Abstract
Ozone (O3) remains a prevalent air pollutant and public health concern. Inf2 is a significant quantitative trait locus (QTL) on murine chromosome 17 that contributes to susceptibility to O3-induced infiltration of polymorphonuclear leukocytes (PMNs) into the lung, but the mechanisms of susceptibility remain unclear. The study objectives were to confirm and restrict Inf2, and to identify and test novel candidate susceptibility gene(s).
Congenic strains of mice that contained overlapping regions of Inf2 and their controls, and mice deficient in either MHC Class II genes or the Tnf cluster were exposed to air or O3. Lung inflammation and gene expression were assessed.
Inf2 was restricted from 16.42 Mbp to 0.96 Mbp, and bioinformatic analysis identified MHC class II, the Tnf cluster, and other genes in this region that contain potentially informative SNPs between the susceptible and resistant mice. Furthermore, O3-induced inflammation was significantly reduced in mice deficient in MHC class II genes, or the Tnf cluster genes, compared to wild-type controls. Gene expression differences were also observed in MHC class II and Tnf cluster genes.
This integrative genetic analysis of Inf2 led to identification of novel O3 susceptibility genes that may provide important, new therapeutic targets in susceptible individuals.
Keywords: inflammation, lymphotoxin α, mouse, major histocompatibility complex, susceptibility, tumor necrosis factor
Introduction
The pollutant ozone (O3) is a highly toxic principal oxidant found in urban environments throughout industrialized cities worldwide. O3 exposure has been associated with many adverse health effects, such as exacerbation of asthma [1, 2]. Identification of susceptibility genes involved in O3-induced pulmonary injury may provide critical information for future risk assessment as well as general international health policies. In 2006, an estimated one-third of U.S. individuals were at an increased risk of adverse effects caused by O3 and 131 million U.S. residents resided in regions that either approached or exceeded the National Ambient Air Quality Standard of 0.08 parts per million (ppm) O3 [3].
Significant intersubject differences in pulmonary function and inflammatory responses to O3 suggest that genetic background contributes to O3 susceptibility in humans and rodents [4–6]. Furthermore, studies in human subjects have suggested that polymorphisms in oxidant defense genes, such as glutathione S transferase M1, and quinone metabolism genes, such as NADPH quinone oxidoreductase 1, associate with differential responsiveness to O3 [7, 8]. Activating polymorphisms in inflammatory genes, such as tumor necrosis factor alpha (TNF), also enhance susceptibility to O3 and asthma [9]. A genome-wide linkage analysis of O3-induced influx of polymorphonuclear leukocytes (PMNs) in an intercross cohort (B6C3F2) derived from susceptible C57BL/6 (B6) and resistant C3H/HeJ (C3) progenitor mouse strains identified a significant susceptibility QTL on chromosome 17 (inflammation 2; Inf2) [5]. Inf2 (33.73–50.15 Mbp; D17Mit16 – D17Mit10) contains the H-2 locus, including major histocompatibility genes and non-MHC genes such as the pro-inflammatory cytokine Tnf. Pre-treatment of B6 mice with a monoclonal antibody to TNF-α and deletion of TNF-α receptors 1 and 2 [5, 10] significantly attenuated the inflammatory response to O3 relative to control mice, supporting the importance of Tnf as a candidate gene in this model. However, these studies did not conclusively identify Tnf as the susceptibility gene in Inf2. In addition, Inf2 is located in the most gene dense, polymorphic region of the entire mouse genome, thus making single candidate gene identification difficult [11].
In the current study we sought to confirm the importance of Inf2 in O3-induced lung inflammation, to reduce Inf2, and to identify candidate susceptibility genes. Using an integrative genomics approach, we utilized congenic mouse strains to limit Inf2 from 16.42 Mbp to 0.96 Mbp, and bioinformatic analysis which identified MHC class II genes and the entire TNF cluster as candidate susceptibility loci. Functional analyses of these genes confirmed novel roles for modulation of the inflammatory response to O3 exposure.
Materials and Methods
Mouse strains and O3 exposure
The following male (6-8 wk) congenic mice were used: B10.A- H2h2/(2R)SgSnJ (2R); B10.A-H2h4/(4R)SgDvEg (4R), B10.A-H2i5H2-T18a/(5R)SgSnJ (5R); and C3.SW-H2b/SnJ (C3H-H2b). The congenic region and haplotype for each of the strains are shown in Table 1 and Figure 1. Control strains for the congenic mice were C57BL/10SnJ (B10), A/WySnJ (A), and C3H/HeSnJ (HeSnJ). The location of the congenic region of the 2R, 4R, and 5R mice with respect to the B10 and A/Wy background strains was identified by genotyping (Malhotra and Travis, data not shown). H2k haplotypes are O3-resistant and the H2b haplotype is O3-susceptible [5] . Additional strains used for the candidate gene studies were B6.129-H2dlAb1-Ea/J (H2-dlAbl-Ea; B6 background strain), B6.129S2-Ltatm1Dch/J (Lta−/−, B6 background), and B6.129P2-Ltb/Tnf/Ltatm1Dvk/J (Ltb/Tnf/Lta−/− mice, B6 background), C57BL/6J (B6) mice, and C3H/HeJ (C3) mice. B6 are O3-susceptible and are H2b, while C3H/HeJ (C3) mice are O3-resistant and are H2k. All mice were purchased from Jackson Laboratory (Bar Harbor, ME). All animal use was conducted in facilities accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care and approved by the NIEHS Animal Care and Use Committee and follows the Helsinki convention for the use and care of animals. For air and O3 exposures, mice were caged in a humidity and temperature-controlled room and provided water and pelleted open-formula rodent diet NIH-07 (Zeigler Brothers, Gardners, PA) ad libitum (see O3 exposure procedures in Supplement Methods).
Table 1.
Nomenclature and characteristics of the congenic and control strains used for investigation.
| Congenic Region | ||||
|---|---|---|---|---|
| Strain Name | Abbreviation | Mbp | cM | H2 Haplotype** |
| Congenic strains | ||||
| B10.A-H2h2/(2R)SgSnJ | 2R | 33.74 – 35.34 | 18.20 – 19.06 | H2h2 |
| B10.A-H2h4/(4R)SgDvEgJ | 4R | 29.09 – 34.38 | 16.30 – 18.64 | H2h4 |
| B10.A-H2i5 H2-T18a/(5R)SgSnJ | 5R | 34.45 – 36.35 | 18.70 – 19.14 | H2i5 |
| C3.SW-H2b/SnJ | C3H-H2b | 33.74 – 43.80 | 18.20 – 23.30 | H2bc |
| Control strains | ||||
| C57BL/10SnJ† | B10 | - - - | - - - | H2b |
| A/WySnJ ‡ | A | - - - | - - - | H2a |
| C3H/HeSnJ++ | HeSnJ | - - - | - - - | H2k |
Mbp and cM were determined using the MGI website:http://www.informatics.jax.org/searches/marker and genotyped by Malhotra and Travis, unpublished).
MHC H2 Haplotypes according to Appendix 2: MHC H2 haplotypes (www.jax.org/jaxmice/search).
background strain for 2R, 4R, and 5R mice ,
donor strain for 2R, 4R, and 5R mice.,
background strain for C3H-H2b mice.
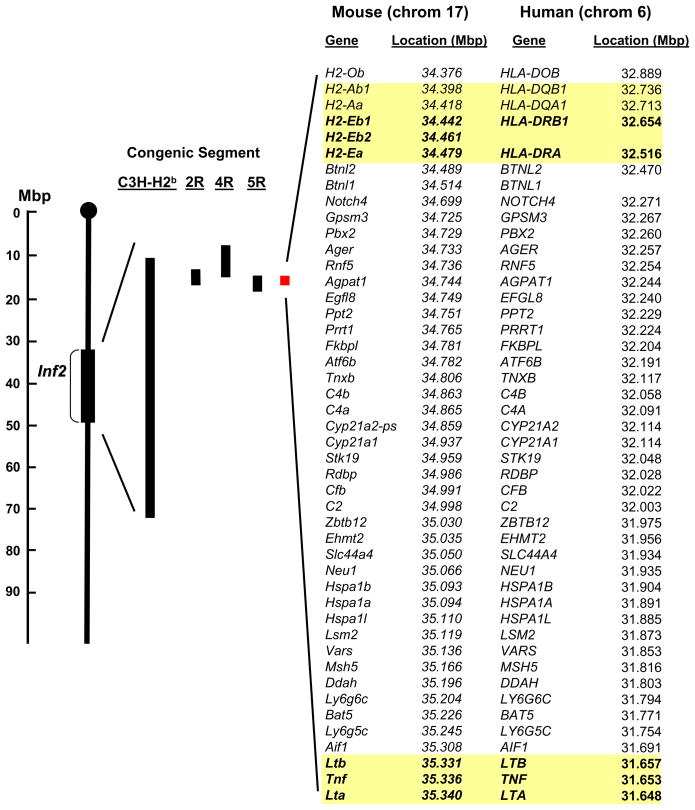
Figure 1. Schematic of the chromosome 17 congenic segments for 2R, 4R, 5R, and C3H-H2b mice that were used to reduce inflammation 2 (Inf2).
The congenic segments are shown with respect to Inf2 (on left). The red rectangle represents the reduced Inf2. Mouse genes and human homologues and their chromosomal locations within the reduced Inf2 are shown at right. While most of the genes are in the same order in both species, some are not. These locations were identified using the MGI website:http://www.informatics.jax.org/searches/marker website and NCBI. The yellow highlighted genes are major histocompatibility complex (MHC) class II genes and TNF cluster genes identified as candidate genes using proof of concept experiments (see Figs. 5 and 6). Abbreviations:Ager, advanced glycosylation end product-specific receptor; Agpat1; 1-acylglycerol-3-phosphate O-acyltransferase 1; Aif1, allograft inflammatory factor 1; Atf6b, activating transcription factor 6 beta; Bat5, HLA-B associated transcript 5; Btnl2, butyrophilin-like 2; Btnl1, butyrophilin-like 1; C2, complement component 2; C4a, complement component 4A; C4b, complement component 4B; Cfb, complement factor B; CREBL1, cAMP responsive element binding protein-like 1; Cyp21a1, cytochrome P450, family 21, subfamily a, polypeptide 1; Cyp21a2-ps, cytochrome P450, family 21, subfamily a, polypeptide 2 pseudogene; Ddah, dimethylarginine dimethylaminohydrolase 2;Egfl8, EGF-like domain 8; Ehmt2, euchromatic histone-lysine N-methyltransferase 2; Fkbpl, FK506 binding protein-like; Gpsm3, G-protein signalling modulator 3; H2-Ab1, class II antigen A, beta 1; H2-Aa, class II antigen A, alpha; H2-Eb1, class II antigen E beta; H2-Eb2, class II antigen E beta 2; H2-Ea, class II antigen E alpha; H2-ob, O region beta locus; Hspa1b, heat shock 70kDa protein 1B; Hspa1a, heat shock 70kDa protein 1A; Lsm2, LSM2 homolog; Lta, lymphotoxin a; Ltb, lymphotoxin b; Ly6g6c, lymphocyte antigen 6 complex, locus G6C; Ly6g5c, lymphocyte antigen 6 complex, locus G5C; Msh5, mutS homolog 5;Neu1, neuraminidase1; Notch4, notch gene homolog 4; Pbx2, pre B-cell leukemia transcription factor 2; Ppt2, palmitoyl-protein thioesterase 2; Prrt1, proline-rich transmembrane protein 1; Psmb8, proteosome subunit, beta type 8; Rdbp, RD RNA-binding protein; Rnf5, ring finger protein 5; Slc44a4; solute carrier family 44, member 4; Stk19, serine/threonine kinase 19;Tap-1, transporter 1, ATP-binding cassette, sub-family B (MDR/TAP); Tap-2, transporter 2, ATP-binding cassette, sub-family B (MDR/TAP); Tnf, tumor necrosis factor; Tnxb, tenascin B; Vars, valyl-tRNA synthetase; Zbtb12, zinc finger and BTB domain containing 12. No human homolog exists for Btnl1.
Bronchoalveolar lavage fluid (BALF) analysis
The procedures used for these techniques have been described previously for right lung lavages and inflammatory cell analysis [5, 10, 12].
Total RNA Isolation and Real-Time Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Mice were sacrificed immediately after air or O3 exposure, and lungs of each animal were snap-frozen in liquid nitrogen. Total RNA isolation, reverse transcription into cDNA and PCR reaction procedures are described in Supplemental Methods.
Statistics
Data are expressed as group means ± standard error of the mean (S.E.M.). Two-way analysis of variance (ANOVA) was used to evaluate the effects of exposure (air vs O3) and strain (B10, 2R, 4R, 5R, A, HeSnJ, C3H-H2b, H2-dlAbl-Ea−/−, H2-dlAbl-Ea+/+, Lta−/−, Lta+/+, Lta/Tnf/Ltb−/−, Lta/Tnf/Ltb+/+, B6, C3) on BAL phenotypes and mRNA expression. The figure legends contain the number of mice used per experiment. Student-Newman-Keuls test was used for a posteriori comparisons of means. All analyses were performed using a commercial statistical analysis package (SigmaStat; Jandel Scientific Software, San Rafael, CA). Statistical significance was accepted at P<0.05.
Results
Restriction of Inf2
Responses to O3 in congenic mice
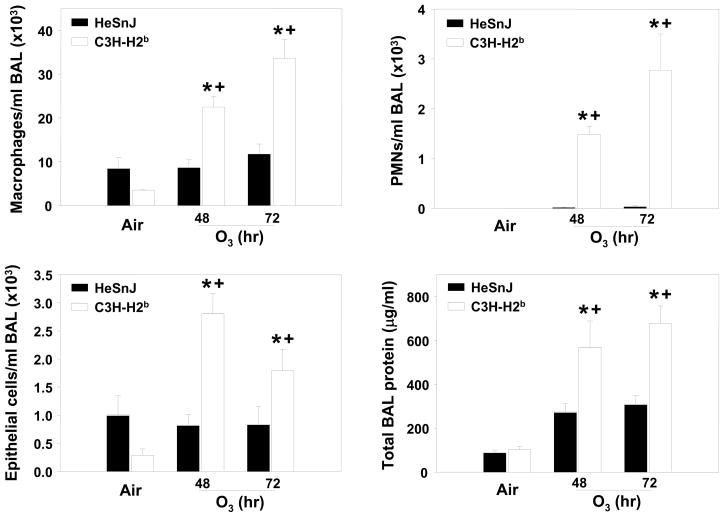
To confirm the significance of Inf2, C3H-H2b mice which contain Inf2 from a susceptible mouse (H2b) on an O3-resistant H2K background [HeSnJ; (see Figure 1)] were exposed to air and O3. Relative to HeSnJ controls, significant increases in mean BAL protein concentration and numbers of macrophages, PMNs, and epithelial cells were found in C3H-H2b mice after 48 and 72 h exposure to O3 (Figure 2); no strain differences were observed in any parameter in the air exposed mice. Results thus confirmed the importance of the region of chromosome 17 encompassed by Inf2 in susceptibility to O3-induced inflammation.
Figure 2. Bronchoalveolar lavage fluid (BALF) inflammatory parameters in the C3H-H2b and HeSnJ mice exposed continuously to 0.3 ppm O3 or air for 48 or 72 h.
A. Macrophages; B. Polymorphonuclear leukocytes (PMNs); C. Epithelial cells; D. Total BALF protein. Data are presented as means ± SEM (n=3-5 per experimental group). *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from HeSnJ mice (P < 0.05).
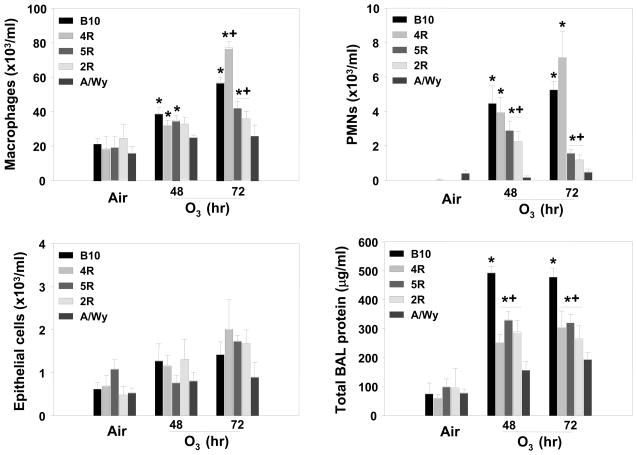
To reduce the length of Inf2, O3-induced inflammatory responses in congenic 2R, 4R, and 5R mice were compared to those of similarly exposed O3-susceptible background B10 and O3-resistant donor A strains. Each congenic strain contains a different region of the Inf2 locus from the A strain mouse on a B10 background (see Table 1). O3 caused significant increases in mean numbers of macrophages and PMNs in B10 but not A mice compared to respective air-exposed animals, and numbers of macrophages and PMNs were greater in B10 mice compared to A mice after O3 (Figure 3). The mean numbers of PMNs and macrophages in 2R and 5R mice were not different from A strain mice after 48 and 72 h O3; however, numbers of PMNs and macrophages in 2R and 5R mice were significantly lower compared to B10 mice (Figure 3). In contrast, the numbers of BALF macrophages and PMNs from O3-exposed 4R mice were not significantly different from those from O3-exposed B10 mice, and were greater than those from A, 2R, and 5R mice (Figure 3). The mean BALF protein concentrations were significantly reduced in 2R, 4R, and 5R mice compared to B10 mice and significantly greater than A mice after 48 and 72 h O3. Comparison of the mean numbers of macrophages and PMNs between congenic strains with respective congenic regions thus suggested that the 34.38–35.34 Mbp [18.64–19.06 centimorgan (cM)] region of chromosome 17 accounted for a major portion of the inflammatory response to O3 in these mice (Figure 1). No differences in mean numbers of BALF epithelial cells were found between the congenic, A, or B10 strains after exposure to O3 (Figure 3C).
Figure 3. Bronchoalveolar lavage fluid (BALF) inflammatory parameters in B10, 4R, 5R, 2R, and A/Wy mice exposed continuously to 0.3 ppm O3 or air for 48 or 72 h.
A. Macrophages; B. Polymorphonuclear leukocytes (PMNs); C. Epithelial cells; D. Total BALF protein. Data are presented as means ± SEM (n=3-12 per experimental group). *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from B10 mice (P < 0.05).
Candidate Gene Analysis
Database search for polymorphisms
Genes within the reduced region of Inf2 (34.38–35.34 Mbp) were searched for known exon (non-synonymous), intron (including splice sites) and UTR polymorphisms using the public Mouse Phenome Database [13] (Supplemental Table 1). H2-Aa and H2-Ab1 genes are haplotype H2k in the 2R and 4R congenic strains, thus excluding these as candidate genes [14]. Additionally, the H2-Eb1 gene is divided between resistant and susceptible haplotypes and therefore remains a candidate gene, as well as H2-Eb2 [15]. The genes that contain known non-synonymous polymorphisms between the B6 (susceptible) and C3 (resistant) mice in the reduced Inf2 in order of location on chromosome 17 are H2-Eb1, H2-Eb2, Btnl1, Notch4, Fkbpl, Crebl1, Tnxb, C4b, C4a, and Stk19. Two genes (Hspa1b and Hspa1a) may have exonic polymorphisms between the B6 and C3, but no details are currently available for the B6 strain at these sites. Genes with intronic polymorphisms between the B6 (susceptible) and C3 (resistant) mice in the reduced Inf2 are: H2-Eb1, H2-Eb2, H2-Ea, Btln2, Btln1, Btln7, Notch4, Ppt2, Crebl1, Tnxb, C4b, Cyp21a1, C4a, Stk19, Cfb, C2, Msh5, Ddah2, Ly6g6c, Bat5, Ly6g5c, Csnk2b, Aif1, Tnf, and Lta (Supplemental Table 1). UTR polymorphisms between the B6 and C3 mice were found in H2-Eb2, Btnl2, Ppt2, C4b, C4a, G6b, Bat5, Ly6g5b, and Tnf (Supplemental Table 1). Information is absent for Ltb and Tnf UTR and intronic polymorphisms in the C3 strain, thus it is possible that more exist between the B6 and C3 strains.
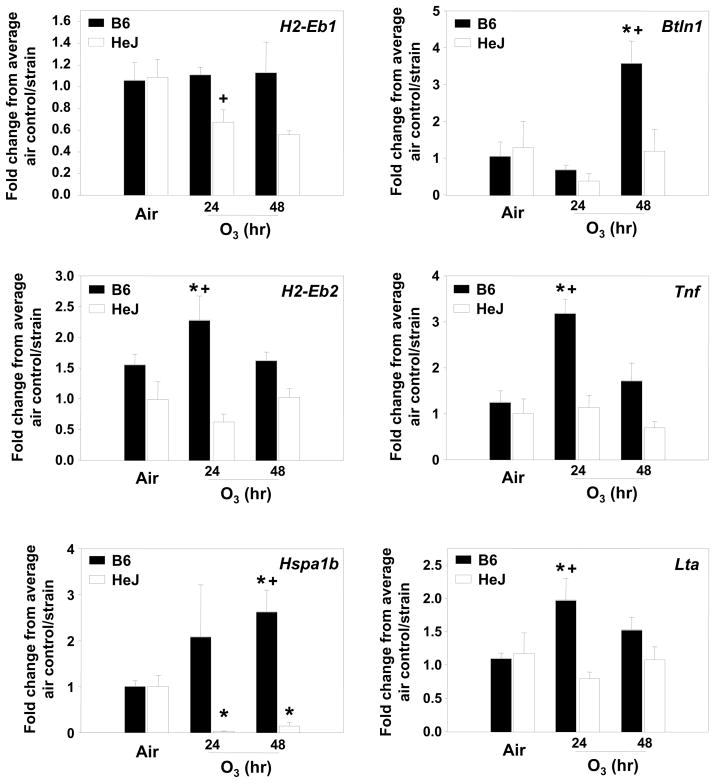
We then asked if mRNA expression differences between B6 and C3 mice existed for several candidate genes within the reduced Inf2, and with either known or possible SNPs (Fig. 4). H2-Eb1 expression was reduced after 24 h of O3 exposure in the C3 mice, but unchanged in the B6 mice; H2-Eb2 expression was significantly increased only in B6 mice at 24 h. Expression of Hspa1b and Btnl1 was significantly increased after 72 h O3 in B6 mice only. Tnf and Lta expression was significantly increased in B6 mice after 24 h O3 compared to no increases observed in C3 mice.
Figure 4. Gene expression for candidate genes identified within the reduced Inf2 region of B6 (susceptible) and C3 (resistant) mice.
Six genes were analyzed using quantitative RT-PCR (H2Eb1, H2Eb2, Hspa1b, Btnl1, Tnf, Lta). Data are presented as means ± SEM (n=3–7 per experimental group) and were determined using the comparative CT method (see Supplemental Materials and Methods section). Y-axis represents the gene of interest normalized first to 18S followed by determination of the fold-change relative to the average air control value for each strain. *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from B6 mice (P < 0.05).
MHC class II deficient mice
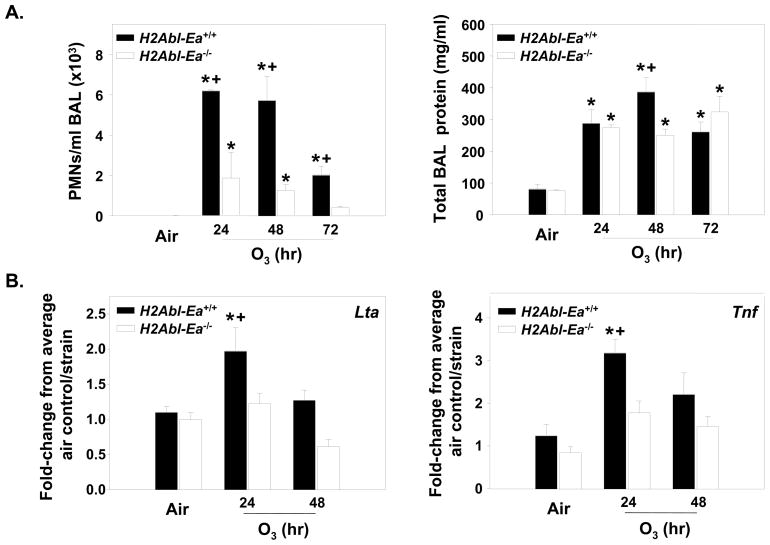
Because of the prevalence of MHC class II genes in Inf2 and the non-synonymous, intronic, and UTR polymorphisms in H2-Eb1 and -Eb2 between the susceptible (B6) and resistant strains (C3) (Supplemental Table 1), we hypothesized that they were candidate susceptibility genes for O3-induced inflammation. H2Abl-Ea−/− mice are deficient in MHC class II genes H2-Ab1, -Aa, -Eb1, -Eb2, and -Ea compared to the H2Abl-Ea+/+ mice. The B6 background of these mice is null at H2-Ea, therefore H2Abl-Ea−/− mice only test the importance of the H2-Ab1 through H2-Eb2 MHC class II region. Relative to respective air controls, O3 caused significant (P < 0.05) increases in mean numbers of BALF PMNs and macrophages in H2Abl-Ea+/+ mice after 24, 48 and 72 h, and after 24 and 48 h O3 in H2Abl-Ea−/− mice (Fig. 5A and Table 2). However, significantly greater numbers of PMNs and macrophages were found in H2Abl-Ea+/+ mice compared to H2Abl-Ea−/− mice after 24–72 h and 48 and 72 h exposures, respectively. Significant strain and O3 effects on BAL epithelial cells were found after 48 h O3 (Table 2). Significant strain differences in BAL protein content were also found after 48 h O3 (Fig. 5A). mRNA expression of Tnf and Lta was significantly increased in the H2Abl-Ea+/+ mice after 24 h O3; however no increase was observed in the H2Abl-Ea−/− mice (Fig. 5B).
Figure 5. Deficiency in MHC class II genes significantly reduced O3-induced responses.
A. Bronchoalveolar lavage fluid (BALF) polymorphonuclear leukocytes (PMNs) and total protein in H2Ab1-Ea−/− and H2Ab1-Ea+/+ mice exposed continuously to 0.3 ppm O3 or air for 48 or 72 h. Data are presented as means ± SEM (n=3-10 per experimental group). *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from H2Ab1-Ea+/+ mice (P < 0.05). B. Gene expression for Lta and Tnf genes in H2Ab1-Ea deficient mice compared to wildtype mice. Data are presented as means ± SEM (n=3-6 per experimental group) and were determined using the comparative CT method (see Supplemental Materials and Methods section). Y-axis represents either Lta or Tnf normalized first to 18S followed by determination of the fold-change relative to the average air control value for each strain. *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from H2Ab1-Ea+/+ mice (P < 0.05).
Table 2.
Effect of deletion of H2-Abl-Ea, Lta, or Lta/Tnf/Ltb on macrophages and epithelial cells recovered from BAL following O3 exposure.
| No. of cells (X 103 ml/BAL) |
|||||
|---|---|---|---|---|---|
| O3 exposed |
|||||
| Cell type | Genotype | Air* | 24 h | 48 h | 72 h |
| Macrophages | H2Abl-Ea+/+ | 19.6 ± 0.1 | 33.6 ± 3.9** | 48.1 ± 0.4**§ | 37.9 ± 3.7**§ |
| H2Abl-Ea −/− | 19.0 ±0.1 | 38.5 ± 6.3** | 33.2 ± 0.4** | 22.5 ± 1.3 | |
| Lta+/+ | 12.1 ± 1.2 | 22.2 ± 3.9**§ | 29.7 ± 7.7**§ | 37.9 ± 3.7** | |
| Lta−/− | 13.8 ±1.9 | 7.6 ± 2.2 | 21.2 ±2.7 | 32.0 ± 2.5** | |
| Lta/Tnf/Ltb+/+ | 18.3 ± 3.4 | 26.7 ± 3.0 | 36.8 ± 3.9**§ | 38.5 ± 3.4**§ | |
| Lta/Tnf/Ltb−/− | 26.6 ± 3.2 | 32.7 ± 3.7 | 25.2 ± 3.0 | 24.4 ± 3.9 | |
| Epithelial cells | MHC+/+ | 1.4 ± 0.3 | 1.0 ± 0.2 | 2.6 ± 0.7**§ | 1.0 ± 0.3 |
| MHC−/− | 0.1 ± 0.2 | 1.8 ± 0.6 | 0.9 ± 0.1 | 0.9 ± 0.2 | |
| Lta+/+ | 0.7 ± 0.2 | 0.85 ± 0.3 | 1.4 ± 0.2 | 1.0 ± 0.3 | |
| Lta−/− | 0.4 ± 0.2 | 0.5 ± 0.2 | 1.2 ± 0.3 | 1.7 ± 0.2 | |
| *Lta/Tnf/Ltb+/+ | 1.0 ± 0.5 | 0.6 ± 0.04 | 1.5 ±0.3§ | 1.8 ± 1.5 | |
| *Lta/Tnf/Ltb−/− | 1.8 ± 1.5 | 1.0 ± 0.3 | 0.6 ± 0.05 | 1.7 ± 0.3 | |
Data are presented as mean ± SEM, n = 3 – 10 mice per treatment group.
Air controls shown are for the 72 hr time point except for the Tnf +/+ and Tnf −/− mice (48hr time point shown). Air controls for each time point were not significantly different from one another.
p < 0.05, significantly elevated numbers of cells compared to air controls.
p < 0.05, significantly elevated numbers of cells compared to the transgenic strain.
Mice deficient in Lta or the Tnf cluster
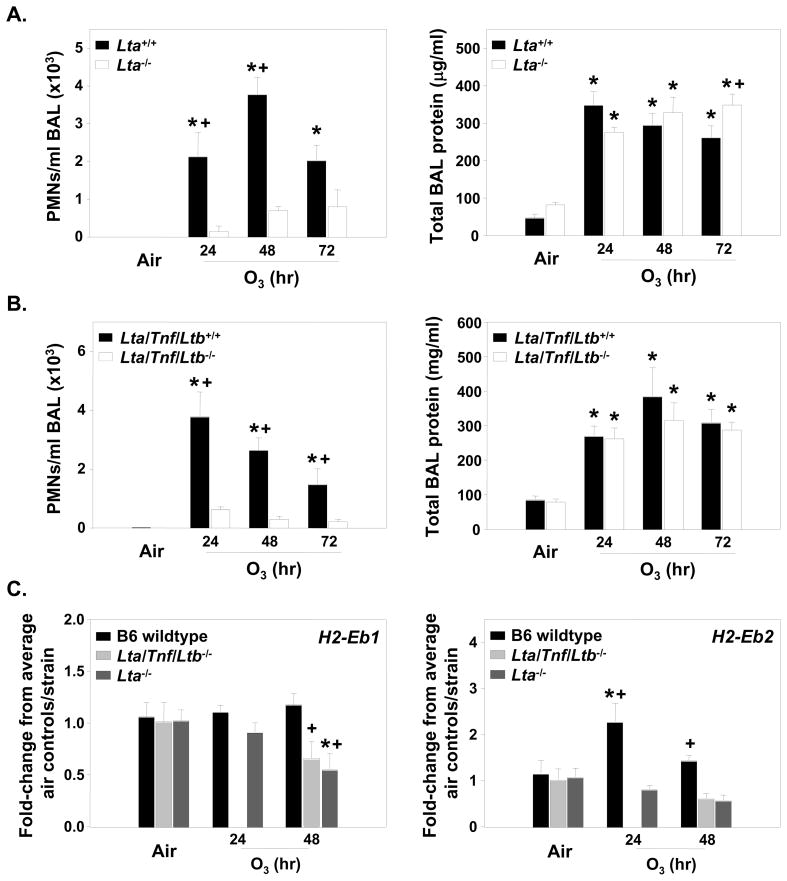
Based on the existing evidence to support a role for the TNF cluster in the O3 response [10], as well as the known intronic and UTR polymorphisms in Tnf and Lta genes between the B6 and HeJ mice, we tested the role of these three genes in the model. Lta−/−, Lta/Tnf/Ltb−/−, and wildtype mice were exposed continuously to air or 0.3 ppm O3 for 24, 48 or 72 h. PMNs were significantly elevated in the Lta+/+ and Lta/Tnf/Ltb+/+ mice compared to mice deficient in Lta−/− or the Lta/Tnf/Ltb cluster following 24 and 48 h O3 for both strains, and 72 h exposure for Lta/Tnf/Ltb deficient mice compared to air controls (p<0.05; Fig. 6A, B). Mean numbers of BALF macrophages were also significantly elevated in wildtype mice compared to Lta−/− and Lta/Tnf/Ltb−/− mice following 24–72 h O3 compared to air controls (p<0.05; Table 2). Epithelial cell numbers were not different between exposure or strains (Table 2). The BAL protein content was not affected by these genes, except for 72 h exposure in the Lta−/− mice, supporting the hypothesis that different genes regulate these specific phenotypes [16] (Fig. 6A, B).
Figure 6. O3-induced responses in mice deficient in Lta or the Tnf cluster (Lta, Tnf, Ltb) genes.
A) BALF PMNs and total protein for mice deficient in Lta and their wildtype controls. Data are presented as means ± SEM (n=3–10 per experimental group). *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from Lta+/+ mice (P < 0.05). B. BALF inflammatory parameters for the Lta/Tnf/Ltb deficient mice in response to O3 compared to wildtype mice. Data are presented as means ± SEM (n=3–10 per experimental group). *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from Lta/Tnf/Ltb+/+ mice (P < 0.05). C) Gene expression for the MHC class II genes (H2-Eb1 and H2-Eb2) in the mice deficient in either Lta or Lta/Tnf/Ltb compared to the wildtype mice. B6 are the wildtype for both strains and the numbers were combined since they were not significantly different from one another. No 24 h samples for Lta/Tnf/Ltb−/− mice were done for gene expression analysis. Data are presented as means ± SEM (n=3–6 per experimental group) and were determined using the comparative CT method (see Supplemental Materials and Methods section). Y-axis represents either H2-Eb1 or H2-Eb2 normalized first to 18S followed by determination of the fold-change relative to the average air control value for each strain. *, Significantly different from air exposed mice (P < 0.05). +, Significantly different from wildtype mice (P < 0.05).
MHC class II gene expression was also significantly different between the strains deficient in the TNF cluster. H2-Eb1 mRNA was unchanged in B6 wildtype mice, however, this gene was significantly decreased after 48 h O3 in the Lta−/− and Lta/Tnf/Ltb−/− mice compared to controls (Fig. 6C). H2-Eb2 mRNA was significantly increased in B6 mice after 24 and 48 h O3, and was significantly greater after O3 than mRNA expression in Lta−/− and Lta/Tnf/Ltb−/− mice at these time points.
Discussion
The overall objective of this investigation was two-fold. We first sought to restrict the length of Inf2 to elucidate candidate susceptibility genes for O3-induced inflammation. The second objective was to validate the role(s) of identified gene candidates. Significant differences in O3-induced inflammation between C3.SW-H2b/SJ and C3H/HeSnJ strains confirmed the role of Inf2 inasmuch as the only difference between these two strains is a congenic region of chromosome 17 that encompasses Inf2. Comparison of inflammatory responses in 2R, 4R, and 5R congenic mice with respective background strains further confirmed the importance of Inf2, and more importantly reduced Inf2 to 0.96 Mbp. The reduced Inf2 includes MHC class I, II, and III genes, and non-MHC genes, some of which have previously been identified as candidate genes in other lung injury models [e.g. C4a; [17]. In the current study, gene expression and sequence analyses of the reduced Inf2 suggested that MHC class II genes and the TNF cluster may be important in O3-induced inflammation, and significantly different phenotypes between O3-exposed H2Abl-Ea+/+ and H2Abl-Ea−/− mice and the mice deficient in Lta alone or the entire Tnf cluster confirmed a role for these genes. To our knowledge, these are the first studies to conclusively demonstrate a role for MHC class II genes and the entire TNF cluster in oxidant-induced lung inflammation and provide evidence supporting a susceptibility “superlocus”.
Genetic association studies have implicated several gene categories, such as pro-inflammatory cytokine genes [9], metabolism genes [18], and innate immunity genes in responses to environmental stimuli in human populations [19]. In inbred mouse models, positional cloning approaches identified QTLs for a number of lung diseases [20]. However, while identification of QTLs is an important initial step to understanding the genetic determinants of disease, QTLs can contain hundreds of genes. It is therefore necessary to refine the disease QTLs to a limited set of genes that can be evaluated [21]. In our integrative genomics approach to reduce Inf2 and identify candidate genes, we used congenic strains that were developed initially for histo-incompatiblity studies by repeated backcrossing of regions of chromosome 17 from the donor strain onto the recipient background strain [22]. One advantage of these strains is that the allelic designations are well characterized and, after phenotyping these strains for their inflammatory response to O3, it enabled restriction of the previously described Inf2 QTL to a limited chromosomal interval to identify gene candidates.
We queried the Mouse Phenome Database to identify genes located within the narrowed Inf2 that are polymorphic between susceptible B6 and resistant C3 mice. Several of the MHC class II and III genes (and those non-MHC genes located within these regions) have non-synonymous polymorphisms in exons, many have intronic polymorphisms, and several have UTR polymorphisms. However, not all of the genes have been assessed for polymorphisms between B6 and C3 mice (e.g. Hspa1b). It is important to note that functional polymorphisms may also exist in intronic regions, such as with K-ras and mouse lung cancer [23]. Future studies will delineate the functionality of polymorphic regions between the B6 and C3 mice in those genes in which the functionality is unknown, such as Ltb.
Organization of the H2 locus
Genes in the mouse H-2 locus and the homologous human MHC locus are organized in the same relative gene order [24] and span several Mbp. The H2 locus contains class I, class II, and class III genes. Class I and class II genes are important in fulfilling immunologic functions. Class I molecules are expressed by most cells, and present endogenous antigens (cytosolic-derived) to CD8+ cytotoxic T cells. Class II molecules (e.g. H2-Eb2, Btnl1) are expressed in antigen presenting cells (APC) such as dendritic cells, and present exogenous, endocytically derived antigens to CD4+ helper T-cells [25]. Class III genes are diverse in functionality, are located between the class I and class II genes and include the complement component genes (C2 and C4), heat shock proteins (Hspa1a, Hspa1b, and Hsc70t), and the Tnf cluster.
Candidate gene analysis
Deficiency in MHC class II genes and the TNF cluster decreased the O3-induced PMN and macrophage phenotypes, similar to those reductions seen in the 2R and 5R mice. Previous studies as well as the data presented here support a clear role for the TNF cluster in these O3-induced phenotypes. The H2-Ea gene (which is deficient in B6 mice), H2-Ab1, and H2-Aa (as described earlier) can be excluded as candidate susceptibility genes.
Chen et al. [26] demonstrated that inhibition of the CD4+ T-cell population using a monoclonal CD4 antibody in vivo greatly reduced the O3-induced phenotypes (including PMNs, lymphocytes, and epithelial cells) in B6 mice after 72 h continuous O3 exposure. These studies support our findings in the H2Abl-Ea−/− mice and suggest that in O3 susceptible strains, CD4+ T- cells are critical to O3-induced inflammation and injury. A recent study in human airway monocytes isolated from exposed individuals suggests that O3 primes the airway monocytes for innate immune responses, increases the capacity of the monocytes to present antigen to the CD4+ T-cells, and increases the overall population of antigen presenting cells in the lungs [19].
Candidate susceptibility genes in Inf2 and human disease
The MHC class II genes and the TNF cluster in the reduced Inf2 are all located in close proximity on chromosome 17 in mice and chromosome 6 in humans (Fig. 1, Supplement Table 1). Several of these homologous genes have been implicated in human association studies. For example, polymorphisms in HLA-DRB1 (H2-Eb1), TNF, and LTA have been associated with sarcoidosis [27], a chronic granulomatous disease of unknown etiology, supporting a role for a susceptibility “superlocus”. In addition, TNF is a susceptibility gene for O3-induced changes in lung function in humans [28]. Thus, it is possible that a cluster of genes in the MHC class II region (H2-Eb1 and H2-Eb2) interacts with the Tnf cluster (Tnf, Lta and Ltb) to promote O3-induced lung inflammation and injury. In support of this notion, the MHC (specifically HLA genes) has the strongest influence on susceptibility to human autoimmune diseases, but recent evidence suggests that TNF may also be involved in autoimmune diseases [29].
In summary, this integrative genomics investigation utilized congenic mouse lines to narrow the chromosome 17 QTL for susceptibility to O3-induced inflammation in the mouse. We used sequence analysis to identify candidate genes in the narrowed QTL, which were then tested for proof of concept. The novel role of MHC Class II genes and the TNF cluster in susceptibility to O3-induced inflammation provides unique insight to the mechanisms of O3 effects in the lung and may lead to alternative means to prevent oxidant injury to lung tissues.
Supplementary Material
Acknowledgments
Ozone exposures were conducted at the NIEHS Inhalation Facility under contract to Alion Science and Technology, Inc. The authors thank Dr. Daniel Morgan and Mr. Herman Price for coordinating the inhalation exposures. We also thank Dr. Donald Cook for reviewing the manuscript and providing excellent suggestions. This research was supported by the Intramural Research program of the National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services (SRK, LMD, CW, ST, WG) and Michigan State University (AKB, EAR).
References
- 1.Delfino RJ, Murphy-Moulton AM, Burnett RT, Brook JR, Becklake MR. Effects of air pollution on emergency room visits for respiratory illnesses in Montreal, Quebec. American journal of respiratory and critical care medicine. 1997;155(2):568–576. doi: 10.1164/ajrccm.155.2.9032196. [DOI] [PubMed] [Google Scholar]
- 2.Romieu I, Meneses F, Ramirez M, Ruiz S, Perez Padilla R, Sienra JJ, Gerber M, Grievink L, Dekker R, Walda I, Brunekreef B. Antioxidant supplementation and respiratory functions among workers exposed to high levels of ozone. Am J Respir Crit Care Med. 1998;158(1):226–232. doi: 10.1164/ajrccm.158.1.9712053. [DOI] [PubMed] [Google Scholar]
- 3.Air quality criteria for ozone and related photochemical oxidants. U.S. Environmental Protection Agency; 2006. [Google Scholar]
- 4.Goldstein BD, Lai LY, Ross SR, Cuzzi-Spada R. Susceptibility of inbred mouse strains to ozone. Arch Environ Health. 1973;27(6):412–413. doi: 10.1080/00039896.1973.10666416. [DOI] [PubMed] [Google Scholar]
- 5.Kleeberger SR, Levitt RC, Zhang LY, Longphre M, Harkema J, Jedlicka A, Eleff SM, DiSilvestre D, Holroyd KJ. Linkage analysis of susceptibility to ozone-induced lung inflammation in inbred mice. Nat Genet. 1997;17(4):475–478. doi: 10.1038/ng1297-475. [DOI] [PubMed] [Google Scholar]
- 6.Prows DR, Shertzer HG, Daly MJ, Sidman CL, Leikauf GD. Genetic analysis of ozone-induced acute lung injury in sensitive and resistant strains of mice. Nat Genet. 1997;17(4):471–474. doi: 10.1038/ng1297-471. [DOI] [PubMed] [Google Scholar]
- 7.Alexis NE, Zhou H, Lay JC, Harris B, Hernandez ML, Lu TS, Bromberg PA, Diaz-Sanchez D, Devlin RB, Kleeberger SR, Peden DB. The glutathione-S-transferase Mu 1 null genotype modulates ozone-induced airway inflammation in human subjects. The Journal of allergy and clinical immunology. 2009 Sept 29; doi: 10.1016/j.jaci.2009.07.036. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergamaschi E, De Palma G, Mozzoni P, Vanni S, Vettori MV, Broeckaert F, Bernard A, Mutti A. Polymorphism of quinone-metabolizing enzymes and susceptibility to ozone-induced acute effects. American journal of respiratory and critical care medicine. 2001;163(6):1426–1431. doi: 10.1164/ajrccm.163.6.2006056. [DOI] [PubMed] [Google Scholar]
- 9.Lee YL, McConnell R, Berhane K, Gilliland FD. Ambient ozone modifies the effect of tumor necrosis factor G-308A on bronchitic symptoms among children with asthma. Allergy. 2009 doi: 10.1111/j.1398-9995.2009.02014.x. [DOI] [PubMed] [Google Scholar]
- 10.Cho HY, Zhang LY, Kleeberger SR. Ozone-induced lung inflammation and hyperreactivity are mediated via tumor necrosis factor-alpha receptors. Am J Physiol Lung Cell Mol Physiol. 2001;280(3):L537–546. doi: 10.1152/ajplung.2001.280.3.L537. [DOI] [PubMed] [Google Scholar]
- 11.Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14(6):1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer AK, Dixon D, DeGraff LM, Cho HY, Walker CR, Malkinson AM, Kleeberger SR. Toll-like receptor 4 in butylated hydroxytoluene-induced mouse pulmonary inflammation and tumorigenesis. Journal of the National Cancer Institute. 2005;97(23):1778–1781. doi: 10.1093/jnci/dji403. [DOI] [PubMed] [Google Scholar]
- 13.Laboratory MPPatJ. Mouse Phenome Database. http://wwwjaxorg/phenome/SNP [cited 2009 2009]; Available from.
- 14.Laboratories J. H2 Haplotype Appendix 2. [cited; Available from: www.jax.org/jaxmice/search.
- 15.Hood L, Steinmetz M, Malissen B. Genes of the major histocompatibility complex of the mouse. Annual review of immunology. 1983;1:529–568. doi: 10.1146/annurev.iy.01.040183.002525. [DOI] [PubMed] [Google Scholar]
- 16.Kleeberger SR, Reddy S, Zhang LY, Jedlicka AE. Genetic susceptibility to ozone-induced lung hyperpermeability: role of toll-like receptor 4. American journal of respiratory cell and molecular biology. 2000;22(5):620–627. doi: 10.1165/ajrcmb.22.5.3912. [DOI] [PubMed] [Google Scholar]
- 17.Haston CK, Tomko TG, Godin N, Kerckhoff L, Hallett MT. Murine candidate bleomycin induced pulmonary fibrosis susceptibility genes identified by gene expression and sequence analysis of linkage regions. J Med Genet. 2005;42(6):464–473. doi: 10.1136/jmg.2004.027938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Islam T, Berhane K, McConnell R, Gauderman WJ, Avol E, Peters JM, Gilliland FD. Glutathione-S-transferase (GST) P1, GSTM1, exercise, ozone and asthma incidence in school children. Thorax. 2009;64(3):197–202. doi: 10.1136/thx.2008.099366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lay JC, Alexis NE, Kleeberger SR, Roubey RA, Harris BD, Bromberg PA, Hazucha MJ, Devlin RB, Peden DB. Ozone enhances markers of innate immunity and antigen presentation on airway monocytes in healthy individuals. The Journal of allergy and clinical immunology. 2007;120(3):719–722. doi: 10.1016/j.jaci.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Bauer AK, Malkinson AM, Kleeberger SR. Susceptibility to neoplastic and non-neoplastic pulmonary diseases in mice: genetic similarities. Am J Physiol Lung Cell Mol Physiol. 2004;287(4):L685–703. doi: 10.1152/ajplung.00223.2003. [DOI] [PubMed] [Google Scholar]
- 21.Kleeberger SR, Schwartz DA. From quantitative trait locus to gene: a work in progress. American journal of respiratory and critical care medicine. 2005;171(8):804–805. doi: 10.1164/rccm.2501002. [DOI] [PubMed] [Google Scholar]
- 22.Silver L. Concepts and applications. Oxford Univ Press; New York: 1995. Mouse genetics. [Google Scholar]
- 23.Chen B, Johanson L, Wiest JS, Anderson MW, You M. The second intron of the K-ras gene contains regulatory elements associated with mouse lung tumor susceptibility. ProcNatlAcadSciUSA. 1994;91(4):1589–1593. doi: 10.1073/pnas.91.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snoek M, Groot PC, Spies T, Campbell RD, Demant P. Fine mapping of the crossover-sites in the C4-H-2D region of H-2 recombinant mouse strains. Immunogenetics. 1991;34(6):409–412. doi: 10.1007/BF01787492. [DOI] [PubMed] [Google Scholar]
- 25.Snoek M, Teuscher C, van Vugt H. Molecular analysis of the major MHC recombinational hot spot located within the G7c gene of the murine class III region that is involved in disease susceptibility. J Immunol. 1998;160(1):266–272. [PubMed] [Google Scholar]
- 26.Chen X, Gavett SH, Wills-Karp M. CD4+ T lymphocyte modulation of ozone-induced murine pulmonary inflammation. American journal of respiratory cell and molecular biology. 1995;12(4):396–403. doi: 10.1165/ajrcmb.12.4.7695918. [DOI] [PubMed] [Google Scholar]
- 27.Mrazek F, Holla LI, Hutyrova B, Znojil V, Vasku A, Kolek V, Welsh KI, Vacha J, du Bois RM, Petrek M. Association of tumour necrosis factor-alpha, lymphotoxin-alpha and HLA-DRB1 gene polymorphisms with Lofgren's syndrome in Czech patients with sarcoidosis. Tissue antigens. 2005;65(2):163–171. doi: 10.1111/j.1399-0039.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 28.Yang IA, Holz O, Jorres RA, Magnussen H, Barton SJ, Rodriguez S, Cakebread JA, Holloway JW, Holgate ST. Association of tumor necrosis factor-alpha polymorphisms and ozone-induced change in lung function. American journal of respiratory and critical care medicine. 2005;171(2):171–176. doi: 10.1164/rccm.200402-194OC. [DOI] [PubMed] [Google Scholar]
- 29.Camarena A, Juarez A, Mejia M, Estrada A, Carrillo G, Falfan R, Zuniga J, Navarro C, Granados J, Selman M. Major histocompatibility complex and tumor necrosis factor-alpha polymorphisms in pigeon breeder's disease. American journal of respiratory and critical care medicine. 2001;163(7):1528–1533. doi: 10.1164/ajrccm.163.7.2004023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.