Abstract
Surveillance for Plasmodium falciparum drug resistance mutations is becoming an established tool for assessing antimalarial treatment effectiveness. We used an extended version of a high-throughput post-PCR multiplexed ligase detection reaction fluorescent microsphere assay (LDR-FMA) to detect single-nucleotide P. falciparum drug resistance polymorphisms in 402 isolates from children in Papua New Guinea (PNG) participating in an antimalarial treatment trial. There was a fixation of P. falciparum crt (pfcrt) K76T, pfdhfr C59R and S108N, and pfmdr1 mutations (92%, 93%, 95%, and 91%, respectively). Multiple mutations were frequent. Eighty-eight percent of isolates possessed a quintuple mutation (underlined), SVMNT, NRNI, KAA, and YYSND, in codons 72 to 76 for pfcrt; 51, 59, 108, and 164 for pfdhfr; 540, 581, and 613 for pfdhps; and 86, 184, 1034, 1042, and 1246 for pfmdr1, and four of these carried the K540E pfdhps allele. The pfmdr1 D1246Y mutation was associated with PCR-corrected day 42 in vivo treatment failure in children allocated piperaquine-dihydroartemisinin (P = 0.004). Although the pfmdr1 NFSDD haplotype was found in only four isolates, it has been associated with artemether-lumefantrine treatment failure in Africa. LDR-FMA allows the large-scale assessment of resistance-associated single-nucleotide polymorphisms (SNPs). Our findings reflect previous heavy 4-aminoquinoline/sulfadoxine-pyrimethamine use in PNG. Since artemether-lumefantrine and piperaquine-dihydroartemisinin will become first- and second-line treatments, respectively, the monitoring of pfmdr1 SNPs appears to be a high priority.
Resistance of Plasmodium species to 4-aminoquinoline drugs emerged in Papua New Guinea (PNG) in 1976 and then spread across the country (18, 24). In addition, mass pyrimethamine dosing in the 1960s led to high-level resistance, and in vitro (38) and in vivo (11, 21) chloroquine (CQ) or amodiaquine (AQ) monotherapy was retained as the first-line treatment for uncomplicated malaria until 2000, when sulfadoxine-pyrimethamine (SP) was added to improve clinical efficacy (5). Despite initial successes, cure rates have since declined (21, 24).
Single-nucleotide polymorphisms (SNPs) in parasite genes determining drug effects can underlie resistance. This includes mutations in the Plasmodium falciparum CQ transporter (pfcrt) gene (3, 15), but higher-level CQ resistance results from other SNPs and is inversely associated with the copy number of the P. falciparum multidrug resistance 1 (pfmdr1) gene (16, 35, 37). pfmdr1 polymorphisms also confer resistance to other antimalarial drugs, including mefloquine, lumefantrine, and quinine (6, 33, 37). Of particular concern are the results of a previously reported pfmdr1 gene allelic replacement study in which various polymorphisms reduced artemisinin susceptibility in cloned parasite lines (37). Polymorphic changes in the genes encoding dihydrofolate reductase (dhfr) and dihydropteroate synthetase (dhps) underlie parasite resistance to pyrimethamine (7, 34) and sulfadoxine (44, 45), respectively. Almost all strains of P. falciparum from patients from the Madang Province in PNG who fail CQ-SP treatment carry pfcrt K76T and pfmdr1 N86Y mutations, while the pfdhfr C59R and S108N mutations are also found at moderate or high levels (4, 5).
Current techniques for SNP analysis are based on PCR-restriction fragment length polymorphism (RFLP) of selected mutations or hybridization. However, most such methods identify a small number of candidate SNPs (36). Mutations not directly involved in resistance but with compensatory or modulating effects on the overall parasite phenotype are often omitted. An approach based on DNA microarrays allows the parallel detection of multiple SNPs (8) but remains relatively expensive. An alternative technique is a post-PCR ligase detection reaction fluorescent microsphere assay (LDR-FMA) that enables the cost-effective evaluation of 28 SNPs (4). We have adapted this system to detect an additional 10 different pfmdr1 allelic variants in a study of key drug resistance mutations in P. falciparum field isolates from clinical studies conducted in PNG. We also examined associations between these mutations and treatment outcomes.
(The major findings of this study were presented at the 14th International Congress on Infectious Diseases, Miami, FL, 12 March 2010 [46a].)
MATERIALS AND METHODS
Field studies, Plasmodium falciparum isolates, and genomic DNA.
The present study utilized a subset of 402 blood samples available for the identification of Plasmodium isolates to the species level from a large-scale treatment trial with children aged 6 months to 5 years (mean age, 36 months) with uncompli- cated malaria (21) (Australian New Zealand Clinical Trials Registry ACTRN12605000550606). The study was conducted between 2005 and 2007 in the Madang and East Sepik Provinces of PNG. Participants were randomized into CQ-SP, artesunate (ART)-SP, piperaquine-dihydroartemisinin (PQ-DHA), and artemether-lumefantrine (AL) treatment groups. Children who had been treated with antimalarial drugs within the previous 14 days were excluded. The samples used in the present study were those collected at baseline, prior to treatment allocation. Full details of the trial protocol were reported previously (21).
The numbers of samples that were assayed for pfcrt, pfdhps, and pfdhfr genotypes from the CQ-SP, ART-SP, PQ-DHA, and AL treatment groups were 81, 86, 94, and 90, respectively (total of 351), and the numbers of samples that were assayed for pfmdr1 were 63, 65, 79, and 72, respectively (total of 279), due to a limited sample volume. Efficacy was assessed over 42 days by using World Health Organization definitions (47) with correction for reinfections by PCR genotyping (21), specifically, adequate clinical and parasitological response (ACPR), early treatment failure (ETF) (an inadequate parasitological response and/or worsening of clinical signs by day 3), late parasitological failure (LPF) (emergent parasitemia between days 4 and 42), or late clinical failure (LCF) (LPF associated with fever). Informed consent was obtained from parents or guardians before recruitment. Scientific and/or ethical approvals for the main study and present substudy were obtained from the Medical Research and Advisory Committee of the Ministry of Health of PNG, the University of Western Australia Human Research Ethics Committee, and the University Hospitals Case Medical Center.
Laboratory-adapted P. falciparum strains, including 3D7, Dd2, K1, 7G8, and HB3, were provided by the Malaria Research and Reference Reagent Resource Center (MR4), American Type Culture Collection. DNA was extracted from 200 μl whole blood (field samples) or hemolysate (laboratory-adapted strains) using the QIAmp 96 DNA blood kit or the DNeasy blood and tissue kit (Qiagen, CA) according to the manufacturer's protocol.
PCR amplification of Plasmodium species rRNA genes and Plasmodium falciparum resistance-associated sequences.
The detection of Plasmodium species was done by the amplification of small-subunit (SSU) rRNA genes by a modified multiplex LDR-FMA (26). All PCRs (25-μl mixtures) were performed by using a Peltier PTC-225 thermal cycler (MJ Research, MA), and the mixture consisted of 3 μl genomic DNA in a master mix containing 3 pmol appropriate upstream and downstream primers. The amplification of target sequences for P. falciparum dhps, dhfr, pfcrt (4), and pfmdr1 was achieved by using the oligonucleotide primers and conditions described in Table 1. Target sequences in the 3′ and 5′ fragments of the pfmdr1 gene encode alleles at codons 86 and 184 and codons 1034, 1042, and 1246, respectively. DNA samples from laboratory-adapted P. falciparum strains HB3, Dd2, K1, 3D7, and 7G8 were included as haplotype controls for pfmdr1 polymorphisms.
TABLE 1.
PCR oligonucleotide primer sequences and thermocycling conditions for target sequences in pfcrt, pfdhps, pfdhfr, and pfmdr1g
| Gene fragment | PCR primer sequence | Thermocycling conditionsf |
|---|---|---|
| pfcrta | 5′-TAATACGACTCACTATAGGGCCGTTA-3′ | 35 cycles of 95°C for 30 s, 56°C for 30 s, 60°C for 1 min |
| 5′-ATTAACCCTCACTAAAGGGACGGATG-3′ | ||
| pfdhfrb | 5′-TAACTACACATTTAGAGGTCTA-3′ | 35 cycles of 95°C for 30 s, 56°C for 30 s, 72°C for 1 min |
| 5′-GTTGTATTGTTACTAGTATATAC-3′ | ||
| pfdhpsc | 5′-AATGATAAATGAAGGTGCTAGT-3′ | 35 cycles of 95°C for 30 s, 56°C for 30 s, 60°C for 1 min |
| 5′-ATGTAATTTTTGTTGTGTATTTA-3′ | ||
| pfmdr1 3′ fragmentd | 5′-TGTATGTGCTGTATTATCAG-3′ | 32 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s |
| 5′-CTTATTACATATGACACCACA-3′ | ||
| pfmdr1 5′ fragmente | 5′-TAGAAGATTATTTCTGTAATTTG-3′ | 40 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 1 min |
| 5′-CAATGTTGCATCTTCTCTTCCA-3′ |
pfcrt for SNPs at codons 72 to 76.
pfdhfr fragment for SNPs at codons 51, 59, 108, and 164.
pfdhps gene fragment for SNPs at codons 540, 581, and 613.
pfmdr1 3′ fragment for SNPs at codons 86 and 184.
pfmdr1 5′ fragment for SNPs at codons 1034, 1042, and 1246.
PCR programs were preceded by an initial denaturation step at 95°C for 2 min.
Conditions for pfdhfr and pfcrt were optimized to obviate the need for nested reactions.
To evaluate the amplification efficiency, 5-μl PCR products were mixed with loading dye buffer and loaded onto 2% agarose I gels. For a 96-well gel, electrophoresis was performed at 290 V for 30 min. The gel was subsequently stained with SYBR gold (Molecular Probes, Eugene, OR) diluted 1:10,000 in 1× Tris-borate-EDTA (TBE) buffer, and DNA products were visualized with a Storm 860 imaging system coupled with Image-Quant (version 5.2) software (Molecular Dynamics, CA).
LDR-FMA evaluation of Plasmodium species and Plasmodium falciparum resistance-associated sequences.
Details of the LDR-FMA single-tube diagnosis of Plasmodium species and the simultaneous analysis of parasite drug resistance-associated SNPs in dhfr, dhps, and pfcrt were described previously (4, 26). The technique was adapted to include 10 different pfmdr1 allelic variants reported previously to confer resistance to CQ (16, 17), amino-alcohols, and quinine (6, 33, 37).
Following PCR amplification of the target sequences carrying drug resistance-associated pfmdr1 SNPs, the products were subjected to a ligase detection reaction (LDR) in which allele-specific upstream primers ligate to conserved-sequence downstream primers. The LDR upstream allele-specific primers have unique 5′ TAG sequences of 24 nucleotides that enable subsequent SNP identification. Downstream conserved-sequence oligonucleotides were 5′ phosphorylated and 3′ biotinylated (Table 2). LDRs were performed with a solution containing 10 nM each LDR primer, 1 μl PCR product, and 2 units of Taq DNA ligase (New England Biolabs, MA). For pfmdr1 region 1, reaction mixtures were initially heated at 95°C for 1 min, followed by 32 cycles of denaturation at 95°C for 15 s and annealing/ligation at 60°C for 2 min. Similar thermocycling conditions were used for LDR of pfmdr1 region 2, apart from the final step (65°C for 2 min).
TABLE 2.
Ligase detection reaction with tagged allele-specific upstream primers ligated to conserved sequence downstream primers
| Gene | LDR primerc | Sequencea | FlexMAP microsphere set IDb |
|---|---|---|---|
| pfmdr1 (region 1) | 86N | 5′-tacactttctttctttctttctttTTGGTGTAATATTAAAGAACATGA-3′ | 10 |
| 86Y | 5′-atcatacatacatacaaatctacaTTGGTGTAATATTAAAGAACATGT-3′ | 12 | |
| Com 86 | 5′-phosphate-ATTTAGGTGATGATATTAATCCTA-biotin-3′ | ||
| 184Y | 5′-tcaaaatctcaaatactcaaatcaGCCAGTTCCTTTTTAGGTTTATA-3′ | 18 | |
| 184F | 5′-ctacaaacaaacaaacattatcaaGCCAGTTCCTTTTTAGGTTTATT-3′ | 28 | |
| Com 184 | 5′-phosphate-TATTTGGTCATTAATAAAAAATGCA-biotin-3′ | ||
| pfmdr1 (region 2) | 1034S | 5′-ttacctttatacctttctttttacATGCAGCTTTATGGGGATTCA-3′ | 30 |
| 1034C | 5′-caatttcatcattcattcatttcaATGCAGCTTTATGGGGATTCT-3′ | 35 | |
| Com 1034 | 5′-phosphate-GTCAAAGCGCTCAATTATTTATT-biotin-3′ | ||
| 1042N | 5′-cttttcatcttttcatctttcaatCCAAACCAATAGGCAAAACTATT-3′ | 37 | |
| 1042D | 5′-ctttttcaatcactttcaattcatCAAACCAATAGGCAAAACTATC-3′ | 54 | |
| Com 1042 | 5′-phosphate-AATAAATAATTGAGCGCTTTGAC-biotin-3′ | ||
| 1246D | 5′-tcatcaatcaatctttttcactttAATATATGTGATTATAACTTAAGAG-3′ | 59 | |
| 1246Y | 5′-tcataatctcaacaatctttctttTAATATATGTGATTATAACTTAAGAT-3′ | 68 | |
| Com 1246 | 5′-phosphate-ATCTTAGAAACTTATTTTCAATAG-biotin-3′ |
Nucleotides in lowercase type represent TAG sequences added to the 5′ ends of each allele-specific LDR primer.
ID, identification. Luminex microsphere sets are synthesized to exhibit unique fluorescence. Each microsphere set is coupled to different anti-TAG sequences that are complementary to allele-specific TAG sequences.
Com, common (conserved) sequence primer positioned immediately downstream from the allele-specific primer.
The second stage of the reaction initiates the specific classification of the LDR products, where hybridization occurs between anti-TAG oligonucleotides probes that are bound to fluorescent microspheres (Luminex Corporation, TX) specific for the various TAG sequences at the 5′ ends of the LDR products. This required combining 2.5 μl of LDR products from both pfmdr1 regions with 60 μl of prewarmed hybridization solution containing 250 Luminex FlexMAP microspheres from each allelic set (n = 10). The reaction mixture was heated to 95°C for 90 s and incubated at 37°C for 35 to 45 min. Reporter labeling of the conjugate followed, with the addition of streptavidin-R-phycoerythrin dye (Molecular Probes, OR) diluted 1:50 in tetramethylammonium chloride (TMAC), as it binds to the 3′ biotin on the conserved-sequence primers, at 37°C for 35 to 45 min in Costar 6511 M 96-well plates (Corning Inc., Corning, NY). SNP-specific LDR products with microsphere-labeled anti-TAG probes were detected by dual-fluorescence flow cytometry with a Bio-Plex array reader (Bio-Rad Laboratories, CA) set at 37°C, with reporter signals collected into allele-specific classification bins via Bio-Plex Manager 3.0 software.
Data analysis.
Statistical analysis was performed by using GraphPad PRISM version 4.0 (GraphPad Software, CA). Fluorescence signals from the field samples were classified as being positive or negative for drug susceptibility markers according to thresholds determined by standardized procedures. Fluorescence signals were first normalized to a mean of 10,000 arbitrary units and a standard deviation (SD) of 1,000 arbitrary units for each codon by subtracting the calculated mean from every signal within the corresponding codon and then multiplying by 1,000/codon-specific SD and finally adding 10,000. The same procedure was applied to fluorescence signals from cultured-adapted strains with known genotypes, thus providing controls within each SNP assay. Once adjusted, codon-specific cut points that applied to all control strains were derived with a value that predicted the highest number of true positives as a conservative cut point for distinguishing positive signals from background fluorescence.
A cut point of >9,600 had 97.5%, 98.8%, and 98.6% accuracies for predicting true positive alleles for codons 540, 581, and 613, respectively, in the pfdhps gene in control strains. The cut point of >9,600 also applied to codons 1042 and 1246 in the pfmdr1 gene, while a cut point of >9,800 accurately predicted known alleles at codon 86 in the pfmdr1 gene; at codons 51, 59, 108, and 164 in the pfdhfr gene; and in the CVMNK, SVMNT, and CVIET pfcrt haplotypes. A threshold of >10,000 was applied for pfmdr1 codons 184 and 1034. By reversing the normalization process, the cut points were made specific to each drug resistance marker. A similar approach that uses polar coordinates was also used previously to determine thresholds for the LDR-FMA system (9).
Mixed-strain infections can be identified when fluorescence signals from both alleles (i.e., wild type and mutated) from the same codon occur above calculated cut points. Previous experiments have shown that strain-specific allele fluorescence signals are in direct proportion to the ratio of the parasite strain densities within the sample (4). We have also assayed day 28 and day 42 posttreatment blood samples from patients from the clinical trial (21) that were parasite negative by both microscopy and PCR and found very low fluorescence signals (<200). These observations indicate that multiple P. falciparum strains and non-P. falciparum DNA such as that from the human host do not interfere with SNP detection by LDR-FMA. While haplotypes were assigned based on the dominant allele signals at each locus, they have masked the presence of a minor clone in the case of a multiple-strain infection.
Associations between parasite mutations and measures of treatment outcome were assessed by using Fisher's exact test or analysis of variance (ANOVA) with Bonferroni post hoc adjustment for multiple comparisons (SPSS v16.0).
RESULTS
Identification of drug resistance alleles.
The specificities of the pfcrt, pfdhfr, and pfdhps assays were reported previously, with 100% concordance with haplotypes from a variety of laboratory-adapted strains (4). The specificities of the pfmdr1 LDR-FMA probes are shown in Table 3. The results for the pfmdr1-specific LDR-FMAs show that allele-specific background median fluorescence intensity (MFI) signals ranged from 168 to 6,464 and that positive allele-specific signals ranged from 1,421 to 22,008. Strain-specific LDR-FMA results were 100% concordant with previously reported haplotypes (29).
TABLE 3.
LDR-FMA evaluation of pfmdr1 SNPs in laboratory-adapted Plasmodium falciparum strainsb
| Strain | MFI for pfmdr1 codona: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 86 |
184 |
1034 |
1042 |
1246 |
||||||
| Y | N | Y | F | S | C | D | N | D | Y | |
| 3D7 | 2,657 | 15,306 | 17,956 | 6,464 | 21,903 | 5,568 | 225 | 21,182 | 5,243 | 327 |
| 7G8 | 1,724 | 11,351 | 1,869 | 21,092 | 1,475 | 16,556 | 6,022 | 436 | 746 | 1,421 |
| Dd2 | 12,684 | 3,418 | 15,465 | 5,891 | 22,008 | 5,061 | 234 | 18,333 | 4,357 | 359 |
| HB3 | 555 | 6,381 | 530 | 10,036 | 17,881 | 1,555 | 8,238 | 271 | 2,686 | 168 |
| K1 | 19,971 | 3,482 | 17,393 | 6,055 | 21,501 | 4,519 | 199 | 18,265 | 3,476 | 543 |
pfmdr1 haplotypes for P. falciparum strains are NYSND for 3D7, NFCDY for 7G8, YYSND for Dd2, and NFSDD for HB3.
The fluorescence signal is expressed as median fluorescence intensity (MFI) units. Boldface type indicates positive allele-specific signals.
Identification to the species level and drug resistance genes of field isolates.
For 402 samples with microscopy-confirmed P. falciparum, the Plasmodium species LDR-FMA identified 28 patients with P. vivax coinfections and 13 with Plasmodium malariae coinfections. No Plasmodium ovale coinfections were found. The sensitivity and specificity of LDR-FMA compared to the reference RFLP method for the identification of P. falciparum isolates to the species level were 96.6% and 100%, respectively. If a sample is positive for P. falciparum by LDR-FMA, then the pfcrt, pfdhps, pfdhfr, and pfmdr1 genes should also be detectable. There were 304 samples for which identification to the species level and all four specific gene assays were performed by using LDR-FMA. For 241 (79%) samples, each of the pfcrt, pfdhps, pfdhfr, and pfmdr1 genes were identified. For discordant samples, the pfdhps and pfdhfr assays were more often PCR negative in the amplification of Plasmodium genome. The maximum median fluorescence intensity obtained for field isolates and thresholds used to determine the presence of an allele are shown in Table 4.
TABLE 4.
Fluorescence intensity detection thresholds and maxima for Plasmodium falciparum pfdhps, pfdhfr, pfcrt, and pfmdr1 in field samples
| Gene and allelea | Fluorescence intensity signal |
|
|---|---|---|
| Threshold | Maximum | |
| pfdhps | ||
| 540K | 1,213 | 20,400 |
| 540E | 9,774 | |
| 581A | 1,940 | 21,876 |
| 581S* | ||
| 613A | 2,374 | 21,403 |
| 613S* | ||
| pfdhfr | ||
| 51I* | 4,147 | |
| 51N | 23,429 | |
| 59R | 6,087 | 23,368 |
| 59C | 23,490 | |
| 108T* | 3,348 | |
| 108S | 16,258 | |
| 108N | 22,198 | |
| 164I | 3,914 | 19,695 |
| 164L* | ||
| pfcrt | ||
| CVIET* | 4,791 | |
| CVMNK | 20,657 | |
| SVMNT | 22,523 | |
| pfmdr1 | ||
| 86Y | 6,722 | 21,994 |
| 86N | 18,189 | |
| 184Y | 7,899 | 21,145 |
| 184F | 20,412 | |
| 1034S | 11,166 | 25,792 |
| 1034C* | ||
| 1042D | 2,836 | 10,973 |
| 1042N | 23,200 | |
| 1246D | 1,410 | 9,921 |
| 1246Y | 3,174 | |
Alleles not detected in the field isolates are indicated with an asterisk.
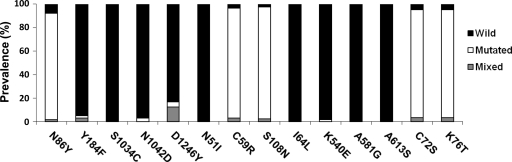
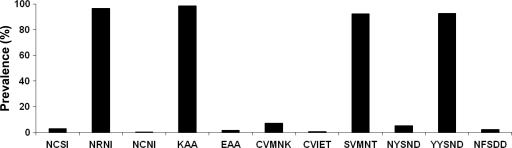
Overall, 9 mutant alleles were detected. These alleles were at pfmdr1 codons N86Y (91%), Y184F (2%), N1042D (2%), and D1246Y (4%); pfcrt codons C72S and K76T (both 92%); pfdhfr codons C59R (93%) and S108N (95%); and pfdhps codon K540E (1.5%) (Fig. 1). We found the pfcrt SVMNT haplotype to be at fixation in the samples (92%), with only 7% of P. falciparum strains retaining the CQ-sensitive CVMNK haplotype. Two children were infected with mixed strains carrying CVIET and SVMNT mutations. The majority of the P. falciparum isolates carried the CQ resistance-associated YYSND (93%) haplotype of the pfmdr1 gene (Fig. 2).
FIG. 1.
Prevalence of pfcrt, pfmdr1, pfdhfr, and pfdhps alleles in Plasmodium falciparum-infected individuals from the Madang and East Sepik Provinces, Papua New Guinea.
FIG. 2.
Frequency distributions of pfcrt, pfdhps, pfdhfr, and pfmdr1 alleles in Plasmodium falciparum-infected Papua New Guinean children. The pfdhfr NCSI, NRNI, and NCNI haplotypes denote codons 51, 59, 108, and 164. The pfdhps KAA and EAA haplotypes denote codons 540, 581, and 613. The pfcrt CVMNK, CVIET, and SVMNT haplotypes denote codons 72 to 76. The pfmdr1 NYSND, YYSND, and NFSDD haplotypes denote codons 86, 184, 1034, 1042, and 1246. The numbers of samples demonstrating complete haplotypes for each gene were 376, 254, 273, and 195, respectively.
One patient was infected with an isolate carrying a single pfdhfr S108N mutation. However, most children (97%) had parasites with double mutations in the NRNI haplotype corresponding to amino acids at codons 51, 59, 108, and 164. A few isolates (3%) retained the wild haplotype NCSI. No isolates carried the pfdhfr I164L allele. The pfdhps KAA haplotype, which harbors a single mutation at codon 613, was predominant (98%). Four isolates carried the pfdhps K540E mutation.
When the isolates with completely defined haplotypes were assessed for multiple-gene mutations, we found that 88% (100/113) of children were infected with P. falciparum carrying quintuple mutations across the pfcrt, pfdhfr, pfmdr1, and pfdhps genes that were characterized by the SVMNT, NRNI, YYSND, and KAA haplotypes, respectively. Four patients (one per treatment group) were infected with parasites carrying the 6-fold-mutated SVMNT, NRNI, YYSND, and EAA haplotypes. One of these patients was treated with PQ-DHA and had LPF, while the other three had ACPR. Table 5 details the number of P. falciparum isolates with multiple combinations of mutations across drug resistance genes.
TABLE 5.
Occurrence of Plasmodium falciparum isolates carrying multiple mutations across 4 genes associated with drug resistanced
| pfcrt haplotype | pfdhfr haplotype | No. of isolates with: |
|||
|---|---|---|---|---|---|
| Double gene mutation (pfcrt + pfdhfr only)a | Triple gene mutation (+pfmdr1) |
||||
| NYSND | YYSND | NFSDD | |||
| CVMNK | NCSI | 1 | 0 | 1 | 0 |
| NRNI | 14 | 1 | 1 | 0 | |
| NCNIa | 0 | 0 | 0 | 0 | |
| CVIET | NCSI | 0 | 0 | 0 | 0 |
| NRNI | 1b | 0 | 1b | 0 | |
| NCNIa | |||||
| SVMNT | NCSI | 2 | 0 | 2 | 1 |
| NRNI | 114 | 3 | 100c | 3 | |
| NCNI | 1 | 0 | 0 | 0 | |
Assayed only for pfcrt and pfdhfr due to an inadequate sample volume.
Sample with P. falciparum carrying both the CVIET and SVMNT mutations.
Four of these isolates were also carrying the K540E mutation, while all other P. falciparum samples carried the 540K allele of the pfdhps gene.
Wild-type alleles of pfcrt, pfdhfr, and pfmdr1 are those in boldface type. Allele mutations are underlined. the pfcrt CVMNK, CVIET, and SVMNT haplotypes correspond to codons 72 to 76. The pfdhfr NCSI, NRNI, and NCNI haplotypes correspond to codons 51, 59, 108, and 164. The pfmdr1 NYSND, YYSND, and NFSDD haplotypes correspond to codons 86, 184, 1034, 1042, and 1246.
Parasite drug resistance mutations and treatment outcomes.
In evaluating the association of parasite genes with treatment outcome, we first compared the numbers of parasite mutations in the ACPR, ETF, LPF, and LCF groups for a total of 351 cases. Overall, there were 308 cases of ACPR, 3 of ETF, 3 of LPF, and 37 of LCF. There were 4 mutations in 41.6% of samples, with 0.6%, 6.8%, 16%, 27.1%, 7.1%, and 0.9% carrying no mutation and single, double, triple, quintuple, and sextuple mutations, respectively. There was no significant difference between the numbers of mutations in the ACPR group (mean, 3.3 [95% confidence interval {CI}, 3.1 to 3.4]) and those in the ETF, LCF, and LPF groups (means, 2.3 [95% CI, 0 to 6.1], 3.0 [95% CI, 0.5 to 5.5], and 3.4 [95% CI, 3.0 to 3.8], respectively; P > 0.05 in each case). For the purposes of subsequent analyses, we combined the ETF, LCF, and LPF groups into a single treatment failure group.
The pfdhfr N51I, C59R, and S108N and pfdhps K540E polymorphisms did not predict treatment failure for 81 children given CQ-SP and for 86 children given ART-SP (P > 0.05). In analyses of pfmdr1 N86Y, Y184F, and N1042D and pfcrt K76T mutations, the pfmdr1 D1246Y mutation was a significant predictor of treatment failure for 79 children treated with PQ-DHA therapy; 4 of 10 (40%) of such children carried D1246Y strains, while only 3 of 69 (4%) who responded to treatment harbored parasites with this mutation (P = 0.004). For none of the other three groups was there a significant association between the presence of the D1246Y mutation and treatment failure (P ≥ 0.10). Similar analyses did not reveal any associations between haplotypes of pfcrt, pfdhfr, and pfdhps and treatment failure (data not shown).
DISCUSSION
The consequences of past antimalarial treatment policies in PNG were evident in our data. Only 7% of the parasites carried the wild-type pfcrt CVMNK mutation (codons 72 to 76) associated with a CQ-sensitive phenotype. This high prevalence of the resistant K76T polymorphism concurred with data from studies conducted between 2000 and 2005 in Madang and East Sepik Provinces (5, 28, 41). The SVMNT haplotype (codons 72 to 76) is at fixation in the PNG parasite population, with an increase in prevalence from 83% from the early 1990s to 92.3% in 2003 to 2005 (25, 27, 42). Another CQ-resistant haplotype, CVIET (codons 72 to 76), found commonly in Africa and Southeast Asia (22, 23, 32, 48), was detected in two isolates as a mixed infection with the SVMNT strain, confirming its recent emergence in PNG (10). Differences between the pfcrt intronic microsatellite diversity haplotypes of the SVMNT and CVIET parasites from the province adjacent to Madang suggest that they have been imported (10), presumably by economic migrants from nearby Asian countries (27-29).
Although assessed with limited numbers of isolates (n = 195) due to an inadequate sample volume, the CQ-sensitive pfmdr1 NYSND mutation (codons 86, 184, 1034, 1042, and 1246) was present in 5.1% of isolates. Most others carried the N86Y polymorphism with the YYSND haplotype (codons 86, 184, 1034, 1042, and 1246), associated with CQ resistance. Our results reflect an increase in the frequency of this haplotype since the mid-1990s (28, 29). We also observed four isolates (2%) that carried the Y184F and N1042D double point mutations, confirming the recent emergence of these pfmdr1 polymorphisms in PNG (25). Although none of the four patients with parasites carrying the pfmdr1 NFSDD haplotype (codons 86, 184, 1034, 1042, and 1246) were assigned to the AL treatment group in our study, the presence of these SNPs with the wild-type 86N allele was significantly associated with AL treatment failure in a previously reported Nigerian pediatric study (19). Since the widespread introduction of this treatment in PNG is imminent, the monitoring of changes in the NFSDD pfmdr1 haplotype should be a high priority.
We found that the signal intensities for the pfmdr1 codon 1246 alleles were relatively weak even in the controls. This was unlikely to reflect poor PCR amplification, as signal intensities for codons 1034 and 1042 derived from the same region were unaffected. Poor hybridization with the LDR probe is possible. The D1246Y allele, present in 12% of our isolates in association with the wild-type 1246D allele, has not been previously detected in PNG (25, 29). A recent African study observed an increased prevalence of the pfmdr1 N86Y and D1246Y alleles post-AQ exposure (31), but sporadic mutations at codon 1246 have also been documented in Thailand (39), and it may also have been imported into PNG (28). Both the D1246Y and N86Y mutations have been associated with diminished in vitro sensitivity to CQ and AQ (35, 37).
We did not detect the pfmdr1 NFCDD, NFSND, and YFSND haplotypes (codons 86, 184, 1034, 1042, and 1246) found previously in Thailand (35) or the NFCDY and NFSDY haplotypes found in Brazil and Colombia (29). Previously reported in vitro studies by Sa et al. showed that 7G8 and hybrid crosses with its pfmdr1 haplotype (NFCDY) resulted in higher 50% growth-inhibitory concentrations (IC50s) for AQ and its metabolite (40). However, the “CDY” (S1034C, N1042D, and D1246Y) mutation of the pfmdr1 gene has not been detected in PNG. Collective polymorphisms in the 3′ coding region (N86Y and Y184F) that are found in South America, Africa, and Asia can confer resistance to quinine, mefloquine, and halofantrine and modulate parasite sensitivity to artemisinin drugs (37). Their geographic distribution suggests that drug pressure is required for their maintenance and spread (16), as may occur with the future use of artemisinin combination therapies (ACTs) in PNG. However, the frequency of wild-type 86N may increase with the adoption of ACT, as has been reported by previous studies from Africa (12, 20).
We found that 97% of the isolates reported here carried both the C59R and S108N SNPs, which constitute the major pfdhfr resistance alleles. Since the prevalence of this double mutant in the East Sepik Province was 72% to 91% in 2001 to 2003 (4, 30), this suggests that there has been continuing selection pressure for alleles conferring pyrimethamine resistance in northern PNG. Carnevale et al. noted previously that a number of isolates from this area carried a single mutation at codon 108 with the NCNI (n = 13) and NCTI (n = 1) haplotypes (4). In the present study, only one isolate had a single pfdhfr mutation with the NCNI haplotype (codons 51, 59, 108, and 164), and, consistent with data reported previously by Marfurt et al. (25), we did not detect the S108T mutant that was previously reported to be present in PNG in 1996 (38).
As we evaluated pfdhps polymorphisms at codons 540, 581, and 613, we detected only wild-type alleles at loci at codons 581 and 613. Four isolates were found to carry the K540E variant first detected in studies in East Sepik and Simbu Provinces in 2002 to 2004 (25, 30). Most of the isolates studied here (98.4%) carried the KAA haplotype, and 1.6% carried the EAA haplotype. We were unable to evaluate SNPs at codons 436 and 437, as the respective LDR probes were unavailable at the time of study.
The lack of an association between well-known mutations (pfcrt K76T, pfdhfr S108N, pfdhfr C59R, and pfmdr1 N86Y mutations) and treatment failure is due largely to fixation levels of these mutations in the Madang Province. Although carriers of multiple mutations were observed more often for treatment failure cases, this did not reach statistical significance. We had limited statistical power because of the relatively small number of treatment failures (43 of 351, or 12.3%), but the high baseline number of mutations per isolate and the emergence of immunity in this age group in a high-transmission setting may also have attenuated the relationship. Although the pfdhfr K540E mutation was shown previously to predict SP treatment failure (2, 43), there was a low prevalence of this allele in our isolates, and it was unrelated to outcome. Our data are consistent with those reported previously by Marfurt et al., who showed that the pfcrt K76T mutation and the pfdhfr C59R and S108N mutations did not predict treatment failure after CQ-SP treatment in a region northwest of Madang (25), reflecting a fixation of these mutations. Interestingly, the association of the pfmdr1 D1246Y allele with PQ-DHA treatment failure highlights the increasingly recognized role of this transporter gene in modulating resistance to antimalarial drugs from different classes (6, 33, 37).
The LDR-FMA system is a novel approach to determine the presence or absence of resistance-associated mutations in P. falciparum. In determining signal positivity, we tested various statistical models, but in the absence of valid malaria-negative samples, we found that bimodal and gamma distributions were inappropriate. The subsequent use of allelic signal ratios within each codon appeared robust when applied to controls with known haplotypes, especially in cases where strong positive signals were accompanied by high background levels. However, this approach was based on the assumption of single-strain infections, which can be circumvented only by establishing a specific threshold. We found that through normalization of the data, the application of gene-specific cut points from the control strains, and reversing the process to obtain codon-specific cutoffs, the prediction of positivity was relatively accurate. Unless a better resolution of positive from negative signals is possible, we suggest that this method or related approaches (9) be adopted for future LDR-FMA studies.
The multiplex LDR-FMA technique proved to be a cost-effective tool for epidemiological studies. Taking into consideration the reagents required for each step (DNA extraction, plasticware, PCR and LDR primers, and Fleximap microspheres), the total cost per sample for the analysis of 28 SNPs was US$4.14 ($0.15 per SNP), with over 60% of the cost attributable to DNA extraction. This amounts to less than half of the cost of a recently described microarray SNP detection approach (8). Compared to PCR-based approaches that have been used to evaluate polymorphisms in the pfdhfr, pfdhps, pfcrt, and pfmdr1 genes, most of which involve DNA probe hybridization and post-PCR RFLP methods (1, 5, 13, 14, 46), the LDR-FMA system offers greater efficiency and objectivity when analyzing multiple mutations. Given these considerations and the increase in the frequency of key parasite mutations in resource-poor countries where malaria is endemic, such as PNG, the LDR-FMA SNP assay represents an excellent tool for molecular surveillance.
The present LDR-FMA platform enables the assessment of 18 SNPs in the pfcrt, pfdhps, and pfdhfr genes in a single-tube multiplex assay, a task that is beyond the capabilities of existing real-time PCR and RFLP methodologies. The inclusion of 10 additional SNPs in the pfmdr1 gene can be easily accommodated through a modified microsphere set selection with all 28 SNPs having their own unique classification codes. Monitoring of the spread of resistance to CQ, sulfadoxine, and pyrimethamine using this or an equivalent methodology is a high priority in countries, such as PNG, where these drugs are still used. In addition, there will be an increasing need for the monitoring of pfmdr1 SNPs, since polymorphisms encompass diverse effects on parasite sensitivity to a range of antimalarial drugs, including those used in ACT (37).
Acknowledgments
We thank patients and staff at Alexishafen Health Centre and the staff at the PNG Institute of Medical Research for their kind cooperation and logistic assistance; Laurie Gray, Jeana DaRe, and Eric Carnevale from Case Western Reserve University for laboratory and technical support during assay development; Rajeev Mehlotra for technical advice and critical review of the manuscript; and Martin Firth, Wendy Davis, and Shih Ching Fu from the University of Western Australia for statistical advice and mathematical modeling.
This project was supported by grants from the WHO Western Pacific Region, the National Health and Medical Research Council of Australia (grant 353663), and the U.S. National Institutes of Health (grants AI52312 and TW007872). R.P.M.W. was supported by a University of Western Australia graduate research travel award, and T.M.E.D. is the recipient of a National Health and Medical Research Council of Australia practitioner fellowship.
Footnotes
Published ahead of print on 15 November 2010.
REFERENCES
- 1.Alifrangis, M., et al. 2005. A simple, high-throughput method to detect Plasmodium falciparum single nucleotide polymorphisms in the dihydrofolate reductase, dihydropteroate synthase, and P. falciparum chloroquine resistance transporter genes using polymerase chain reaction- and enzyme-linked immunosorbent assay-based technology. Am. J. Trop. Med. Hyg. 72:155-162. [PubMed] [Google Scholar]
- 2.Bacon, D. J., et al. 2009. Effects of point mutations in Plasmodium falciparum dihydrofolate reductase and dihydropterate synthase genes on clinical outcomes and in vitro susceptibility to sulfadoxine and pyrimethamine. PLoS One 4:e6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basco, L. K., M. Ndounga, V. F. Ngane, and G. Soula. 2002. Molecular epidemiology of malaria in Cameroon. XIV. Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene sequences of isolates before and after chloroquine treatment. Am. J. Trop. Med. Hyg. 67:392-395. [DOI] [PubMed] [Google Scholar]
- 4.Carnevale, E. P., et al. 2007. A multiplex ligase detection reaction-fluorescent microsphere assay for simultaneous detection of single nucleotide polymorphisms associated with Plasmodium falciparum drug resistance. J. Clin. Microbiol. 45:752-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, G. J., et al. 2004. Molecular analysis of Plasmodium falciparum from drug treatment failure patients in Papua New Guinea. Am. J. Trop. Med. Hyg. 70:251-255. [PubMed] [Google Scholar]
- 6.Cowman, A. F., D. Galatis, and K. J. Thompson. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. U. S. A. 91:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowman, A. F., M. J. Morry, B. A. Biggs, G. A. Cross, and S. J. Foote. 1988. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reducatase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 85:9109-9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crameri, A., et al. 2007. Rapid microarray-based method for monitoring of all currently known single-nucleotide polymorphisms associated with parasite resistance to antimalarial drugs. J. Clin. Microbiol. 45:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DaRe, J. T., D. P. Kouri, P. A. Zimmerman, and P. J. Thomas. 2010. Differentiating Plasmodium falciparum alleles by transforming Cartesian X,Y data to polar coordinates. BMC Genet. 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DaRe, J. T., et al. 2007. Microsatellite polymorphism within pfcrt provides evidence of continuing evolution of chloroquine-resistant alleles in Papua New Guinea. Malar. J. 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlow, B., H. Vrbova, J. Stace, P. Heywood, and M. P. Alpers. 1980. Fansidar-resistant falciparum malaria in Papua New Guinea. Lancet ii:1243. [DOI] [PubMed] [Google Scholar]
- 12.Dokomajilar, C., S. L. Nsobya, B. Greenhouse, P. J. Rosenthal, and G. Dorsey. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 50:1893-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duraisingh, M. T., J. Curtis, and D. C. Warhurst. 1998. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp. Parasitol. 89:1-8. [DOI] [PubMed] [Google Scholar]
- 14.Farcas, G. A., R. Soeller, K. Zhong, A. Zahirieh, and K. C. Kain. 2006. Real-time polymerase chain reaction assay for the rapid detection and characterization of chloroquine-resistant Plasmodium falciparum malaria in returned travelers. Clin. Infect. Dis. 42:622-627. [DOI] [PubMed] [Google Scholar]
- 15.Fidock, D. A., et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein pfcrt and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foote, S. J., et al. 1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255-258. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Saladin, E., et al. 1999. Plasmodium falciparum mdr1 mutations and in vivo chloroquine resistance in Indonesia. Am. J. Trop. Med. Hyg. 61:240-244. [DOI] [PubMed] [Google Scholar]
- 18.Grimmond, T. R., K. O. Donovan, and I. D. Riley. 1976. Chloroquine resistant malaria in Papua New Guinea. P. N. G. Med. J. 19:184-185. [PubMed] [Google Scholar]
- 19.Happi, C. T., et al. 2008. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 53:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphreys, G. S., et al. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 51:991-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karunajeewa, H. A., et al. 2008. A trial of combination antimalarial therapies in children from Papua New Guinea. N. Engl. J. Med. 359:2545-2557. [DOI] [PubMed] [Google Scholar]
- 22.Keen, J., et al. 2007. Real-time PCR assay for rapid detection and analysis of pfcrt haplotypes of chloroquine-resistant Plasmodium falciparum isolates from India. J. Clin. Microbiol. 45:2889-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, P., et al. 2003. pfcrt polymorphism and chloroquine resistance in Plasmodium falciparum strains isolated in Cambodia. Antimicrob. Agents Chemother. 47:87-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marfurt, J., et al. 2007. Low efficacy of amodiaquine or chloroquine plus sulfadoxine-pyrimethamine against Plasmodium falciparum and P. vivax malaria in Papua New Guinea. Am. J. Trop. Med. Hyg. 77:947-954. [PubMed] [Google Scholar]
- 25.Marfurt, J., et al. 2008. The usefulness of twenty-four molecular markers in predicting treatment outcome with combination therapy of amodiaquine plus sulphadoxine-pyrimethamine against falciparum malaria in Papua New Guinea. Malar. J. 7:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNamara, D. T., et al. 2006. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am. J. Trop. Med. Hyg. 74:413-421. [PMC free article] [PubMed] [Google Scholar]
- 27.Mehlotra, R. K., et al. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. U. S. A. 98:12689-12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehlotra, R. K., et al. 2005. Insight into the early spread of chloroquine-resistant Plasmodium falciparum infections in Papua New Guinea. J. Infect. Dis. 192:2174-2179. [DOI] [PubMed] [Google Scholar]
- 29.Mehlotra, R. K., et al. 2008. Disconcordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 52:2212-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mita, T., et al. 2006. Rapid selection of dhfr mutant allele in Plasmodium falciparum isolates after the introduction of sulfadoxine/pyrimethamine in combination with 4-aminoquinolines in Papua New Guinea. Infect. Genet. Evol. 6:447-452. [DOI] [PubMed] [Google Scholar]
- 31.Nawaz, F., S. L. Nsobya, M. Kiggundu, M. Joloba, and P. J. Rosenthal. 2009. Selection of parasites with diminished drug susceptibility by amodiaquine-containing antimalarial regimens in Uganda. J. Infect. Dis. 200:1650-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nsobya, S. L., C. Dokomajilar, M. Joloba, G. Dorsey, and P. J. Rosenthal. 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob. Agents Chemother. 51:3023-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peel, S. A., P. Bright, B. Yount, J. Handy, and R. S. Baric. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648-658. [DOI] [PubMed] [Google Scholar]
- 34.Peterson, D. S., D. Walliker, and T. E. Wellems. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 85:9114-9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickard, A. L., et al. 2003. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob. Agents Chemother. 47:2418-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranford-Cartwright, L. C., K. L. Johnston, A. M. Abdel-Muhsin, B. K. Khan, and H. A. Babiker. 2002. Critical comparison of molecular genotyping methods for detection of drug-resistant Plasmodium falciparum. Trans. R. Soc. Trop. Med. Hyg. 96:568-572. [DOI] [PubMed] [Google Scholar]
- 37.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 38.Reeder, J., et al. 1996. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am. J. Trop. Med. Hyg. 55:209-213. [DOI] [PubMed] [Google Scholar]
- 39.Rungsihirunrat, K., W. Chaijareonkul, A. Seugorn, K. Na-Bangchang, and S. Thaithong. 2009. Association between chloroquine resistance phenotypes and point mutations in pfcrt and pfmdr1 in Plasmodium falciparum isolates from Thailand. Acta Trop. 109:37-40. [DOI] [PubMed] [Google Scholar]
- 40.Sa, J. M., et al. 2009. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. U. S. A. 106:18883-18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoepflin, S., et al. 2008. Heterogeneous distribution of Plasmodium falciparum drug resistance haplotypes in subsets of the host population. Malar. J. 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoepflin, S., et al. 2009. Comparison of Plasmodium falciparum allelic frequency distribution in different endemic settings by high-resolution genotyping. Malar. J. 8:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talisuna, A. O., et al. 2004. Efficacy of sulphadoxine-pyrimethamine alone or combined with amodiaquine or chloroquine for the treatment of uncomplicated falciparum malaria in Ugandan children. Trop. Med. Int. Health 9:222-229. [DOI] [PubMed] [Google Scholar]
- 44.Triglia, T., and A. F. Cowman. 1999. The mechanism of resistance to sulfa drugs in Plasmodium falciparum. Drug Resist. 2:15-19. [DOI] [PubMed] [Google Scholar]
- 45.Triglia, T., and A. F. Cowman. 1994. Primary structure and expression of dihydropteroate synthetase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 91:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veiga, M. I., P. E. Ferreira, A. Bjorkman, and J. P. Gil. 2006. Multiplex PCR-RFLP methods for pfcrt, pfmdr1 and pfdhfr mutations in Plasmodium falciparum. Mol. Cell. Probes 20:100-104. [DOI] [PubMed] [Google Scholar]
- 46a.Wong, R. P. M., H. Karunajeewa, I. Mueller, P. Siba, E. P. Carnevale, P. A. Zimmerman, and T. M. E. Davis. 2010. Novel molecular detection of drug resistance markers in Plasmodium falciparum from Papua New Guinean children with uncomplicated malaria, abstr. 72.005. Proceedings of the 14th International Congress on Infectious Diseases. International Society for Infectious Diseases, Brookline, MA.
- 47.World Health Organization. 2003. Assessment and monitoring of antimalarial drug efficacy for the treatment of uncomplicated falciparum malaria, vol. WHO/HTM/RBM/2003.50. World Health Organization, Geneva, Switzerland.
- 48.Yang, Z., et al. 2007. Molecular analysis of chloroquine resistance in Plasmodium falciparum in Yunnan Province, China. Trop. Med. Int. Health 12:1051-1060. [DOI] [PubMed] [Google Scholar]