Abstract
The Campylobacter jejuni pgl gene cluster encodes a complete N-linked protein glycosylation pathway that can be functionally transferred into Escherichia coli. In this system, we analyzed the interplay between N-linked glycosylation, membrane translocation and folding of acceptor proteins in bacteria. We developed a recombinant N-glycan acceptor peptide tag that permits N-linked glycosylation of diverse recombinant proteins expressed in the periplasm of glycosylation-competent E. coli cells. With this “glycosylation tag,” a clear difference was observed in the glycosylation patterns found on periplasmic proteins depending on their mode of inner membrane translocation (i.e., Sec, signal recognition particle [SRP], or twin-arginine translocation [Tat] export), indicating that the mode of protein export can influence N-glycosylation efficiency. We also established that engineered substrate proteins targeted to environments beyond the periplasm, such as the outer membrane, the membrane vesicles, and the extracellular medium, could serve as substrates for N-linked glycosylation. Taken together, our results demonstrate that the C. jejuni N-glycosylation machinery is compatible with distinct secretory mechanisms in E. coli, effectively expanding the N-linked glycome of recombinant E. coli. Moreover, this simple glycosylation tag strategy expands the glycoengineering toolbox and opens the door to bacterial synthesis of a wide array of recombinant glycoprotein conjugates.
Asparagine-linked (N-linked) protein glycosylation is essential and conserved in eukaryotic organisms. It is the most prevalent of all posttranslational protein modifications, affecting nearly 70% of the eukaryotic proteome (2). The attachment of N-glycans to eukaryotic secretory and membrane proteins can influence their folding and stability, oligomerization, resistance to proteolysis, sorting, and transport (23, 24). N-linked glycosylation occurs in the endoplasmic reticulum (ER) and involves the assembly of glycans on a lipid carrier in the ER membrane followed by transfer to specific asparagine residues of target polypeptides.
Initially, it was believed that N-linked glycosylation was unique to eukaryotes. However, N-glycoproteins have now been described for all domains of life, including archaea and more recently bacteria, of which the best-characterized example is the human gastroenteric pathogen Campylobacter jejuni (52). In C. jejuni, the genes for this pathway comprise a 17-kb locus named pgl for protein glycosylation (53). To date, more than 40 periplasmic and membrane glycoproteins have been identified in C. jejuni (34, 61), and most of these bind to the N-acetyl galactosamine (GalNAc)-specific lectin soybean agglutinin (SBA) (39). Mass spectrometry and nuclear magnetic resonance (NMR) studies revealed that the N-linked glycan is GlcGalNAc5Bac, where Bac is bacillosamine (2,4-diacetamido-2,4,6-trideoxyglucose) (61). This branched heptasaccharide is synthesized by sequential addition of nucleotide-activated sugars on the lipid carrier undecaprenyl pyrophosphate on the cytoplasmic face of the inner membrane (16). Once assembled, the lipid-linked heptasaccharide is flipped across the membrane by the putative ATP-binding cassette (ABC) transporter PglK (1, 29). Transfer of the heptasaccharide to periplasmic substrate proteins is catalyzed by an oligosaccharyltransferase (OST) called PglB, a single-integral-membrane protein with significant sequence similarity to the catalytic subunit of the eukaryotic OST STT3 (61). PglB attaches the heptasaccharide to asparagine in the motif D/E-X1-N-X2-S/T (where X1 and X2 are any residues except proline), a sequon similar to that of eukaryotes (34).
Recently, Wacker and coworkers transferred the entire C. jejuni pgl locus into Escherichia coli, conferring upon these cells the ability to N glycosylate proteins (57). Native C. jejuni glycoproteins such as Peb3 and AcrA, which are localized to the periplasm by the Sec pathway, can be N glycosylated in glycosylation-competent E. coli (57). AcrA can also become N glycosylated when transported via the twin-arginine translocation (Tat) pathway, which is well known for its ability to export folded proteins across the inner membrane (33). In addition to periplasmic proteins, some native C. jejuni N-glycoproteins are predicted to be integral membrane proteins based on bioinformatic analysis (34). Collectively, these earlier studies suggest that the C. jejuni N-linked glycosylation machinery is compatible with diverse secretory mechanisms and can tolerate an array of structures ranging from unfolded polypeptides to completely folded (albeit highly flexible and solvent-exposed) protein domains. However, the influence of membrane translocation and folding of acceptor proteins on bacterial N-linked glycosylation has not been thoroughly addressed. It is also not known whether proteins destined for locations beyond the periplasm, such as the outer membrane or the extracellular medium, are compatible with N-linked glycosylation.
Therefore, the goal of this study was to investigate the extent to which different secretory and extracellular protein substrates could be N glycosylated in E. coli cells carrying the pgl locus. To address this issue, we developed a genetically encoded N-glycan acceptor peptide tag (GT) that can be appended terminally or inserted at internal locations of recombinant proteins. Numerous recombinant proteins modified with the GT were reliably glycosylated in E. coli strains expressing the pgl genes. When the GT was used in combination with proteins targeted to the periplasm by different export pathways (e.g., Sec, signal recognition particle [SRP], or Tat), we observed a clear difference in the glycosylation patterns on these proteins depending on their mode of inner membrane translocation. In all cases tested, N-glycan attachment via the GT did not have any measurable effect on the protein's activity. Finally, we show that proteins targeted to different locations, including the periplasm, the outer membrane, membrane vesicles, and the extracellular medium, were amenable to N-linked glycosylation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table 1. E. coli strain DH5α was used for cloning of plasmids. E. coli strain CLM24 (16) was used for all glycoprotein expression experiments unless otherwise noted. For labeling of cell surface glycoproteins, E. coli strain BW25113 ΔwaaC::Kan was used (3). For preparation of outer membrane vesicles, E. coli strain CE8032 was created by P1vir phage transduction. Briefly, a kanamycin (Kan)-marked allele derived from BW25113 ΔwaaL::Kan was transduced in recipient JC8031 cells, a tolRA mutant strain that is known to hyper-vesiculate (5). For YebF-mediated secretion studies, strain MC4100 was used because this strain exhibits the least leakage of maltose-binding protein (MBP) of 15 common E. coli strains tested (62), and a derivative strain showed no leakage of MBP during its early growth stage (20). Overnight E. coli cultures were diluted in fresh Luria-Bertani broth (LB) supplemented with antibiotics and 0.2% glucose and grown at 30°C or 37°C. At mid-log phase (optical density at 600 nm [OD600], ∼0.5), the medium was changed to glucose-free LB containing antibiotics and protein expression was induced with either 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for pTrc99A-based expression vectors or 0.2% arabinose for pBAD-based expression vectors. Induction was at 25°C or 30°C for up to 24 h. Antibiotics were used at the following concentrations: 100 μg/ml ampicillin (Amp), 25 μg/ml chloramphenicol (Cm), and 50 μg/ml Kan.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | F−endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG φ80lacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | Laboratory stock |
| MC4100 | F′ araD139 Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | Laboratory stock |
| CLM24 | W3110 ΔwaaL | 16 |
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78, where subscripts indicate strain names | 12 |
| BW25113 waaC::Kan | BW25113 waaC::Kan | 3 |
| BW25113 ΔwaaC | BW25113 waaC::Kan with kanamycin marker removed | This study |
| BW25113 waaL::Kan | BW25113, waaL::Kan | 3 |
| BW25113 ΔwaaL | BW25113 waaL::Kan with kanamycin marker removed | This study |
| JC8031 | 1292 ΔtolRA | 5 |
| CE8032 waaL::Kan | JC8031 waaL::Kan | This study |
| Plasmids | ||
| pACYCpgl | Entire pgl locus from C. jejuni in pACYC184 | 57 |
| pACYCpglmut | pACYCpgl with W458A and Y459A mutations in the highly conserved region of pglB | 57 |
| pACYCpglB::Kan | pACYCpgl with a Kan insertion in pglB | 40 |
| pBAD18 | Cloning vector, arabinose inducible, Ampr | 22 |
| pBAD24 | Cloning vector, arabinose inducible, Ampr | 22 |
| pCP20 | Ampr and Cmr plasmid with temp-sensitive replication and thermal induction of the yeast Flp recombinase | 9 |
| pTrc99A | Cloning vector, IPTG inducible, Ampr | Amersham Pharmacia |
| pTrc-GT-6×-His | GT acceptor site sequence with C-terminal 6×-His tag in pTrc99A | This study |
| pTrc-MBP-GT | malE gene from E. coli in plasmid pTrc-GT-6×-His | This study |
| pET-AcrA-4×-His | acrA gene from C. jejuni with 2 additional glycan acceptor sites in pET22b | This study |
| pTrc-ΔspMBP-GT | malE lacking the native signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spDsbA-MBP-GT | malE with the native signal peptide replaced by the DsbA signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spTorA-MBP-GT | malE with the native signal peptide replaced by the TorA signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spMBP-GT-MBP | malE with GT after the native signal peptide in plasmid pTrc-6×-His | This study |
| pTrc-spMBP-GT-MBP-GT | malE with GT after the native signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spDsbA-TOP7-GT | top7 gene with the DsbA signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spTorA-GFP-GT | gfpmut2 gene with the TorA signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spDsbA-26.10LC-spPelB-26.10HC-GT | 26.10 light chain with the DsbA signal peptide followed by the 26.10 heavy chain with the PelB signal peptide in plasmid pTrc-GT-6×-His | This study |
| pTrc-spDsbA-Fc | IgG1 Fc domain with the DsbA signal peptide in pTrc99A | This study |
| pTrc-spDsbA-FcDQNAT | IgG1 Fc domain with Q295D, Y296Q, and S298A substitutions and the DsbA signal peptide in pTrc99A | This study |
| pTrc-spDsbA-AcrA-4× | acrA gene from C. jejuni with 2 additional glycan acceptor sites and the native export signal replaced with the signal peptide from E. coli DsbA in pTrc99A | This study |
| pTrc-spTorA-AcrA-4× | acrA gene from C. jejuni with 2 additional glycan acceptor sites and the native export signal replaced with the signal peptide from E. coli TorA in pTrc99A | This study |
| pBAD18-CjaA | cjaA gene from C. jejuni with a C-terminal Flag epitope in pBAD18 | This study |
| pBAD24-OmpX*-His | ompX gene from E. coli with a C-terminal 6×-His tag in pBAD24; contains unique KpnI and SpeI restriction sites between S53 and S54 of extracellular loop 2 of OmpX | 44 |
| pBAD24-OmpX-GT | GT acceptor site sequence in pBAD24-OmpX*-His | This study |
| pBAD24-OmpX-GT::PglB | Bicistronic expression of OmpX-GT and C. jejuni PglB in pBAD24 | This study |
| pBAD18-ClyA-GT | clyA gene from E. coli with a C-terminal GT and 6×-His tag in pBAD18 | This study |
| pTrc-YebF-GT | yebF gene from E. coli with a C-terminal GT and 6×-His tag in pTrc99A | This study |
| pTrc-ΔspYebF-GT | yebF gene lacking the N-terminal signal peptide with a C-terminal GT and 6×-His tag in pTrc99A | This study |
| pTrc-spYebF-GT | N-terminal signal peptide from yebF with a C-terminal GT and 6×-His tag in pTrc99A | This study |
| pTrc-YebF-ΔspMBP-GT | malE gene from E. coli lacking the N-terminal signal peptide in pTrc-YebF-GT | This study |
| pTrc-ΔspYebF-ΔspMBP-GT | malE gene from E. coli lacking the N-terminal signal peptide in pTrc- ΔspYebF-GT | This study |
| pTrc-spYebF-ΔspMBP-GT | malE gene from E. coli lacking the N-terminal signal peptide in pTrc-spYebF-GT | This study |
Strr, streptomycin resistant.
Plasmid construction.
The plasmid pTrc-GT-6×-His was cloned by inserting synthesized DNA encoding the GT (Integrated DNA Technologies [IDT] DNA) and a six-His (6×-His) motif between XbaI and HindIII of pTRC99A. DNA encoding malE, ΔspmalE (where sp indicates the signal sequence/peptide), sptorA-malE, spdsbA-malE, and spmalE was inserted between SacI and XhoI of pTrc-GT-6×-His to create pTrc-MBP-GT, pTrc-ΔspMBP-GT, pTrc-spTorA-MBP-GT, pTrc-spDsbA-MBP-GT, and pTrc-spMBP-GT, respectively. DNA encoding ΔspmalE was then cloned into the SalI site of pTrc-spMBP-GT to create pTrc-spMBP-GT-MBP. DNA encoding GT-MBP was then cloned back into pTrc-MBP-GT between SacI and AfeI to create pTrc-spMBP-GT-MBP-GT. DNA encoding Top7 (36) was cloned into XbaI and XhoI of pTrc-spDsbA-MBP-GT to create pTrc-spDsbA-TOP7-GT. DNA encoding sptorA-gfpmut2 was cloned into SacI and BamHI of pTrc-GT-6×-His to create pTrc-spTorA-GFP-GT. DNA encoding a bicistronic version of the heavy and light chains of the 26.10 IgG was PCR amplified from pMAZ360-26.10 (41) and digested with AvrII and SpeI. The PCR product was then cloned into pTrc-spDsbA-TOP7-GT that had been cut with XbaI and SpeI to remove the gene encoding Top7. The resulting plasmid, pTrc-spDsbA-26.10LC-spPelB-26.10HC-GT, expressed the light chain with a DsbA signal peptide and the heavy chain with a PelB signal peptide and a C-terminal GT. Plasmid pTrc-spDsbA-Fc was created by cloning the Fc region of human IgG1 into XbaI and HindIII of pTrc-spDsbA-MBP-GT. The pTrc-spDsbA-Fc DQNAT plasmid was created in an identical way, except that an Fc gene encoding point mutations Q295D, Y296Q, and S298A was initially cloned into pMQ70 via homologous recombination in Saccharomyces cerevisiae (49) and then subcloned into the XbaI and HindIII sites of pTrc-spDsbA-MBP-GT. Plasmid pBAD18-CjaA was created by amplifying the cjaA gene from C. jejuni genomic DNA (kindly provided by Brendan Wren). PCR-amplified cjaA was cloned between NcoI and NotI of plasmid pBAD18, containing a Flag epitope tag between NotI and PsiI. The plasmid pBAD24-OmpX-GT was created by inserting the GT sequence between the KpnI and SpeI sites of plasmid pBAD24-OmpX*-His (where * indicates a mutated form of OmpX containing a cloning site for peptide insertion into loop 2) (44) such that the GT peptide tag was positioned between the serine residues at positions 53 and 54 of extracellular loop 2 of OmpX. This transmembrane loop is able to tolerate short peptide insertions without affecting the surface expression of OmpX (44). A GQSGQ linker that flanked the KpnI and SpeI sites was also introduced. A bicistronic construct for coexpression of OmpX-GT and PglB was created by amplifying pglB from C. jejuni genomic DNA and inserting the resulting PCR product between XbaI and SbfI in plasmid pBAD24. Next, OmpX-GT was inserted between NcoI and XmaI in the same plasmid but with its own ribosome-binding site identical to the one upstream of pglB. Plasmid pBAD18-ClyA-GT was constructed by first inserting the PCR-amplified clyA gene into pTrc-GT-6×-His between SacI and XhoI. The entire ClyA-GT-6×-His construct was amplified by PCR and inserted between the SacI and HindIII sites in pBAD18. All YebF plasmids were derivatives of pTrc99A. For these, E. coli yebF was PCR amplified and cloned between SacI and XbaI in pTrc99A. Controls were similarly constructed by cloning either the YebF N-terminal signal peptide (spYebF) or the mature domain of YebF lacking the N-terminal signal peptide (ΔspYebF) between SacI and XbaI in pTrc99A. The GT or MBP-GT fusion was then added to each YebF vector between the XbaI and SalI sites. Lastly, a 6×-His affinity tag was inserted between the SalI and HindIII sites. The sequences of all plasmids constructed in this study were confirmed by DNA sequencing.
In-solution digestion of engineered glycoproteins.
The in-solution digestion for MBP-GT was performed as described previously (63) without reduction and alkylation. A protein sample (200 μg) was dissolved in a total of 100 μl denaturing solution containing 6.0 M guanidine-HCl and 50 mM Tris, pH 8.0, and incubated at 56°C for 45 min. Samples were diluted 1:5 in 50 mM ammonium bicarbonate, pH 7.8. Then, either 10 μg trypsin (Promega) or 1 μg LysC (Sigma) was added to the MBP-GT or AcrA-4× (AcrA containing four possible glycosylation sites) solution, respectively, at an enzyme-to-substrate ratio of 1:20 (wt/wt). Digestions were performed at 37°C for 16 h and stopped by the addition of 0.5% (vol/vol) trifluoroacetic acid (TFA). The digests were further desalted by solid-phase extraction using a Sep-Pack cartridge (Waters Corporation), and the eluted tryptic peptides were evaporated to dryness with a Speedvac SC110 (Thermo Savant). The samples were reconstituted in 200 μl of 0.1% formic acid with 2% acetonitrile to get a stock solution of 20 pmol/μl.
Infusion nanoelectrospray MS analysis.
MBP-GT was analyzed by infusion nanoelectrospray mass spectrometry (MS) analysis as follows. The enzymatic digest sample was diluted at a concentration of 2 pmol/μl in 50% acetonitrile with 0.1% formic acid prior to MS analysis. The sample (6 μl) was loaded into a discrete glass tip (New Objective Inc.) and delivered to an LTQ Orbitrap XL mass spectrometer (Thermo) equipped with a nanoscale ion source. The sample was analyzed in tune and positive ion modes for both a survey MS scan and a tandem MS (MS/MS) scan of selected ions using Orbitrap as a mass analyzer at a resolution of 60,000 using collision-induced dissociation (CID) fragmentation. The instrument was operated, and the resulting data analysis was performed with Xcalibur 2.0.7 software. The spray voltage used for all experiments was 1.8 kV. The capillary temperature was set up to 150°C, and collision energy was set to 20% for MS/MS experiments. The maximum scan time was set to 50 ms, and the results of 2 to 3 microscans were summed for each scan. Both the survey MS scan and the MS/MS scan of a singly glycosylated peptide with (M + 5H)5+ were acquired for 20 min over the mass range from m/z 200 to m/z 2,000. Data analysis of acquired raw data was performed using Xcalibur 2.0.7 software. Both MS and MS/MS spectra were summed from the 20-min acquisition.
nanoLC-MS/MS analysis.
The digested AcrA-4× samples (4 μl) were injected using a Famous autosampler onto a C18 column (5 μm, 300 μm by 5 mm; Dionex) for on-line desalting and then separated on a PepMap C18 reversed-phase (RP) nanoscale column (3 μm, 75 μm by 15 cm; Dionex) and eluted in a 60-min gradient of 5% to 45% acetonitrile in 0.1% formic acid at 275 nl/min. The nanoscale liquid chromatography (nanoLC) column was connected in-line to a hybrid triple-quadrupole linear ion trap mass spectrometer (4000 Q Trap equipped with a Micro-ion spray head II ion source; Applied Biosystems/MDS SCIEX).
MS data acquisition was performed using Analyst 1.4.2 software (Applied Biosystems) for precursor-ion (PI) scan-triggered, information-dependent acquisition (IDA). The precursor-ion scan at m/z 204.08 for oxonium ions of N-acetylhexoamine (HexNAc) was monitored at a step size of 0.2 Da across a mass range of m/z 650 to 1,600 for detecting glycopeptides containing N-acetylhexoamine unit. The nanospray voltage was 2.0 kV and was used in positive-ion mode for all experiments. The declustering potential was set at 50 eV, with nitrogen as the collision gas. In the IDA analysis, after each precursor-ion scan and enhanced-resolution scan, the two to three highest-intensity ions with multiple charge states were selected for tandem MS (MS/MS), with rolling collision energy applied for detected ions based on different charge states and m/z values. All acquired MS/MS spectra for detected glycopeptide ions by precursor ion scanning were manually inspected and interpreted with BioAnalysis 1.4 software (Applied Biosystems).
Subcellular fractionation and protein purification.
To isolate intracellular glycoproteins, equal numbers of cells were pelleted by centrifugation at 5,000 × g for 15 min at 4°C, resuspended in 1 ml lysis buffer supplemented with 1% (vol/vol) Triton X-100 and 1 mg/ml lysozyme, and incubated on ice for 30 min. Cells were then sonicated 4 times for 30 s with a 1-min rest between each sonication. The sonicated cells were spun down at 10,000 × g at 4°C for 20 min, and the supernatants were collected. For preparation of periplasmic and culture supernatant fractions, equal numbers of cells were harvested by centrifugation at 5,000 × g for 15 min at 4°C. The supernatant fraction was subjected to 0.2-μm filtration and then concentrated with an Amicon Ultra centrifugal filter from Millipore. The cell pellet was washed and then subjected to subcellular fractionation into periplasmic and cytoplasmic fractions using the ice-cold osmotic-shock procedure as described elsewhere (13, 31). Outer membrane vesicles (OMVs) were isolated from cell-free supernatants as described previously (58). Briefly, following centrifugation at 5,000 × g for 15 min at 4°C, the cell-free supernatants were filtered through a 0.2-μm vacuum filter and spun by ultracentrifugation at 141,000 × g for 2 h at 4°C in a 28 Ti rotor (Beckman Instruments, Inc.), and the pellet containing the OMVs was collected in phosphate-buffered saline (PBS; pH 7.0). OMV preparations were plated on LB agar to confirm complete removal of bacterial cells. All 6×-His-tagged proteins were purified using Ni-nitrilotriacetic acid (NTA) spin columns (Qiagen) according to the manufacturer's instructions (Ni-NTA spin kit; Qiagen) under native conditions. Fc domains were purified using NAb protein A/G spin columns according to the manufacturer's instructions (Thermo Scientific).
Protein analysis.
Proteins were separated with SDS-polyacrylamide gels (Bio-Rad), and Western blotting was performed as described previously (13). Briefly, proteins were transferred onto polyvinylidene fluoride (PVDF) membranes, and membranes were probed with one of the following: anti-MBP antibodies conjugated with horseradish peroxidase (HRP) (New England Biolabs), anti-6×-His antibodies conjugated to HRP, polyclonal antibodies directed against OmpX (14) (kindly provided by Jean-Marie Pagès), hR6P antiserum, which is specific for the C. jejuni heptasaccharide (kindly provided by Markus Aebi), anti-human IgG-HRP (Promega), and SBA-HRP (Sigma). In the cases of OmpX and hR6P antiserum, anti-rabbit IgG-HRP (Promega) was used as the secondary antibody.
Enzyme-linked immunosorbent assay (ELISA) was performed using plates coated with 4 μg/ml bovine serum albumin (BSA)-digoxin (Sigma) or Fc domains purified using NAb protein A/G spin columns. BSA-digoxin- or Fc-coated plates were incubated with serial dilutions of IgG 26.10 samples purified using Ni-NTA spin columns (Qiagen) or FcγRI (CD64; R & D Systems), respectively. The substrate for detection was Sigma Fast o-phenylenediamine dihydrochloride (OPD; Sigma), and reactions were tracked for 20 to 30 min at 490 nm in a SpectraMax 190 plate reader (Molecular Devices). For IgG ELISAs, the signals were normalized to the total amount of IgG 26.10, as determined by Western blot imaging.
For measuring green fluorescent protein (GFP) fluorescence activity, fluorescence measurements were made using a fluorescence microplate reader (Molecular Devices). Analysis of fluorescently labeled bacteria was performed using flow cytometry and fluorescence microscopy as described previously (32). Briefly, after induction of protein expression, 100 μl cells was washed with PBS and incubated with 20 μg/ml SBA-Alexa Fluor 488 (Invitrogen) in PBS for 45 min in the dark. Following an additional PBS wash, flow cytometric data were collected on a FACSCalibur system (Becton Dickinson). Mean fluorescence was determined from histograms of the cell fluorescence emitted by 50,000 events collected using the FACSCalibur flow cytometer in scan mode. For microscopy, 15 μl of the cells incubated with Alexa Fluor 488-tagged SBA was placed onto a microscope slide with a coverslip. Microscopy was performed on a Zeiss Axioskop 40 microscope equipped with a Zeiss 100×/1.30 oil Plan-Neofluar lens, an X-Cite light source (EXFO, Mississauga, Ontario), and a Semrock (Rochester, NY) Brightline filter cube for GFP emission. Digital images were obtained with a Spot Flex digital camera (Diagnostic Instruments, Inc.) and controlled with Spot imaging software. All images were captured under bright-field illumination or under UV illumination.
RESULTS
A universal acceptor sequence for N-linked glycosylation of recombinant proteins.
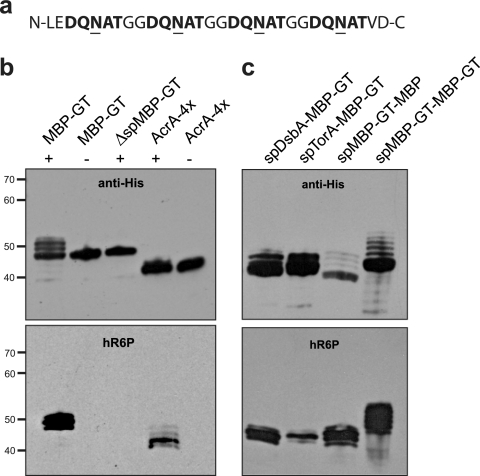
Our overall goal was to systematically evaluate secretion of N-linked glycoproteins to the periplasm and other extracytoplasmic locations in glycosylation-competent E. coli. Toward this goal, we first sought to develop a peptide tag that could be genetically encoded in recombinant proteins of interest, thereby enabling universal glycosylation of these proteins in E. coli. Recent studies by Chen and coworkers revealed that the sequence DQNAT was an optimal acceptor site for in vitro glycosylation by PglB (7). Based on this result, we reasoned that genetic modification of target proteins with an N- or C-terminal peptide containing one or more DQNAT motifs might be sufficient for N-linked glycosylation in E. coli harboring the pgl locus. To test this notion, we designed a C-terminal GT consisting of four consecutive DQNAT motifs separated from one another by consecutive glycine residues (Fig. 1a).
FIG. 1.
Glycosylation tag for making recombinant glycoproteins in E. coli. (a) The GT is comprised of four consecutive D-X1-N-X2-T sequons that are efficiently glycosylated in bacteria. (b) Western blot analysis of (from left to right) MBP with C-terminal GT (MBP-GT), mature MBP lacking its native signal peptide with C-terminal GT (ΔspMBP-GT), and the native C. jejuni glycoprotein AcrA engineered with two additional glycan acceptor sites (AcrA-4×). Proteins were expressed in cells carrying pACYCpgl (+) or pACYCpglmut (−). (c) Western blot analysis of (from left to right) MBP-GT with its native signal peptide replaced with the signal peptide of DsbA or TorA, MBP harboring an N-terminal GT following its native signal sequence (spMBP-GT-MBP), and MBP carrying both N- and C-terminal GT sequences (spMBP-GT-MBP-GT). Each was expressed in cells carrying pACYCpgl. Proteins were purified from cell lysates by Ni-NTA affinity chromatography. The same amount of protein was loaded in each lane. Blots were probed with anti-His (top) or hR6P (bottom) antibodies.
To test the reliability of this tag, we cloned the E. coli malE gene, encoding maltose-binding protein (MBP), in plasmid pTrc99A with a C-terminal GT followed by a hexa-histidine (6×-His) tag to facilitate purification. The resulting plasmid was used to transform E. coli strain BL21(DE3) harboring either plasmid pACYCpgl or plasmid pACYCpglmut, which bears the native (pgl) or a mutant version (pglmut) of the C. jejuni glycosylation locus, respectively (57). Recombinant proteins were purified from cell extracts by Ni-NTA affinity chromatography and analyzed by SDS-PAGE and subsequent immunoblotting. Ni-NTA affinity-purified fractions from cells expressing MBP-GT showed a major protein with a mass of 45 kDa and 3 additional higher-molecular-mass protein bands in the presence of the wild-type pgl locus but not pglmut (Fig. 1b). The three high-molecular-mass bands were absent when MBP-GT was expressed in the cytoplasm by removal of its native Sec pathway export signal (ΔspMBP-GT) (Fig. 1b). Thus, we hypothesized that these higher-molecular-mass bands were multiply glycosylated forms of MBP-GT. To confirm this, we tested the reactivity of hR6P antiserum with the purified proteins. This serum was raised against C. jejuni whole-cell extracts and has been shown to preferentially detect C. jejuni N-glycoproteins (Markus Aebi, personal communication). Only cells carrying the native pgl locus produced MBP-GT that was immunoreactive with hR6P antiserum (Fig. 1b, bottom panel). Neither the MBP-GT expressed in glycosylation-deficient cells nor the cytoplasmically expressed MBP-GT was detected with hR6P antiserum (Fig. 1b, bottom panel). For comparison, we expressed an engineered version of the C. jejuni glycoprotein AcrA that contained 4 possible glycosylation sites at N117, N123, N147, and N273 (AcrA-4×) and a PelB export signal for localization to the periplasm via the Sec pathway (34). Expression of AcrA-4× in glycosylation-competent E. coli cells led to the production of multiple high-molecular-mass proteins that were detected using hR6P antiserum (Fig. 1b), indicating the presence of multiple C. jejuni N-glycans. These glycans were not detected when AcrA-4× was expressed in glycosylation-deficient cells.
Fragmentation analysis of tryptic peptides derived from the purified MBP-GT protein from glycosylation-competent cells by nanoLC-MS/MS confirmed the identity of the characteristic C. jejuni heptasaccharide GlcGalNAc5Bac (see Fig. S1 in the supplemental material). Further, the MS analysis revealed three glycopeptide isoforms detected at the mass and charges expected for the attachment of one (5,162 Da; charges +7, +6, and +5), two (6,568 Da; +7, +6, +5), and three (7,974 Da; +7, +6, +5) C. jejuni N-glycans. Extracted chromatogram peak analysis revealed that the isoforms were found roughly in the proportions 64%, 34%, and 2% for the one-, two-, and three-glycan isoforms, respectively (data not shown), assuming all glycan isoforms have the same ionization efficiency. It should be noted that a four-glycan isoform was not detected (data not shown), consistent with the observation of three distinct bands following Western blot analysis (Fig. 1b and c).
Targeting proteins to the periplasm via the SRP and Tat pathways.
The above-described results demonstrate glycosylation of MBP that was targeted to the periplasm via the Sec pathway by virtue of its native export signal (37). We next determined whether site-specific attachment of C. jejuni glycans to MBP-GT could be similarly achieved following export via alternative protein export strategies, namely, the SRP or Tat pathway. These two export pathways are noteworthy for their starkly contrasting transport mechanisms. The SRP pathway mediates the cotranslational export of unfolded proteins (25, 48), while the Tat pathway is well known for its ability to posttranslationally export folded proteins (13, 45). To target MBP-GT to the SRP and Tat pathways, we replaced its native signal peptide with the SRP-dependent signal from disulfide isomerase I (spDsbA) (48) and the Tat-dependent signal from trimethylamine N-oxide reductase (spTorA) (47), respectively. Cells expressing spDsbA-MBP-GT or spTorA-MBP-GT along with the functional pgl locus triggered a strong signal with anti-His antibodies (Fig. 1c). In the case of spDsbA-MBP-GT, which was targeted to the SRP pathway, cells produced three proteins which were, respectively, mono-, di-, and triglycosylated forms of MBP-GT, as shown by their reactivity with the hR6P antiserum (Fig. 1c). This was reminiscent of the case described above where MBP-GT was targeted to the Sec pathway by its native signal peptide. Interestingly, cells expressing spTorA-MBP-GT produced one dominant protein band corresponding to the diglycosylated form and two much fainter bands corresponding to the mono- and triglycosylated forms (Fig. 1c). Hence, the efficiencies of bacterial N-linked glycosylation appear to be different for proteins routed through the Tat system versus the Sec/SRP export systems, which is not surprising given the fundamental differences in these modes of export (i.e., folded versus unfolded substrates). It is also noteworthy that a difference in glycosylation was observed when the GT sequence was moved from the C to the N terminus of MBP, at a position just after the native signal peptide (compare lanes MBP-GT and spMBP-GT-MBP in Fig. 1b and c, respectively). Expression of spMBP-GT-MBP in glycosylation-competent E. coli cells led to the production of four proteins that reacted with hR6P antiserum, whereas only three MBP-GT proteins were observed to react with this serum (Fig. 1c). This indicates that up to four C. jejuni N-glycans are covalently attached to spMBP-GT-MBP. When a GT was introduced at both the N and C terminus of MBP, seven proteins reactive toward the hR6P antiserum were detected (Fig. 1b and c), giving rise to one of the most extensively glycosylated proteins expressed in bacteria to date.
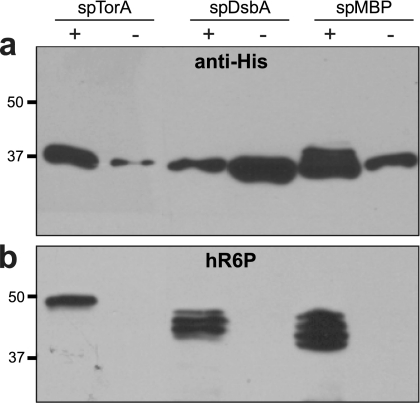
Given the difference in N-linked glycosylation levels observed above for MBP routed through the Sec/SRP and Tat pathways, we next addressed whether this was specific for the GT acceptor peptide or was a more general phenomenon that affected other glycoproteins expressed in E. coli. To test this, we modified the AcrA-4× construct described above with signal peptides for the Tat (spTorA), Sec (spMBP), and SRP (spDsbA) pathways and determined the glycosylation status of each by SDS-PAGE and immunoblotting with anti-His or hR6P antisera. Unlike with MBP-GT, where the acceptor sites are clustered at the C terminus of the protein, the glycosylation sites in AcrA-4× are distributed throughout the protein. Periplasmic extracts from glycosylation-competent cells expressing these constructs all contained anti-His-reactive proteins (Fig. 2a). However, only the spMBP-AcrA-4× construct gave rise to higher-molecular-weight bands following Western blotting with anti-His antibodies. In contrast, the spTorA-AcrA-4× and spDsbA-AcrA-4× constructs each gave rise to only a single band at ∼37 kDa. The lack of higher-molecular-weight bands for spTorA-AcrA-4× and spDsbA-AcrA-4× suggests that the ratio of glycosylated to aglycosylated AcrA in these cases was quite low compared to that for spMBP-AcrA-4×. Nonetheless, export via each of these pathways gave rise to glycosylated AcrA, as confirmed by Western blotting with hR6P antibodies (Fig. 2b). The Sec and SRP pathways triggered the detection of four proteins, indicating a mixture of mono-, di-, tri-, and tetraglycosylated forms of AcrA-4×. Indeed, nanoLC-MS/MS analysis confirmed N-linked glycosylation at all four sites (shown for spMBP-AcrA-4× in Fig. S2 in the supplemental material). A somewhat different result was obtained when AcrA-4× was targeted to the Tat pathway. Cells expressing spTorA-AcrA-4× and the functional pgl locus produced a single slower-migrating protein that cross-reacted with the hR6P antiserum (Fig. 2b). The molecular mass of this band was just under 50 kDa, which likely corresponds to tetraglycosylated AcrA-4× (44.2 kDa) that has retained its N-terminal spTorA signal peptide (4.2 kDa). Indeed, it has been observed that overexpression of Tat substrates and cell stress negatively affect the efficiency of signal peptide removal in E. coli (19, 54). Based on the slow electrophoretic mobility of this protein and its analysis by nanoLC-MS/MS, which revealed N-linked glycosylation at all four sites (data not shown), we conclude that tetraglycosylated AcrA-4× is the predominant glycosylated species produced by the Tat pathway. Taken together, proteins targeted to the Tat, Sec, and SRP pathways all become N glycosylated, with variations in the glycosylation patterns for Tat versus Sec/SRP substrates that may reflect the different substrate conformations (folded versus unfolded) that are presented to the OST.
FIG. 2.
Inner membrane translocation of glycoproteins. Western blot analysis of AcrA-4× targeted to the Tat (spTorA), SRP (spDsbA), or Sec (spMBP) protein export pathway. Proteins were purified from the periplasmic fraction of cells by Ni-NTA affinity chromatography. The same amount of protein was loaded in each lane. Blots were probed with anti-His (a) or hR6P (b) antibodies.
N-linked glycosylation of nonbacterial proteins in the periplasm.
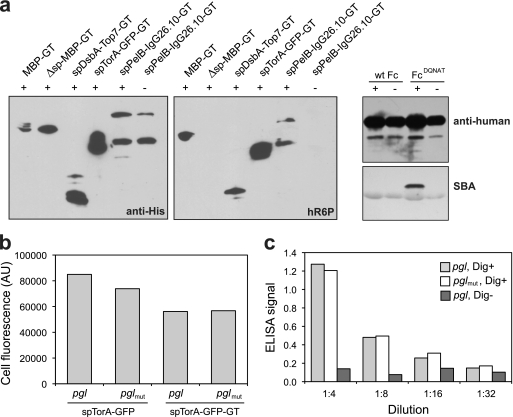
We next determined whether the GT sequence could permit N-linked glycosylation of nonbacterial substrate proteins that were localized in the periplasm of E. coli carrying the pgl gene cluster. As the first target, we chose the Top7 protein, a de novo-designed α/β protein that folds into a structure not observed in nature (36). We modified Top7 with an N-terminal DsbA export signal and C-terminal GT. Following expression in glycosylation-competent cells, spDsbA-Top7-GT reacted with anti-His antibodies and was clearly glycosylated based on its reactivity toward hR6P (Fig. 3a). Next we examined GFPmut2, a well-folded, fluorescence-activated cell sorting (FACS)-optimized variant of GFP (11). GFP is fluorescent only if it is allowed to fold in the cytoplasm of Gram-negative bacteria (15). Due to this peculiar property, export via the Tat pathway is the only way to localize fluorescent GFP to the periplasm (18, 46, 55). Thus, an spTorA-GFP-GT chimera was created and expressed in the presence of the functional pgl locus. Extracts from strains containing this construct elicited a strong signal with the hR6P antiserum (Fig. 3a). Finally, we examined a full-length mammalian antibody, the murine anti-digoxin 26-10 antibody (IgG 26.10) (41). Simmons and colleagues showed previously that expression of aglycosylated IgGs is possible in E. coli via separate expression and targeting of the IgG heavy and light chains to the periplasm (50). In a similar fashion, we targeted the light and heavy chains of IgG 26.10 to the periplasm via a DsbA and a PelB export signal, respectively (41). Additionally, we amended the heavy chain for glycosylation via a C-terminal GT acceptor sequence. Following expression and purification of this IgG, proteins reactive toward hR6P were detected only in the presence of the functional pgl locus (Fig. 3a), indicating the covalent attachment of bacterial glycan.
FIG. 3.
Glycosylation of diverse recombinant proteins in glycoengineered E. coli. (a) Western blot analysis of (from left to right) MBP-GT with or without its native signal peptide, TOP7-GT with the DsbA signal peptide, GFP-GT with the TorA signal peptide, and the murine anti-digoxin IgG 26.10 with a PelB signal peptide on the light chain and a DsbA signal peptide on the heavy chain, which was also appended with the GT. Proteins were expressed in cells carrying pACYCpgl (+) or pACYCpglmut (−). Blots were probed with anti-His (left) or hR6P (right) antibodies. Western blot analysis of wild-type Fc (wt Fc) and FcDQNAT was conducted with anti-human antibodies (top) and SBA (bottom). (b) Fluorescence of cells expressing spTorA-GFP or spTorA-GFP-GT in pgl or pglmut cells as indicated. Data are the averages of results from three replicate experiments, and the standard error was less than 5%. (c) ELISA signals for plates coated with BSA-digoxin conjugate (Dig+) and probed with 26.10 IgG purified from pgl or pglmut cells. Control wells without BSA-digoxin (Dig−) were incubated with 26.10 IgG purified from pgl cells.
To determine if glycosylation affected the functions of these recombinant glycoproteins, we examined the activities of our engineered GFP and IgG 26.10 constructs following their expression in glycosylation-competent cells. In the case of spTorA-GFP-GT, the fluorescence activity of GFP was unaffected by the addition of C. jejuni N-glycans, although the addition of the GT sequence did cause a nearly 30% decrease in spTorA-GFP fluorescence relative to that of the version expressed without the GT (Fig. 3b). Thus, we concluded that while the tag itself moderately decreased the fluorescence of GFP, this was independent of the glycosylation process. We also examined the binding activity of E. coli-expressed IgG 26.10 against its cognate antigen digoxin. The activities of IgG 26.10 proteins produced in cells carrying the active or inactive pgl gene clusters were virtually identical, as measured by ELISA (Fig. 3c), which is similar to the case for GFP, indicating that the process of glycosylation did not affect the antigen binding activity of IgG 26.10.
Encouraged by the results with IgG 26.10, we next determined whether the conserved acceptor site at N297 in the IgG1 Fc receptor domain could be glycosylated in the bacterial system. To address this, we cloned a mutant of the IgG1 Fc domain with triple mutations (Q295D Y296Q S298A) that encoded the preferred DQNAT sequon at N297 for PglB-mediated glycosylation (the FcDQNAT mutant). Following expression in E. coli cells carrying the pgl genes, the FcDQNAT mutant but not wild-type Fc was effectively N glycosylated (Fig. 3a). It is noteworthy that despite this glycosylation at N297, the bacterially glycosylated FcDQNAT was not observed to bind the FcγRI receptor, as determined by ELISA, whereas an aglycosylated Fc variant (FcE382V M428I) that was engineered to bind FcγRI (28) gave a strong ELISA signal (see Fig. S3 in the supplemental material). This indicates either that the bacterial N-glycan is insufficient for creating the “open” conformation needed for Fc receptor binding (35) or that the amino acids surrounding Asn297 (i.e., Q295, Y296, and S298) are needed in conjunction with N-linked glycosylation to restore binding to FcγRI.
Localization of N-linked glycoproteins in the outer membrane.
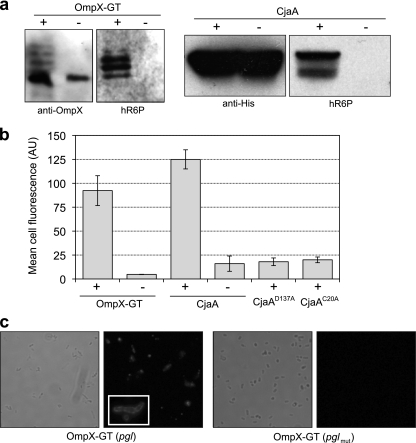
To date, only soluble periplasmic proteins have been glycosylated in E. coli cells carrying the pgl locus. Therefore, we next investigated whether outer membrane proteins could be N glycosylated. In a manner similar to the strategy described above for MBP, we engineered a GT acceptor sequence into the second extracellular loop of the β-barrel outer membrane protein OmpX from E. coli (44). Cells expressing OmpX-GT in the presence of the pgl locus produced glycosylated OmpX, whereas OmpX-GT expressed in cells carrying the pglmut locus showed no detectable glycosylation (Fig. 4a). To confirm that glycosylated OmpX-GT was localized in the outer membrane, we attempted to label intact E. coli cells with a fluorescently labeled version of SBA (SBA-Alexa Fluor 488). Since lectin SBA selectively binds terminal GalNAc residues, such as those of the C. jejuni heptasaccharide, we reasoned that if OmpX-GT was localized in the outer membrane, then N-glycans in extracellular loop 2 would be accessible for SBA binding. Further, since SBA is a 120,000-Da tetramer, it is too large to cross the outer membrane and thus is restricted to binding only those N-glycans that are displayed on the cell surface. To our surprise, E. coli cells carrying the pgl locus in the absence of an acceptor protein gave a strong fluorescent signal following labeling with SBA-Alexa Fluor 488, whereas plasmid-free cells were nonfluorescent (see Fig. S4a and b in the supplemental material). This signal was not dependent on PglB, as labeling of pglmut cells resulted in similarly high cell fluorescence (Fig. S4c in the supplemental material). We suspected that this fluorescence was due to transfer of the C. jejuni N-glycan to lipid A and subsequent localization of this glycolipid conjugate to the outer membrane. The basis for our hypothesis is the observation that undecaprenyl pyrophosphate-linked oligosaccharides are the substrates for the E. coli WaaL ligase, which transfers oligosaccharides from the undecaprenol lipid carrier to the lipid A core molecule (43). To test this notion, we created an E. coli strain in which the genomic copies of waaL or waaC were inactivated by allelic-replacement mutagenesis. The waaL gene codes for O-antigen ligase, which catalyzes the transfer of an O-antigen to the lipopolysaccharide (LPS) core, while the waaC gene codes for LPS heptosyl transferase I, which transfers the first heptose residue on the inner core of the LPS (43). Indeed, waaL- and waaC-deficient E. coli strains carrying the pgl locus were nonfluorescent following SBA-Alexa Fluor 488 labeling (as shown for waaL-deficient cells in Fig. S4d in the supplemental material), confirming that N-glycan transfer to the outer membrane occurs by a piggyback mechanism on lipid A. Importantly, inactivation of either waaL or waaC is sufficient to block this lipid A-mediated surface display and provides a suitable host for detecting outer membrane glycoproteins. Hence, we used the waaC mutant strain for investigating the surface display of N-glycosylated OmpX-GT. Expression of OmpX-GT in waaC-deficient cells carrying the pgl locus resulted in a significant increase in cell fluorescence following SBA-Alexa Fluor 488 labeling, whereas control cells expressing the same OmpX-GT construct in the presence of the pglmut locus were nonfluorescent (Fig. 4b). Fluorescence microscopy analysis confirmed the outer membrane localization of glycosylated OmpX-GT in waaC mutant cells carrying the pgl gene cluster (Fig. 4c). In contrast, no detectable staining was observed for waaC mutant cells expressing OmpX-GT in the presence of pglmut.
FIG. 4.
Expression of outer membrane glycoproteins. (a) Western blot analysis of OmpX-GT or CjaA expression in pgl (+) or pglmut (−) cells. Proteins were purified from cell lysates by Ni-NTA affinity chromatography. The same amount of protein was loaded in each lane. Blots were probed with anti-OmpX, anti-His, or hR6P antibodies as indicated. (b) Flow cytometric analysis of waaC-deficient cells expressing OmpX-GT or CjaA in the presence of pACYCpgl (+) or pACYCpglmut (−). Also shown are the results of a flow cytometric analysis of waaC-deficient cells expressing CjaAD137A or CjaAC20A in pgl cells. All cells were labeled with a fluorescently labeled version of SBA (SBA-Alexa Fluor 488). Data are the averages of results from three replicate experiments, and the error bars represent the standard errors of the means. (c) Light and fluorescence microscopy of waaC-deficient cells expressing OmpX-GT in the presence of pACYCpgl (+) or pACYCpglmut (−). The inset depicts an enlarged view of SBA-Alexa Fluor 488-labeled cells.
To determine whether glycosylation of outer membrane proteins with native N-glycan acceptor sites could be achieved in E. coli, we turned our attention to C. jejuni OmpH1 (CjaA). CjaA is an N-glycosylated lipoprotein that contains a single acceptor motif (137DSNIS141) and is expressed on the surfaces of C. jejuni cells (10, 60). Consistent with earlier studies (60), only glycosylation-competent E. coli produced CjaA that immunoreacted with hR6P antiserum (Fig. 4a). As with OmpX-GT, expression of CjaA in waaC-deficient cells carrying the pgl locus, but not pglmut, resulted in strong cell fluorescence following SBA-Alexa Fluor 488 labeling (Fig. 4b). When a D137A substitution was introduced into the CjaA acceptor motif, cell fluorescence in glycosylation-competent waaC mutant cells was lost (Fig. 4b). A similar loss of cell fluorescence was observed when the cysteine located 2 residues downstream of the CjaA signal peptide, which is required for lipoprotein processing and localization (27), was mutated to alanine (C20A).
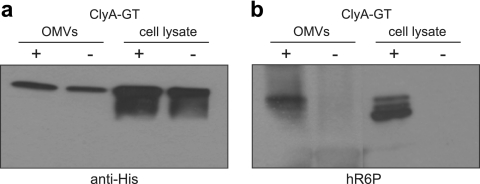
As a final test of outer membrane protein glycosylation, we investigated E. coli cytolysin A (ClyA, HlyE, SheA). ClyA is a hemolytic pore-forming toxin that is localized in the outer membrane of E. coli and subsequently released from the cell surface in outer membrane vesicles (OMVs) (58). Our studies revealed that the introduction of fusion partners to the C terminus of ClyA is well tolerated and does not affect vesicle localization (30). Therefore, to create a version of ClyA that could be N glycosylated, we introduced a C-terminal GT acceptor sequence to the clyA gene in plasmid pTrc99A. ClyA-GT was expressed in hyper-vesiculating E. coli strain CE8032, which also carried either the pgl or the pglmut plasmid (Fig. 5a). Whereas both pgl and pglmut cells produced ClyA-GT, only the glycosylation-competent cells produced ClyA-GT that immunoreacted with hR6P antiserum (Fig. 5b). Similarly, OMVs isolated from pgl and pglmut cells contained equivalent levels of ClyA-GT (Fig. 5a), but only those vesicles produced from glycosylation-competent cells contained N-glycosylated ClyA-GT, on the basis of reactivity with hR6P (Fig. 5b). Collectively, these results indicate that the biogenesis of diverse outer membrane proteins, including a β-barrel protein, a lipoprotein, and a membrane vesicle-associated protein, are compatible with the N-glycosylation process.
FIG. 5.
Localization of N-glycoproteins in OMVs. Western blot analysis of ClyA-GT in OMV fractions or cell lysates isolated from CE8032 cells carrying pACYCpgl (+) or pACYCpglmut (−). For cell lysates, proteins were Ni-NTA affinity purified. The same amount of protein was loaded in each lane. Blots were probed with anti-His (a) or hR6P (b) antibodies.
Extracellular secretion of recombinant N-glycoproteins.
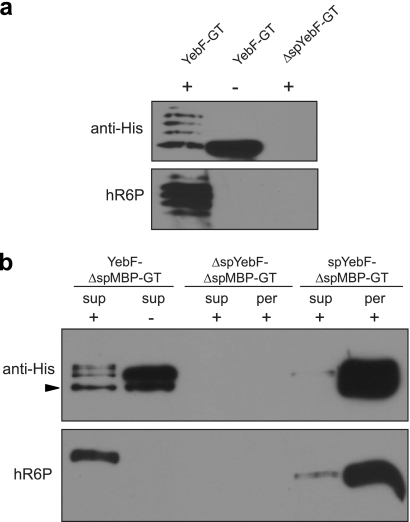
Since vesicle-mediated secretion was compatible with N-linked glycosylation, we next determined whether proteins glycosylated in the periplasm could be directed for secretion across the outer membrane and into the culture medium. We chose the E. coli YebF protein as a carrier to target proteins for extracellular secretion. YebF is a small protein (10.8 kDa in the mature form) that is transported into the bacterial periplasm by the Sec translocase and then delivered across the outer membrane by an unknown mechanism (62). YebF can carry C-terminal fusion partners into the culture medium at high levels (62). To determine if N-glycosylated derivatives of YebF could be secreted into the culture supernatant, we created a YebF-GT chimera in plasmid pTrc99A. Extracellular YebF-GT was secreted from both pgl and pglmut cells and reacted readily with anti-His antibodies (Fig. 6a). The secreted YebF-GT from pgl cells, but not pglmut cells, also reacted with hR6P (Fig. 6a). As expected, YebF-GT lacking its native Sec signal peptide (ΔspYebF-GT) was not detected in the supernatant fraction (Fig. 6a).
FIG. 6.
Secretion of N-glycoproteins in the culture supernatant. (a) Western blot analysis of YebF-GT and ΔspYebF-GT in the culture supernatant of MC4100 cells. (b) Western blot analysis of ΔspMBP-GT fused to full-length YebF, ΔspYebF, and the spYebF signal peptide as indicated. The arrowhead indicates the YebF-ΔspMBP-GT degradation product. The supernatant (sup) and periplasmic (per) fractions were probed with anti-His antibodies or hR6P serum as indicated. Proteins were expressed in cells carrying pACYCpgl (+) or pACYCpglmut (−). All proteins were Ni-NTA affinity purified from the supernatant and periplasmic fractions. The same amount of protein was loaded in each lane. Blots were probed with anti-His or hR6P antibodies as indicated.
To determine if YebF could carry recombinant glycoproteins into the supernatant, we translationally fused ΔspMBP-GT to the C terminus of YebF. The YebF-ΔspMBP-GT fusion was secreted into the culture medium by both pgl and pglmut cells, but only the fusion protein secreted from pgl cells was observed to react with hR6P (Fig. 6b). For secretion controls, we tested ΔspYebF-ΔspMBP-GT, which lacks an N-terminal export signal, and spYebF-ΔspMBP-GT, which modifies ΔspMBP-GT with only the native YebF signal peptide and does not include the mature YebF domain. As expected, ΔspYebF-ΔspMBP-GT was not localized in the periplasm or the supernatant (Fig. 6b) owing to the absence of an N-terminal signal peptide. The spYebF-ΔspMBP-GT construct was exported into the periplasm of glycosylation-competent cells, where it became glycosylated; however, only a faint band corresponding to the fusion was detected in the culture medium (Fig. 6b), indicating that the YebF protein is required for extracellular secretion of the fusion proteins tested here.
DISCUSSION
In this study, we have demonstrated the N-linked glycosylation of diverse secretory and extracellular protein substrates expressed in E. coli cells carrying the C. jejuni pgl locus, thereby expanding the N-linked glycome of recombinant E. coli. This was achieved by the development of a genetically encoded glycosylation tag that promoted reliable N-linked glycosylation when introduced at the termini or at internal locations of recombinant proteins. Using this tag, proteins targeted to different locations, including the periplasm (via the Sec, SRP, and Tat pathways), the outer membrane, membrane vesicles, and the extracellular medium, were efficiently glycosylated. To our knowledge, this is the first report that the C. jejuni N-glycosylation machinery is compatible with cotranslational (i.e., SRP), outer membrane, and extracellular protein targeting mechanisms in E. coli.
These results also illuminate key mechanistic similarities and differences between prokaryotic and eukaryotic N-linked glycosylation. In eukaryotes, N-glycan addition occurs on polypeptides as they emerge through the translocation pore, a process called cotranslocational glycosylation. In this scenario, acceptor proteins are translocated into the ER in an extended conformation, and the OST is thought to recognize polypeptides in an unfolded state (8, 59). In the bacterial system, translocation and glycosylation are apparently uncoupled, and glycosylation sites are located in flexible parts of folded proteins (33). Indeed, we found that protein substrates exported in a folded conformation by the Tat pathway were N glycosylated in the periplasm, consistent with recent findings of Kowarik and coworkers (33), who also showed that completely folded proteins can be modified by PglB in vivo. We also found that the bacterial OST can accommodate unfolded proteins, as well as folded proteins, as substrates. For example, outer membrane proteins, which are exported by the Sec pathway and maintained in an unfolded conformation until insertion in the outer membrane (6), were efficiently glycosylated by PglB. Presumably, glycosylation occurred in the periplasm prior to outer membrane integration, when folding was still incomplete. Likewise, the vesicle-mediated and extracellular secretion mechanisms investigated here, although poorly understood, are likely to involve unfolded periplasmic intermediates that are amenable to glycosylation by PglB. Taken together, our results reveal PglB to be a very versatile enzyme based on its ability to glycosylate both folded and unfolded substrates. While the glycosylation of Tat substrates here and elsewhere (33) clearly indicates that PglB attaches N-glycans posttranslationally, our results do not exclude the possibility that PglB glycosylates some substrates in a cotranslational manner. In support of this possibility, we observed that proteins targeted to the SRP pathway were competent for glycosylation, and we are now determining whether these proteins undergo cotranslational glycosylation. Such a coupling between translocation and glycosylation would have important consequences for bacterial glycoengineering because potential acceptor sites would not be confined to locally flexible structures but instead could be located in structured domains that become glycosylated prior to folding.
An important outcome of our studies is the demonstration that a simple glycosylation tag is sufficient to promote covalent attachment of multiple glycans to a single protein carrier. We anticipate that this tag will be useful in a number of important areas related to glycobiology and glycomedicine. For example, the GT could be used to express and isolate carrier proteins that are hyper-glycosylated with a multiplicity of homogeneous glycans. Such hyper-glycosylated proteins could then be used as features on carbohydrate microarrays (so-called glycoarrays), which are currently very challenging to fabricate due in large part to difficulties associated with the synthesis and isolation of sufficient quantities of naturally occurring oligosaccharides (38). Glycan diversity could be “genetically encoded” by expression of different glycosyltransferases to control the specific glycoform, and covalent transfer onto target proteins could be achieved using PglB, which is relatively promiscuous in its choice of both oligosaccharide (16) and protein (Fig. 3) substrates. Another potential use of the GT is in the development of therapeutic glycoprotein conjugates. For example, bacterial polysaccharides conjugated to proteins have proven effective as vaccines, as evidenced by the Haemophilus influenzae type b conjugate vaccine (56). By expanding the spectrum of recombinant protein carriers that can be N glycosylated via fusion to the GT, more-complex conjugate vaccine candidates may be developed. For example, multiple doses of H. influenzae type b capsular polysaccharide conjugate vaccines are required to induce protective antibody responses in infants. When conjugated to the meningococcal outer membrane protein (OMPC), protective antibody responses are induced after a single dose (42). A recombinant glycosylation tag could allow for N-linked glycosylation of optimized protein carriers in E. coli. Even MBP could serve as a useful carrier for glycoconjugate vaccines since (i) it can be engineered to carry far more glycans than most naturally occurring glycoproteins (Fig. 1b) (4) and (ii) it has demonstrated immunostimulatory properties (17). A final application for the GT is in the development of genetic tools for analyzing glycosylation phenotypes, which at present are severely lacking. A recombinant peptide tag allows glycosylation of proteins that may be amenable to extracellular display (21, 51) and/or two-hybrid systems (26) for the isolation of specific N-glycans. Further, a GT could be fused to a permissive site in a binding protein or enzyme such that glycosylation modulates the activity of the enzyme. Such a scheme would allow for genetic selection or screening dependent on functional N-linked glycosylation. It is our hope that the glycosylation tag strategy presented here allows for a higher level of sophistication and throughput in the emerging field of glycoengineering and opens the door to bacterial synthesis of a wide array of recombinant glycoprotein conjugates.
Supplementary Material
Acknowledgments
We thank Brendan Wren, Jean-Marie Pagès, George Georgiou, and Markus Aebi for providing strains, plasmids, and antiserum used in this work.
This work was supported by National Science Foundation Career Award CBET-0449080 (to M.P.D.), the New York State Office of Science, Technology and Academic Research Distinguished Faculty Award (to M.P.D.), the Department of Energy Great Lakes Bioenergy Research Center (GLBRC) Emerging Opportunities Program (all grants to M.P.D.), and the National Institutes of Health Small Business Innovation Research Awards R43 GM087766 and R43 GM086965 (to A.C.F.).
A.C.F., C.A.R., and J.H.M. are employees of Glycobia, Inc. A.C.F., C.A.R., J.H.M., C.G., and M.P.D. have a financial interest in Glycobia, Inc.
Footnotes
Published ahead of print on 3 December 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alaimo, C., et al. 2006. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apweiler, R., H. Hermjakob, and N. Sharon. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473:4-8. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Dor, S., N. Esterman, E. Rubin, and N. Sharon. 2004. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology 14:95-101. [DOI] [PubMed] [Google Scholar]
- 5.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubes. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos, M. P., V. Robert, and J. Tommassen. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191-214. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M. M., K. J. Glover, and B. Imperiali. 2007. From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46:5579-5585. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W., and A. Helenius. 2000. Role of ribosome and translocon complex during folding of influenza hemagglutinin in the endoplasmic reticulum of living cells. Mol. Biol. Cell 11:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Cordwell, S. J., et al. 2008. Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8:122-139. [DOI] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. U. S. A. 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont, M., C. E. James, J. Chevalier, and J. M. Pages. 2007. An early response to environmental stress involves regulation of OmpX and OmpF, two enterobacterial outer membrane pore-forming proteins. Antimicrob. Agents Chemother. 51:3190-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman, M. F., et al. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102:3016-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez, S., et al. 2007. Potential role for Toll-like receptor 4 in mediating Escherichia coli maltose-binding protein activation of dendritic cells. Infect. Immun. 75:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher, A. C., and M. P. DeLisa. 2008. Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS One 3:e2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher, A. C., et al. 2008. Exploration of twin-arginine translocation for the expression and purification of correctly folded proteins in Escherichia coli. Microb. Biotechnol. 1:403-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francetic, O., D. Belin, C. Badaut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francisco, J. A., and G. Georgiou. 1994. The expression of recombinant proteins on the external surface of Escherichia coli. Biotechnological applications. Ann. N. Y. Acad. Sci. 745:372-382. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 24.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019-1049. [DOI] [PubMed] [Google Scholar]
- 25.Huber, D., et al. 2005. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 187:2983-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joung, J. K., E. I. Ramm, and C. O. Pabo. 2000. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 97:7382-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juncker, A. S., et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, S. T., et al. 2010. Aglycosylated IgG variants expressed in bacteria that selectively bind FcgammaRI potentiate tumor cell killing by monocyte-dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 107:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly, J., et al. 2006. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, J. Y., et al. 2008. Engineered bacterial outer membrane vesicles with enhanced functionality. J. Mol. Biol. 380:51-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. Y., et al. 2005. Twin-arginine translocation of active human tissue plasminogen activator in Escherichia coli. Appl. Environ. Microbiol. 71:8451-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostecki, J. S., H. Li, R. J. Turner, and M. P. DeLisa. 2010. Visualizing interactions along the Escherichia coli twin-arginine translocation pathway using protein fragment complementation. PLoS One 5:e9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowarik, M., et al. 2006. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314:1148-1150. [DOI] [PubMed] [Google Scholar]
- 34.Kowarik, M., et al. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krapp, S., Y. Mimura, R. Jefferis, R. Huber, and P. Sondermann. 2003. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 325:979-989. [DOI] [PubMed] [Google Scholar]
- 36.Kuhlman, B., et al. 2003. Design of a novel globular protein fold with atomic-level accuracy. Science 302:1364-1368. [DOI] [PubMed] [Google Scholar]
- 37.Kumamoto, C. A., and P. M. Gannon. 1988. Effects of Escherichia coli secB mutations on pre-maltose binding protein conformation and export kinetics. J. Biol. Chem. 263:11554-11558. [PubMed] [Google Scholar]
- 38.Laurent, N., J. Voglmeir, and S. L. Flitsch. 2008. Glycoarrays—tools for determining protein-carbohydrate interactions and glycoenzyme specificity. Chem. Commun. (Camb.) 2008:4400-4412. [DOI] [PubMed] [Google Scholar]
- 39.Linton, D., E. Allan, A. V. Karlyshev, A. D. Cronshaw, and B. W. Wren. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497-508. [DOI] [PubMed] [Google Scholar]
- 40.Linton, D., et al. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695-1703. [DOI] [PubMed] [Google Scholar]
- 41.Mazor, Y., T. Van Blarcom, R. Mabry, B. L. Iverson, and G. Georgiou. 2007. Isolation of engineered, full-length antibodies from libraries expressed in Escherichia coli. Nat. Biotechnol. 25:563-565. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Melgosa, M., et al. 2001. Carrier-mediated enhancement of cognate T cell help: the basis for enhanced immunogenicity of meningococcal outer membrane protein polysaccharide conjugate vaccine. Eur. J. Immunol. 31:2373-2381. [DOI] [PubMed] [Google Scholar]
- 43.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocco, M. A., et al. 2009. Site-specific labeling of surface proteins on living cells using genetically encoded peptides that bind fluorescent nanoparticle probes. Bioconjug. Chem. 20:1482-1489. [DOI] [PubMed] [Google Scholar]
- 45.Sanders, C., N. Wethkamp, and H. Lill. 2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41:241-246. [DOI] [PubMed] [Google Scholar]
- 46.Santini, C. L., et al. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 47.Santini, C. L., et al. 1998. A novel sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schierle, C. F., et al. 2003. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185:5706-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmons, L. C., et al. 2002. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J. Immunol. Methods 263:133-147. [DOI] [PubMed] [Google Scholar]
- 51.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 52.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 53.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, G., L. Potter, and J. A. Cole. 1999. The periplasmic nitrate reductase from Escherichia coli: a heterodimeric molybdoprotein with a double-arginine signal sequence and an unusual leader peptide cleavage site. FEMS Microbiol. Lett. 174:167-171. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 56.Verez-Bencomo, V., et al. 2004. A synthetic conjugate polysaccharide vaccine against Haemophilus influenzae type b. Science 305:522-525. [DOI] [PubMed] [Google Scholar]
- 57.Wacker, M., et al. 2002. N-Linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 58.Wai, S. N., et al. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 59.Whitley, P., I. M. Nilsson, and G. von Heijne. 1996. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271:6241-6244. [DOI] [PubMed] [Google Scholar]
- 60.Wyszynska, A., J. Zycka, R. Godlewska, and E. K. Jagusztyn-Krynicka. 2008. The Campylobacter jejuni/coli cjaA (cj0982c) gene encodes an N-glycosylated lipoprotein localized in the inner membrane. Curr. Microbiol. 57:181-188. [DOI] [PubMed] [Google Scholar]
- 61.Young, N. M., et al. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, G., S. Brokx, and J. H. Weiner. 2006. Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nat. Biotechnol. 24:100-104. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, S., C. K. Van Pelt, and J. D. Henion. 2003. Automated chip-based nanoelectrospray-mass spectrometry for rapid identification of proteins separated by two-dimensional gel electrophoresis. Electrophoresis 24:3620-3632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.