex, large-scale, rotational movements of CTs in assemblies rather than isolated movements of individual CTs. Based on this evidence, we propose a new model of chromatin dynamics, which explains the formation of specific long-range DNA interactions in trans10–13 by a combination of large-scale rotational and locally constrained chromatin movements.
Results
Experimental rationale.
To test Boveri's hypotheses with state-of-the-art live cell experiments we stably transfected the human cell-line RPE-1 with constructs for H4 tagged with a photoactivatable paGFP25,26 and H2B tagged with mRFP.27 In selected areas of interphase nuclei and mitotic cells paGFP fluorescence was activated by microirradiation with a 440 nm laser-line. The resulting H4 paGFP fluorescent chromatin patterns were traced through interphase and mitosis. Indirect immunofluorescence tests were negative for the accumulation of γ-H2AX28 or polyadenylated ribose (PAR)29 expected at sites of radiation induced single and double strand DNA breaks indicating that the induction of the fluorescent chromatin patterns did not result in DNA damage (data not shown).
Evidence for stably maintained chromosome territory arrangements during interphase.
Consistent with early evidence for CTs obtained with laser-uv-microirradiation experiments (reviewed in ref. 23) activation of paGFP fluorescence of a single prometaphase chromosome of living RPE-1 cells yielded distinct CTs in the resulting two daughter nuclei (Fig. 2A). The long term stability of a variety of different microbeam induced, fluorescent chromatin proximity patterns during interphase was studied in >100 nuclei by 4D live cell imaging ≥2 hours and 50 nuclei at ≥3 hours. To avoid photoinduced damage, 3D images of nuclei in part of the cells were recorded only twice, i.e., at the beginning and the end of the observation period. From other cells 3D nuclear images were recorded in intervals between 5 and 30 min. In both cases we obtained evidence for the interphase stability of chromatin proximity patterns except for locally constrained movements26 (Fig. 2B–E, Movie S1). Cells often showed pronounced nuclear rotations around an axis perpendicular to the growth surface as exemplified in Figure 2B. Since these nuclear rotations apparently did not result in changes of CT proximity patterns, we refer to them as simple rotations in order to distinguish them from complex rotational movements of CT assemblies described in the next paragraph during which major changes of CT proximity patterns were observed. In order to facilitate pattern comparisons in interphase nuclei recorded at different times we corrected for simple nuclear rotations (Fig. 2C). Figure 2D shows a nucleus with a cross-like fluorescent pattern traced from telophase to the next prophase (see Movie S2). The cross persisted through interphase and into the next prophase, where it became fuzzier reflecting the condensation of prophase chromosomes. In the nucleus shown in Figure 2E a double cross-like fluorescent nuclear pattern was produced in G1 and followed through S-phase till G2 (see Movie S3). In addition to paGFP-H4 this cell was transiently transfected with a construct for PCNA-mRFP to visualize the changing patterns of replication foci when the cell passed through S-phase.30 These experiments indicated that global patterns of CT arrangements were stably maintained during interphase in most RPE-1 cells.
Figure 2.
Stable higher order chromatin arrangements in interphase nuclei of living RPE-1 cells. (A) Activation of paGFP-H4 fluorescence (green) along a prometaphase chromosome yields distinct chromosome-territories in the two daughter nuclei. The whole chromosome complement is visualized mRFP-H2B (red). (B) Simple nuclear rotation along an axis perpendicular to the growth surface indicated by positional changes of the paGFP fluorescent area (for more complex rotational movements compare Fig. 3). (C) Various patterns of paGFP-H4 fluorescence are stably maintained in interphase nuclei during the whole observation period (up to 30 h). Images taken at the end point were corrected for simple rotational nuclear movements to ease the comparison with the pattern recorded at the start point. Signal blurring indicates locally constrained chromatin movements (compare Movie S1). (D) A cross-like paGFP-H4 fluorescence pattern induced in telophase is maintained throughout interphase into the next prophase (36 h). Labeling of neighboring prophase chromosomes indicates that the stripes of paGFP-H4 fluorescence induced at telophase covered neighboring CTs. (E) Fluorescent paGFP-H4 stripes are stably maintained through S-phase. Proliferating Cellular Nuclear Antigen (PCNA) tagged with mRFP reveals the typical change of the PCNA S-phase pattern.
Evidence for large-scale changes of CT proximity during complex, rotational chromatin movements.
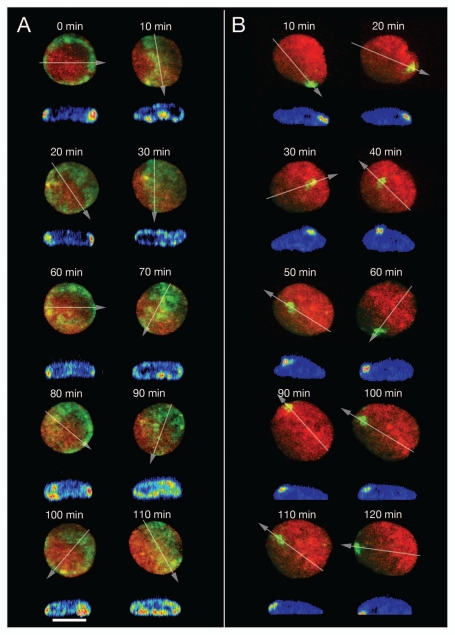
Figure 3 shows two exceptional cases which were detected during our 4D studies of living RPE-1 interphase cells. Figure 3A depicts a nucleus of a living RPE-1 cell, where we activated paGFP-H4 around the entire nuclear rim as seen from the top 2D perspective. After 10 min the fluorescent chromatin ring had seemingly disappeared. Instead a broad band of fluorescent chromatin expanded from one site of the nucleus to the other. Virtual sections made in x, z through the 3D image reconstruction of the flatly shaped RPE-1 nucleus revealed that the fluorescent chromatin was still associated with the nuclear envelope, although the CT proximity pattern had largely changed within 10 min enabling widely separated CTs to move into close proximity. Another 10 min later, the fluorescent chromatin ring reappeared in the 2D image projection. This drastic, transient change was repeated four times during the total observation period of 110 minutes. Another pertinent case is shown in Figure 3B. Here, fluorescence was activated in an area close to the nuclear rim. During the subsequent observation period of 120 min we noted an apparent movement of fluorescent chromatin away from the nuclear rim and back to it. This nucleus demonstrates the necessity of a full 4D analysis in order to understand the complexities of possible chromatin movements during interphase. Whereas a simple 2D analysis could lead to the erroneous conclusion that fluorescent marker chromatin underwent directed movements away from the nuclear rim and back to it, x, z-cross-sections demonstrated that the fluorescent chromatin segment always stayed at the nuclear periphery. Based on the analysis of 3D sequences (Fig. S1) the two cases presented in Figure 3 were recognized as examples of complex large-scale, rotational chromatin movements along axes positioned perpendicular and parallel to the growth surface. Notably, complex rotational chromatin movements with major changes of CT proximity pattern depended on the movement of CT assemblies rather than of isolated, individual CTs (see Discussion). This phenomenon was observed in 4 out of 150 interphase cells (ca. 2.5%) studied by time lapse imaging (see above). While it seems unlikely that this small percentage of cells plays an important functional role in RPE-1 cell cultures, the observation that sudden, complex rotational chromatin movements occur at all justifies speculations about a major role in tissue cells (see Discussion).
Figure 3.
Major changes of CT proximity patterns in RPE-1 interphase nuclei during complex large-scale rotational movements of chromatin assemblies. (A) A ring of paGFP fluorescent chromatin was induced at the 2D rim of a flat-ellipsoidal RPE-1 cell nucleus (time 0). 3D images were taken every 10 min for a total period of 110 min observation time. Note the drastic, repetitive changes of the fluorescent patterns noted in the 2D projections of this image sequence from ring-shaped to broad bands etc., indicating repeated rotations along an perpendicular axis. Since the flat-ellipsoidal shape of the nucleus was maintained during these rotations of entire CT assemblies, proximity patterns changed accordingly (compare Fig. 7D). Virtual cross-sections performed through 3D nuclear reconstructions along the lines indicated by arrows demonstrate that the mass of fluorescent chromatin remained at the nuclear envelope during these rotations indicating that radial interphase chromatin arrangements were maiantained during complex nuclear rotation. (B) Photoactivation of paGFP in a small area at the nuclear rim as a marker for major chromatin movements at the nuclear periphery. At face value 2D projections of x, y-nuclear images recorded at different times after photoactivation indicate movements of marker chromatin away from the nuclear rim (40–60 min) and back to it (70–130 min); virtual x, z-cross-sections through 3D nuclear reconstructions along lines indicated in the 2D projections by arrows demonstrate that the marker chromatin always stayed at the nuclear periphery. During the whole observation period the nucleus performed a counter-clockwise rotation of about 630° around an axis perpendicular to the growth surface and simultaneously another rotation around an axis parallel to it.
Proximity patterns of chromatin differ extensively between mother and daughter nuclei.
In order to test whether higher order chromatin arrangements in the mother cell nucleus are transmitted to daughter nuclei, we produced different fluorescent chromatin patterns in >130 RPE-1 interphase or prophase nuclei (Fig. 4, upper row) and studied the pattern in the resulting daughter nuclei (Fig. 4 and bottom row). In about ∼80 interphase cell nuclei 2D images were recorded at the beginning (prophase) and the end of the observation period (G1). In another ∼50 cells 3D images were recorded throughout mitosis into G1. In agreement with previous studies14,16 patterns differed strongly from the pattern produced in the mother nuclei. Pattern changes were in particular obvious in 18 cases, where we activated paGFP-H4 fluorescence in small nuclear areas of the mother nucleus (about 5% of the total nuclear area) either close to the nuclear rim or in central nuclear regions. In all cases both daughter nuclei revealed patches of chromatin marked with fluorescent paGFP-H4 intermixed with patches of unmarked chromatin. This finding was confirmed for HeLa cells transiently transfected with constructs for the expression of mRFP-H2B and paGFP-H4, as well as NRK cells kindly provided by Jan Ellenberg (EMBL, Heidelberg) (Fig. S2). These data indicate that marked chromatin segments in trans, which are intimately associated in the mother nucleus, are typically separated in daughter nuclei.
Figure 4.
Chromatin proximity patterns are not preserved through mitosis. (A–F) Upper row: different patterns of paGFP-H4 fluorescence produced in prophase nuclei of living RPE-1 cells. Bottom row: patterns observed in pairs of daughter nuclei. Note the strong discordance of the patterns of fluorescent paGFP between the mother nucleus and its daughters. Fluorescent chromatin activated at the nuclear rim in mother nuclei is distributed over the whole nuclear area in 2D projections of the daughter nuclei (E and F). As exemplified in (F, bottom) virtual z, x-cross-sections reveal fluorescent chromatin patches maintained their association with the nuclear envelope.
Major changes of chromosome proximity patterns during prometaphase are transmitted to daughter nuclei.
In order to explore the dynamics of positional chromosome changes during mitosis and the contribution of different mitotic stages to the drastic change of CT proximity patterns, we induced paGFP fluorescence within a circular field covering about 5% of prophase nuclei. Chromosomes or segments thereof located inside this area showed paGFP-H4 fluorescence, as well as mRFP-H2B fluorescence, while non-irradiated prophase chromosomes outside this area showed red fluorescence only. Figure 5A–C show 2D projections of light optical serial sections scanned at every 10 min through the entire subsequent mitosis into G1. In order to simplify the follow-up of individual paGFP labeled chromosomes, we coded each chromosome by a different false color. The discrimination was based on the complete 4D sequence from the mother cell at prophase to the two daughter cells at G1 shown in Figure S3A, which provides interactively rotatable images of individual 3D reconstructions made at all time points. Movie S4 presents this and a second cell as another pertinent example. Already within the first few minutes after entry of the cell into prometaphase we noted that labeled and unlabeled prometaphase chromosomes started to intermix, whereas in established metaphase plates chromosome movements became strongly constrained (Figs. 5 and S3B). Activation of paGFP-H4 in one half of the metaphase plate yielded daughter nuclei, which exhibited contiguous paGFP fluorescence over one half of their nuclear areas (Fig. 5D). Note that this experiment per se does not rule out major changes between the proximity patterns of metaphase chromosomes and the proximity patterns of CTs in the resulting daughter nuclei. Evidence for the global maintenance of proximity patterns established during metaphase through anaphase and telophase and into G1, however, is provided by the stable, mirror-like proximity patterns of color coded, paGFP-labeled chromosome segments (Fig. 5B).
Figure 5.
Chromosome neighborhood arrangements change during prometaphase. (A–C) Live cell sequence recorded from a RPE-1 prophase nucleus (0 min) through mitosis into G1 (150 min). A small area of fluorescent paGFP-H4 (∼5%) was marked at the nuclear border (0 min). Light optical serial sections were recorded every ten minutes (images recorded at 30–70 min not shown). Left column: Maximum intensity projections show chromatin labeled with fluorescent paGFP-H4 (green) within total chromatin labeled exclusively with mRFP-H2B (red-only). At early prometaphase (10 min) green-labeled segments of chromosomes derived from the marked area of the prophase nucleus were already separated by red-only chromosomes. The proximity pattern established at metaphase (80 min) was largely maintained through anaphase-telophase (90 min) and into G1 (100–150 min). Middle and right columns: In order to follow individual movements, labeled chromosomes were color-coded. Panels display images from 3D reconstructions recorded as top views (middle, 0–120 min; right 130–150 min) and side views (right, 0–120 min; compare Fig. S3 for fully rotatable 3D reconstructions). Note the mirror like symmetry of color-coded chromosomes during the transition from anaphase rosettes into G1 nuclei.
Spindle position does not affect the extent of positional changes of chromosomes during prometaphase.
Gerlich et al.15 showed that the distribution of chromosomes in the metaphase plate was starkly different depending on a parallel or perpendicular position of the spindle with respect to the labeling boundary (see Introduction). The following experiments were performed to test a possible impact of spindle position on global transmission of chromosome proximity patterns from one cell generation to the next in RPE-1 cells. We transiently transfected RPE-1 cells with a construct for the expression of GFP-FOP (Fibroblast Growth Factor Receptor-1 Oncogene Partner, a component of the centrosome) and activated paGFP-H4 fluorescence in extended areas of prophase nuclei in a way that the labeling boundary between green and red fluorescent chromatin was either roughly parallel to the connecting line between the two centrosomes (Fig. 6A) or perpendicular to it (Fig. 6B). Parallel activation of paGFP-H4 fluorescence resulted in daughter nuclei with a contiguous area of fluorescent chromatin, although additional green fluorescent chunks separated from this contiguous mass were also observed (Fig. 6A). In contrast, a perpendicular activation yielded daughter nuclei with a scattered distribution of green fluorescent chromatin patches intermixed with exclusively red fluorescent patches (Fig. 6B). Figures S4A and B present corresponding, rotatable 3D reconstructions. Figures S4C and D show additional examples. At face value these experiments seem to argue that the extent of positional changes of chromosomes during mitosis is strongly affected by the spindle position. The following experiments, however, provide evidence against this conclusion. We bleached the mRFP-H2B fluorescence in approximately half of a prophase nucleus along the short nuclear axis (Fig. 6C, upper) and paGFP-H4 fluorescence along the long axis (Fig. 6D, upper). As expected, in the resulting daughter nuclei we found contiguous nuclear areas of mRFP-H2B fluorescence, while patches of GFP-H4 fluorescence were scattered over the two nuclei (Fig. 6C and D, lower). An overlay of these images (Fig. 6E) shows green fluorescent patches distributed within the red fluorescent contiguous area. This finding demonstrates that the transmission of a contiguous fluorescent area from a mother nucleus to its daughters was observed despite massive changes of chromatin neighborhood arrangements within this area. This conclusion was confirmed in experiments, where paGFP-H4 activation was restricted to one quarter of a RPE-1 prophase nucleus (Fig. 6F, upper). The resulting daughter nuclei revealed green fluorescent chromatin patches scattered over one half of the nucleus (Fig. 6F, lower). In line with previous studies,14,16 however, this finding argues against a complete randomization of CT proximity patterns present in the mother nucleus already after a single mitotic event. In such a case we would have expected scattering of fluorescent chromatin patches throughout the entire nuclear space in all experiments where we activated paGFP-H4 in a half or a quarter of the nuclear area.
Figure 6.
Changes of chromatin proximity patterns during mitosis are not affected by the spindle topography. (A and B) Centrosomes in living RPE-1 cells were visualized by GFP-FOP. The spindle axis was defined by a line through the centrosomes. Activation of paGFP-H4 fluorescence was performed at prophase either parallel (A) or perpendicular (B) to the spindle axis. Although the fidelity of transmission of global chromosome positions appeared higher in (A) than in (B), additional experiments (C–F) provide evidence against this impression. (C–E, upper row) In roughly half of a prophase nucleus photobleaching of mRFP-H2B (red) was performed along its short axis (C), whereas photoactivation of paGFP-H4 (green) was induced along its long axis (D). (C–E, lower row) The resulting daughter nuclei show coherent regions with mRFP fluorescence, but scattered patches of paGFP fluorescence. Of particular interest are the zones of overlap between red and green fluorescence in the daughter nuclei (E). These zones demonstrate patches of green fluorescence within contiguous areas of red fluorescence. This result provides direct evidence that chromatin proximity patterns changed strongly independently of whether fluorescent labeling was performed parallel or perpendicular to the spindle axis. (F) Photoactivation of paGFP-H4 fluorescence in a quarter of a prophase nucleus resulted in daughter nuclei with patches of green fluorescent chromatin scattered over one nuclear half. (G–I) Schemes explaining the outcome of experiments with induction of paGFP-H4 fluorescence in half of prophase nuclei, either parallel to the spindle axis (G) or perpendicular to it (H), as well as in a quarter of a prophase nucleus (I). Top: Arrows point at the centrosomes and define the spindle axis. Filled symbols represent chromosomes after activation of paGFP-H4 fluorescence, open symbols chromosomes with mRFP-H2B fluorescence only. Bottom: Schemes of the metaphase plate with daughter nuclei. In (H and I) the upper pair of daughter nuclei represents mirror like symmetry, the lower pair translational symmetry (for further explanation see text).
Figure 6G–I provide schemes for the outcome of experiments, in which chromatin fluorescence labeling was induced in half of the nucleus parallel (Fig. 6G) or perpendicular (Fig. 6H) to the spindle axis or in a quarter of the nucleus (Fig. 6I). It should be emphasized that the schemes presented in Figure 6 do not reflect the complex events, which take place during the first part of mitosis from prophase to metaphase (Fig. S5). Whereas in this scheme the two centrosomes are always placed on opposite sites of the prophase nucleus, the actual placement of centrosomes in RPE-1 cells varied strongly. At prophase they were often still placed at one side of the nucleus in invaginations of the nuclear envelope. During or briefly after breakdown of the nuclear envelope the spindle was typically deeply immersed within a tunnel like space surrounded by prometaphase chromosomes and kinetochores exposed towards the tunnel31 (see rotatable 3D reconstructions provided in Fig. S5C, E, F). These complexities, however, do not affect the following conclusions. At face value a comparison of Figure 6G and H argues for a higher fidelity of the global transmission of CT order in experiments where the labeling boundary is chosen parallel to the spindle axis. This conclusion, however, is based on the erroneous assumption that the recurrence of a contiguous fluorescent area produced in the mother nucleus in its daughters demonstrates a high fidelity of global positional transmission of chromosomes through mitosis. Figure 6I summarizes the evidence obtained after paGFP-H4 activation of a quarter (Fig. 6F) or of still smaller nuclear areas in RPE-1 cells (Figs. 4 and S3). This evidence, as well evidence for other cell lines, including NRK and HeLa cells (Fig. S2) support the conclusion that major changes of CT neighborhood arrangements occurred independent of the topography of the spindle apparatus. Changes of chromosome proximity patterns during prometaphase were not restored during anaphase (Figs. 5, S3A, S4B and S4D). Note that CT arrangements in pairs of daughter nuclei show either a mirror like symmetry (upper pair of daughter nuclei in Fig. 6H and I) or a translational symmetry (lower pair of daughter nuclei). In cultured cells, which grow attached on a surface, the spindle is typically arranged parallel to this surface, whereas both the metaphase plate and the anaphase rosettes are typically arranged perpendicular to it.32 At the end of anaphase the two anaphase rosettes fall over either in the same direction or in opposite directions. The first case yields a pair of daughter nuclei with a direct symmetry of their chromatin proximity patterns, the second case yields mirror like patterns.
Discussion
Evidence for Boveri's hypotheses.
Our live cell study of cycling RPE-1 cells presents direct experimental support for three hypotheses first proposed by Theodor Boveri with respect to the dynamics of chromosome territory arrangements in cycling blastomeres of Parascaris equorum19 (see Introduction). In support of hypothesis I we provide evidence for persistent proximity patterns of CTs during interphase in most RPE-1 cell nuclei (Fig. 2). Unexpectedly, however, we observed occasional cells undergoing complex nuclear rotations, which led to at least transient, major changes of chromatin proximity patterns. Radial chromatin arrangements, however, were maintained, i.e., chromatin with fluorescent paGFP retained its location at the nuclear periphery (Fig. 3). Possible implications of this finding for the formation of specific long-range DNA interactions in trans11–13 are discussed below. In support of hypothesis II we demonstrate profound changes of chromosome proximity patterns during prometaphase (Figs. 5, S3, S4B and D). A randomizing effect of prometaphase on chromosome proximity patterns was well established in previous studies and can be explained by a random order of spindle attachment to the kinetochores of individual prometaphase chromosomes.31,33–35 Due to the stochastic nature of the spindle attachment, widely separated kinetochores can become attached at an earlier time point than juxtaposed kinetochores. Prometaphase chromosomes connected to the spindle apparatus immediately start to move and thus are driven away from originally juxtaposed, but still unconnected neighbors. The varying positions of centrosomes introduce another stochastic component (Fig. S5A, C–F). In agreement with previous studies14,16 the patterns observed in daughter nuclei differed considerably and often strikingly from the pattern originally induced in the mother nucleus (Fig. 4). In support of hypothesis III we demonstrate that the new proximity patterns of chromosomes established in the metaphase plate persisted largely through anaphase and telophase yielding similar CT proximity patterns in RPE-1 daughter cell nuclei (Figs. 5, S3A, S4).
The case for long-range chromatin movements.
Several groups reported chromatin loops carrying specific clusters of genes expanding up to several micrometers away from the surface of their home CTs.36–39 Such an extrusion of a gene locus from a CT is not necessarily indicative of transcriptional activity, but also can reflect a poised state for activation.37 Notably, the compaction level of one such giant loop studied in detail was about one order of magnitude higher than that of an extended 30 nm thick chromatin loop.40 Live-cell microscopy performed with cultured mammalian and Drosophila cells directly demonstrated constrained movements (<2 µm) of subchromosomal domains,41–43 as well as long-range movements (<2 µm).42,44 This distinction is based on the assumption that constrained Brownian motions may suffice for interactions between DNA segments with distances <2 µm, while movements >2 µm require more elaborate mechanisms, including energy dependent directed movements45–47 and/or rotational movements (see below).
Chromosome conformation capture, first introduced by Dekker et al.48 has provided compelling evidence for specific DNA-DNA interactions in cis and trans in cycling cells.10,12,49–60 The combination of circular chromosome conformation capture (4C) with DNA microarrays13,61 or massively parallel sequencing11 has allowed for the first time mapping of DNA-DNA interactions in cis and trans at a genome wide level. Although DNA interactions in cis were found to abound, significant interactions in trans were detected in agreement with other studies cited above. This wealth of data, often substantiated by 3D FISH assays, has provided strong support for long-range interactions in cis, i.e., between genes located many Mbs apart on the same chromosomes, or trans, i.e., between genes located on different chromosomes.
We agree with Gerlich et al.15 and Cvacková et al.14 that the CT neighborhood pattern present in the mother nucleus is not entirely lost after a single mitotic event. In an additional study performed by the T. Cremer group in collaboration with the group of Roland Eils (Heidelberg) landmark-based registration methods were employed to measure the similarity of CT arrangements visualized by multicolor 3D FISH in cell clones from different human cell types (Koehler D, Mattes J, Gao J, Joffe B, Cremer T, Eils R and Solovei I, unpublished data). Two mitotic events were typically sufficient to dilute the global transmission of chromosome proximity patterns to the extent that the similarity within a given clone was not significantly higher than between unrelated clones from the same culture. Our data strongly argue against an anaphase mechanism, which restores the loss of order during prometaphase. Based on this evidence, we will consider below possible mechanisms for the interactions of genes in trans, which act during interphase or in postmitotic cell nuclei.
Implications of variable CT proximity patterns for large-scale, non-random DNA-DNA interactions.
In search for a mechanism, which explains the formation of specific, long-range DNA-DNA interactions in trans, the evidence discussed above for probabilistic CT proximity patterns in cycling cells, which result from the randomizing effect of mitosis, must be considered. A mechanism for such interactions requires long-range movements (>2 µm) of the respective CTs and/or of chromatin loops carrying the genes in question. In an attempt to reconcile present evidence for long-range gene “kissing” events in trans during interphase with evidence for probabilistic CT neighborhood arrangements we consider four gene “kissing” scenarios (Fig. 7A–D). In scenarios A and B we consider the demanding problems of directed long-range chromatin movements for gene “kissing” events in trans in such nuclei. It is utterly speculative at present how the topographical information required for directed movements in trans can be gained and used. Scenario C considers a possibility, where constrained Brownian chromatin motions may suffice for the formation of functionally relevant DNA-DNA interactions in trans. In order to explain “kissing” events between primarily widely separated genes (>2 µm)45,62 scenario D assumes a combination of large-scale rotational chromatin movements and short-scale constrained Brownian chromatin motions.
Figure 7.
Four model scenarios for large-scale chromatin interactions in trans. (A) “Contact first” scenario. It assumes a functional necessity for long-range movements of giant chromatin loops carrying two genes in order to achieve a “gene-kissing” event. At the “kissing” site these genes initiate the formation of a specialized expression hub with specific factors serving the particular needs for the co-regulation of these genes. (B) “Expression hub/transcription factory first” scenario. It assumes that a specialized expression hub/transcription factory already exists at a specific site in the nucleus. Genes in need for co-regulation by this unique expression hub must congress towards this site. (C) This scenario argues that numerous specialized expression hubs/transcription factories, which already exist at different nuclear sites, are responsible for co-regulated expression of a set of genes present in CTs with variable positions in different nuclei. Driven by constrained Brownian motions these genes may reach the closest hub/factory serving their special needs and stick to this hub/factory, but associate only briefly with other hubs/factories specialized for the transcription of other genes. (D) Upper: example of clockwise rotational movements of CT assemblies in a flat-shaped nucleus around an axis perpendicular to the z-axis. This rotation brings widely separated CTs located at the rim of the nucleus (left) in a position close to each other (right). At this stage the distance between the two CTs may be close enough to allow the generation of specific DNA-DNA interactions in trans by additional Brownian chromatin motions. Complex rotational movements along various axes have the potential to achieve a close neighborhood of any pair of heterologous or homologous CTs starting from random CT proximity patterns. In the lower panel this potential is exemplified by group of square dancers rotating together in a clock-wise fashion.
Scenario A (Fig. 7A) describes a “contact first” model, which considers the possibility that widely separated genes initially need to come together and then form a hub or specialized transcription factory at their meeting point;7–9 (reviewed in ref. 63). To some extent non-random radial chromatin arrangements may favor the de novo formation of preferential proximity patterns, since genes positioned in the nuclear interior are on average located closer to each other than genes located in the nuclear periphery and thus should have a better chance to be positioned close enough for a kiss enabled by constrained Brownian motions. Taking into account that the relative positions of the CTs carrying the genes involved in the requested “kissing” event can change strongly from nucleus to nucleus, the mechanism for large-scale movements requires information about the direction into which the CTs and/or giant loops expanding from the respective CT surfaces should move. The congression of two widely separated CTs necessitates major rearrangements of other CTs as well, since CTs located in-between need to move aside affecting in turn other neighboring CTs. The demand for major chromatin rearrangements could be minimized by the congression of giant chromatin loops, which carry the genes of interest (reviewed in ref. 49). In this case physical constraints need to be overcome that hinder long distance passages of giant loops through the nuclear space. On their route giant loops must penetrate through or pass around one or several other CTs forming obstacles between the site of departure and the site of arrival of genes traveling with giant loops. Although interchromatin channels could serve as routes for expanding loops to remote nuclear sites,40,64,65 we need to take into account that the IC is crowded with macromolecules and non-chromatin domains, which provide physical obstacles in addition to chromatin clusters for expanding and retracting giant chromatin loops.66–68 At the onset of mitosis the undisturbed retraction of a giant loop towards its corresponding chromosome generates a further potential problem.
Scenario B (Fig. 7B) presents a “factory first” model, where a specialized expression hub/transcription factory already exists and recruits specific genes for co-regulated expression at such a hub or factory. The same topographical problems discussed for scenario A hold for scenario B. To use a more martial comparison than kissing, the problem resembles the task of duelists with pistols in hand (representing the CTs and the respective genes). To know where the opponent stands is a minimum requirement for each duelist. In the nucleus this requirement would be served best by strictly deterministic relative positions of the two CTs carrying genes involved in a kissing event. In case that the relative positions change strongly from nucleus to nucleus, the mechanism for a successful kissing event requires information about the direction into which a given CT and/or a giant loop expanding from these CTs should move.
Movements of chromatin loops expanding from or retracting towards a CT36,38,40 may serve functional demands by re-positioning genes from a compartment of silent chromatin into a nearby compartment of active chromatin (and vice versa). Such movements are less complex than movements necessary for long-range “kissing” events between genes in trans. Long-range movements of chromatin away from the nuclear periphery into the nuclear interior (and vice versa)32,44,69–71 apparently require a choreography different from the example presented in Figure 7D. Using live cell microscopy, Chuang et al.44 observed long-range directional movements of a chromosome locus undergoing an inducible repositioning from the nuclear periphery to the interior 1–2 hr after targeting a transcriptional activator to this site. Extended periods of chromosome immobility were interspersed with several minute periods in which chromosomes moved unidirectionally along curvilinear paths oriented roughly perpendicular to the nuclear envelope at velocities of 0.1–0.9 µm/min over distances of 1–5 µm. We suggest that this movement implied movements of higher order chromatin assemblies rather than the isolated movement of a single locus (compare Fig. 3).
Scenario C (Fig. 7C) assumes random CT proximity patterns and argues that DNA-DNA interactions in trans are limited to CTs, which by chance are located sufficiently close to each other. This chance increases with the number of pre-existing special expression hubs/transcription factories (reviewed in ref. 13). A set of genes involved in a distinct gene network functions as long as each gene is able to get access to one of multiple specialized hubs/factories, the more the better, which may be widely dispersed in the nuclear space. Probabilistic CT proximity patterns should yield a fraction of cells where two or even more genes, which are involved in the same regulatory gene network, but located on different CTs, are by chance located near enough to each other to explore their immediate nuclear environment by constrained Brownian motions and attach to the same hub/factory. Genes located far away from each other will explore different nuclear sub-volumes and attach to different hubs/factories suitable for their special needs of regulatory factors.
Scenario D (Fig. 7D) attempts to explain long-range gene “kissing” events starting with random CT proximity patterns by a two-step mechanism. Clockwise and anti-clockwise rotational movements of CT assemblies provide the means to bring widely separated CTs carrying genes involved in a required “kissing” event into proximity close enough to interact thereafter by constrained Brownian movements. While simple rotational movements of whole spherical nuclei would not change the relative positions of CTs, the situation is different for complex rotational movements of flatly shaped nuclei and/or CT assemblies within such nuclei. In this case global, large-scale, rotational chromatin movements around an axis parallel to the growth surface resembles a moving device built up from an endless belt (representing the CTs) rotating around sprocket wheels (representing the unknown molecular mechanism) (Fig. 7D, upper). Importantly, in contrast to scenarios denoted in Figure 7A and B, scenarios C and D do not require a priori information on the topography of two CTs involved in a “gene-kissing” event. It is possible with any starting assembly of the individuals to bring a desired pair of dancers into a directly opposite position by a clockwise or anti-clockwise rotation of the whole assembly.
To illustrate this point, we compare the dance of CTs with a group of square dancers (Fig. 7D, lower). Consider for example dancers 1 and 4. In the starting configuration they stand widely apart from each other, whereas in the later configuration they occupy directly opposite positions. Figure 7D is just meant as an example to illustrate one of many possibilities of changes of CT arrangements brought about by choreography of CTs dancing in assemblies. The larger the diploid number of chromosomes, the more obvious is the need for choreography in order to enable positional changes without chaos. Such choreography seems essential for an ordered mechanism of homologous alignment during meiotic prophase and it is tempting to speculate that some part of the meiotic pairing mechanism72 is also used in somatic cells to achieve homologous chromosome pairing during Drosophila melanogaster embryogenesis,73 as well as homologous and heterologous chromatin alignments necessary for intranuclear DNA-DNA interactions in trans. Since the necessary dance of CTs in assemblies takes place within a 3D nuclear space, which can change shape transiently or permanently, the possibilities for choreographies may be exceedingly more complex than the simple choreographic example of some square dancers.
Simple and complex nuclear rotation (NR) with and without changes of CT proximity patterns has been observed in various cell types, including neurons.74,75 De Boni and co-workers described saltatory, rotational movements with changes in direction including reversals. To our knowledge these authors were the first, who argued for a link between NR and the positioning of specific chromatin domains into cytotypic, nonrandom chromosome pattern in cycling and even in terminal differentiated cells. In multinucleolated neurons studied by time-lapse imaging they observed examples of nucleolar fusion, where nucleoli moved along curvilinear trajectories within the 3D nuclear space prior to the fusion event. Since large-scale movements of nucleoli require concomitant large-scale movements of NOR-bearing CTs and likely also of other CTs carrying chromatin domains specifically associated with perinucleolar heterochromatin, they provide a case in point for the necessity of a choreography, in which CTs move in assemblies rather than as independent individuals.
An important question with respect to the mechanism(s) involved in NR is whether NR represents motion of nuclei in toto, including the nuclear envelope, or independent motion of subnuclear structures, relative to each other, while the nuclear envelope stays fixed. For rotations of whole nuclei a motor fiber system located in the cytoplasm and attached to the nuclear envelope is required. For intranuclear chromatin rotations such a motor fiber system may either pass from the cytoplasm through the nuclear envelope in order to attach to chromatin sites or the system may be located within the nucleus. In both cases interactions of chromatin with lamin receptors would have to be highly dynamic in order to free peripheral chromatin transiently for rotational movements. Reports on significant changes of chromatin domains relative to stationary cytoplasmic structures with a juxtanuclear position have provided circumstantial evidence that the interface for NR may lie on the karyoplasmic side of the nuclear envelope.76
Complex rotational movements of CTs in assemblies, followed by constrained Brownian motions of chromatin domains or loops harboring specific genes provide an experimentally testable hypothesis how log-range DNA-DNA interactions in trans can be established in a population of cells starting with random CT proximity patterns. The stabilization of useful DNA-DNA interactions requests a molecular part for the mutual recognition and stabilization of aligned chromatin segments. This task may involve connecting filaments (see below) analogous to meiotic transverse filaments77 or an IC channel specifically connecting two CTs or loci of interest. Although both assumptions are presently not supported by experimental data, they are fully testable by state-of-the-art methods. In case that CTs connected in trans by such filaments move apart from each other because of continued rotational CT movements, connected giant loops expanding from different CTs would be the consequence rather than the cause of “gene kissing” events. If such connecting filaments become sufficiently long, they could even help to permanently widely separated genes once connected by a “kissing” event and trigger directed movements of such genes towards each other upon a stimulus provided by a signaling pathway.12 Connecting fibers could even provide a possibility to re-establish functionally important patterns of co-localized genes present in a mother nucleus in its daughters.
Numerous molecular components are likely expected to participate in mechanism(s) for long-range DNA-DNA interactions in trans and remain to be identified except for a few candidates. In addition to cytoplasmic and/or nuclear actin and myosin,10,44,78–80 other proteins, including dynein,81 should be considered. A disturbance in the connection between the nucleus and the cytoskeleton and a concomitant loss of nuclear rotation was observed in A-type lamin-deficient (lmna−/−) fibroblasts isolated from lmna knockout mice, as well as in 3T3 cells with RNAi induced reduction of lmna expression.82
Concluding Remarks: Mapping and Understanding the Dynamic Nuclear Architecture Requires a Systems Biological Approach
The above considerations encourage a new perspective of dynamic chromatin arrangements in somatic cell types. The hypothesis that chromosome territories dance in assemblies in order to establish transient or permanent cell type specific DNA-DNA interactions implies that mapping the dynamic nuclear architecture from the fertilized egg to a multitude of cell types becomes an exceedingly demanding undertaking. The envisaged complexity of the mechanism(s) responsible for dynamic chromatin arrangements in somatic cell nuclei involves a great number of proteins assembled in molecular machines, potentially including proteins also involved in alignment and pairing events in meiotic prophase. Genome wide mapping of DNA-DNA interactions is only the beginning paving the way to even more demanding problems. How are dynamic DNA-DNA interactions in cis and trans related to the topography of protein machineries involved in transcription, splicing, DNA replication and repair? What do these interactions mean in functional terms? What molecular mechanisms are required to establish, maintain and disassemble such interactions?
Materials and Methods
Cell lines and constructs.
H-tert immortalized retina pigment epithelium 1 cells (RPE-1) originally established by Clonetech (California) were kindly provided by Friederike Eckhardt-Schupp (Helmholtz Zentrum, Munich, Germany). Although these cells were considered to have a normal karyotype (46,XX), M-FISH revealed that our batch of cells was subtetraploid with a mean of 73 chromosomes (counted from 15 metaphase spreads) and carrying a few chromosomal rearrangements (data not shown). Using Fugene HD (Roche) cells were simultaneously transfected with a plasmid-construct for paGFP-H4 carrying a CMV promotor and a resistance marker against G418 (a kind gift from Roeland W. Dirks, University of Leiden26) and a second plasmid-construct for mRFP-H2B with a SV40 promotor (kind gift from Ruth Brack-Werner, Helmholtz Zentrum, München, Germany27). After selection with G418 clones expressing both paGFP and mRFP were picked manually. For some experiments cells were transiently transfected with mRFP-PCNA kindly provided by Heinrich Leonhardt (LMU München). After further cultivation clones were subjected to fluorescence activated cell sorting (FACS). Clonal cell populations exhibiting both green and red fluorescence cells were stored in liquid nitrogen until use. After thawing >85% of all cell-nuclei showed both colors. The cells were grown in a density dependent manner with a generation time of ∼30 hours (data not shown). For live cell observations of centrosomes cells were transiently transfected with a plasmid-construct coding for a GFP tagged FOP (Fibroblast Growth Factor Receptor-1 Oncogene Partner) kindly provided by Erich Nigg (Biozentrum Basel).
Cell culture conditions.
RPE-1(paGFP-H4; mRFP-H2B or mRFP-PCNA) cells were cultured in DMEM-HEM-F12 medium (1:1) containing 15 mM HEPES (Sigma), 250 µM Trolox (Sigma) added 12 h prior to imaging, 20% FCS and 5% Penicillin/Streptomycin. Cells were grown at 37°C in a humidified incubator at 5% CO2 and seeded into LabTek-Chambers (Nunc) for in vivo observations. During in vivo imaging cells were kept at 37°C and pH 7.2.
Live cell microscopy combined with photoactivation of paGFP-H4 and bleaching of mRFP-H2B.
Live-cell imaging was performed with a spinning disc microscope. A Perkin Elmer Ultra View Vox Confocal Imaging System was connected to a Zeiss Axio Observer D1 fluorescence microscope. A FRAP unit was used to perform photoactivation and/or photobleaching experiments through a 63x Plan Apochromat (C. Zeiss) objective with laser beams focused to the mid nuclear plane. Photoactivation of paGFP-H4 in selected nuclear areas was performed by 10x repeated scanning with a 440 nm laser beam of a diode laser at 4% (100% = 40 mW). Photobleaching of mRFP was performed in the same way using a diode laser with a 651-nm laser line at 100% (75 mW) with 100 times repetition. Image stacks were acquired automatically with a z-resolution between 0.3 and 0.75 µm covering a total stack height of 15 µm (voxel size 109 × 109 × 300 up to 750 nm).
Image acquisition software.
Live-cell image acquisition was performed with the Volocity v5.2.1 Software (Improvision/Perkin Elmer), acquisition of fixed cells with the LAS-Software (Leica). Image processing was performed in Volocity 5.2.1, ImageJ 1.42 and Photoshop CS4. 3D reconstructions were made with Amira 5.2.2. Rotatable 3D reconstructions were achieved with Adobe Acrobat 3D reviewer.83
Immunofluorescence and 3D imaging of fixed cells.
For immunofluorescence cells studied in the Lab-Tek-chamber were fixed 10 min in 1× PBS with 4% paraformaldehyde, permeabilized 20 min in 1X PBS with 0.4% Triton X-100 and blocked in 1x PBS with 4% BSA. The following, commercial primary and secondary antibodies were used: polyclonal-rabbit-anti-pericentin (Abcam; catalog-#ab4448), monoclonal-mouse-anti-γ-H2AX (Upstate; catalog-#06-636), monoclonal-mouse-anti-tubulin (Sigma; catalog-#T4026), monoclonal-mouse-anti-PAR (Trevigen; catalog-# 4335-MC-100), human antiserum CREST (Euroimmun, catalog-# CA 1611-0101), polyclonal goat-anti-mouse-fab-cy3 and TexasRed (Jackson Immuno Research; catalog-# 115-165-072 and #111-076-045 respectively), polyclonal goat-anti-rabbit-alexa 633 (Molecular Probes; catalog-# A-21071) and polyclonal goat-anti-human-FITC (Jackson Immuno Research; catalog-# 109-095-006). Primary antibodies were incubated over night at 4°C in a humidified chamber. Secondary antibodies were incubated for 1 h at 37°C. Thereafter cell samples were embedded with Vectashield (Vector Labs) and 3D image stacks with a voxel size of 50 × 50 × 200 nm were recorded with a Leica SP5 confocal microscope.
Acknowledgements
This study was supported by grants awarded to Thomas Cremer by the Human Science Frontier Program (HSFP), the Deutsche Forschungsgemeinschaft (DFG) and the Munich Center of integrated Protein Science (CIPSM). The authors are indebted to the following colleagues, who have provided materials necessary for this study: Heinrich Leonhardt (LMU Biocenter, Munich; mRFP-PCNA, PAR antibodies), Erich Nigg (Biocenter University of Basel; FOP-GFP), Roeland W. Dirks (University of Leiden; paGFP-H4), Ruth Brack-Werner (Helmholtz Center, Munich; mRFP-H2B), Friederike Eckhardt-Schupp (Helmhotz Center, Munich; RPE-1 cells), Jan Ellenberg (EMBL, Heidelberg; NRK-cells).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11969
Supplementary Material
References
- 1.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Pederson T. The spatial organization of the genome in mammalian cells. Curr Opin Genet Dev. 2004;14:203–209. doi: 10.1016/j.gde.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 7.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 8.Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 9.Kosak ST, Groudine M. Gene order and dynamic domains. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- 10.Hu Q, Kwon Y-S, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C, Yang L, Tanasa B, Hutt K, Ju B-g, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cvacková Z, Masata M, Staněek D, Fidlerová H, Raska I. Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. J Struct Biol. 2009;165:107–117. doi: 10.1016/j.jsb.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 16.Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essers J, van Cappellen WA, Theil AF, van Drunen E, Jaspers NGJ, Hoeijmakers JHJ, et al. Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell. 2005;16:769–775. doi: 10.1091/mbc.E04-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 19.Boveri T. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforsch. 1909;3:181–268. (Ger). [Google Scholar]
- 20.Rabl C. Über Zelltheilung. Morph Jb. 1885;10:214–330. (Ger). [Google Scholar]
- 21.Boveri T. Zellen Studien. Jenaische Zeitschrift für Naturwissenschaft. 1888;22:687–882. (Ger). [Google Scholar]
- 22.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem. 2006;50:161–176. [PubMed] [Google Scholar]
- 23.Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem. 2006;50:223–272. [PubMed] [Google Scholar]
- 24.Gerlich D, Ellenberg J. Dynamics of chromosome positioning during the cell cycle. Curr Opin Cell Biol. 2003;15:664–671. doi: 10.1016/j.ceb.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 26.Wiesmeijer K, Krouwels IM, Tanke HJ, Dirks RW. Chromatin movement visualized with photoactivable GFP-labeled histone H4. Differentiation. 2007;76:83–90. doi: 10.1111/j.1432-0436.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- 27.Wolff H, Hadian K, Ziegler M, Weierich C, Kramer-Hammerle S, Kleinschmidt A, et al. Analysis of the influence of subcellular localization of the HIV Rev protein on Rev-dependent gene expression by multi-fluorescence live-cell imaging. Exper Cell Res. 2006;312:443–456. doi: 10.1016/j.yexcr.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 28.Ismail IH, Hendzel MJ. The gamma-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ Mol Mutagen. 2008;49:73–82. doi: 10.1002/em.20358. [DOI] [PubMed] [Google Scholar]
- 29.Mortusewicz O, Amé J-C, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berezney R, Dubey DD, Huberman JA. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. doi: 10.1007/s004120050399. [DOI] [PubMed] [Google Scholar]
- 31.Chaly N, Brown DL. The prometaphase configuration and chromosome order in early mitosis. J Cell Sci. 1988;91:325–335. doi: 10.1242/jcs.91.3.325. [DOI] [PubMed] [Google Scholar]
- 32.Solovei I, Schermelleh L, Düring K, Engelhardt A, Stein S, Cremer C, et al. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 2004;112:410–423. doi: 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]
- 33.Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. The J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 35.Manders EM, Kimura H, Cook PR. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J Cell Biol. 1999;144:813–821. doi: 10.1083/jcb.144.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- 38.Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- 39.Williams RRE, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exper Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- 40.Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 41.Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol. 1997;137:1459–1468. doi: 10.1083/jcb.137.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77:2871–2886. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 44.Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 45.Chuang C-H, Belmont AS. Moving chromatin within the interphase nucleus-controlled transitions? Semin Cell Dev Biol. 2007;18:698–706. doi: 10.1016/j.semcdb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C. Morphology and dynamics of chromosome territories in living cells. Biochim Biophys Acta. 2001;1551:29–39. doi: 10.1016/s0304-419x(01)00023-3. [DOI] [PubMed] [Google Scholar]
- 47.Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 49.Göndör A, Ohlsson R. Chromosome crosstalk in three dimensions. Nature. 2009;461:212–217. doi: 10.1038/nature08453. [DOI] [PubMed] [Google Scholar]
- 50.Ling JQ, Hoffman AR. Epigenetics of long-range chromatin interactions. Pediatr Res. 2007;61:11–16. doi: 10.1203/pdr.0b013e31804575db. [DOI] [PubMed] [Google Scholar]
- 51.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 52.Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, et al. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008;4:1000016. doi: 10.1371/journal.pgen.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunez E, Fu X-D, Rosenfeld MG. Nuclear organization in the 3D space of the nucleus—cause or consequence? Curr Opin Genet Dev. 2009;19:424–436. doi: 10.1016/j.gde.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexton T, Bantignies F, Cavalli G. Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Simonis M, De Laat W. FISH-eyed and genome-wide views on the spatial organisation of gene expression. Biochim Biophys Acta. 2008;1783:2052–2060. doi: 10.1016/j.bbamcr.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 57.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 58.Tiwari VK, Cope L, McGarvey KM, Ohm JE, Baylin SB. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18:1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams A, Flavell RA. The role of CTCF in regulating nuclear organization. J Exp Med. 2008;205:747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Z, Tavoosidana G, Sjölinder M, öndör A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 61.Göndör A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat Protoc. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- 62.Wachsmuth M, Caudron-Herger M, Rippe K. Genome organization: balancing stability and plasticity. Biochim Biophys Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 63.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. Fifth Edition 2008. [Google Scholar]
- 64.Bouchet-Marquis C, Dubochet J, Fakan S. Cryoelectron microscopy of vitrified sections: a new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol. 2006;125:43–51. doi: 10.1007/s00418-005-0093-x. [DOI] [PubMed] [Google Scholar]
- 65.Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009;17:801–810. doi: 10.1007/s10577-009-9070-x. [DOI] [PubMed] [Google Scholar]
- 66.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hancock R. Packing of the polynucleosome chain in interphase chromosomes: evidence for a contribution of crowding and entropic forces. Semin Cell Dev Biol. 2007;18:668–675. doi: 10.1016/j.semcdb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta. 2008;1783:2100–2117. doi: 10.1016/j.bbamcr.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 69.Barr ML, Bertram EG. The behaviour of nuclear structures during depletion and restoration of Nissl material in motor neurons. J Anat. 1951;85:171–181. [PMC free article] [PubMed] [Google Scholar]
- 70.Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- 71.Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 72.Moore G, Shaw P. Improving the chances of finding the right partner. Curr Opin Genet Dev. 2009;19:99–104. doi: 10.1016/j.gde.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Hiraoka Y, Dernburg AF, Parmelee SJ, Rykowski MC, Agard DA, Sedat JW. The onset of homologous chromosome pairing during Drosophila melanogaster embryogenesis. J Cell Biol. 1993;120:591–600. doi: 10.1083/jcb.120.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Boni U, Mintz AH. Curvilinear, three-dimensional motion of chromatin domains and nucleoli in neuronal interphase nuclei. Science. 1986;234:863–866. doi: 10.1126/science.3775367. [DOI] [PubMed] [Google Scholar]
- 75.Park PC, De Boni U. Dynamics of nucleolar fusion in neuronal interphase nuclei in vitro: association with nuclear rotation. Exp Cell Res. 1991;197:213–221. doi: 10.1016/0014-4827(91)90425-t. [DOI] [PubMed] [Google Scholar]
- 76.De Boni U. Chromatin motion in interphase nuclei, its modulation and its potential role in gene expression. Anticancer Res. 1988;8:885–898. [PubMed] [Google Scholar]
- 77.Heyting C. Meiotic transverse filament proteins: essential for crossing over. Transgenic Res. 2005;14:547–550. doi: 10.1007/s11248-005-8925-y. [DOI] [PubMed] [Google Scholar]
- 78.Mehta IS, Elcock LS, Amira M, Kill IR, Bridger JM. Nuclear motors and nuclear structures containing A-type lamins and emerin: is there a functional link? Biochem Soc Trans. 2008;36:1384–1388. doi: 10.1042/BST0361384. [DOI] [PubMed] [Google Scholar]
- 79.Pederson T. As functional nuclear actin comes into view, is it globular, filamentous or both? J Cell Biol. 2008;180:1061–1064. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pederson T, Aebi U. Nuclear actin extends, with no contraction in sight. Mol Biol Cell. 2005;16:5055–5060. doi: 10.1091/mbc.E05-07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levy JR, Holzbaur ELF. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Houben F, Willems CHMP, Declercq ILJ, Hochstenbach K, Kamps MA, Snoeckx LHEH, et al. Disturbed nuclear orientation and cellular migration in A-type lamin deficient cells. Biochim Biophys Acta. 2009;1793:312–324. doi: 10.1016/j.bbamcr.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Ruthensteiner B, Heß M. Embedding 3D models of biological specimens in PDF publications. Microsc Res Tech. 2008;71:778–786. doi: 10.1002/jemt.20618. [DOI] [PubMed] [Google Scholar]
- 84.van Beneden E. Recherches sur la Maturation de L'Oeuf, la Fécondation et la Division Cellulaire. Paris: Gand, Leipzig; 1883. (Ger). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.