This work analyzes the localization, phloem transport, and metal binding properties of SODIUM POTASSIUM ROOT DEFECTIVE1. The results provide evidence for (1) Na+ recirculation in the phloem, (2) a function for the phloem in root meristem maintenance, and (3) a previously undiscovered symplasmic domain in the root tip.
Abstract
SODIUM POTASSIUM ROOT DEFECTIVE1 (NaKR1; previously called NPCC6) encodes a soluble metal binding protein that is specifically expressed in companion cells of the phloem. The nakr1-1 mutant phenotype includes high Na+, K+, Rb+, and starch accumulation in leaves, short roots, late flowering, and decreased long-distance transport of sucrose. Using traditional and DNA microarray-based deletion mapping, a 7-bp deletion was found in an exon of NaKR1 that introduced a premature stop codon. The mutant phenotypes were complemented by transformation with the native gene or NaKR1-GFP (green fluorescent protein) and NaKR1-β-glucuronidase fusions driven by the native promoter. NAKR1-GFP was mobile in the phloem; it moved from companion cells into sieve elements and into a previously undiscovered symplasmic domain in the root meristem. Grafting experiments revealed that the high Na+ accumulation was due mainly to loss of NaKR1 function in the leaves. This supports a role for the phloem in recirculating Na+ to the roots to limit Na+ accumulation in leaves. The onset of root phenotypes coincided with NaKR1 expression after germination. The nakr1-1 short root phenotype was due primarily to a decreased cell division rate in the root meristem, indicating a role in root meristem maintenance for NaKR1 expression in the phloem.
INTRODUCTION
Plant vascular tissue consists of two parallel pathways for long-distance transport: the xylem and phloem. The xylem transports water and mineral nutrients from roots to the shoot. The phloem transports sugars and other metabolites, proteins, and RNA from source (photosynthetic or storage) tissues to sinks, such as roots, developing leaves, flowers, seeds, and storage tissues (reviewed in Lough and Lucas, 2006). In angiosperms, the main unit of the phloem is the companion cell/sieve element complex. Sieve elements that form the conduits for transport are highly differentiated cells that lack many of the normal intracellular organelles, such as mitochondria and a nucleus. Companion cells are connected to sieve elements via plasmodesmata; they provide metabolic support and synthesize RNAs and proteins that are transported in sieve elements. In addition to the basic function of the phloem in transporting reduced carbon to sink tissues, the phloem performs a variety of important roles in plants. Genes specifically expressed in the phloem have been identified (Brady et al., 2007; Zhang et al., 2008), but the functions of relatively few phloem-expressed genes have been analyzed.
The classic theory that the phloem in leaves has a significant function in promoting Na+ tolerance (Greenway and Munns, 1980) does not have much support from molecular genetic studies. Based on the phenotype of Arabidopsis thaliana sodium overaccumulation in shoot2 (sas2 allele of HKT1) mutants, Na+ transporter HKT1 was proposed to function in the phloem (Berthomieu et al., 2003). However, the current consensus is that HKT1 mainly functions to remove Na+ from the xylem, thus limiting Na+ accumulation in leaves (Mäser et al., 2002; Sunarpi et al., 2005; Rus et al., 2006). The extent and mechanism of Na+ translocation in the phloem remains unknown and has been identified as a current research topic that is important for understanding Na+ tolerance (Munns and Tester, 2008). More is known about the function of K+ in the phloem. K+, along with sugars, serves as one of the major osmotica in phloem sap. K+ efflux via AKT2/3 also serves to repolarize the membrane potential during sucrose uptake (Deeken et al., 2002).
One main function of the phloem is the transport of fixed carbon from photosynthetic tissues to sink tissues, such as growing leaves, roots, flowers, and seeds. Arabidopsis suc2 mutants are specifically defective in sucrose transport into the phloem and have a severe phenotype that includes dwarf stature, starch accumulation in leaves, and anthocyanin accumulation (Gottwald et al., 2000). The severity of the suc2 phenotype indicates the importance of companion cell function but provides limited information about other roles of the phloem, such as in the control of flowering, K+ and Na+ translocation, and meristem functions.
Plasmodesmata (PD) are essential for phloem function. Companion cells and sieve elements are connected by specialized PD with a size exclusion limit of at least 67 kD (Stadler et al., 2005). Observing cell-to-cell movement of green fluorescent protein (GFP) fusion proteins has been instrumental in identifying symplasmic domains (groups of cells connected by PD). GFP fusions of 36 to 67 kD expressed under the control of the companion cell–specific SUC2 promoter were translocated in the phloem and entered the protophloem but did not move throughout the root tip, although free GFP (27 kD) was able to move throughout the root tip (Stadler et al., 2005). This sets a size exclusion limit of 27 to 36 kD for PD connecting cells of the root tip with protophloem cells. Little is known concerning the specificity or sequence requirements for protein transit through PD (Lucas et al., 2009).
In this article, the cloning of the SODIUM POTASSIUM ROOT DEFECTIVE1 (NaKR1) gene (which was previously called NPCC6; Zhang et al., 2008), the analysis of the nakr1-1 mutant phenotype, and the localization, phloem transport, and metal binding properties of the NaKR1 protein are presented. The results provide evidence for Na+ recirculation in the phloem, a function for the phloem in root meristem maintenance, and a previously undiscovered symplasmic domain in the root tip.
RESULTS
nakr1-1 Has High K+, Na+, and Rb+ in Leaves
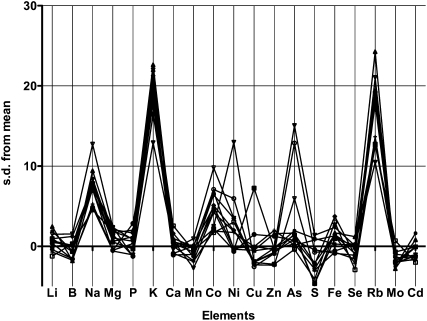
In previous work, an Arabidopsis fast neutron-induced mutant (136:31) was identified based on differences in elemental composition in leaves as determined by inductively coupled plasma–mass spectrometry (ICP-MS) analysis (Lahner et al., 2003). When first isolated, the mutant showed a complex phenotype with high Li, Mg, K, As, and low Mn compared with the wild type (Columbia-0 [Col-0]). After backcrossing twice to Col-0, a single recessive allele was resolved that resulted in high Na, K, and Rb (a chemical analog of K) (Figure 1) accumulation in leaf tissue. The mutant and Col-0 plants were grown under various soil conditions, and the high Na+ and K+ phenotypes were consistently observed. Figures 2A to 2C show results from a typical experiment in which plants were grown under conditions described by Lahner et al. (2003). The mutant accumulated around threefold higher Na+ and twofold higher K+ and Rb+ compared with Col-0. The mutant also showed defects in primary root growth (Figure 2D). Therefore, we named the mutant nakr1-1 for sodium potassium root defective 1-1.
Figure 1.
Leaves of nakr1-1 Contain Higher Na+, K+, and Rb+ Than Col-0.
Plant growth conditions and ICP-MS analysis were as described by Lahner et al. (2003). Rosette leaves were sampled after 5 to 6 weeks of growth. Differences in content of the mutant compared with Col-0 for each element analyzed are plotted as z-values (sd) for individual plants. The zero line indicates the average for Col-0 grown in the same tray.
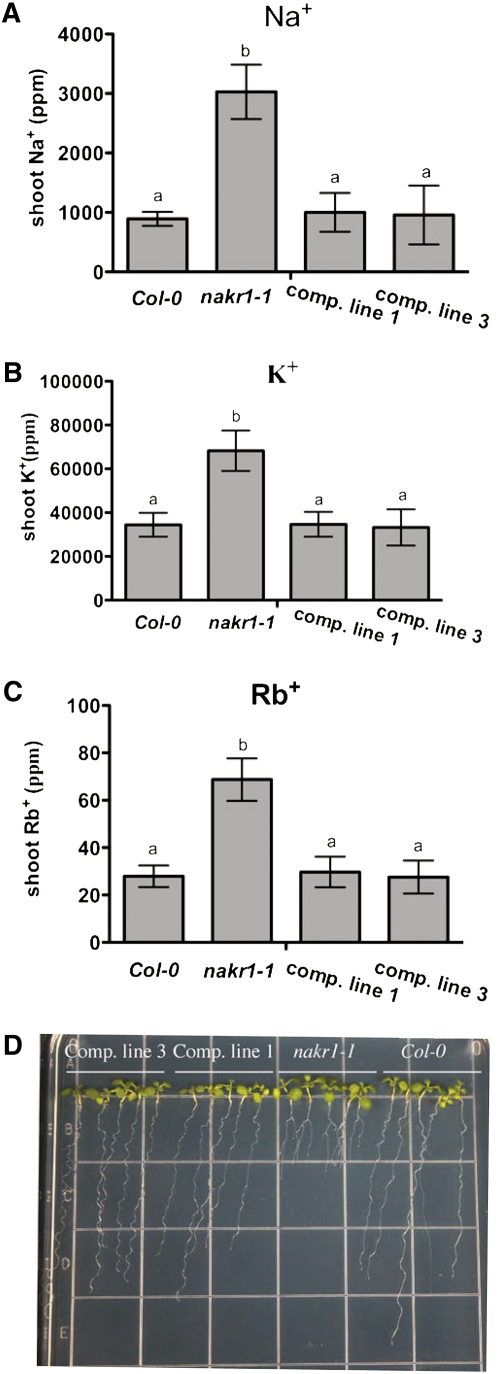
Figure 2.
nakr1-1 Na, K, and Rb Accumulation Phenotypes and Root Growth Defects Were Complemented by a NaKR1 Whole-Gene Construct.
(A) to (C) Concentrations of shoot Na+, K+, and Rb+ in Col-0, nakr1-1, and complemented lines 1 and 3 (T2 generation) are presented as mean ± sd (n = 12). Student’s t test was performed, and the different letters indicate significant difference with P < 0.05. Plant growth, sample collection, and ICP-MS analysis were according to Lahner et al. (2003).
(D) Eight day-old seedlings grown vertically on ATS media. From left to right are two complementation lines, nakr1-1, and Col-0. Note that nakr1-1 had reduced primary root growth.
[See online article for color version of this figure.]
NaKR1 Encodes a Heavy Metal Coordinating Protein
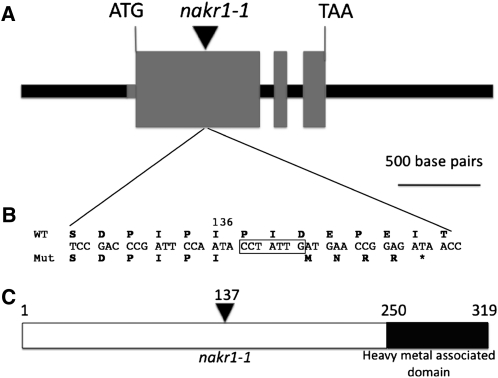
A mapping population was created after backcrossing nakr1-1 once to Col-0; the mutant was crossed with Landsberg erecta (Ler-0), and 300 F2 progeny with short primary roots were selected. The primary root growth defect in nakr1-1 is illustrated in Figure 2D. Using PCR-based mapping, NaKR1 was localized within a region of 90 kb (514.5 to 604.6 kb) at the top of chromosome 5. To identify the mutation, DNA microarray-based deletion mapping was used. Genomic DNA was prepared from two populations (nakr1-1 mutants and Col-0) and hybridized to Arabidopsis ATTILE 1.0R arrays. A single deletion of 7 bp was identified within At5g02600 (see Supplemental Figure 1 online), which caused a frame shift mutation (Figures 3A and 3B) and premature truncation of the C-terminal 183 amino acids (Figure 3C). Transformation of nakr1-1 with a 2817-bp genomic construct containing a 689-bp promoter and 5′ untranslated region (UTR), all the exons and introns, and 1005 nucleotide sequence after the stop codon (Figure 3A) complemented the root growth defects (Figure 2D) as well as the Na+ (Figure 2A), K+ (Figure 2B), and Rb+ (Figure 2C) overaccumulation phenotypes. This confirmed the identity of the NaKR1 gene. Four T-DNA insertion lines, with insertions in the NaKR1 promoter region (SALK_022426) or downstream of the coding region (SALK_033325, SALK_049519, and FLAG_633FO3) were examined; however, none displayed a phenotype.
Figure 3.
NaKR1 Gene and Protein Structure.
(A) The gene structure of NaKR1 (used for complementation), with the positions of the initiation and stop codon highlighted. From left to right are the promoter region and 5′ UTR (the black and gray box in front of ATG), three exons, two introns, and 3′ UTR. The black arrowhead indicates the position of nakr1-1 mutation.
(B) A deletion of 7 bp in the first exon of nakr1-1, indicated by the boxed sequence, caused a frame shift and truncation of C-terminal amino acid sequences. Mut., mutant; WT, wild type.
(C) The protein structure of NaKR1 with a C-terminal heavy metal–associated domain and an N-terminal region with little similarity with other functional domains. Amino acid position 137 indicates the first changed position in the mutant protein.
NaKR1 is predicted to encode a soluble protein of 319 amino acids, with a heavy metal–associated domain (HMA domain, pfam 00403.6) of 59 amino acids at the C-terminal end (Figure 3C). No similarity was found between the N-terminal region of NaKR1 and proteins of known function. Two Arabidopsis proteins of unknown function share high sequence similarity with NaKR1 in the C-terminal domain; NaKR2 (At2g37390) is 79% identical and NaKR3 (At3g53530) is 72% identical at the amino acid level. These show lower similarity to NaKR1 in the N-terminal domain (NaKR2 is 36% identical and NaKR3 is 32% identical). The HMA domain is found in all organisms, in proteins that transfer/bind heavy metals, including small metallochaperones, heavy metal transporters, and enzymes that use heavy metals as cofactors. The conserved sequence L(M)XCXXC is believed to be a Cu binding site, in which the two Cys residues coordinate metal directly; this motif was also observed in transport proteins that specifically transport Zn2+, Ni+, and Hg+ (Bull and Cox, 1994; Dykema et al., 1999; Suzuki et al., 2002).
The HMA domain of NaKR1 shares 47 and 45% identity with Arabidopsis Cu chaperones ATX1 and CCH, respectively. The homolog in yeast (Saccharomyces cerevisiae) ATX1 has been well characterized (Lin and Culotta, 1995; Lin et al., 1997; Pufahl et al., 1997) and found to be involved both in Cu homeostasis by specifically delivering Cu to heavy metal P-type ATPases and in defense against oxidative stress. When overexpressed in a yeast strain lacking the superoxide dismutase gene SOD1, both At-ATX1 and At-CCH can protect the mutant from active oxygen toxicity in a Cu-dependent manner (Himelblau et al., 1998; Puig et al., 2007). In our experiments, expression of NaKR1 or the C-terminal region alone (containing the HMA domain) failed to complement the reactive oxygen toxicity phenotype of yeast sod1Δ mutant (see Supplemental Figure 2 online), indicating that the HMA domain of NaKR1 functions differently than At-ATX1/At-CCH when expressed in yeast.
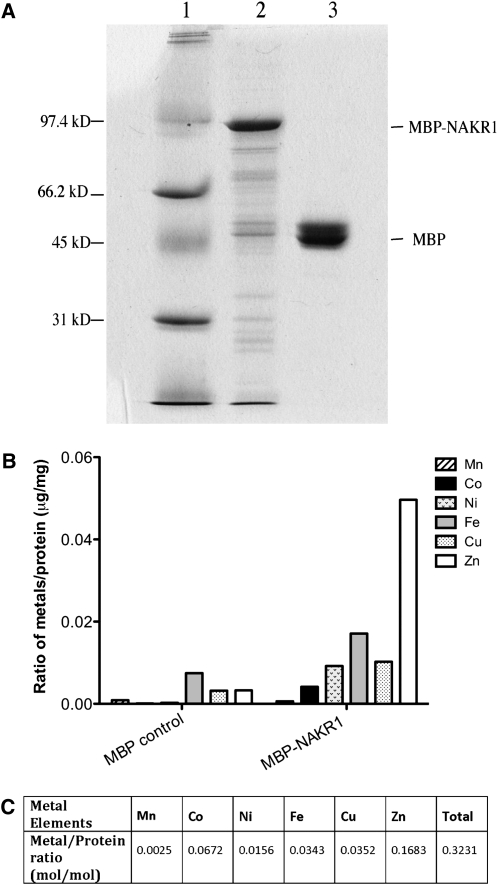
To test for possible metal binding, NaKR1 was expressed in Escherichia coli as a fusion to maltose binding protein (MBP). The purified MBP-NaKR1 fusion protein is shown in Figure 4A. The identity of the purified MBP-NaKR1 fusion protein was confirmed by MS analysis. Forty unique peptides were detected, covering 48.5% of the fusion protein sequence (see Supplemental Figure 3 online). Several heavy metals were detected in the purified protein sample using ICP-MS analysis (Figure 4B). Higher Zn, Cu, Fe, Ni, and Co was associated with the MBP-NaKR1 fusion protein compared with the MBP control. The molar ratio of protein to metals was around 3:1 (Figure 4C), indicating that NaKR1 is a metal binding protein. A mutated version of the NaKR1 whole-gene construct in which two conserved Cys residues in the HMA domain were changed to Gly failed to complement the nakr1-1 short root and late flowering phenotypes (data not shown). This indicates that metal binding is essential for NaKR1 function in the plant.
Figure 4.
Purified MBP-NaKR1 Fusion Protein Binds Heavy Metals.
MBP-NaKR1 or MBP (control) were expressed in E. coli BL21-AI and purified using amylose resin.
(A) Coomassie blue–stained polyacrylamide gel (12%) showing 3 μg purified MBP-NaKR1 (lane 2) and 3 μg MBP (lane 3). Lane 1 contained low-range SDS-PAGE standards (Bio-Rad).
(B) Metal content of the purified protein samples MBP-NaKR1 and MBP. Average metal concentration from two independent experiments is presented. The metal concentrations were determined by ICP-MS analysis.
(C) Molar ratio of heavy metal to protein in purified MBP-NaKR1 samples. The metal concentrations were determined by ICP-MS analysis and protein concentrations by amino acid analysis. The values represent the average of results from two independent experiments.
NaKR1 Is Specifically Expressed in the Phloem
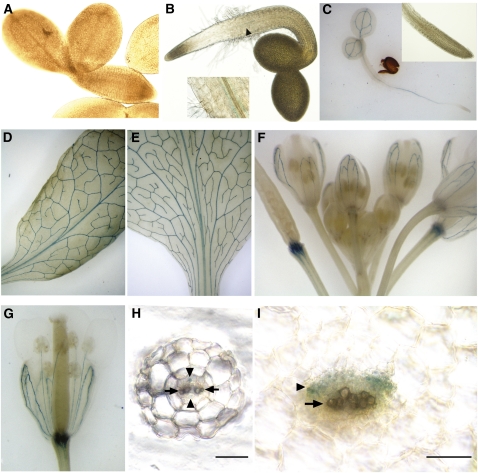
To study the expression pattern, a whole-gene β-glucuronidase (GUS) fusion (NaKR1pro:NaKR1-GUS) was expressed in Col-0, and 10 independent transformants were analyzed. No GUS staining was found in embryos or imbibed (but not germinated) seeds (Figure 5A). At 1 d after germination (DAG), GUS staining was observed in 10% of the seedlings. GUS activity was found in the vasculature of the root-hypocotyl junction and to a lesser extent in the hypocotyl (Figure 5B). At 2 DAG, GUS activity was found in the vascular tissue throughout the seedling (Figure 5C). In more mature plants, strong GUS staining was found in vascular tissue in rosette (Figure 5D) and cauline leaves (Figure 5E). In floral tissue, GUS staining was mainly localized to the sepals and the filaments of the flowers and the pedicel of the siliques (Figures 5F and 5G). Cross sections of roots (Figure 5H) and leaf petioles (Figure 5I) revealed expression in the phloem region of the vasculature, but not in the xylem.
Figure 5.
Expression Pattern of NaKR1pro:NaKR1-GUS Specifically in the Phloem Region of the Vasculature.
(A) No GUS expression was detected in imbibed seeds prior to germination. Seed coat was removed to facilitate observation.
(B) GUS staining was visible in the vasculature of a 1-DAG seedling in the region of root-hypocotyl boundary (indicated by arrow and shown at higher magnification in the inset).
(C) A 2-DAG seedling. Inset shows root tip at higher magnification.
(D) Mature rosette leaf.
(E) Cauline leaf.
(F) Floral tissue. Note that the strongest staining is in the pedicel of developing siliques and in the sepals.
(G) Sepals and stamen filaments in an opening flower.
(H) Cross section of a young root after GUS staining. The positions of the phloem and xylem region are indicated (by the arrowheads and arrows, respectively). Note that GUS expression was found in the phloem cells.
(I) Cross section of a rosette leaf petiole after GUS staining. The position of the phloem and the xylem regions of the main vasculature are indicated as in (H). Note that GUS expression was limited to the phloem tissue. Bars in (H) and (I) = 50 μm.
NaKR1 (At5g02600) expression was previously reported to be companion cell specific. This is based on microarray results and cDNA library analysis of mRNA transcripts isolated from purified companion cell protoplasts, nuclei, or isolated phloem tissue (Brady et al., 2007; Deeken et al., 2008; Zhang et al., 2008). In the work by Zhang et al. (2008), NaKR1 (called NPCC6 in that study) was among the top 12 transcripts most enriched in companion cell nuclei. Furthermore, NaKR1pro:histoneB2-GFP was expressed in Arabidopsis and shown to label companion cells specifically.
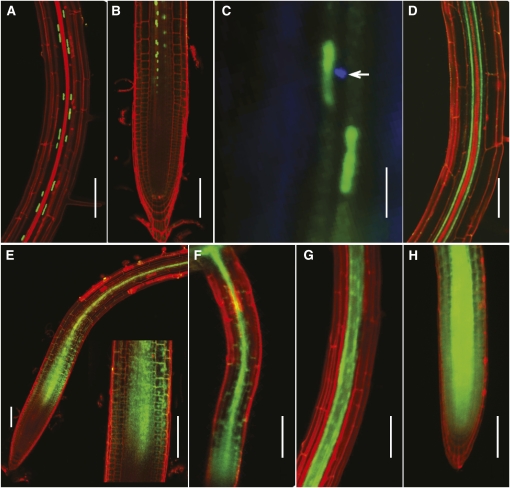
In our experiments, expression of histoneB2-GFP driven by the NaKR1 promoter labeled nuclei within the phloem of roots (Figure 6A), confirming the results of Zhang et al. (2008). In the root tip, histoneB2-GFP fluorescence was found only within the phloem (Figure 6B). Sieve plates stained with aniline blue were identified in the sieve element adjacent to cells containing histoneB2-GFP fluorescence in nuclei (Figure 6C). This confirms that NaKR1 is expressed in companion cells. A whole-gene GFP fusion (NaKR1pro:NaKR1-GFP) complemented the nakr1-1 mutant phenotypes (see Supplemental Figure 4 online), indicating that the NaKR1-GFP fusion protein is functional. Confocal microscopy was performed on the T2 generation seedlings. Within the root, GFP fluorescence was found in two parallel cell files on either side of the xylem within the vasculature, indicating phloem localization (Figure 6D). At the root tip, NaKR1-GFP fluorescence was observed in cells proximal to the zone of cell division in both primary (Figure 6E) and lateral (Figure 6F) roots. Fluorescence was apparent in both the cytosol and nucleus (Figure 6E, inset). Since expression of the NaKR1 gene is specific to companion cells (Figures 6A to 6C; Zhang et al., 2008), our results indicate that the NaKR1-GFP fusion protein is mobile through PD into sieve elements and is unloaded into some cells of the root meristem. As a control, GFP fluorescence in seedlings of the same age transformed with SUC2pro:GFP was analyzed. SUC2 is specifically expressed in companion cells, and GFP fluorescence was observed in the phloem and within two cell layers surrounding the phloem in the mature region of roots (Figure 6G) as previously reported (Stadler et al., 2005). Free GFP diffused through the symplast into the whole root meristem (Figure 6H) (Stadler et al., 2005; Benitez-Alfonso et al., 2009). Compared with free GFP (27 kD), NaKR1-GFP (61 kD) was more restricted to the phloem in mature roots. This is similar to results for multiple soluble GFP fusions of 36 to 67 kD analyzed by Stadler et al. (2005). Compared with free GFP, NaKR1-GFP mobility in the root tip was more restricted.
Figure 6.
NaKR1 Is Expressed in Companion Cells, and NaKR1-GFP Is Phloem Mobile.
Confocal images (except in [C]) of Arabidopsis tissue stained briefly with propidium iodide.
(A) NaKR1pro:histone2B-GFP localization in companion cell nuclei of Col-0 primary root.
(B) NaKR1pro:histone2B-GFP localization in the proximal meristem region of the primary root meristem.
(C) Epifluorescence image of NaKR1pro:histone2B-GFP expression in Col-0 root. Aniline blue staining shows sieve plate (indicated by arrow) of the adjacent sieve element.
(D) NaKR1pro:NaKR1-GFP expression in a mature, complemented nakr1-1 root.
(E) NaKR1pro:NaKR1-GFP fluorescence in the proximal meristem region of a primary root of complemented nakr1-1. Inset is magnified meristem region.
(F) NaKR1pro:NaKR1-GFP expression in the lateral root of complemented nakr1-1.
(G) Free GFP expressed using the SUC2 promoter showing fluorescence in the mature primary root of Col-0.
(H) Expression of free GFP under control of the SUC2 promoter in a primary root meristem of Col-0. Scale bars indicate 100 μm except for C in which the bar is 10 μm.
To test whether NaKR1 protein mobility was essential for its function, NaKR1pro:NaKR1-GUS was used to transform nakr1-1. The larger size of NaKR1-GUS (102 kD) compared with NaKR1-GFP (61 kD) was expected to limit NaKR1-GUS mobility through PD. Of 24 primary transformants, 20 showed full complementation of both root defects and the late flowering phenotype (see Supplemental Figure 5 online). The high Na+, K+, and Rb+ phenotype of the mutant was also complemented (see Supplemental Figure 5F online). GUS staining patterns in the complemented lines were similar to the expression pattern described in Col-0 background plants (Figure 5). This indicated that the postphloem trafficking of NaKR1 protein within the root meristem may not be essential for the function of NaKR1 in root growth.
Based on the result that NaKR1 was expressed specifically within the phloem region, a detailed characterization of the vasculature of Col-0 and the nakr1-1 mutant was performed. The general pattern of cell layers within the vasculature of nakr1-1 was normal. Based on the results of transmission electron microscopy, no difference in subcellular structure was detected for nakr1-1 compared with Col-0 in the mature phloem region (see Supplemental Figures 6A to 6F online). The phloem-specific gene ALTERED PHLOEM DEVELOPMENT (APL) is essential for maintaining phloem cell identity. The expression of APLpro:GFP-APL was used to study phloem cell identity as the fusion protein specifically labels companion cells and developing protophloem/metaphloem cells (Bonke et al., 2003). No difference was detected in the expression of GFP-APL in Col-0 and nakr1-1 plants, indicating that the phloem differentiation process proceeded normally in the nakr1-1 mutant (see Supplemental Figures 7A to 7D online).
nakr1-1 Is Defective in Phloem Loading or Translocation
The accumulation of starch in leaves of the mutant was analyzed as a test for phloem function. Previous work has shown that reduced phloem loading leads to starch accumulation in leaves (Riesmeier et al., 1994; Gottwald et al., 2000; Ma et al., 2009). Leaves of short-day-grown plants were collected at the beginning and end of the day and stained with I2/KI solution. In the nakr1-1 mutant, excess starch was retained at the end of the night (Figure 7A). Starch was quantified during the 8-h/16-h light/dark cycle (see Supplemental Figure 8A online), showing that nakr1-1 accumulated around fourfold more starch in leaves compared with the wild type during the day. By the end of the following dark period, 30% of the starch remained in nakr1-1 compared with undetectable amounts of starch in Col-0. When the plants were kept in continuous darkness for 3 d, leaf starch in the wild type decreased completely, while nakr1-1 retained leaf starch even after 88 h of darkness (see Supplemental Figure 8B online). When plants were illuminated for 8 h following 88 h of continuous darkness, the starch accumulation rate was twice as high in the mutant compared with Col-0 (see Supplemental Figure 8B online). This indicated that the starch overaccumulation phenotype was caused by a higher accumulation rate during the day and a decreased ability to deplete leaf starch during the night.
Figure 7.
nakr1-1 Mutants Are Defective in Phloem Function.
(A) I2/KI staining of Col-0 and nakr1-1 leaves, sampled at the beginning and end of the day. Col-0 and nakr1-1 plants were grown in the same tray under short-day conditions for 12 weeks. Mature healthy leaves were sampled for I2/KI staining and starch quantification.
(B) to (E) Autoradiography of individual Col-0 ([B] and [C]) and nakr1-1 ([D] and [E]) plants exposed to 14C-sucrose. Plants were grown in Turface. One fully expanded rosette leaf was labeled with 2 μL of 14C-sucrose (indicated by arrows). After 30 min, the application site was rinsed, and plants were dissected, dried overnight at 20°C, and exposed to film for 5 d ([C] and [E]). Arrowhead in (C) indicates appearance of radioactivity in the root of Col-0.
Considering that NaKR1 is expressed in companion cells, it was likely that the starch excess phenotype of nakr1-1 was caused by a defect in phloem loading or translocation. To test this possibility, sucrose translocation from source to sink was analyzed in nakr1-1 and Col-0. Plants were grown in Turface (Turface Athletics) to facilitate access to the root system. A 2-μL aliquot of 14C- sucrose was applied to wounded, fully expanded rosette leaves (Figures 7B and 7D). Plants were then incubated for 30 min to allow translocation. After dissection and drying, the plants were exposed to x-ray film for 5 d. As shown in the autoradiographs, in both nakr1-1 and Col-0, radioactivity is present in the labeled leaf. In Col-0, radioactivity was translocated to the root system (Figure 7C). However, no radioactivity was observed in the root system of the nakr1-1 mutant (Figure 7E). The results indicate that the nakr1-1 mutant is defective in phloem loading or translocation.
The K+ and Na+ Overaccumulation Phenotype Is Caused Mainly by Shoot Defects
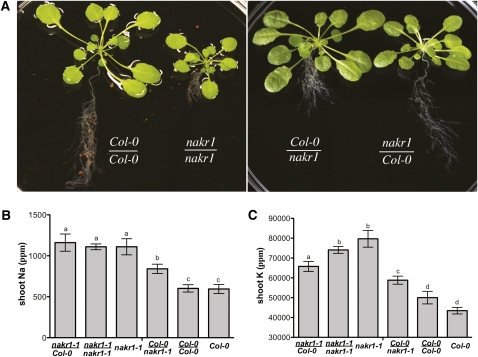
To begin to analyze how the nakr1-1 defect in phloem translocation was related to the Na+ and K+ accumulation phenotypes, reciprocal grafts were made between nakr1-1 and wild-type plants. The working hypothesis was that if a defect in phloem loading caused Na+ and K+ accumulation in leaves, then plants with mutant shoots grafted to wild-type roots would show high Na+ and K+ accumulation similar to the mutant. Grafted and control plants were grown in Turface. Self-grafting did not affect the visible phenotype of the plants; nakr1-1 shoots grafted to nakr1-1 roots produced plants with small root and shoot structures (Figure 8A), similar to nongrafted nakr1-1 mutants growing in the same conditions (data not shown). When Col-0 shoots were grafted to nakr1-1 roots, the plants produced larger shoot architecture with short roots (Figure 8A), indicating that the defect in root growth was caused by loss of NaKR1 function in the roots. Leaf K+ and Na+ levels were tested for all the grafted combinations using ICP-MS analysis. The self-grafted nakr1-1 and Col-0 plants displayed K+ and Na+ accumulation similar to the control nakr1-1 mutants and Col-0, respectively (Figures 8B and 8C). This indicates the grafting per se had little effect on K+ and Na+ accumulation. Mutant shoots grafted to wild-type roots showed leaf Na+ levels similar to that of the mutant. Plants with wild-type shoots grafted to mutant roots also showed Na+ content in leaves higher than the wild type, although lower than in mutant plants (Figure 8B). The results indicate that high Na+ in nakr1-1 is caused mainly by loss of NaKR1 function in the shoot and that loss of NaKR1 in the roots also contributes.
Figure 8.
Grafting Followed by ICP-MS Analysis Indicates That Loss of Function of NaKR1 in the Shoot Tissue Is the Main Cause of the nakr1-1 Na+/K+ Overaccumulation Phenotype.
(A) The different grafting types. From left to right are Col-0 shoot-Col-0 root, nakr1-1 shoot-nakr1-1 root, Col-0 shoot-nark1-1 root, and nakr1-1 shoot-Col-0 root.
(B) Shoot Na+ content in different grafting types and control plants. Data are presented as mean ± se.
(C) Shoot K+ content presented as mean ± se; n = 9 to 12 plants for grafting between Col-0 and nakr1-1, and n = 5 for self-grafting types and control plants (Col-0 and nakr1-1). Student’s t test was performed, and different letters in (B) and (C) indicate P values < 0.01.
[See online article for color version of this figure.]
The K+ levels of mutant shoot/wild-type root plants and wild-type shoot/mutant root plants were intermediate between the self-grafted mutant and wild-type plants. However, mutant shoot/wild-type root plants still had higher K+ in leaves than did the wild-type shoot/mutant root plants. These results suggest that the K+ accumulation phenotype was also caused by both root and shoot defects; however, the effect of loss of NaKR1 in the shoot predominated (Figure 8C). Similar results were obtained when the different grafted types and control plants were grown in soil (see Supplemental Figure 9 online).
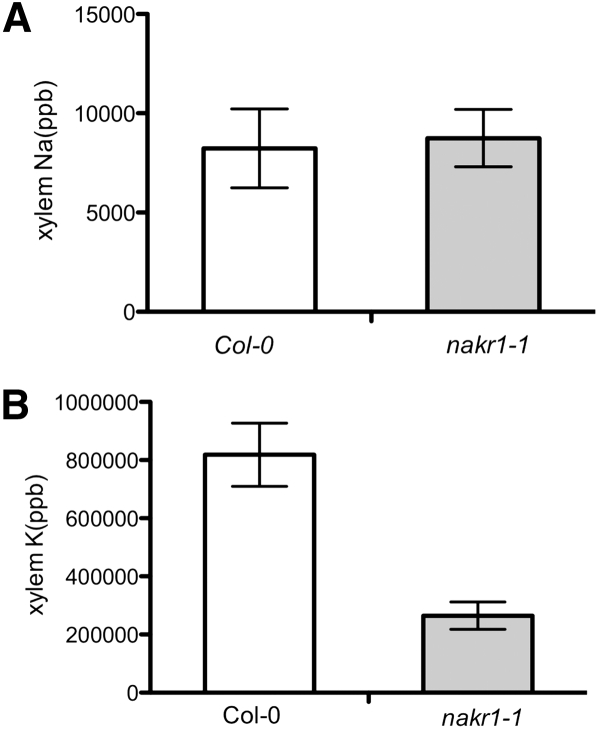
To test the effect of root K+/Na+ uptake on their accumulation in the shoot, xylem sap was collected from Col-0 and nakr1-1 mutant plants grown in the same soil conditions, and K+ and Na+ concentrations were analyzed by ICP-MS. Similar Na+ concentrations were detected in nakr1-1 and Col-0 xylem samples (Figure 9A), and lower K+ concentration was detected in the xylem sap samples of nakr1-1 plants than in those of Col-0 plants (Figure 9B). These results indicated K+/Na+ overaccumulation in the shoot tissue of nakr1-1 was not caused by increased Na+/K+ uptake from the mutant roots or increased K+/Na+ concentration within the xylem sap.
Figure 9.
nakr1-1 Plants Displayed Similar Na+ Content and Less K+ in Xylem Sap Than Did Col-0 Plants.
(A) Na+ content (ppb) in the xylem sap of nakr1-1 and Col-0 plants.
(B) K+ content (ppb) in the xylem sap of nakr1-1 and Col-0 plants. Na+ and K+ concentrations were determined by ICP-MS analysis of xylem sap collected as inflorescence stem exudates from plants grown under the same conditions. Each value was represented as mean ± se; n = 7 to 12 plants.
NaKR1 Is Necessary for Root Meristem Maintenance after Germination
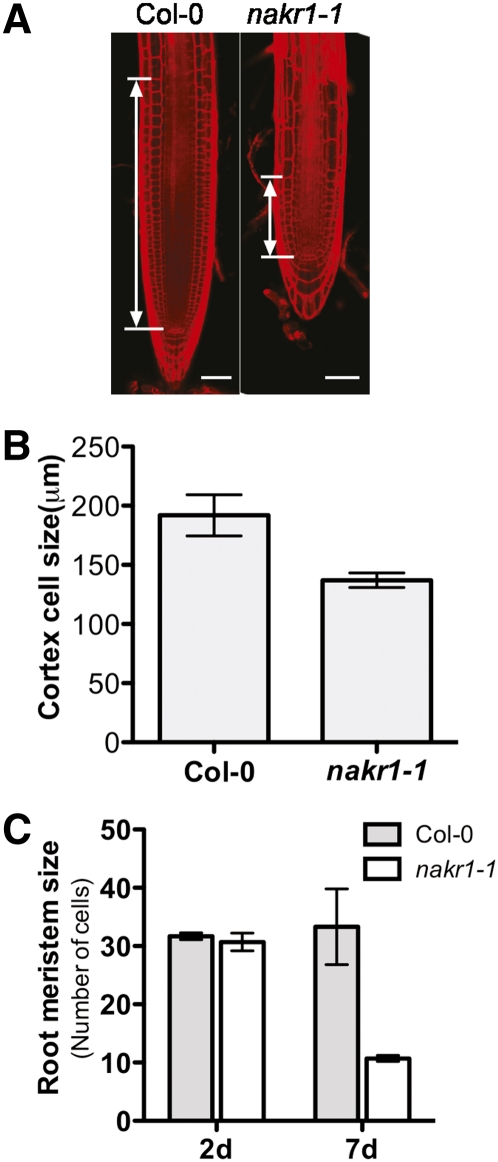
nakr1-1 showed a severe root growth phenotype shortly after germination that involved short primary roots and increased lateral root growth (Figure 2D). When grown on normal ATS plates for 1 week, the primary root length of nakr1-1 seedlings was 50% of that of the wild-type roots (Figure 2D). The size of the nakr1-1 root meristem region decreased dramatically during the first 7 DAG as determined by analysis of propidium iodide–stained roots using confocal microscopy (Figures 10A and 10C). In comparison, meristem size remained more stable in wild-type seedlings (Figures 10A and 10C), and a decrease in the number of meristematic cells does not occur in wild-type Arabidopsis until 3 to 4 weeks after germination (Baum et al., 2002). The elongation zone of nakr1-1 seedlings at 7 DAG was typically reduced to three cells in length (Figure 10A). The length of fully elongated cortex cells was also measured (Figure 10B). nakr1-1 had a smaller cell length than the wild type. Thus, the short roots of nakr1-1 were the result of both fewer cells in the root and a smaller cell size.
Figure 10.
Root Phenotypes of the nakr1-1 Mutant.
(A) Confocal images of Col-0 and nakr1-1 primary roots at 7 DAG stained with propidium iodide. The smaller root meristematic zone of nakr1-1 compared with Col-0 is indicated. Bars = 50 μm.
(B) The lengths of fully elongated cortex cells were measured in Col-0 and nakr1-1. Data are presented as mean ± sd (n = 11 for Col-0 seedlings, and n = 7 for nakr1-1 seedlings).
(C) The size of the root meristem in Col-0 and nakr1-1 seedlings at 2 and 7 DAG was measured by counting the number of cells within the cortex cell files from the quiescent center to the first elongated cortex cell. Compared with Col-0, the size of the root meristem in nakr1-1 was reduced at 7 DAG. Data are presented as mean ± sd (n = 7 to 10 for seedlings analyzed at each time point).
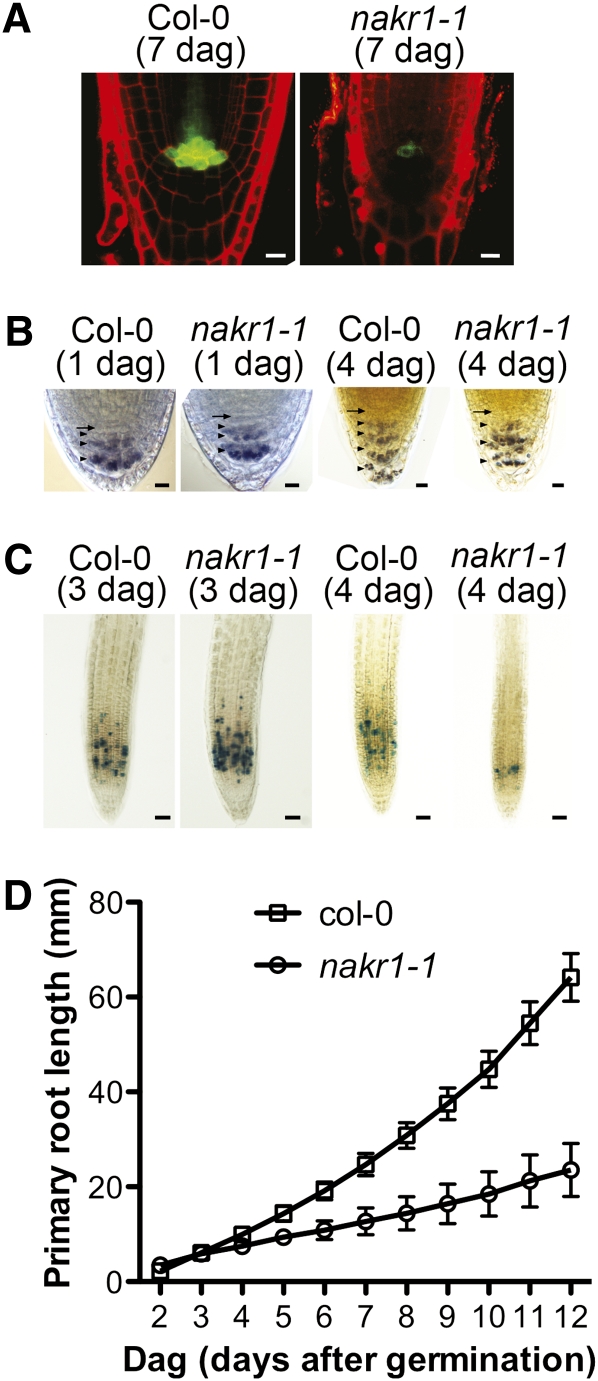
To determine whether the root meristem defects originated from an abnormality in embryo development, embryos of different stages were dissected from developing seeds of nakr1-1 and Col-0. No obvious differences between the wild type and nakr1-1 were found in early embryo stages (data not shown). This result was supported by the finding that nakr1-1 seeds can germinate normally on both plates and soil, as well as by GUS staining results showing that NaKR1 is expressed only after germination (Figure 5A). The root meristem defect in seedlings was investigated using a quiescent center marker, starch accumulation in root columella cells, and the expression of cyclin-GUS (CycB1;1:uida; Colón-Carmona et al., 1999) in the root apical meristem. Compared with Col-0, nakr1-1 seedlings showed much lower expression of quiescent center marker JYB1234 at 7 DAG (Figure 11A). At 3 DAG, nakr1-1 seedlings showed a similar cyclin-GUS expression pattern as the wild type (Figure 11C); however, by 4 DAG, the number of cells expressing cyclin-GUS in the root apical meristem decreased significantly in some nakr1-1 roots compared with the wild type (Figure 11C). Within the first 3 DAG, no difference was detected in starch accumulation within root columella cells (Figure 11B). From 4 DAG, starch accumulation in the outermost columulla cell layer disappeared in nakr1-1 roots; as a result, only three layers of columella cells exhibited starch accumulation compared with four layers in the wild-type roots (Figure 11B). Abnormality of starch accumulation in nakr1-1 root columella cells is consistent with defects in quiescent center cells. However, no sign of differentiation in nakr1-1 columella stem cells was detected, as revealed by the absence of stainable starch granules within the columella stem cell layer in nakr1-1 (Figure 11B). The growth of primary roots within Col-0 and nakr1-1 mutants was recorded for the first 2 weeks after germination (Figure 11D). Clear differences between Col-0 and nakr1-1 root growth was not observed until 4 DAG when primary roots grew at a slower but constant rate. These results suggest that defects in the nakr1-1 root apical meristem occurred at 3 to 4 DAG.
Figure 11.
Root Defects in nakr1-1 Appeared after Germination.
(A) Expression of quiescent center marker JYB1234 (At5g17800pro:ER-GFP) in Col-0 and nakr1-1 primary roots at 7 DAG. Images were collected by confocal microscopy. Bars = 10 μm.
(B) Starch accumulation pattern in root columella cells of Col-0 and nakr1-1 seedlings at 1 and 4 DAG. The position of columella cell layers and columella stem cells were labeled with arrowheads and arrows, respectively. Note, at 4 DAG, Col-0 roots showed starch staining in four layers of columella cells, while nakr1-1 roots showed starch in only three layers. Bars = 10 μm.
(C) Cyclin:GUS expression in Col-0 and nakr1-1 roots at 3 and 4 DAG. Bars = 40 μm.
(D) Primary root growth of nakr1-1 and Col-0 within the first 2 weeks after germination. Within the first week, 20% of nakr1-1 primary roots stopped growing, and their primary roots were not measured in the second week. Col-0 and nakr1-1 seedlings were grown vertically on ATS plates under long-day conditions (16-h day/8-h night). Experiments were repeated three times under the same conditions. The y axis value at each time point represents mean ± sd.
nakr1-1 Is a Late-Flowering Mutant Especially in Long-Day Conditions
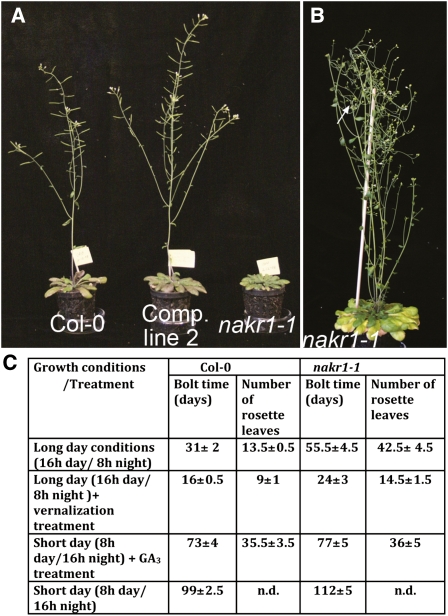
In the early stage of vegetative growth, nakr1-1 rosette leaves appeared at the same rate as in the wild type. nakr1-1 leaves were slightly smaller and had shorter petioles than those of Col-0. After the wild-type plants bolted, nakr1-1 mutants continued to form new rosette leaves, displaying a late flowering phenotype (Figure 12A). In Arabidopsis, several different pathways promote floral induction. The autonomous pathway responds to endogenous signals at specific developmental stages. The photoperiod and vernalization pathways respond to environmental stimuli, such as light and temperature. The gibberellin (GA) pathway plays a relatively independent role in floral transition under noninductive photoperiods (Araki, 2001). Among these pathways, the photoperiod pathway is largely dependent on phloem activity, since the signals induced by photoperiod, CONSTANS and its downstream target FLOWERING LOCUS T (FT) are expressed in the vasculature and the mobile flowering signal FT needs to be transported through the phloem to the meristem (Corbesier et al., 2007; Mathieu et al., 2007). The floral transition times of nakr1-1 and Col-0 were measured under different floral promotion conditions. In long-day conditions (16-h day/8-h night, 55 μmol photons m−2 s−1), wild-type plants began to bolt at 31 ± 2 DAG, after forming 13.5 ± 0.5 rosette leaves; nakr1-1 mutants required 55.5 ± 4.5 d to bolt, and by that time, 42.5 ± 4.5 rosette leaves had developed (Figure 12C). nakr1-1 and Col-0 bolted at similar times in short-day conditions (8-h day/16-h night, 55 μmol photons m−2 sec−1 light intensity; Figure 12C). Wild-type plants took 99 ± 2.5 d to bolt, whereas nakr1-1 plants took 112 ± 5 d to bolt. The effects of vernalization and GA3 treatment were also tested. Both treatments accelerated flowering in nakr1-1, although in both conditions the mutant plants still required slightly more time to bolt (Figure 12C). Therefore, nakr1-1 mutants were affected mainly in the photoperiod pathway. This is consistent with a phloem defect in the nakr1-1 mutant.
Figure 12.
nakr1-1 Displayed a Strong Late Flowering Phenotype, Most Notably in Long-Day Growth Conditions.
(A) The late-flowering phenotype of nakr1-1 was complemented by the NaKR1 whole-gene construct. Col-0, complementation line 2 (T2 generation), and nakr1-1 were grown in long-day conditions within the same tray. The image was taken 45 d after sowing.
(B) A flowering nakr1-1 mutant after 90 d of growth in long-day conditions. The white arrow indicates the position of the primary shoot meristem. Note that the mutant showed a loss of apical dominance and set seeds poorly.
(C) Bolting time of nakr1-1 and Col-0 under different growth conditions. Days after sowing and the number of rosette leaves are expressed as mean ± sd (n = 17 to 25 Col-0 or nakr1-1 plants counted for each growth condition).
[See online article for color version of this figure.]
In addition to extended vegetative growth, nakr1-1 also showed elongated reproductive growth and late senescence phenotypes. Abundant flowers were made in this period, but the early flowers set seeds poorly. The main stem displayed loss of apical dominance (Figure 12B) in both short- and long-day conditions, and the floral meristem in the primary shoot died before producing any flowers. The same phenomenon also appeared in the floral meristems of lateral branches.
DISCUSSION
The nakr1-1 mutant phenotype included elevated Na+, K+, and Rb+ in leaves, short roots, elevated starch accumulation during the day, decreased long-distance transport of sucrose, and late flowering under long-day conditions. The NPCC6/NaKR1 gene was identified and encodes a soluble protein of 34.5 kD with a conserved HMA domain in the C-terminal half. NPCC6/NaKR1 expression was specific to companion cells as previously reported (Zhang et al., 2008). NaKR1-GFP was mobile in the phloem and was detected in the proximal root meristem region.
The specificity of NaKR1 expression in companion cells and the observation that long-distance sucrose transport in the phloem is affected indicated that elevated Na+ and K+ in nakr1-1 leaves is likely to be due to a defect in phloem transport of these ions in the mutant. However, the cause of this decrease in ion translocation is not clear. Transport of sucrose in the phloem is dependent on K+ channel activity, as evidenced by the decreased sucrose concentration in the phloem of mutants of the K+ channel AKT2/3 (Deeken et al., 2002). In theory, any defect in companion cells that limits ATP production or plasma membrane H+-ATPase activity would be expected to decrease both sucrose and K+ loading into the phloem.
The nakr1-1 phenotypes cannot be fully explained by a decrease in phloem loading in leaves, as demonstrated in reciprocal grafting experiments. Reduced root growth, as well as elevated Na+ and K+ in leaves, was found when the root lacked NaKR1. Whereas NaKR1-GFP was found to be mobile in the phloem, a NaKR1-GUS fusion that complemented all nakr1-1 phenotypes did not translocate outside of the phloem. Therefore, it is possible that all phenotypes observed for the nakr1-1 mutant are due to a lack of NAKR1 in the phloem. This does not rule out additional functions that require postphloem NaKR1 mobility, functions that may be redundant with other metal binding domain proteins.
These results contribute to our understanding of phloem function in long-distance transport of K+, Na+, and sugars, flowering time, and maintenance of the root meristem. They raise questions regarding (1) cell-to-cell movement of protein, (2) the significance of Na+ and K+ recirculation in the phloem from leaves to roots, and (3) the function of the phloem in influencing root growth.
NaKR1 Movement through PD
NaKR1 expression was reported to be specific to companion cells in microarray experiments performed with fluorescently labeled protoplasts (Brady et al., 2007). When companion cell nuclei were labeled with histoneB2-GFP, NaKR1 was identified as one of the 12 most companion cell-specific transcripts (Zhang et al., 2008). HistoneB2-GFP targets GFP to nuclei in cells where translation occurs, and when driven by the NaKR1 promoter, was specific to companion cells (Zhang et al., 2008). We confirmed the companion cell localization of histoneB2-GFP driven by the NaKR1 promoter (Figures 6A and 6B). This localization was also supported by NaKR1pro:NAKR1-GUS expression in Col-0, which was restricted to vascular tissue (Figure 5). Interestingly, NaKR1-GFP driven by the native promoter produced a different localization, consistent with transport through PD. In mature parts of the root, NaKR1-GFP fluorescence was observed in files of cells consistent with sieve elements and appeared similar to other soluble GFP fusions that move from companion cells into sieve elements (Stadler et al., 2005). In mature regions of the root, NaKR1-GFP was more restricted to sieve elements compared with free GFP, which is able to move one to two cell layers laterally (Figure 6G; Stadler et al., 2005). Free GFP, expressed in companion cells driven by the SUC2 promoter, can move out of the transport phloem in mature regions of the root, is unloaded from the protophloem and moves throughout the root tip, indicating that these cells are connected by PD (Figure 6H; Stadler et al., 2005). NaKR1-GFP movement was restricted to an area of the root tip below the transition zone of the root meristem. Since NaKR1-GFP was translocated to only a portion of the root tip, in contrast with free GFP, this identifies a new symplasmic domain in the root tip. In a previous study, the smallest GFP protein fusions (ubiquitin-GFP is 36 kD) did not move from the protophloem into cells of the root tip (Stadler et al., 2005).
Maize KNOTTED1 (KN1) was the first protein identified to function non-cell-autonomously (Lucas et al., 1995). Trafficking of KN1 through PD requires an internal signal contained in the homeodomain (Kim et al., 2005), and the trafficking is essential for KN1 function in maintaining stem cell populations in plant shoots (Kim et al., 2003). NaKR1 shares no homology with the KN1 homeobox domain, and the presence of a signal sequence in NaKR1 remains to be tested.
Phloem and Na+/K+ Homeostasis
The nakr1-1 mutant overaccumulates Na+ and K+ in leaves. In theory, the amount of K+ and Na+ in leaves is determined by several distinct processes: Na+/K+ uptake from the soil, long-distance transport to the shoot via the xylem, and redistribution through the phloem. The function of xylem-localized HKT1 in limiting Na+ translocation to the shoot has been established (Mäser et al., 2002; Sunarpi et al., 2005; Rus et al., 2006; Davenport et al., 2007). In hkt1 mutants, xylem Na+ concentration is higher than in the wild type (Sunarpi et al., 2005). In the nakr1-1 mutant, xylem sap Na+ concentration was the same as in wild-type plants (Figure 9A). This indicates that the nakr1-1 mutation does not cause high Na+ in leaves by downregulation of HKT1. Compared with the xylem, less is known about the possible function of the phloem regulating Na+ concentration in leaves (Horie et al., 2009). As shown here, the specificity of NAKR1 expression in companion cells combined with the nakr1-1 high K+ and Na+ phenotypes indicates a role for the phloem in determining leaf K+ and Na+ accumulation.
NaKR1 is expressed in the phloem throughout the plant. We hypothesized that K+ and Na+ are loaded into the phloem in leaves and that NaKR1 is required. To test this, reciprocal grafting experiments were done with nakr1-1 and Col-0. The results (Figure 8) showed that lack of NaKR1 in the shoot resulted in a Na+ concentration in leaves as high as in nakr1-1. However, a lack of NaKR1 in roots also resulted in leaf Na+ that was higher than in Col-0. The results indicate that companion cell function in both the root and shoot contributes to the limitation of Na+ accumulation in shoots.
The effect of the nakr1-1 mutation on K+ accumulation was similar. In grafting experiments, the lack of NaKR1 in either the root or shoot led to K+ accumulation in leaves. Differences in K+ concentration between source and sink tissues have been detected and are thought to contribute to the turgor gradient (Fischer, 1987; Fromm and Eschrich, 1989; Hayashi and Chino, 1990) that drives phloem sap movement (Mengel and Haeder, 1977; Lang, 1983). The K+ channel AKT2/3 is specifically expressed in the phloem and functions in regulating the membrane potential (Marten et al., 1999; Deeken et al., 2000; Lacombe et al., 2000). The akt2/3-1 mutant exhibited reduced sucrose concentration in the phloem sap, but K+ concentration was not significantly affected (Deeken et al., 2002). Transporters functioning in loading K+ into the phloem remain to be identified. The grafting experiments indicated that NaKR1 is required in both the shoot and the root for K+ recycling via the phloem. The requirement in the shoot may be related to phloem loading of K+. In the root, NaKR1 may be required for phloem unloading. The lower K+ concentration observed in the xylem sap in nakr1-1 is consistent with this hypothesis.
nakr1-1 Affects Phloem Translocation, Starch Accumulation, and Flowering
nakr1-1 accumulated more starch in leaves during the day. This is consistent with a decrease in phloem loading of sucrose as exemplified by the effects of antisense inhibition of SUT1 in potato (Solanum tuberosum; Riesmeier et al., 1994), SUC2 insertional mutants in Arabidopsis (Gottwald et al., 2000), and tdy1 mutants of maize (Zea mays; Ma et al., 2009). The ability of nakr1-1 to translocate 14C sucrose was tested in intact plants. When 14C-sucrose was applied to a Col-0 leaf, radioactivity was detected in roots within 30 min. With nakr1-1, no radioactivity was detected in roots, indicating that phloem loading or translocation is defective in the mutant.
Consistent with a defect in phloem function, the nakr1-1 mutant displayed a large delay in flowering under long-day conditions. This is consistent because the phloem functions in producing and translocating FT protein (Corbesier et al., 2007), a mobile signal that induces flowering. By contrast, nakr1-1 responded normally to other conditions that promote flowering, such as GA3 treatment or vernalization and to conditions that delay flowering such as short days.
NaKR1 Expression and Root Phenotypes
Considering the short root phenotype of nakr1-1, it was important to compare the timing of NaKR1 expression and onset of the phenotype to determine if NaKR1 is important for root development. Using a NaKR1-GUS fusion driven by the NaKR1 promoter, the earliest expression was identified at 1 DAG in the vascular tissue at the root-hypocotyl junction. No expression was detected in developing embryos or in seeds before germination. Therefore, NaKR1 is not expressed in the earliest protophloem cells that differentiate in mature embryos (Bauby et al., 2007). By 2 DAG, NaKR1 expression was observed in vascular tissue throughout the seedlings. The earliest expression of NaKR1 coincides well with the onset of phenotypes in the root. By 3 DAG, root growth was measurably slower. By 4 DAG, the cell division rate in the meristematic region was lower, and differences were observed in starch accumulation in the root distal region. Overall, the results indicate that NaKR1 is important for root meristem maintenance rather than root development.
The NaKR1pro:NaKR1-GUS construct complemented the nakr1-1 mutant phenotypes. This construct produced GUS staining only in the phloem. This suggests that NaKR1 functions at a distance from areas of the root displaying a phenotype, for example, in the quiescent center and in the zone of cell division. However, we cannot rule out partial proteolysis of the NaKR1-GUS fusion resulting in phloem transport of a truncated protein. The root phenotypes in nakr1-1 could be due to a defect in supply of photosynthate to the roots but cannot be attributed solely to decreased phloem loading. Results from reciprocal grafting experiments (Figure 8) indicated that a lack of NaKR1 in the roots was responsible for the short root phenotype. This indicates a specific function for companion cells in the root in maintenance of the root meristem.
NaKR1 Function as a Metal Binding Protein
A NaKR1 fusion protein was expressed in E. coli, purified, and shown to bind Zn, Cu, Fe, Ni, and Co. A mutated version of NaKR1 in which the two metal-coordinating Cys residues were changed to Gly failed to complement nakr1-1 phenotypes. This indicates that metal binding is important for NaKR1 function. HMA domain proteins have been identified in the phloem sap of Brassica napus and Cucurbita maxima (Giavalisco et al., 2006; Lin et al., 2009). In Arabidopsis, HMA domain protein CCH is found in the phloem and is thought to transport Cu out of senescing tissues (Himelblau et al., 1998; Mira et al., 2001). nakr1-1 seedling growth (for 2 weeks) was not affected more than the wild type when Zn2+ (up to 15 μM) or Cu2+ (up to 25 μM) was included in the growth medium (data not shown). Overall, the results suggest that metal binding is important for NaKR1 function in companion cells. We cannot rule out additional NaKR1 functions that require NaKR1 mobility in the phloem that are redundant with other related HMA domain-containing proteins also known to be present in the phloem.
Conclusions
The NaKR1 gene encodes a soluble, cytoplasmic heavy metal binding protein that is specifically expressed in companion cells of the phloem in Arabidopsis. The nakr1-1 mutant displayed a number of phenotypes that are consistent with defects in phloem function: higher starch accumulation in the mesophyll, defective long-distance transport of 14C-sucrose, accumulation of K+ and Na+ in leaves, and late flowering under long-day conditions. The nakr1-1 mutation also caused a short root phenotype, primarily due to a lower rate of cell division. In addition, NaKR1-GFP was found to move cell to cell through PD, which may indicate that it functions non-cell autonomously.
METHODS
Plant Material
Arabidopsis thaliana ecotypes Col-0 and Ler-0 seeds were obtained from the ABRC. The mutant line 136:31 (Lahner et al., 2003) was backcrossed twice to Col-0 and renamed nakr1-1. Cyclin-GUS (Colón-Carmona et al., 1999) (in Col-0) was obtained from William Gray (University of Minnesota). The SUC2pro:GFP marker (Benitez-Alfonso et al., 2009) (in Col-0) was obtained from David Jackson (Cold Spring Harbor Laboratory). APLpro:APL:GFP (in Ler-0) (Bonke et al., 2003) was obtained from Ykä Helariutta (University of Helsinki, Finland). The JYB1234 marker line was described previously (Benitez-Alfonso et al., 2009). These marker lines were crossed with nakr1-1 mutants, and the F2 progeny with short roots and containing these constructs (selected by the expression of GUS or GFP) were selected. NaKR1pro:HTB2:GFP (NPCC6) (Zhang et al., 2008) was provided by David W. Galbraith (University of Arizona). T-DNA insertion lines SALK_022426, SALK_033325, SALK_049519, and FLAG_633FO3 were obtained from the ABRC.
Plant Growth on Vertical Plates
Seeds were surface sterilized, stratified for 2 d in the dark at 4°C, and placed on square Petri dishes containing ATS medium (Lincoln et al., 1990) supplemented with 0.05% MES (w/v), 0.5% (w/v) sucrose, and 0.8% (w/v) Phytoagar (Caisson Laboratory), pH 5.7. The Petri dishes were placed vertically in a growth room under controlled conditions (16-h/8-h day/night cycle, 22°C, 60 μmol·m−2·s−1).
Plant Growth in Soil
Plants were grown in plastic trays with sterilized LG3 (Sun Gro Horticulture) or BM2 (Berger). Arabidopsis seeds were sowed on the top of the soil. To facilitate seed germination, the trays were kept for 4 d at 4°C before transferred to the growth room. Plants were grown at long-day (16-h/8-h day/night cycle, 22°C, 55 to 70 μmol·m−2·s−1) or short-day (8-h/16-h day/ night cycle, 22°C, 55 to 70 μmol·m−2·s−1) conditions. Trays were regularly rotated and watered twice a week from the bottom of the tray with deionized water and fertilized once with All Purpose Plant Food (Vigoro). For vernalization experiments, seeds were plated on ATS medium and incubated at 4°C for 6 weeks before being transferred to soil and grown under long-day conditions until flowering. To test GA3 effects, plants were grown under short-day conditions for 1 month before they were sprayed with 100 μM GA3 once per week until flowering.
Identification of the nakr1-1 Mutation
For map-based cloning, nakr1-1 mutants were crossed with Ler-0, and F2 progeny with short primary roots were selected after 3 weeks of growth on ATS plates. Polymorphism information used for mapping was provided by Monsanto SNP and Ler collections from The Arabidopsis Information Resource website (http://www.Arabidopsis.org). A genomic DNA pool from 30 mutants was created for bulk segregant analysis, and the mutation was revealed to be at the top of chromosome 5. Three hundred mutant plants were used for fine mapping. Progeny were collected from those F2 mutants with recombination sites close to the mutation region and grown on ATS plates to confirm they were homozygous mutants.
DNA microarray-based deletion mapping was performed to identify the mutation. Three genomic DNA pools were created from the Col-0 plants and three from a nakr1-1 mutant population using the DNeasy plant mini kit (Qiagen). For each pool, the genomic DNA was from 10 to 15 plants. Genomic DNA was labeled and hybridized to Affymetrix ATTILE 1.0R arrays. Probes with no sequence differences should show no difference in hybridization between nakr1-1 and Col-0. Deletions linked to NaKR1 were identified by decreased hybridization signals that were consistent in all three nakr1-1 hybridized samples. Microarray data have been submitted to the Gene Expression Omnibus database with accession number GSE24385.
The nakr1-1 mutation was confirmed by sequencing the PCR products amplified from nakr1-1 genomic DNA using primer pairs F (5′-CACTCCTCCACCTTCCCCAAACCTTAAT-3′) and R (5′-GTCTCCCGTCACGGTAACCTTCTTTGCT-3′). A derived cleaved-amplified polymorphic sequence marker (Neff et al., 1998) was designed to discriminate nakr1-1 and wild-type Col-0 genotype using PCR primer pairs F (5′-CGGTTAGCGAAGAGGAAGAGCAAGAAAGG-3′) and R (5′-TTAAATCATCAGCTTTGGTTATCTCCGGTCCAT-3′). The amplified Col-0 genomic DNA could be specifically digested by the enzyme BccI; the amplified nakr1-1 genomic DNA was not recognized by BccI. Cosegregation of the mutation sequence with mutant phenotypes was further confirmed by genotype analysis of a segregating population containing 108 plants using the derived cleaved-amplified polymorphic sequence marker.
Plasmid Construction and Plant Transformation
The NaKR1 whole-gene sequence (including promoter, exons, introns, and 3′ UTR) was amplified from wild-type Col-0 genomic DNA using primer pairs F (5′-GCCTTTGTGAAATCGATTTGAGTTAGAA-3′) and R (5′-GGTCATCTGTAAAGGTAAGTCTATATGG-3′). The resulting 2424-bp PCR product (NaKR1;1) was cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen). An LR Gateway recombination reaction (Invitrogen) between the vectors pCR8/NaKR1;1 and pMDC123 (Curtis and Grossniklaus, 2003) was performed to subclone NaKR1;1 into the binary vector. To make the constructs NaKR1pro:NaKR1-GUS and NaKR1pro:NaKR1-GFP, the NaKR1 gene sequence (including the promoter, coding, and noncoding region, without TAA stop codon) was PCR amplified using primer pairs F (5′-GCCTTTGTGAAATCGATTTGAGTTAGAA-3′) and R (5′-CTTCTGAATAATCTCAGGCCAAAACTGA-3′). The 1809-bp PCR product (NaKR1;2) was cloned into pCR8/GW/TOPO and by LR recombination reaction into the binary vectors pMDC164 and pMDC107 (Curtis and Grossniklaus, 2003).
Agrobacterium tumefaciens was transformed by electroporation (Micropulser; Bio-Rad). Col-0 and nakr1-1 plants were transformed using the floral dip method (Weigel and Glazebrook, 2002). Transformants containing the construct pMDC123/NaKR1;1 were selected based on Basta resistance. Transformants expressing NaKR1pro:NaKR1-GUS and NaKR1pro:NaKR1-GFP were hygromycin resistant and were selected by growing seeds on half-strength Murashige and Skoog medium (Caisson Laboratories) supplemented with 1% phytoagar, 0.05% MES, 2% sucrose, and hygromycin (30 mg/L for seeds of Col-0 background and 10 mg/L for seeds of nakr1-1 background), pH 5.7.
A NaKR1 cDNA clone in the Gateway entry vector (pENTR223.1) was obtained from the Salk Institute Genomic Analysis Laboratory. For expression in Escherichia coli cells, the cDNA fragment was recombined with the expression vector pRARE_MAL_DEST via LR reaction (Invitrogen), and pRARE MAL DEST/NaKR1 was introduced into E. coli strain BL21-AI by electroporation. A control plasmid was made by LR recombination between an empty pCR8/GW/TOPO vector and pRARE_MAL_DEST to remove the CcdB gene and kanamycin resistance marker between the AttR1/AttR2 sites.
GUS Staining
GUS staining was performed according to the method described by Quaedvlieg et al. (1998). Plant tissues were vacuum infiltrated in X-Gluc solution (Research Products International) twice for 5 min each, followed by incubation at 37°C for 16 h and cleared with 70% ethanol at 65°C for 1 h before samples were whole mounted in 70% ethanol for microscopy. For cross sections, the stained samples (without clearing) were embedded in 4% agarose, and 50-μm sections were made using a Vibratome 1000 Plus sectioning system.
Microscopy
Confocal laser microscopy was performed using C1 spectral imaging confocal microscope (Nikon) and EZ-C1 acquisition and analysis software. For propidium iodide staining of cell walls, seedlings were removed from the growth medium and stained briefly in 10 μg/mL propidium iodide, and the whole plants were mounted in water. The excitation wavelength for propidium iodide was 561 nm and emission wavelength collected at 570 to 610 nm. For GFP fluorescence, the excitation wavelength was 488 nm and the emission wavelength collected at 505 to 545 nm. A z-series of scans was collected to obtain a transverse section view.
GUS expression images were collected using the MZFLIII dissection microscope (Leica), Hoffman modulation contrast microscope (Nikon), and Qcapture acquisition software. Differential interference contrast images of root meristems and starch accumulation in root columella cells and epifluorescence images of GFP and aniline blue staining were captured using a DM5000B microscope (Leica) and Leica Applicaion Suite software (version 2.4.0 R1).
For starch staining of columella cells, whole seedlings were fixed overnight in FAA solution (50% ethanol, 5% formaldehyde, and 10% acetic acid), rinsed briefly in 50% ethanol twice, and stained in a mixture of glycerol:chloral hydrate:I2/KI (containing 30% glycerol [v/v], 8% chloral hydrate [w/v], 0.04% I2 [w/v], and 0.2% KI [w/v]) for 1 h and mounted in 50% glycerol for DIC imaging. For aniline blue staining, seedlings were incubated in 2.5% aniline blue (Fisher Scientific) dissolved in 0.5 M K3PO4 solution, pH 9.5, for 5 min before being mounted in 0.5 M K3PO4 for observation using a fluorescence microscope with UV filter.
For electron microscopy, mature roots were sampled from 8-d-old Col-0 and nakr1-1 seedlings, fixed, embedded, and stained according to the method of Haritatos et al. (2000). The sections were imaged at 60 kV with a Phillips CM12 transmission electron microscope. Images were captured with Maxim DL digital image program and viewed with Adobe Photoshop CS3 program.
NaKR1 cDNA Expression in E. coli and Protein Purification and ICP-MS Analysis
E. coli strain BL21-AI containing the construct pRARE_MAL_DEST/NaKR1 or pRARE_MAL_DEST (with CcdB gene and KAN resistance gene removed) was inoculated into small Luria-Bertani liquid culture containing 100 mg/L ampicillin and incubated overnight at 37°C at 200 rpm. Overnight cultures (10 mL) were inoculated into 1 liter of Luria-Bertani medium for further amplification. When the culture reached OD600 of 0.5 to 0.7, isopropyl β-D-1-thiogalactopyranoside was added (to a final concentration of 0.5 mM), and the culture was allowed to grow for another 3 to 4 h before harvest. Protein purification was according to the pMAL protein purification protocol (New England BioLabs) with minor modifications. The cell culture was disrupted by sonication, and the MBP-NaKR1 fusion protein and MBP were purified by mixing cell supernatant with amylose resin (NEB) at a ratio of 20:1 (v:v) and incubated at 4°C for 0.5 h. The resin was washed with PBS (0.8% NaCl, 0.02% KCl, 0.144% Na2HPO4, and 0.024% KH2PO4, pH7.4) and eluted with 30 mM maltose in PBS. The protein products collected from three independent experiments were combined for one ICP-MS analysis. Protein samples were digested with 50% HNO3 (Mallinckrodt AR Select grade) at 110°C for 1 h prior to ICP-MS analysis. Protein concentrations used to calculate the molar ratio of purified MBP-NaKR1 protein to metal was based on amino acid analysis.
The purified protein sample was digested with trypsin (50:1 ratio in 25 to 100 mM ammonium bicarbonate at 37°C overnight) followed by reduction with 10 mM DTT and alkylated with iodoacetamide. The sample was analyzed by liquid chromatography–tandem mass spectrometry (MS/MS) on a ThermoFinnigan LTQ ion trap mass spectrometer (Applied Biosystems). Liquid chromatography was performed on a Michrom BioResources Paradigm AS1 with an inline C18 12 cm column, packed in house (Michrom BioResources). All MS/MS samples were analyzed using Sequest (ThermoFinnigan) and X! Tandem (www.thegpm.org). The search used a FASTA file for Arabidopsis containing more than 174,668 entries, including known contaminants as well as the sequence of the MBP-NAKR1 fusion protein. Scaffold (version Scaffold-01_05_14; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptides were manually confirmed to NaKR1 to ensure that all peaks were assigned.
Xylem Sap Collection
Col-0 and nakr1-1 plants were grown in soil under long-day conditions until bolting and the inflorescence stems reached 7 to 10 cm in length. Rosette leaves were removed with scissors and inflorescence stems cut with a razor blade. The plants were covered to maintain humidity. The first two drops of xylem sap were discarded, and subsequent drops of xylem sap were collected from each plant with micropipettes (Drummond) (Shi et al., 2002; Sunarpi et al., 2005). Samples were dried overnight at 95°C and dissolved in 5% HNO3 solution. Sodium and potassium content was measured by ICP-MS.
Grafting Analysis
Reciprocal grafting between the shoot and root from different Arabidopsis seedlings was performed. Medium and grafting method were according to Rus et al. (2006). Seeds were germinated and grown vertically on plates containing half-strength Murashige and Skoog macro- and micronutrients, vitamins (Caisson Laboratories), benomyl, indole-3-acetic acid, 6-benzylaminopurine, and 1.2% phytoagar in controlled conditions (16-h/8-h light/dark cycle, 22°C, 55 μmol·m−2·s−1). Four- to five-day-old seedlings were grafted on the plate under a dissection microscope. The grafted plants were grown vertically on the same plate for 1 week, and successful graft unions were transferred to LG3 soil or Turface (Turface Athletics) and grown under short-day conditions (8-h/16-h light/dark cycle, 22°C, 70 μmol·m−2·s−1) for another 5 weeks. Grafted unions were checked 2 weeks after transfer to the soil/Turface as well as after harvesting the rosette leaves. The individuals with adventitious roots formed above the grafted union were discarded. Two to three fully expanded rosette leaves of similar age were sampled from each plant, rinsed three times in deionized water, and dried in the oven (90°C) overnight, and dry weight of each sample was recorded. Plants grown in Turface were watered twice per week with 0.25× Hoagland solution (Sigma-Aldrich) and iron supplement (4.15 mg/L [4.5 μM] sodium ferric EDDHA) from the bottom.
Long-Distance Transport of 14C Sucrose
Phloem loading and long-distance transport of sucrose was analyzed essentially as described (Srivastava et al., 2008). Before the start of the experiment, [U-14C] sucrose stock (21.8 GBq/mmol; MP Biomedicals) was dried to remove the ethanol and then resuspended at a final solution of 0.5 mM [U-14C] sucrose in 10 mM MES, pH 5.5, and 1 mM CaCl2. Eleven-week-old Col-0 and nakr1-1 plants were grown in Turface under short-day conditions, and single, representative Col-0 and nakr1-1 plants were tested for [U-14C] sucrose translocation. On each plant, the tip of a fully expanded rosette leaf was crimped with forceps and 2 μL of the [U-14C] sucrose applied. Plants were maintained at 20°C under high moisture conditions for the duration of the experiment. After 30 min, the remaining sucrose drop was removed and the application site rinsed with buffer (10 mM MES, pH 5.5, and 1 mM CaCl2). The labeled plants were dissected immediately, and rosette leaves, stem, and roots from each plant were pressed flat between Whatman 3M filter paper until dry. The dried plant parts were attached to card stock using double-sided tape and then exposed to Kodak BioMax MR film for 5 d.
Leaf Starch Analysis
Arabidopsis plants were grown for 3 months in short-day conditions, and fully expanded rosette leaves were sampled. For starch staining, leaves were cleared in 70% ethanol at 70°C and stained in I2/KI solution (2 g I2 and 10g KI per liter) for 15 min at room temperature. Starch quantification was according to the method of Smith and Zeeman (2006) with minor modifications. Fresh leaves (0.2 g) were sampled from each plant, boiled in 80% ethanol (five times × 3 min) to remove soluble sugar. The remained pellet was ground in water and adjusted to a final volume of 1.5 to 2 mL. From each sample, 0.1 mL (×4) of homogenate was collected and boiled in a water bath for 20 min, and the gelatinized samples were digested with 6 units of α-amyloglucosidase (dissolved in 200 mM Na acetate, pH 5.5) and 1.2 units of α-amylase (dissolved in 3.2 M ammonium sulfate, pH 7.0) at 37°C for 4 h. The glucose content of each tube was measured by glucose Trinder assay (Sigma Diagnotics). The glucose concentration of each sample was determined based on the absorbance monitored at 495 nm using a microplate scanning spectrophotometer (Power Wave 340; BIO-TER) and the standard curve produced using a series of glucose standards (0.011 mg/mL, 0.051 mg/mL, 0.11 mg/mL, 0.51 mg/mL, and 1 mg/mL).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: At5g02600 (NPCC6/NaKR1), At2g37390 (NaKR2), At3g53530 (NaKR3), At1g22710 (SUC2), At5g17800 (JBY1234), At1g66240 (ATX1), At3g56240 (CCH), At1g79430 (APL), At4g37490 (CYCB1;1), and At4g22200 (AKT2/3). Microarray data were submitted to the Gene Expression Omnibus database with accession number GSE24385.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of a 7-bp Deletion in At5g02600.
Supplemental Figure 2. At-NaKR1 and the C-Terminal HMA Domain Failed to Rescue the Lys Auxotrophy Phenotype of the Yeast sod1Δ Mutant.
Supplemental Figure 3. The Identity of the Purified MBP-NaKR1 Fusion Protein Was Confirmed by Mass Spectrometry Analysis.
Supplemental Figure 4. nakr1-1 Developmental and Na+/K+/Rb+ Accumulation Defects Were Complemented with NaKR1pro:NaKR1-GFP.
Supplemental Figure 5. nakr1-1 Developmental and Na+/K+/Rb+ Accumulation Defects Were Complemented with NaKR1pro:NaKR1-GUS.
Supplemental Figure 6. Cellular Structures of SE/CC Complexes Were Not Affected by the nakr1-1 Mutation as Shown by Transmission Electron Microscopy.
Supplemental Figure 7. The Differentiation of Protophloem/Metaphloem Cells and Companion Cells Was Not Affected by the nakr1-1 Mutation.
Supplemental Figure 8. Quantification of Starch in Col-0 and nakr1-1.
Supplemental Figure 9. Reciprocal Grafting of Col-0 and nakr1-1 Was Used to Test the Contribution of NaKR1 in Root and Shoot on Leaf Na+ and K+ Content of Plants Grown on Soil.
Supplementary Material
Acknowledgments
Mark Sanders and Gail Celio (University of Minnesota) are gratefully acknowledged for assistance with confocal microscopy and electron microscopy, respectively. We thank the following scientists for sharing biological materials or protocols: William Gray (University of Minnesota) provided Cyclin-GUS (in Col-0); David Jackson (Cold Spring Harbor Laboratory) provided SUC2pro:GFP (in Col-0); Ykä Helariutta (University of Helsinki, Finland) provided transgenic seeds containing APLpro:APL:GFP; Jan Dettmer (Helariutta lab) provided the aniline blue staining procedure; David W. Galbraith (University of Arizona) provided NPCC6pro:HTB2:GFP; and Philip Benfey (Duke University) provided quiescent center marker (JYB1234, At5g17800). This research was supported by National Science Foundation Arabidopsis 2010 (Grant IOS 0419695 to J.M.W. and D.E.S.).
References
- Araki T. (2001). Transition from vegetative to reproductive phase. Curr. Opin. Plant Biol. 4: 63–68 [DOI] [PubMed] [Google Scholar]
- Bauby H., Divol F., Truernit E., Grandjean O., Palauqui J.C. (2007). Protophloem differentiation in early Arabidopsis thaliana development. Plant Cell Physiol. 48: 97–109 [DOI] [PubMed] [Google Scholar]
- Baum S.F., Dubrovsky J.G., Rost T.L. (2002). Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am. J. Bot. 89: 908–920 [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Cilia M., San Roman A., Thomas C., Maule A., Hearn S., Jackson D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Bull P.C., Cox D.W. (1994). Wilson disease and Menkes disease: New handles on heavy-metal transport. Trends Genet. 10: 246–252 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R.J., Muñoz-Mayor A., Jha D., Essah P.A., Rus A., Tester M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30: 497–507 [DOI] [PubMed] [Google Scholar]
- Deeken R., Ache P., Kajahn I., Klinkenberg J., Bringmann G., Hedrich R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Deeken R., Geiger D., Fromm J., Koroleva O., Ache P., Langenfeld-Heyser R., Sauer N., May S.T., Hedrich R. (2002). Loss of the AKT2/3 potassium channel affects sugar loading into the phloem of Arabidopsis. Planta 216: 334–344 [DOI] [PubMed] [Google Scholar]
- Deeken R., Sanders C., Ache P., Hedrich R. (2000). Developmental and light-dependent regulation of a phloem-localised K+ channel of Arabidopsis thaliana. Plant J. 23: 285–290 [DOI] [PubMed] [Google Scholar]
- Dykema P.E., Sipes P.R., Marie A., Biermann B.J., Crowell D.N., Randall S.K. (1999). A new class of proteins capable of binding transition metals. Plant Mol. Biol. 41: 139–150 [DOI] [PubMed] [Google Scholar]
- Fischer D.B. (1987). Changes in the concentration and composition of peduncle sieve tube sap during grain filling in normal and phosphate-deficient wheat plants. Aust. J. Plant Physiol. 14: 147–156 [Google Scholar]
- Fromm J., Eschrich W. (1989). Correlation of ionic movements with phloem unloading and loading in barley leaves. Plant Physiol. Biochem. 27: 577–585 [Google Scholar]
- Giavalisco P., Kapitza K., Kolasa A., Buhtz A., Kehr J. (2006). Towards the proteome of Brassica napus phloem sap. Proteomics 6: 896–909 [DOI] [PubMed] [Google Scholar]
- Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway H., Munns R. (1980). Mechanisms of salt tolerance in nonhalophytes. Annu. Rev. Plant Physiol. 31: 149–190 [Google Scholar]
- Haritatos E., Medville R., Turgeon R. (2000). Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 211: 105–111 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Chino M. (1990). Chemical composition of phloem sap from the uppermost internode of the rice plant. Plant Cell Physiol. 31: 247–251 [Google Scholar]
- Himelblau E., Mira H., Lin S.J., Culotta V.C., Peñarrubia L., Amasino R.M. (1998). Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol. 117: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Hauser F., Schroeder J.I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14: 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Rim Y., Wang J., Jackson D. (2005). A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev. 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.Y., Yuan Z., Jackson D. (2003). Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Lacombe B., Pilot G., Michard E., Gaymard F., Sentenac H., Thibaud J.B. (2000). A shaker-like K(+) channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 12: 837–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E.L., Abid K.B., Rogers E.E., Guerinot M.L., Harper J.F., Ward J.M., McIntyre L., Schroeder J.I., Salt D.E. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Lang A. (1983). Turgor-related translocation. Plant Cell Environ. 6: 683–689 [Google Scholar]
- Lin M.K., Lee Y.J., Lough T.J., Phinney B.S., Lucas W.J. (2009). Analysis of the pumpkin phloem proteome provides insights into angiosperm sieve tube function. Mol. Cell. Proteomics 8: 343–356 [DOI] [PubMed] [Google Scholar]
- Lin S.J., Culotta V.C. (1995). The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc. Natl. Acad. Sci. USA 92: 3784–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.J., Pufahl R.A., Dancis A., O’Halloran T.V., Culotta V.C. (1997). A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J. Biol. Chem. 272: 9215–9220 [PubMed] [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough T.J., Lucas W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57: 203–232 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Ham B.K., Kim J.Y. (2009). Plasmodesmata - Bridging the gap between neighboring plant cells. Trends Cell Biol. 19: 495–503 [DOI] [PubMed] [Google Scholar]
- Ma Y., Slewinski T.L., Baker R.F., Braun D.M. (2009). Tie-dyed1 encodes a novel, phloem-expressed transmembrane protein that functions in carbohydrate partitioning. Plant Physiol. 149: 181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten I., Hoth S., Deeken R., Ache P., Ketchum K.A., Hoshi T., Hedrich R. (1999). AKT3, a phloem-localized K+ channel, is blocked by protons. Proc. Natl. Acad. Sci. USA 96: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mengel K., Haeder H.E. (1977). Effect of potassium supply on the rate of phloem sap exudation and the composition of phloem sap of Ricinus communis. Plant Physiol. 59: 282–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira H., Martínez-García F., Peñarrubia L. (2001). Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J. 25: 521–528 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Pufahl R.A., Singer C.P., Peariso K.L., Lin S.J., Schmidt P.J., Fahrni C.J., Culotta V.C., Penner-Hahn J.E., O’Halloran T.V. (1997). Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278: 853–856 [DOI] [PubMed] [Google Scholar]
- Puig S., et al. (2007). Higher plants possess two different types of ATX1-like copper chaperones. Biochem. Biophys. Res. Commun. 354: 385–390 [DOI] [PubMed] [Google Scholar]
- Quaedvlieg N.E., Schlaman H.R., Admiraal P.C., Wijting S.E., Stougaard J., Spaink H.P. (1998). Fusions between green fluorescent protein and beta-glucuronidase as sensitive and vital bifunctional reporters in plants. Plant Mol. Biol. 38: 861–873 [DOI] [PubMed] [Google Scholar]
- Riesmeier J.W., Willmitzer L., Frommer W.B. (1994). Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., Salt D.E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Quintero F.J., Pardo J.M., Zhu J.K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G. (2008). Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R., Wright K.M., Lauterbach C., Amon G., Gahrtz M., Feuerstein A., Oparka K.J., Sauer N. (2005). Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 41: 319–331 [DOI] [PubMed] [Google Scholar]
- Sunarpi H., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Yamaguchi Y., Koizumi N., Sano H. (2002). Functional characterization of a heavy metal binding protein CdI19 from Arabidopsis. Plant J. 32: 165–173 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Zhang C., Barthelson R.A., Lambert G.M., Galbraith D.W. (2008). Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 147: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.