Abstract
The large subunit of the U2 auxiliary factor (U2AF) recognizes the polypyrimidine tract (Py-tract) located adjacent to the 3′ splice site to facilitate U2 snRNP recruitment. While U2AF is considered essential for pre-mRNA splicing, its requirement for splicing on a genome-wide level has not been analyzed. Using Solexa sequencing, we performed mRNA profiling for splicing in the Schizosaccharomyces pombe U2AF59 (prp2.1) temperature-sensitive mutant. Surprisingly, our analysis revealed that introns show a range of splicing defects in the mutant strain. While U2AF59 inactivation (nonpermissive) conditions inhibit splicing of some introns, others are spliced apparently normally. Bioinformatics analysis indicated that U2AF59-insensitive introns have stronger 5′ splice sites and higher A/U content. Most importantly, features that contribute to U2AF59 insensitivity of an intron unexpectedly reside in its 5′-most 30 nucleotides. These include the 5′ splice site, a guanosine at position 7, and the 5′ splice site-to-branch point sequence context. A differential requirement (similar to U2AF59) for introns may also apply to other general splicing factors (e.g., prp10). Our combined results indicate that U2AF insensitivity is a common phenomenon and that varied intron features support the existence of unrecognized aspects of spliceosome assembly.
Pre-mRNA splicing plays a major role in gene regulation (23). Furthermore, alternative splicing serves as an important mechanism to generate molecular diversity (5). In the initial stages of splicing, the U1 snRNP recognizes the 5′ splice site (5′ SS), and the U2 snRNP auxiliary factor (U2AF) recognizes the polypyrimidine tract (Py-tract)/3′ splice site (3′ SS), leading to U2 snRNP recruitment to the branch point sequence (BPS) (27).
In humans, the essential splicing factor U2AF, which has been extensively studied, is a heterodimeric protein containing a large subunit (U2AF65) and a small subunit (U2AF35). These subunits bind to the Py-tract and the 3′ SS, respectively (37, 65, 68, 71). Both subunits of U2AF are essential for viability in many model organisms, such as zebrafish, the fruit fly, the nematode worm, and fission yeast Schizosaccharomyces pombe (U2AF59) (17, 28, 42, 46, 47, 59, 72). However, in the budding yeast Saccharomyces cerevisiae, the large subunit is dispensable and the small subunit is absent (1). In humans, U2AF65 interacts with several splicing factors (BBP/SF1, UAP56, SAP155 [or SF3b155], and p54) (1, 14, 19, 43, 45, 70) and facilitates branch point recognition (2, 4). The mammalian U2AF65 is found to be dispensable for splicing of some introns in vitro in the presence of the SR protein SC35 (36). Microarray analyses revealed an unexpected role of the Drosophila melanogaster large subunit (dU2AF50) in the nuclear export of intronless mRNAs (8). Several splicing regulators, such as SXL, PTB, hnRNP A1, ASF/SF2, SC35, and TRA, can facilitate or antagonize U2AF activity for splicing regulation (5, 53).
Biochemical and structural studies indicate that the essential splicing factor U2AF65 has a bipartite structure (26, 52, 68): the C terminus contains three RNA recognition motifs (RRM1, -2, and -3), and the N terminus contains an arginine-serine-rich (RS) activation domain. Mechanistically, the binding of U2AF to the Py-tract via RRM1 and RRM2 is the key to U2AF recruitment to pre-mRNA. This interaction allows the RS domain to facilitate multiple RNA-RNA interactions during spliceosome assembly, including the base-pairing between the BPS and the U2 snRNA (51, 57). The conserved RRM3 domain of U2AF interacts with BBP/SF1, SAP155 (or S. pombe prp10), and other splicing factors (1, 19, 25, 32). RRM3 is dispensable for RNA binding and in vitro splicing of model introns. However, it is required for viability in S. pombe and may be important for the splicing of only a subset of introns in vivo (3, 54).
We propose that there is a large diversity of features within introns that are relevant for intron definition or splicing and this could influence the requirement for U2AF59 in ways not captured so far by detailed analyses of model pre-mRNAs in metazoans. Despite extensive studies of U2AF, its requirement on a genome-wide level has not been analyzed. Relative to Saccharomyces cerevisiae, S. pombe shares many more features of pre-mRNA splicing with mammals: they both have degenerate splicing signals, their splicing factors (snRNAs and proteins) are similar to one another, and both require the two U2AF subunits (U2AF59 and U2AF23) (60, 64). A temperature-sensitive allele of the S. pombe large U2AF subunit is lethal (42, 46) and has been used to study splicing of specific transcripts in vivo (54, 58). Thus, S. pombe offers an excellent model system to analyze the role of U2AF59 on a genome-wide level.
We report here that when cellular mRNAs are profiled, introns show a range of splicing defects in the temperature-sensitive mutant of U2AF59 (prp2.1). Furthermore, many introns are apparently insensitive to U2AF59 inactivation, and many unanticipated intronic features contribute to U2AF59 insensitivity.
MATERIALS AND METHODS
Hosts/plasmids and RNA preparation and analysis.
The prp2.1 mutant (42) and the prp10-YH03 mutant (21) were previously described. The protocols for RNA preparation and reverse transcription-PCR (RT-PCR) (59) were as previously described and used gene-specific primers (54). For the RT-PCR assay from total RNA, PCR products after 30 cycles were stained with Sybr green. Sequences of gene-specific primers are given in Table S1 of the supplemental material.
Generation of mutants.
Mutants were generated using mutagenic primers (see Table S1 in the supplemental material) and inverse PCR. The clones were confirmed by sequencing.
Sequencing data processing and analysis.
The poly(A)+ RNA from the prp2.1 mutant grown at nonpermissive temperature (37°C) for 1 h (54) was subjected to the RNA-seq protocol and sequenced at the Genome Technology Core Facility at Vanderbilt University Medical Center. The annotated S. pombe genome sequence was obtained from Sanger GeneDB (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/). By using BLAT (tile size of 8, oneoff of 1) (31), sequence reads were mapped to the S. pombe genome and to the spliced gene products generated using the exon coordinates file. The sequence reads confirmed the presence of the TAT codon, reflecting the C387Y mutation of the prp2.1 mutant (42). Sequence reads that mapped to one unique location in the genome (or spliced products) and matched at least 34 of 36 nucleotides (nt) were used for subsequent analysis. The splicing efficiency (SE) was calculated as the fraction of spliced reads to total (spliced and unspliced) reads {SE = SR/[SR + (UR5′ + UR3′)/2]} (61) for introns that showed at least five total reads (spliced reads [SR] plus unspliced reads [UR]); UR5′ and UR3′ are 5′ and 3′ intron-exon junction reads. We used only unambiguous reads corresponding to either the spliced product (SR) or the unspliced product (UR). We included 3,114 of the total 4,878 annotated introns for prp2.1 (37°C) and 3,069 introns for the wild type (32°C); the wild-type data sets, s30_s7 and s30_s8, used for comparison were previously published (62). This study also contributed to the most recent intron-exon coordinates for the S. pombe genome that were used for mapping sequence reads (62). Using the sequence reads from the entire intron also yielded similar overall results for splicing efficiency. There were only 14 sequence reads in the prp2.1 mutant RNA sample that supported exon skipping, but these were not pursued further. The Solexa sequence reads are available upon request.
Sequence analysis.
The 106 introns which had a splicing efficiency of at least 90% under nonpermissive conditions and had between 10 and 100 unspliced plus spliced reads were selected for the U2AF59-insensitive group. Of the 243 introns with a splicing efficiency of <10% and between 10 and 100 unspliced plus spliced reads, 106 were randomly selected for the U2AF59-sensitive group to keep the number of introns in each set the same (the statistical conclusions were comparable when all 243 introns were used [data not shown]). Forty-three introns that were 90 to 100% unspliced in the wild-type data set (62) were excluded from our analysis. The 5′ splice site and branch site consensus strength was calculated using a weight matrix derived from all annotated introns in S. pombe. The sum of the log-odds score for each position in the weight matrix was taken as the overall splice site strength, as described previously (24). Calculation of the intron A/U percentage in Fig. 4E, below, excluded the 5′ and 3′ splice sites. The Py-tract length was determined as the longest contiguous C/U stretch downstream of the annotated branch site, excluding the 3′ splice site. A/U tract length was determined similarly, except we used the entire intron, excluding the 5′ and 3′ splice sites. The exon/intron A/U percentage was calculated using only introns at least 30 nt in length from each group.
RESULTS
Solexa sequencing allows identification of pre-mRNA splicing defects in the U2AF59 mutant.
To address the role of U2AF59 in splicing from a genome-wide perspective, we isolated poly(A)+ RNA from the prp2.1 mutant (C387Y), which was described previously (42), and subjected it to Solexa/Illumina sequencing. We obtained 22,727,544 sequence reads of a 36-nucleotide length. By using BLAT (Fig. 1A), about 79% (17,999,569) of the sequences could be mapped to the S. pombe genome. About 61% (11,059,027) sequence reads were unambiguously mapped to only one location in the genome and were used for subsequent analysis. About 96% of genes were represented in this group. About 80% of genes had over-5-fold coverage (number of nucleotides in sequencing reads normalized to gene length), 47% had 5- to 25-fold coverage, and 33% had over-25-fold coverage. About 18% of reads were from 12 high-expressing genes. For example, the hsp9 gene had 122,618 reads, or 21,324-fold coverage. U2AF59 had 1,545 sequence reads, or 36-fold coverage. Analysis of sequence read distributions across genes showed a 3′ bias (see Fig. S1 in the supplemental material).
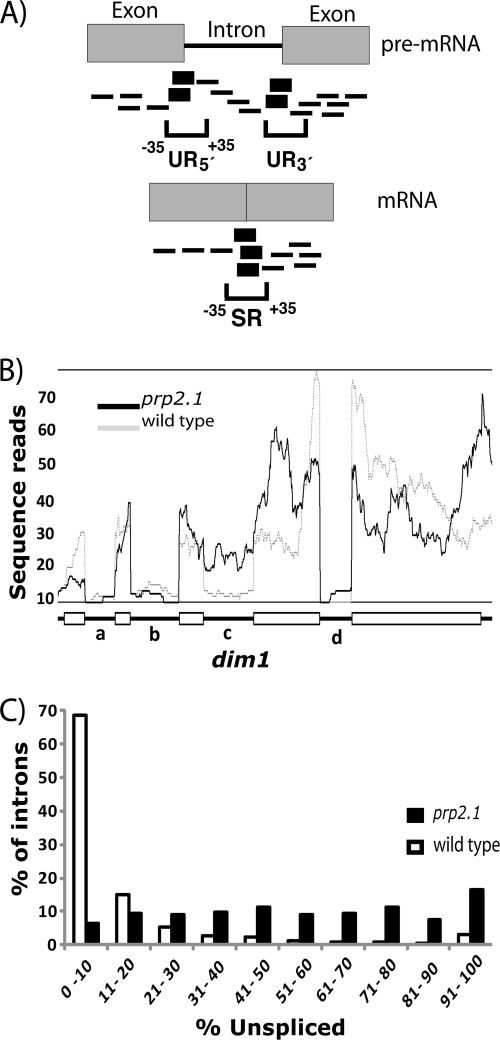
FIG. 1.
High-throughput sequencing by Solexa identified splicing defects in prp2.1. (A) Intron-exon junction sequence reads at the 5′ (UR5′) and the 3′ (UR3′) ends of the intron represent the unspliced mRNA. Mature mRNA sequence reads represent the spliced product. The 36-nt sequence reads that traverse from nt −35 to +35 positions of the exon-intron or exon-exon junctions are shown in bold. The sequence reads (outside the −35 to +35 splice junction positions) that cannot distinguish between the spliced and unspliced mRNAs are shown as thin lines. (B) Single-nucleotide sequence read distribution over exon and intron regions of the dim1 gene. Exons are shown as rectangles, and introns are shown as the lines connecting them. Solid lines and dashes indicate prp2.1 (37°C) and wild-type (prp2+ at 32°C) levels, respectively. The wild-type data set used for comparison was previously published (62). (C) Solexa sequencing revealed a range of splicing defects in prp2.1. The histogram shows percentages of introns (y axis), grouped according to the range of percent pre-mRNA levels (unspliced) (x axis) for introns that showed at least five total reads.
Figure 1B shows an example of the sequence read distribution at single-nucleotide resolution. As expected for the U2AF59 mutant, intron c of dim1 showed efficient splicing in the wild-type data set (62) but splicing inhibition in the prp2.1 mutant data set under nonpermissive conditions (37°C, 1 h). In contrast, introns a, b, and d were efficiently spliced in both the wild type and, surprisingly, U2AF59 mutant. We concluded that Solexa sequencing should allow genome-wide analysis of the effect of a general splicing factor mutation on specific splicing events in vivo.
The range of splicing defects in the U2AF59 mutant.
To analyze splicing efficiency at the genome-wide level in the prp2.1 mutant, we calculated the fraction of spliced reads to total reads (spliced and unspliced) for introns that showed at least five total reads (Fig. 1A) (see Materials and Methods for details). About 91% of introns were represented. About 64% of introns had at least 5 reads, and 41% of introns had between 10 and 100 reads.
In the prp2.1 mutant under nonpermissive conditions, considering at least five total reads, about 15% of introns showed splicing defects of >90% (494 introns), about 8% (189 introns) of introns remained mostly spliced (0 to 10% unspliced), and the remaining introns had a range of unspliced levels (Fig. 1C). When we eliminated introns with very low or high sequence reads, there remained 106 introns in the mostly spliced group with total reads between 10 and 100, which were used for further analysis. Although U2AF59 is considered an essential splicing factor, we concluded that endogenous introns show a range of splicing defects. Most importantly, a significantly large fraction (∼8%) of introns are efficiently spliced in prp2.1 in vivo under conditions that inhibit the splicing of many other introns.
A large number of cellular introns remain efficiently spliced upon U2AF59 inactivation.
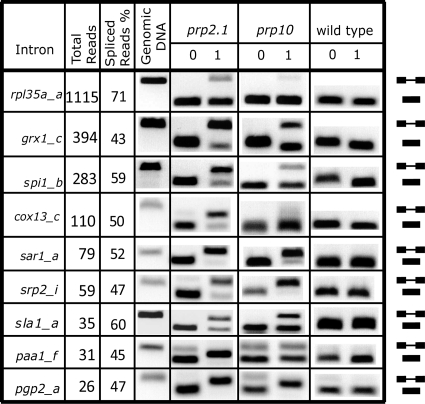
To validate the above predictions from the analysis of Solexa reads, we tested by RT-PCR the introns that were either partially spliced (40% to 60% unspliced) (Fig. 2) or efficiently spliced (<10% unspliced) (Fig. 3). Of nine randomly selected introns from the partially spliced category, we found using RT-PCR that eight introns were completely or mostly unspliced, sla1_a was partially spliced, and rpl35a_a was efficiently spliced. These results indicate that Solexa analysis, in comparison to RT-PCR, underestimated the splicing defect for this category.
FIG. 2.
Solexa analysis, compared to RT-PCR, underestimates splicing defects for the partially spliced introns. The total (unspliced and spliced) reads and percent spliced reads are shown for each intron. The splicing patterns were determined using RT-PCR for introns (indicated on the left) in prp2.1 and prp10 mutant strains and the wild type (prp2+), incubated for 0 and 1 h at 37°C. The genomic DNA PCR product indicates the unspliced product position in the gel. Positions of spliced and unspliced RNAs are indicated on the right.
FIG. 3.
A large fraction of cellular introns are U2AF59 insensitive. (A) Solexa and RT-PCR analyses independently confirmed the presence of U2AF59-insensitive introns. The splicing patterns for each intron are as per the details in the legend for Fig. 2. SPBC32H8.04c_a and SPAC6F6.05_a introns are abbreviated as SPBC32H and SPAC6F6, respectively. (B) Different introns within the same transcript behave differently. (C) The splicing behavior of plasmid-expressed introns mimics the behavior of endogenous introns. Plasmid DNA PCR product indicates the position of the unspliced product. (D) The splicing behavior of introns is similar in another prp2 mutant, rrm3-ts (54).
Next, we analyzed the group of efficiently spliced introns based on Solexa reads (Fig. 3A and B). We found that 17 of the 18 randomly selected introns were fully or mostly spliced, and vma3_c was partially spliced (∼50%); dim1_c was not identified with the spliced group. Our results indicate that RT-PCR and Solexa analysis agree for introns that have a sufficient number of Solexa sequence reads (10 to 100) and >90% splicing efficiency. We concluded, given the high correlation between Solexa and RT-PCR analyses results (17 of 18), that the majority of introns that are predicted to be efficiently spliced under nonpermissive conditions by Solexa sequencing are genuinely efficiently spliced. In other words, U2AF59 insensitivity is a widespread phenomenon in vivo.
U2AF59 insensitivity is not influenced by neighboring introns or expression levels.
An important question arises as to whether neighboring introns within the same gene behave similarly to U2AF59 inactivation. We found that genes with multiple introns had differential splicing patterns. For example, while dim1_c was not spliced, dim1_a, -b, and -d were all efficiently spliced in prp2.1 (Fig. 1B and B). The rhb1_a intron was fully spliced, and the rhb1_b intron was mostly spliced (Fig. 3B). We inferred from these findings that introns on the same transcript can behave differently with respect to U2AF59 insensitivity.
It is possible that the high expression level could have somehow influenced the splicing behavior (high splicing efficiency) of some introns based on both Solexa sequencing and RT-PCR analysis. To address this issue we cloned several introns along with their flanking exons into a plasmid vector, transformed them into the prp2.1 host, and analyzed the splicing of plasmid-encoded introns expressed from the nmt1 (no message in thiamine) promoter. Figure 3C shows that the control U2AF59-sensitive cdc2_b intron, as expected, was spliced under the permissive condition but unspliced under the nonpermissive condition. Thus, under these mRNA expression conditions a splicing defect is observable. Figure 3C also shows that all of the other relevant introns expressed from the plasmid were efficiently spliced, recapitulating the behavior of the corresponding endogenous introns. Another U2AF59 mutant, rrm3-ts (54), showed a similar splicing pattern for these introns (Fig. 3D), indicating that the effect is not specific to the prp2.1 mutation. We concluded that certain introns are inherently U2AF59 insensitive.
The 5′ splice site and base composition bias in U2AF59-sensitive versus -insensitive introns.
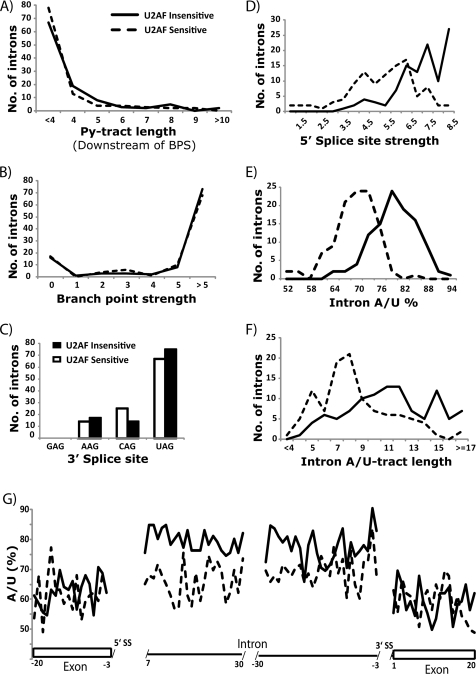
To determine an intronic feature(s) that may contribute to U2AF59 insensitivity, we analyzed the 106 U2AF59-insensitive introns (<10% unspliced) and an equal number of randomly selected U2AF59-sensitive introns (>90% unspliced) of a comparable average expression level. We found no statistically significant difference between the two intron pools for the length of the Py-tract either between the BPS and the 3′ SS (Fig. 4A) or in the entire intron (see Fig. S2 in the supplemental material). Similarly, no difference was observed in the strength of the BPS (Fig. 4B) or the nature of the 3′ SS (Fig. 4C). These findings may seem surprising given that the Py-tract/3′ SS and the BPS are the intron features most relevant to U2AF function.
FIG. 4.
Strengths of the 5′ SS and base composition distinguish the U2AF59-sensitive (>90% unspliced) and U2AF59-insensitive introns (<10% unspliced), based on the findings shown in Fig. 1C. Histograms show comparisons between U2AF59-sensitive and -insensitive introns with respect to the following features: Py-tract length downstream of the branch site (t test, P = 0.12) (A); strength of the BPS based on consensus weight matrix scores (t test, P = 0.84) (B); the 3′ splice site sequences (P = 0.38) (C); strength of the 5′ splice site based on consensus weight matrix scores (t test, P = 1.8 × 10−17) (D); base composition (percent A/U) (t test, P = 1.8 × 10−29) (E); (F) A/U tract length throughout the intron (t test, P = 4.2 × 10−9) (F); single-nucleotide distribution of the percent A/U (y axis) in the intron and flanking exon sequences, indicated by the coordinates in relation to the splice sites (G). Rectangles are exons, and the lines along the x axes are the intron.
Next, we analyzed the 5′ splice site and the base composition of the U2AF59-sensitive and -insensitive introns. Importantly, the U2AF59-insensitive introns had stronger 5′ splice sites in terms of consensus (Fig. 4D) and in terms of complementarity to the U1 snRNA (see Table S2 in the supplemental material). In addition, the U2AF59-insensitive intron pool also had both a higher A/U percentage (Fig. 4E) and longer A/U tracts (Fig. 4F). Furthermore, we analyzed the nucleotide preference at each position both upstream and downstream of 5′ and 3′ splice sites. We found that there was a pronounced preference for A/U residues in the U2AF59-insensitive introns relative to -sensitive introns (Fig. 4G); there was no significant difference in the flanking exons, and the difference became smaller toward the 3′ end of introns (see Fig. S3 in the supplemental material). Our combined results indicate that the strength of the 5′ SS and base composition (A/U tracts and/or percent A/U) in the 5′ intronic region could provide distinguishing features between the U2AF59-insensitive and -sensitive introns.
The 5′-most 30 nucleotides of bpb1_a confer U2AF59 insensitivity to heterologous introns.
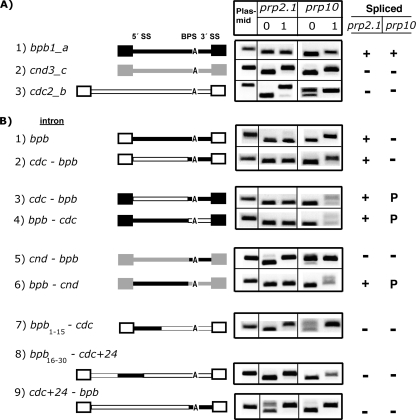
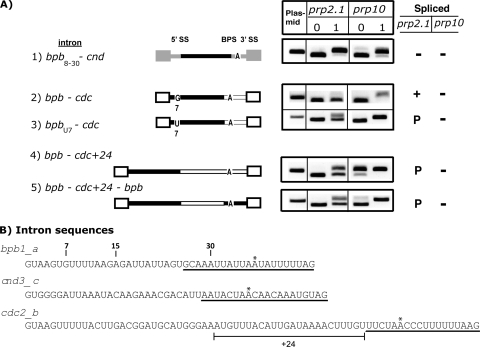
Next, we investigated what sequence feature(s) in a given intron contributes to U2AF59 insensitivity. We performed numerous domain swap experiments between the bpb1_a intron (U2AF insensitive) (Fig. 5A, construct 1) and the cnd3_c and cdc2_b introns (both U2AF59 sensitive) (Fig. 5A, constructs 2 and 3). First, we inserted the bpb1_a intron between the cdc2_b flanking exons. The bpb1_a intron was fully spliced, indicating that the sequence responsible for U2AF59 insensitivity is present entirely within the bpb1_a intron (Fig. 5B, construct 1). Second, we replaced the 3′-half (the sequence underlined in Fig. 6B, below) of the cdc2_b or cnd3_c intron with that of bpb1_a. The 3′-half of bpb1_a had a context-dependent effect: it supported efficient splicing in the cdc2_b intron (Fig. 5B, constructs 2 and 3) but not in the cnd3_c or cdc + 24 introns (Fig. 5B, constructs 5 and 9). Third, the 5′-half (nucleotides 1 to 30) of bpb1_a was similarly analyzed in these backgrounds. The 5′-half of bpb1_a supported efficient splicing in the three sequence contexts tested (Fig. 5B, constructs 4 and 6 and 6A, construct 2). Fourth, while nucleotides 1 to 30 of bpb1_a supported splicing, neither nucleotides 1 to 15 (Fig. 5B, construct 7) nor nucleotides 16 to 30 (Fig. 5B, construct 8) supported splicing in prp2.1. We concluded that the 30-nucleotide sequence is important for U2AF59 insensitivity.
FIG. 5.
The 5′-most 30 nucleotides of bpb1_a confer U2AF59 insensitivity onto heterologous introns. (A) Plasmid-expressed introns (bpb1_a, cnd3_c, and cdc2_b) are spliced, as expected, in the prp2.1 and prp10 mutants at 0 and 1 h under nonpermissive conditions. Schematics of the three introns (lines) and their flanking exons (boxes) are shown in different shades. (B) Schematics of various chimeras between different portions of introns and exons from the three introns are shown, using the same exon-intron shading scheme as in for panel A. For chimeric introns, unless otherwise indicated the 5′ portions of bpb1_a, cnd3_c, and cdc2_b introns are nucleotides 1 to 30 and the 3′ portions are shown below (underlined) in Fig. 6B. cdc+24 refers to the presence of the cdc2_b intron sequence indicated below in Fig. 6B. For construct 7, nt 16 to 30 are from cdc2_b. For construct 8, nt 1 to 15 are from cdc2_b. Splicing patterns for each construct are shown for the two mutants. +, efficiently spliced; −, largely unspliced; P, partially spliced.
FIG. 6.
Mutations that affect the 5′ SS, the guanosine-7 position, the sequence context, or the 5′ SS-to-BPS distance confer conditional U2AF59 sensitivity. (A) Schematics of various mutants and chimeras between different portions of introns and exons (see also Fig. 5). Splicing patterns for each construct are shown for the two mutants. (B) Sequences of the bpb1_a, cnd3_c, and cdc2_b introns. As reference points, positions 7, 15, and 30 are indicated. The 3′ portions used for the chimeras are underlined. The 24-nucleotide sequence of cdc2_b used in the cdc + 24 constructs is highlighted. The asterisk indicates a predicted branch point. +, efficiently spliced; −, largely unspliced; P, partially spliced.
bpb1_a mutations affecting the guanosine-7 (G7) position, the sequence context, or the 5′ SS-to-BPS distance confer conditional U2AF59 sensitivity.
Since the lengths of the bpb1_a and cdc2_b introns were different, we asked if the spacing between the 5′ SS and the BPS was relevant to the U2AF59 requirement. Increasing the spacing between the 5′ and 3′ halves of the bpb1_a intron with 24 nucleotides from the cdc2_b intron had no effect on splicing under permissive conditions. However, this expansion resulted in a partially unspliced intron under nonpermissive conditions (Fig. 6A, constructs 4 and 5), indicating that spacing between the 5′ SS and the BPS confers conditional sensitivity to the U2AF requirement. Moreover, we noticed that the 5′ SS (nucleotides 1 to 6, GUAAGU) was identical between the bpb1_a and cdc2_b introns (Fig. 6B). In fact, the first 10 nucleotides had only one nucleotide difference between the two introns: position 7 had a guanosine in bpb1_a and a uridine in cdc2_b. A guanosine allows complete splicing, while a uridine allows partial splicing (Fig. 6A, constructs 2 and 3). Finally, nucleotides 8 to 30 of bpb1_a were insufficient for U2AF59 insensitivity, because substitution of nucleotides 1 to 7 (or the 5′ SS) of bpb1_a with those from cnd3_c rendered the splicing of this chimera U2AF59 sensitive (Fig. 6A, construct 1 versus 5B, construct 6).
Our combined mutagenesis and sequence swap experiments indicated that nucleotides 1 to 30 of bpb1_a contribute to U2AF59-insensitive splicing and that the 5′ SS is important. Furthermore, there is a conditional requirement of U2AF59 that depends on the G7 position and the spacing (or sequence context) between the 5′SS and the BPS.
Various effects of the U2 snRNP component SAP155 (or SF3b155) on splicing.
Our primary focus in the present study was on the large subunit of U2AF. However, we were interested in whether the U2AF59-insensitive introns also failed to respond to loss of a different component of the general splicing machinery. We previously used the prp10 (also known as SAP155) temperature-sensitive mutant (21) and showed, as expected, that it failed to splice several introns at 37°C (54). We argued earlier that a lack of pre-mRNA accumulation in U2AF59 mutants may not be due simply to preferential pre-mRNA turnover. This is because the spliceosomal factor SAP155 is a core component of the U2 snRNP that interacts with the RRM3 domain of the large subunit of U2AF (19) and results in the accumulation of pre-mRNA (54). While the splicing of cdc2_b and cnd3_c was disrupted under nonpermissive conditions in the prp10 mutant, the splicing of the endogenous bpb1_a was unaffected (Fig. 5A). However, some of the sequence swap chimeras became sensitive to the prp10 mutation. Our analysis of numerous other introns (Fig. 2 and 3) revealed that splicing of many introns was not affected by the prp10 mutation under conditions that inactivated the splicing of some introns. We note that for some chimeras bpb1_a intron sequences show pre-mRNA accumulation in prp10 (Fig. 5B, construct 1, and 6A) and that the pre-mRNA with dim1_c intron accumulates in prp2.1 under nonpermissive conditions (Fig. 1B and 3B), indicating that preferential pre-mRNA instability in the prp2.1 mutant may not contribute to the efficient splicing behavior of these pre-mRNAs. We note that there may be slightly less efficient splicing in the mutant strains at the permissive temperature for some of the substrates.
We conclude that the effect of prp10 is also variable, that this mutation does not always disrupt splicing, and that certain mutations can alter sensitivity to the prp10 mutation (Fig. 5B and 6A). It appears that responses to loss of U2AF59 and SAP155 are quite similar, but not identical, for these introns.
DISCUSSION
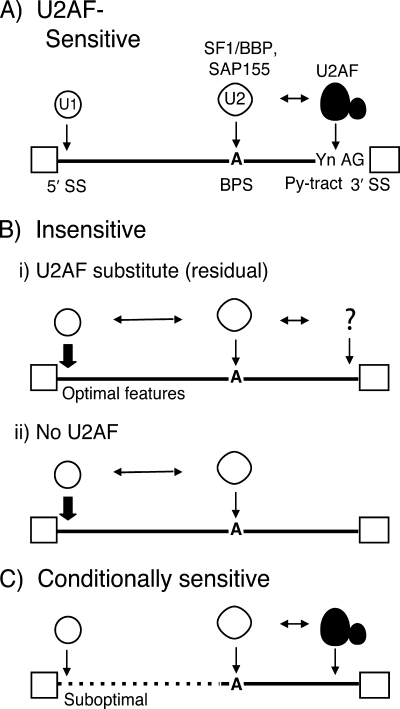
The key finding of this study was that cellular introns show a range of splicing defects in the prp2.1 mutant and that numerous introns are relatively insensitive to U2AF59 inactivation. While U2AF59 is known to function by binding to sequences between the BPS and the 3′ SS, we found that additional distant sequence features near the 5′ SS either eliminate or minimize the apparent need for the essential splicing factor U2AF59. Here, we provide a model of U2AF59 function that accounts for these unexpected findings from our genome-wide analysis for the requirement of U2AF59 in splicing (Fig. 7).
FIG. 7.
A proposed model for U2AF59-insensitive splicing. (A) The U1 snRNP interacts with the 5′ SS and U2AF interacts with the Py-tract/3′ SS. The SF1/BBP, SAP155, and U2 snRNPs interact with the BPS. RNA-RNA and RNA-protein interactions are shown by downward arrows. Horizontal two-sided arrows represent interactions between splicing factors. The question mark in panel B represents residual activity of the U2AF large subunit or another factor that may functionally substitute for it. The dashed line in panel C represents suboptimal features or mutations/alterations in the 5′ SS, the G7 position, spacing between the 5′ SS and the BPS, or portions of nucleotides 1 to 30 of bpb1_a that render this intron U2AF59 sensitive. Exons are represented as boxes, and introns are shown as lines.
Compensatory mechanisms influence the requirement for the splicing factor U2AF59.
For this discussion of the U2AF59 requirement(s) for splicing, we have classified cellular introns as U2AF59 sensitive, U2AF59 insensitive, and conditionally sensitive, as suggested by previous studies of specific introns (54, 58). Similar to the behavior of well-studied model introns, the U2AF59-sensitive cdc2_b intron showed the conventional U2AF59 requirement (Fig. 7A) (51). The second category of introns showed no detectable splicing defect in the prp2.1 mutant at 37°C. There are three possible explanations for the splicing of the newly identified U2AF59-insensitive introns, which constitute a significant fraction of introns in S. pombe (Fig. 3). First, consistent with the prevalent view that U2AF59 is a general or essential splicing factor, these introns may actually require the U2AF59 function. Perhaps the temperature-sensitive prp2.1 mutant protein is not completely inactive and possesses residual activity that is sufficient to support the splicing of introns that are better able to recruit U2AF59 but fails to support viability, under nonpermissive conditions (Fig. 7B, panel i). We found no detectable difference in the level of U2AF59 protein in the prp2.1 mutant under nonpermissive conditions (see Fig. S4 in the supplemental material), suggesting that U2AF59 inactivation rather than degradation likely contributes to loss of function. The second possibility is that some other gene(s) provides an activity that substitutes for U2AF59 for these introns (represented by the question mark in the figure). Finally, U2AF59 is dispensable (partially or fully) for some introns (Fig. 7B, panel ii). According to this view, under nonpermissive conditions the prp2.1 mutant protein shows a complete loss of function for all practical purposes, i.e., the residual activity, if any, is below the threshold level necessary to support U2AF59 function under physiological conditions, especially those lacking a significant Py-tract (bpb1_a or bpb-cnd3_c chimeras) (Fig. 5A, construct 1, and B, construct 6). Thus, the U2AF59-insensitive introns may be truly U2AF59 independent. For the third category, conditionally sensitive introns (Fig. 7C), we discovered that the bpb1_a intron is U2AF59 insensitive but can become sensitive (partially or fully) with any of several changes: the G7U substitution, which increases the spacing between its 5′ SS and the BPS, or when the 5′-most 30-nucleotide sequence is not intact (nucleotides 1 to 7, 1 to 15, or 16 to 30) (Fig. 5 and 6). Our simplest explanation is that several optimal sequence features together allow splicing with little or no U2AF59 activity.
Possible basis for relative U2AF59 insensitivity.
The discovery of U2AF59-insensitive introns does not contradict what we have learned from studies of model introns that showed that U2AF is required for splicing (48, 66). Nonetheless, our genome-wide analysis emphasizes that the notion that U2AF59 is required may not be tenable for a large fraction of introns in S. pombe (and possibly in metazoans). This alternative view is similar to the exceptional situation in S. cerevisiae, where both Mud2p and the Py-tract are generally dispensable for splicing (1, 39), and it is consistent with in vitro studies from mammalian introns (30, 35, 36, 63). Furthermore, it has been argued that different pre-mRNAs likely have distinct rate-limiting steps (3, 12, 36, 55, 56). The splicing factor SF1/BBP, which interacts with the RRM3 domain of U2AF59, plays a kinetic rather than an essential role in splicing and U2 snRNP binding in S. cerevisiae and humans (20, 49). Similarly, the small subunit and the RRM3 domain (which interacts with SF1/BBP) of the human U2AF65 are dispensable for the splicing of some introns in vitro (3, 67) and in vivo (54). Thus, it is possible that certain introns in S. pombe are indeed U2AF59 independent.
We favor the possibility that different U2AF59-insensitive introns may involve distinct compensatory mechanisms for splicing. In previous studies, no correlation between conventional Py-tract strength and U2AF59 requirement for the splicing of specific introns was observed (46). However, the 5′ end of the U2AF59-insensitive ulp2_c intron (54) contains an unusually long (30-nucleotide) uridine tract, which may facilitate splicing by involving either the putative residual activity of the prp2.1 mutant protein or the activity of factors such as p54, TIA-1-like, SMN, or Nam-8p-like protein (9, 15, 16, 29, 44). For the bpb1_a intron, which lacks a Py-tract within the 5′-most 30 nucleotides, different features (Fig. 5 and 6) may support splicing with little or no U2AF59 activity. For example, although the G7 position has not been linked to any known RNA-RNA or RNA-protein interactions (13), the U1C protein of the U1 snRNP could interact with the G7 position. The bridging interactions across introns involving the U1 snRNP at the 5′ SS and factors at the BPS/3′SS (2, 11, 34, 50, 69) could provide a likely mechanism for U2AF large subunit insensitivity. For certain introns, such alternative interactions could provide the major contribution for early events during spliceosome assembly. A previous study on limited introns found no correlation between U2AF59 requirement and the strength of the 5′ splice site or other obvious features of pre-mRNAs; sequence features of the U2AF59-insensitive cdc16-I1 intron remained uncharacterized (58). Our study indicates the presence of a stronger 5′ SS and a higher A/U percentage among the favorable features in U2AF59-insensitive introns (Fig. 4; see also Table S2 in the supplemental material). One mechanism for bypassing the U2AF requirement could be the presence of a stronger 5′ SS sequence in some introns. In other organisms, it has been shown that base composition (A/U richness) in plants and C. elegans contributes to defining the intron-exon boundary (see Fig. S3 in the supplemental material) (10, 18). It is possible that the A/U richness of the U2AF59-insensitive introns may make a similar favorable contribution to splicing in S. pombe (Fig. 4). Our findings raise the intriguing question whether different subsets of interactions between cis elements and general splicing factors are sufficient for splicing. We propose that supplementary or alternative mechanisms exist for early spliceosome assembly to accommodate various intronic features (Fig. 7).
Genomic view of splicing: intron-specific requirement for general splicing factors.
Undoubtedly, characterizations of specific model pre-mRNA substrates using in vitro and in vivo splicing systems have contributed greatly to our understanding of splicing signals, factors, and mechanisms (5, 23, 33). Findings from model substrates, however, may not explain splicing behavior of all cellular introns. The question arises whether general splicing factors show intron-specific requirements. Both prp2.1 and prp10 or SAP155(SF3b) mutations appear to have variable, intron-specific effects (Fig. 2, 3, 5, and 6). Such an intron-specific requirement may also be relevant to other general splicing factors. We believe that the next wave of progress on splicing in particular and gene regulation in general would involve a shift from intron- or gene-centric studies to genome-wide studies for hypothesis generation. This shift is necessary to accommodate the complex landscape of cis- and trans-acting factors from a genomic perspective, which is not captured by model introns or genes. Recent RNA interference and microarray analyses in Drosophila and S. cerevisiae pointed to the possibility of a rich diversity of requirements for splicing factors/regulators (6, 7, 22, 38, 40, 41).
The present study underscores the importance of how weakening (or strengthening) one or more factors in a system can influence the need for others. As a corollary, the requirement for RNA sequences and trans-acting factors is dynamic rather than static. In other words, a mutation could convert a U2AF59-insensitive intron into a sensitive (partially or fully) intron and vice versa. Such a phenomenon is relevant for splicing as well as for many steps along the gene expression pathway (transcription, RNA processing, translation, and signaling) in many organisms. This type of analysis, performed on a genome-wide level, can reveal alternative/compensatory mechanisms that might otherwise be missed.
Supplementary Material
Acknowledgments
We thank Dick McIntosh and his laboratory for help with the yeast work, Judith Potashkin, and Tokio Tani for plasmids, strains, and reagents/antibodies, and Tom Cech, Tom Blumenthal, Mark Winey, and Rajesh Gaur for helpful discussions and critical reading of the manuscript.
This work was supported in part by the American Cancer Society and a Butcher award to R.S.
Footnotes
Published ahead of print on 13 December 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, H., et al. 2004. The conserved RNA recognition motif 3 of U2 snRNA auxiliary factor (U2AF65) is essential in vivo but dispensable for activity in vitro. RNA 10:240-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 6.Blanchette, M., R. E. Green, S. E. Brenner, and D. C. Rio. 2005. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 19:1306-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette, M., et al. 2009. Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members. Mol. Cell 33:438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette, M., E. Labourier, R. E. Green, S. E. Brenner, and D. C. Rio. 2004. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell 14:775-786. [DOI] [PubMed] [Google Scholar]
- 9.Campion, Y., H. Neel, T. Gostan, J. Soret, and R. Bordonne. 2010. Specific splicing defects in S. pombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 29:1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad, R., K. Lea, and T. Blumenthal. 1995. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA 1:164-170. [PMC free article] [PubMed] [Google Scholar]
- 11.Cote, J., J. Beaudoin, R. Tacke, and B. Chabot. 1995. The U1 small nuclear ribonucleoprotein/5′ splice site interaction affects U2AF65 binding to the downstream 3′ splice site. J. Biol. Chem. 270:4031-4036. [DOI] [PubMed] [Google Scholar]
- 12.Crispino, J. D., B. J. Blencowe, and P. A. Sharp. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265:1866-1869. [DOI] [PubMed] [Google Scholar]
- 13.Du, H., and M. Rosbash. 2002. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature 419:86-90. [DOI] [PubMed] [Google Scholar]
- 14.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 15.Forch, P., L. Merendino, C. Martinez, and J. Valcarcel. 2003. U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor of 65 kDa, U2AF65, can promote U1 snRNP recruitment to 5′ splice sites. Biochem. J. 372:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forch, P., et al. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 17.Golling, G., et al. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31:135-140. [DOI] [PubMed] [Google Scholar]
- 18.Goodall, G. J., and W. Filipowicz. 1989. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58:473-483. [DOI] [PubMed] [Google Scholar]
- 19.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guth, S., and J. Valcarcel. 2000. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 275:38059-38066. [DOI] [PubMed] [Google Scholar]
- 21.Habara, Y., S. Urushiyama, T. Tani, and Y. Ohshima. 1998. The fission yeast prp10(+) gene involved in pre-mRNA splicing encodes a homologue of highly conserved splicing factor, SAP155. Nucleic Acids Res. 26:5662-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann, B., et al. 2009. Global analysis of alternative splicing regulation by insulin and wingless signaling in Drosophila cells. Genome Biol. 10:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 24.Hertz, G. Z., G. W. Hartzell III, and G. D. Stormo. 1990. Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput. Appl. Biosci. 6:81-92. [DOI] [PubMed] [Google Scholar]
- 25.Huang, T., J. Vilardell, and C. C. Query. 2002. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23). EMBO J. 21:5516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, J. L., H. Shen, M. R. Green, and C. L. Kielkopf. 2008. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J. Biol. Chem. 283:33641-33649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 28.Kanaar, R., S. E. Roche, E. L. Beall, M. R. Green, and D. C. Rio. 1993. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science 262:569-573. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy, C. F., A. Kramer, and S. M. Berget. 1998. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol. Cell. Biol. 18:5425-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent, O. A., D. B. Ritchie, and A. M. Macmillan. 2005. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 25:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent, W. J. 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konarska, M. M., J. Vilardell, and C. C. Query. 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell 21:543-553. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., and B. J. Blencowe. 1999. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 274:35074-35079. [DOI] [PubMed] [Google Scholar]
- 35.Lutzelberger, M., E. Backstrom, and G. Akusjarvi. 2005. Substrate-dependent differences in U2AF requirement for splicing in adenovirus-infected cell extracts. J. Biol. Chem. 280:25478-25484. [DOI] [PubMed] [Google Scholar]
- 36.MacMillan, A. M., P. S. McCaw, J. D. Crispino, and P. A. Sharp. 1997. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. U. S. A. 94:133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 38.Park, J. W., K. Parisky, A. M. Celotto, R. A. Reenan, and B. R. Graveley. 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 101:15974-15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson, B., and C. Guthrie. 1991. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell 64:181-187. [DOI] [PubMed] [Google Scholar]
- 40.Pleiss, J. A., G. B. Whitworth, M. Bergkessel, and C. Guthrie. 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell 27:928-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pleiss, J. A., G. B. Whitworth, M. Bergkessel, and C. Guthrie. 2007. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 5:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Potashkin, J., K. Naik, and K. Wentz-Hunter. 1993. U2AF homolog required for splicing in vivo. Science 262:573-575. [DOI] [PubMed] [Google Scholar]
- 43.Prigge, J. R., S. V. Iverson, A. M. Siders, and E. E. Schmidt. 2009. Interactome for auxiliary splicing factor U2AF(65) suggests diverse roles. Biochim. Biophys. Acta 1789:487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rain, J. C., Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer. 1998. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romfo, C. M., S. Lakhe-Reddy, and J. A. Wise. 1999. Molecular genetic analysis of U2AF59 in Schizosaccharomyces pombe: differential sensitivity of introns to mutational inactivation. RNA 5:49-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudner, D. Z., R. Kanaar, K. S. Breger, and D. C. Rio. 1996. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. U. S. A. 93:10333-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52:207-219. [DOI] [PubMed] [Google Scholar]
- 49.Rutz, B., and B. Seraphin. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19:1873-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 51.Shen, H., and M. R. Green. 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16:363-373. [DOI] [PubMed] [Google Scholar]
- 52.Sickmier, E. A., et al. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23:49-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh, R., and J. Valcarcel. 2005. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 12:645-653. [DOI] [PubMed] [Google Scholar]
- 54.Sridharan, V., and R. Singh. 2007. A conditional role of U2AF in splicing of introns with unconventional polypyrimidine tracts. Mol. Cell. Biol. 27:7334-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarn, W. Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8:2704-2717. [DOI] [PubMed] [Google Scholar]
- 57.Valcarcel, J., R. K. Gaur, R. Singh, and M. R. Green. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273:1706-1709. [DOI] [PubMed] [Google Scholar]
- 58.Webb, C. J., S. Lakhe-Reddy, C. M. Romfo, and J. A. Wise. 2005. Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell 16:584-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Webb, C. J., and J. A. Wise. 2004. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol. Cell. Biol. 24:4229-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wentz-Hunter, K., and J. Potashkin. 1995. The evolutionary conservation of the splicing apparatus between fission yeast and man. Nucleic Acids Symp. Ser. 33:226-228. [PubMed] [Google Scholar]
- 61.Wilhelm, B. T., S. Marguerat, I. Goodhead, and J. Bahler. 2010. Defining transcribed regions using RNA-seq. Nat. Protoc. 5:255-266. [DOI] [PubMed] [Google Scholar]
- 62.Wilhelm, B. T., et al. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239-1243. [DOI] [PubMed] [Google Scholar]
- 63.Will, C. L., C. Schneider, R. Reed, and R. Luhrmann. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284:2003-2005. [DOI] [PubMed] [Google Scholar]
- 64.Wood, V. R., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 65.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 66.Zamore, P. D., and M. R. Green. 1991. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 10:207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zamore, P. D., and M. R. Green. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. U. S. A. 86:9243-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355:609-614. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, W. J., and J. Y. Wu. 1996. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell. Biol. 16:5400-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]
- 72.Zorio, D. A., and T. Blumenthal. 1999. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.