Abstract
Many Gram-positive pathogens link the expression of virulence genes to the presence of carbon source substrates using overlapping pathways for global control of carbon catabolite regulation. However, how these pathways are integrated to control the behavior of the transcriptome in time- and compartment-specific patterns is typically not well understood. In the present study, global transcriptome profiling was used to determine the extent to which glucose alters gene expression in Streptococcus pyogenes (group A streptococcus) and the contributions of the CcpA and LacD.1 catabolite control pathways to the regulation of this response in vitro. This analysis revealed that the expression of as many as 15% of the genes examined was regulated and that CcpA and LacD.1 together contribute to the regulation of 60% of this subset. However, numerous patterns were observed, including both CcpA- and LacD.1-specific and independent regulation, coregulation, and regulation of genes by these pathways independently of glucose. In addition, CcpA and LacD.1 had antagonistic effects on most coregulated genes. To resolve the roles of these regulators during infection, the expression of selected transcripts representative of different regulatory patterns was examined in a murine model of soft tissue infection. This revealed distinct patterns of misregulation with respect to time in CcpA− versus LacD.1− mutants. Taken together, these data support an important role for carbohydrate in the regulation of the transcriptome in tissue and suggest that the CcpA and LacD.1 pathways are organized to function at different times during the course of an infection.
Carbon catabolite control is a conserved regulatory phenomenon that couples the presence of preferred growth substrates to the behavior of the transcriptome. An important function of this response is to match the cell's metabolic potential to those substrates that can be used most efficiently. Typically, the response includes the up- and/or downregulation of a large number of different genes, including those that are not necessarily involved in metabolic activities. For pathogenic bacteria, the latter genes can include multiple virulence factors, suggesting that substrate availability can be used as a cue to distinguish between specific stages of the infectious process. In the low-G+C Gram-positive bacteria, a major regulator of carbon catabolite control is the transcription factor CcpA, which regulates gene expression mainly in response to glucose and other carbohydrates (for reviews, see references 15 and 41). Numerous studies have shown that CcpA coordinates virulence gene regulation for a broad range of different pathogens (21, 22, 25, 36, 38, 45). However, CcpA may regulate only a subset of a subset of the virulence genes subject to carbon catabolite control (4, 21, 31). This suggests that pathogenesis requires the integration of multiple catabolite-sensing pathways.
Carbon catabolite control is emerging as a major mechanism for the global regulation of virulence genes in Streptococcus pyogenes (group A streptococcus). This human pathogen can cause numerous diseases that range from largely superficial infections of the skin and mucous membranes (impetigo, pharyngitis) to highly invasive and life-threatening diseases (necrotizing fasciitis), as well as serious postinfection sequelae (rheumatic fever, glomerulonephritis [reviewed in reference 13]). Global regulators of amino acid catabolism, including RelA and CodY, have been implicated in the regulation of virulence genes (30, 40). Also, the RopB (Rgg) transcription factor has been shown to coordinate amino acid catabolism with the expression of virulence factors (10). More recently, glucose levels have been shown to regulate the expression of numerous virulence genes, including those encoding the CAMP factor, streptolysin S, and the secreted SpeB cysteine protease (22, 28). Along with this, comparison of CcpA− mutant and wild-type bacteria during growth in vitro has suggested that CcpA regulates numerous virulence genes (22, 25, 38, 39). Taken together, these data suggest that carbohydrate availability is a major cue used by S. pyogenes for the regulation of virulence and that CcpA is a central regulator of this response. However, the contribution that CcpA makes to global regulation of virulence in response to glucose, and the extent to which other regulators of carbohydrate utilization may contribute to this response, has not been clearly established.
There is evidence to suggest that other carbohydrate regulators do contribute to global carbon catabolite regulation. For example, the “stand-alone” virulence gene transcription factor Mga both is regulated by CcpA (1) and has been noted to contain several putative phosphotransferase system (PTS) regulatory domains, suggesting that phosphorylation by the PTS phosphorelay may link virulence gene expression and carbohydrate transport (20). Analysis of the repression of the SpeB cysteine protease in response to glucose revealed that repression could be relieved by mutation of LacD.1, a tagatose 1,6-bisphosphate aldolase (28). Interestingly, LacD.1 appears to have arisen from a duplication of the catabolic LacD.2 aldolase that was then adapted for a role as a regulator (27). Regulation by LacD.1 functions independently of its catalytic activity, although it does apparently require an ability to bind to its substrates (28). The mechanism by which LacD.1 regulates speB transcription is not known. However, the fact that the products of its aldolase reaction, glyceraldehyde-3-phosphate and dihydroxyacetone phosphate, are also important intermediates in the Embden-Meyerhof-Parnas pathway of glycolysis suggests that LacD.1 functions as a global sensor of carbon flow through the central metabolism (28). However, the contribution of LacD.1 to global carbon catabolite repression has not been examined.
While both LacD.1 and CcpA are involved in the regulation of speB, comparison of S. pyogenes infection in murine models has suggested that CcpA and LacD.1 may regulate distinct subsets of the transcriptome. When examined in a subcutaneous ulcer model, a CcpA− mutant, but not a LacD.1− mutant, was attenuated and produced lesions with areas significantly smaller than those of lesions produced by the wild type (22). Furthermore, when examined in vitro, LacD.1− and CcpA− mutants were found to have different patterns of speB expression. Inactivation of LacD.1 results in derepression of speB expression in the presence of glucose, while inactivation of CcpA results in reduced expression of speB under these conditions (22). When examined during infection of murine subcutaneous tissue, the LacD.1− mutant expressed high levels of speB, indistinguishable from those expressed by the wild type, while the CcpA− mutant expressed reduced levels (22). These data suggest that even though these two pathways sense similar cues, they may be organized so as to function at different stages of the infectious process.
In this study we have expanded our analysis of the CcpA and LacD.1 pathways in order to better understand how catabolite-responsive pathways are organized to promote virulence. Using transcriptional profiles derived from global in vitro analyses, we identify different classes of genes regulated by CcpA, LacD.1, and glucose. The abundance of selected messages representative of genes from each of these classes was probed over time during soft tissue infection. Analysis of expression patterns derived from this panel of candidate genes supported the idea that these two catabolite-sensitive pathways are organized to function at different times in this environment. Since catabolite availability and virulence are linked in many pathogens, and since many other bacterial species also possess multiple mechanisms for catabolite control, this analysis contributes to our understanding of how pathogens utilize multiple pathways to sense a central environmental signal.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type S. pyogenes strain HSC5 (M-type 14) (19) and isogenic mutants with in-frame deletions in ccpA (22) or lacD.1 (28) were grown in Todd-Hewitt broth (Difco) with 0.2% yeast extract (Difco) (THYB) or in C medium supplemented as described previously (29). Routine growth was carried out in sealed culture tubes at 37°C under static conditions. Streptococci grown on THYB medium solidified with 1.4% Bacto agar (Difco) were cultured anaerobically in sealed jars with commercially available gas-generating sachets (GasPak; catalog no. 70304; BBL).
Computational techniques.
All references to genomic loci are based on the genome of strain SF370 (14). Analyses of homology were conducted using BLAST (2, 16). Signal peptides were predicted with the SignalP 3.0 program (http://www.cbs.dtu.dk/services/SignalP/) (5, 33) set for Gram-positive bacteria and using the hidden Markov model. Predicted gene products were assigned to clusters of orthologous groups (COGs) as defined in references 43 and 44.
Isolation of RNA and analysis using real-time RT-PCR.
Total cellular RNA was isolated from streptococcal strains grown in the indicated medium at the times indicated in the text (see Table 1) as previously described (7). RNA was subjected to reverse transcription (RT) using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time RT-PCR analysis of the resulting cDNA samples was performed using iQ SYBR green Supermix (Bio-Rad) and the primers listed in Table S1 in the supplemental material. Relative transcript levels were determined by the ΔΔCT method using the recA (SPy_2116) transcript as a standard as described previously (6). The data presented are means derived from triplicate determinations for three samples per experimental group prepared from at least two independent experiments. The differences between the means for different experimental groups were tested for significance using the paired Student t test as described previously (6).
TABLE 1.
Genes whose expression is altered by glucose or inactivation of CcpA or LacD.1
| Gene code | Functional categorya (no. of genes in categoryb) | No. (%) of genes with differential expressionc |
|||||
|---|---|---|---|---|---|---|---|
| Glucose |
CcpA |
LacD.1 |
|||||
| Increased | Decreased | Increased | Decreased | Increased | Decreased | ||
| J | Translation (131) | 15 (11.5) | 5 (3.8) | 4 (3.1) | 0 | 1 (0.8) | 1 (0.8) |
| K | Transcription (65) | 3 (4.6) | 9 (13.8) | 8 (12.3) | 1 (1.5) | 0 | 1 (1.5) |
| L | Replication, recombination, repair (93) | 3 (3.2) | 2 (2.2) | 5 (5.4) | 0 | 2 (2.2) | 0 |
| D | Cell cycle, cell division, chromosome partitioning (14) | 1 (7.1) | 0 | 0 | 0 | 0 | 0 |
| V | Defense mechanisms (15) | 1 (6.7) | 4 (26.7) | 2 (13.3) | 1 (6.7) | 0 | 0 |
| T | Signal transduction mechanisms (30) | 0 | 5 (16.7) | 6 (20.0) | 0 | 0 | 0 |
| M | Cell wall, membrane, envelope biogenesis (51) | 4 (7.8) | 2 (3.9) | 0 | 0 | 0 | 0 |
| N | Cell motility (6) | 1 (16.7) | 1 (16.7) | 0 | 0 | 1 (16.7) | 0 |
| O | Posttranslational modification, protein turnover, chaperones (42) | 2 (4.8) | 3 (7.1) | 4 (9.5) | 0 | 0 | 0 |
| C | Energy production and conversion (50) | 2 (4.0) | 18 (36.0) | 19 (38.0) | 1 (2.0) | 0 | 5 (10.0) |
| G | Carbohydrate transport and metabolism (78) | 7 (9.0) | 58 (74.4) | 49 (62.8) | 5 (6.4) | 1 (1.3) | 3 (3.8) |
| E | Amino acid transport and metabolism (76) | 5 (6.6) | 10 (13.2) | 14 (18.4) | 5 (6.6) | 5 (6.6) | 6 (7.9) |
| F | Nucleotide transport and metabolism (53) | 6 (11.3) | 7 (13.2) | 8 (15.1) | 2 (3.8) | 1 (1.9) | 2 (3.8) |
| H | Coenzyme transport and metabolism (36) | 0 | 4 (11.1) | 3 (8.3) | 0 | 1 (2.8) | 0 |
| I | Lipid transport and metabolism (34) | 0 | 2 (5.9) | 2 (5.9) | 0 | 0 | 6 (17.6) |
| P | Inorganic ion transport and metabolism (47) | 2 (4.3) | 4 (8.5) | 4 (8.5) | 1 (2.1) | 1 (2.1) | 0 |
| Q | Secondary-metabolite biosynthesis, transport, and catabolism (10) | 0 | 0 | 1 (10) | 0 | 0 | 0 |
| R | General function prediction only (127) | 6 (4.7) | 15 (11.8) | 12 (9.4) | 2 (1.6) | 0 | 1 (1.6) |
| S | Unknown function (122) | 4 (3.3) | 8 (6.6) | 7 (5.7) | 5 (4.1) | 2 (1.6) | 1 (0.8) |
| Uncategorized (hypothetical) | 8 | 25 | 26 | 2 | 22 | 1 | |
| Total | 70 | 182 | 174 | 25 | 37 | 27 | |
Genes were categorized according to a functional classification system, COGs (clusters of orthologous groups) (43, 44).
Number of genes in the assigned category in the S. pyogenes SF370 genome.
Genes displaying a change in expression of >2-fold that was statistically significant (P < 0.05) by ANOVA across all experiments. Gene expression was compared as follows: for glucose, wild-type strain (HSC5) in C medium supplemented with 1% glucose/wild-type strain in unsupplemented C medium in exponential growth; for CcpA, CcpA− strain (CKB206)/wild-type strain in C medium supplemented with glucose in exponential growth; for LacD.1, LacD.1− strain (JL151)/wild-type strain in C medium supplemented with glucose in stationary-phase growth.
Microarray analysis.
Total RNA was isolated at the times indicated in the text from the various strains grown under the conditions described herein. Transcript abundance was then assessed by microarray analysis conducted as described previously (12) using the services provided by the Microarray Core of the Genome Center of Washington University (http://gtac.wustl.edu/). For consistency with prior studies (12, 26), the genomic loci reported are numbered in accordance with the genome of the S. pyogenes serotype M1 strain SF370 (14). Expression ratios were derived from at least two biological replicates, each of which was analyzed a minimum of 3 times, including at least one experiment with reciprocal fluorescent labeling. Fluorescence values from each array were measured and normalized as described previously (12), and for each probe, the null hypothesis of equal binding of experimental and reference cDNA was tested for significance computationally by using the Partek Genomics Suite (Partek Incorporated). Genes were considered differentially expressed when the average ratio between experimental and reference probe intensities was 2-fold or greater and analysis of variance (ANOVA) yielded a P value of <0.05.
Subcutaneous infection of mice.
Five- to 6-week-old female SKH1 mice (Charles River Laboratories) were injected subcutaneously with 107 CFU of the various streptococcal strains indicated in the text by the method of Bunce et al. (9) as described in detail elsewhere (7). Each experimental group consisted of at least 5 mice, and the data presented are pooled from two independent experiments. The differences in the areas of the resulting lesions between experimental groups were tested for significance using the Mann-Whitney U test (17). Total RNA was isolated from lesions as previously described (8) and was subjected to analyses by real-time RT-PCR as described above using the primers listed in Table S1 in the supplemental material. The data are presented as the means and standard errors of the means derived from samples obtained from at least 3 mice per experimental group, each analyzed in triplicate and combined from two independent experiments. Differences in the mean values between experimental groups were tested for significance using Student's t test.
Microarray data accession number.
The microarray data obtained in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) site (www.ncbi.nlm.nih.gov/geo/) under accession number GSE25253.
RESULTS
Effect of glucose on the S. pyogenes transcriptome.
To gauge the global effect of glucose on the behavior of the S. pyogenes transcriptome, wild-type strain HSC5 was grown in C medium with or without the addition of glucose (1.0%), and global transcript abundances were compared at the mid-exponential phase of growth. This analysis revealed that 252 genes (approximately 15% of the 1,696 genes analyzed) were differentially regulated by the presence of glucose (Table 1; see also Table S2 in the supplemental material). Most of these were repressed (182 genes) rather than activated (70 genes) by glucose (Table 1; see also Table S2). When these genes were categorized according to their COG (cluster of orthologous groups) designations (43, 44), it was found that a large fraction (119 of the 252 genes) were predicted to be involved in metabolic processes (for which the COG gene codes are given in parentheses), including energy conversion (C) and carbohydrate (G), amino acid (E), nucleotide (F), coenzyme (H), and lipid (I) metabolism (Table 1). Reflecting the overall pattern, a majority of these 119 metabolic genes were repressed (83%) as opposed to activated (17%) by glucose. Similarly, of the total subset of 182 repressed genes, a majority (54%) were associated with metabolism, compared to only 28% of the 70 genes in the activated subset. Genes from several COG categories not associated with metabolic processes, including those whose predicted functions are poorly defined, were also affected by glucose (Table 1; see also Table S2), establishing a link between the presence of carbohydrate and the regulation of phenotypes with diverse physiological outcomes.
Influence of glucose on genes encoding virulence factors.
Many virulence factors are secreted proteins, and analysis of the protein-encoding open reading frames predicted from the S. pyogenes SF370 genome sequence (14) revealed 205 open reading frames with predicted signal peptides (data not shown). Of these, 43 were differentially regulated in response to glucose, including 15 genes encoding established or predicted virulence factors (see Fig. S1 and Table S2 in the supplemental material). Of the latter, most were repressed (12 genes), including those encoding the cholesterol-dependent cytolysin (SLO), the NAD glycohydrolase (SPN), and mitogenic factor (MF), as well as those in the streptolysin S (SLS) biogenesis operon. Activated genes include those encoding protein GRAB (protein G-related α2-macroglobulin [α2M]-binding protein) and the SpeG superantigen (see Fig. S1 and Table S2 in the supplemental material). Taking the findings together, while the presence of glucose influences the regulation of 15% of the transcriptome overall (see above), 21% of the genes encoding putative secreted proteins were regulated, and of these, 33% have been shown to contribute to pathogenesis. This enrichment highlights the importance of carbon catabolite control in the regulation of virulence in S. pyogenes.
Contribution of CcpA to regulation in response to glucose.
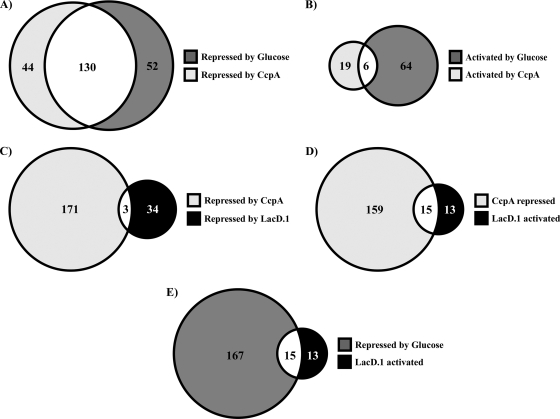
Global transcriptome profiling of a CcpA− mutant was conducted in order to assess the contributions made by this regulator to overall catabolite regulation in response to glucose. The analysis described above, in combination with a prior analysis of a candidate CcpA-repressed gene (lctO), indicated that significant disruption of glucose- and CcpA-mediated regulation was readily detectable at the mid-exponential phase of growth (22). Under this condition, 199 genes (12% of the total) exhibited a significant difference in expression between the wild type and the CcpA− mutant (see Table S3 in the supplemental material); these genes correspond closely with those reported in other studies (25, 38, 39) (For a detailed comparison of CcpA regulons from previous studies with the results of this study, see Table S5 in the supplemental material.) This subset included 33 genes predicted to encode proteins with signal sequences, of which 11 (33%) are known or predicted to be associated with virulence (see Table S3 in the supplemental material). Of the total number of differentially regulated genes, 174 were increased in abundance in the CcpA− mutant, while 25 were decreased (Table 1; see also Table S3 in the supplemental material). There was extensive overlap (approximately 75%) between those transcripts that were more abundant in the absence of CcpA and those whose expression was repressed during growth in glucose (Fig. 1A). However, this analysis also indicates that 29% of the glucose-regulated genes were not regulated by CcpA (Fig. 1A). Similarly, the expression of only about 9% of the transcripts that were more abundant in the presence of glucose was disrupted by the absence of CcpA, and most CcpA-activated genes (76%) were not sensitive to glucose (Fig. 1B). In addition, 24% of the genes regulated by CcpA, including 2 known to be associated with virulence (SPy_0416 and SPy_1273, encoding an interleukin 8 [IL-8] protease and CAMP factor, respectively), were not significantly affected by glucose under these conditions (Fig. 1A; see also Table S3 in the supplemental material). Taken together, these data indicate that under certain conditions, CcpA can function as a regulator independently of glucose and that while CcpA is a major regulator of carbon catabolite repression, other mechanisms for catabolite control exist in S. pyogenes.
FIG. 1.
Overlap between CcpA, LacD.1, and glucose transcriptional profiles. The Venn diagrams show the common and unique transcripts in different modes of regulation by CcpA, LacD.1, and glucose. (A) Numbers of transcripts repressed by glucose and CcpA (decreased abundance in wild-type [HSC5] bacteria grown with glucose; increased abundance in CcpA− [CKB206] bacteria). (B) Numbers of transcripts activated by glucose and CcpA (increased abundance in bacteria grown with glucose; decreased abundance in CcpA− bacteria). (C) Numbers of transcripts repressed by CcpA and LacD.1 (increased abundance in CcpA− and LacD.1− [JL141] bacteria). (D) Numbers of transcripts repressed by CcpA and activated by LacD.1 (increased abundance in CcpA− and decreased abundance in LacD.1− bacteria). (E) Numbers of transcripts repressed by glucose and activated by LacD.1 (decreased abundance in wild-type bacteria grown in glucose; decreased abundance in LacD.1− bacteria). The numbers given are the total numbers of transcripts for which the changes in abundance were found significant (P < 0.05) by ANOVA as described in Materials and Methods.
Contribution of LacD.1 to catabolite regulation.
Global transcription profiles were then compared between a LacD.1− mutant and the wild type during growth in C medium supplemented with glucose. An initial attempt examining cells harvested in mid-exponential phase yielded few genes whose transcript abundance was significantly misregulated (data not shown). However, a comparison of cultures at the early-stationary phase of growth yielded 64 genes that were significantly differentially expressed (see Table S4 in the supplemental material). There was very little overlap between genes repressed by CcpA and LacD.1 (Fig. 1C). In contrast, nearly 50% of the transcripts that were activated by LacD.1 were also repressed by CcpA (Fig. 1D). This shared subset of genes was also regulated by glucose (Fig. 1E). Taken together, these results indicate that there is considerable overlap between regulation by LacD.1 and regulation by CcpA. Furthermore, while this subset of genes is regulated by a common signal, regulation by LacD.1 and regulation by CcpA are apparently antagonistic.
Identification of shared, distinct, and opposing regulatory activities.
In order to resolve the dynamics of regulation by LacD.1 and CcpA during infection, a panel of transcripts was identified whose abundances represented the full spectrum of the regulatory activities of the respective controlling pathways, as revealed by analysis of mutants in vitro. Candidate genes were identified from the global profiling data, and their expression phenotypes were confirmed by real-time RT-PCR. Seven classes of genes were recognized. The first, represented by the gene encoding lactate oxidase (lctO), is repressed by glucose and is derepressed in the absence of CcpA (Table 2). The second, represented by the gene encoding acetate kinase (ackA), is repressed by CcpA but not by glucose (Table 2). Conversely, the gene encoding 4-α-glucanotransferase (malM) was chosen to be representative of those repressed by glucose but not affected by the loss of CcpA (Table 2). Genes regulated by CcpA also included those activated by glucose, represented by xanthine phosphoribosyltransferase (xpt) (Table 2) and those repressed in the presence of glucose (dexS, encoding a putative dextran glucosidase) (Table 2). The next two classes included those activated by LacD.1 but not affected by the presence of glucose (SPy_1507) and those activated by LacD.1 and repressed by CcpA (arcA, encoding a putative arginine deaminase) (Table 2). These patterns of regulation are summarized in Fig. 2.
TABLE 2.
Selected genes responsive to glucose-sensitive pathways
| Genea | Ratio of transcript abundanceb |
|||||
|---|---|---|---|---|---|---|
| Glucose |
CcpA |
LacD.1 |
||||
| Array | RT-PCR | Array | RT-PCR | Array | RT-PCR | |
| lctO | 0.047 | 0.010 | 43.5 | 388.1 | NSc | 1.31 |
| malM | 0.33 | 0.28 | 1.3 | 1.6 | NS | |
| ackA | 0.63 | 0.77 | 2.8 | 3.7 | NS | |
| xpt | 2.5 | 10.7 | 0.361 | 0.31 | 1.8 | |
| dexS | 0.082 | 0.22 | 0.17 | 0.20 | NS | |
| SPy_1507 | NS | 1.4 | 1.76 | 3.7 | 4.6 | |
| arcA | 0.044 | 19.7 | 35.6 | 0.055 | 0.111 | |
The locus tags corresponding to the genes are as follows: for lctO, SPy_0414; for malM, SPy_1292; for ackA, SPy_0109; for xpt, SPy_1136; for dexS, SPy_2096; and for arcA, SPy_1547.
For glucose, the ratio of transcript abundance in the wild-type strain (HSC5) in C medium plus glucose to that in the wild-type strain in C medium alone; for CcpA, the ratio of transcript abundance in the CcpA− strain (CKB206) in C medium plus glucose to that in the wild-type strain in C medium plus glucose; for LacD.1, the ratio of transcript abundance in the LacD.1− strain (JL151) in C medium plus glucose to that in the wild-type strain in C medium plus glucose.
NS, not significant by ANOVA.
FIG. 2.

Summary of the regulatory activities of CcpA, glucose, and LacD.1 on selected genes. Arrows indicate transcriptional activation; bars indicate repression.
Analysis of virulence in tissue over time.
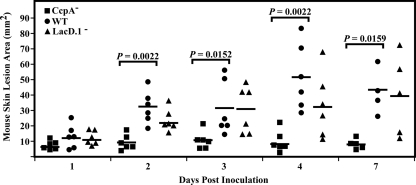
Our previous comparison of the virulence of CcpA− and LacD.1− mutants in the murine subcutaneous infection model revealed that at the time of maximal lesion formation by the wild-type strain (3 days postinfection), the CcpA− mutant, but not the LacD.1− mutant, was significantly attenuated in virulence (22). In order to extend this analysis for the examination of transcriptome dynamics, lesion development was monitored from the time of infection to the point of maximum lesion size (1 to 4 days) and at 7 days postinfection. In this comparison, visible ulcers were present by day 1 after the injection of bacteria into murine subcutaneous tissue, and these ulcers did not differ significantly in size between the wild type, the CcpA− mutant, and the LacD.1− mutant (Fig. 3). The lesions for all strains increased in size by day 2; however, those for the CcpA− mutant, but not those for the LacD.1− mutant, had significantly smaller areas than those for the wild-type strain (Fig. 3). By day 3, the lesions in tissue infected by the CcpA− mutant had reached their maximal size, while those resulting from the other strains continued to increase, reaching their maximal area on day 4 (Fig. 3). Lesions from all 3 strains then remained constant in size until the end of the observation period (day 7) (Fig. 3). Taken together, these data show that neither CcpA nor LacD.1 is essential for the formation of lesions in this model, although the former does influence the degree of lesion formation over time. Furthermore, resolution of lesions caused by any of these strains does not begin by day 7 postinfection.
FIG. 3.
CcpA but not LacD.1 affects virulence after early time points. Hairless SKH1 mice were injected with a wild-type (WT) (HSC5), CcpA− (CKB206), or LacD.1− (JL141) strain in order to determine relative virulence. At the indicated times, the 2-dimensional lesion area from the resulting skin ulceration was quantitated by image analysis. The data presented are plotted lesion areas from mice from combined experiments. Differences in means between mutant and WT lesion areas were tested for significance using the Mann-Whitney U test.
Analysis of genes repressed by glucose and/or CcpA reveals time-dependent variation in regulatory activity.
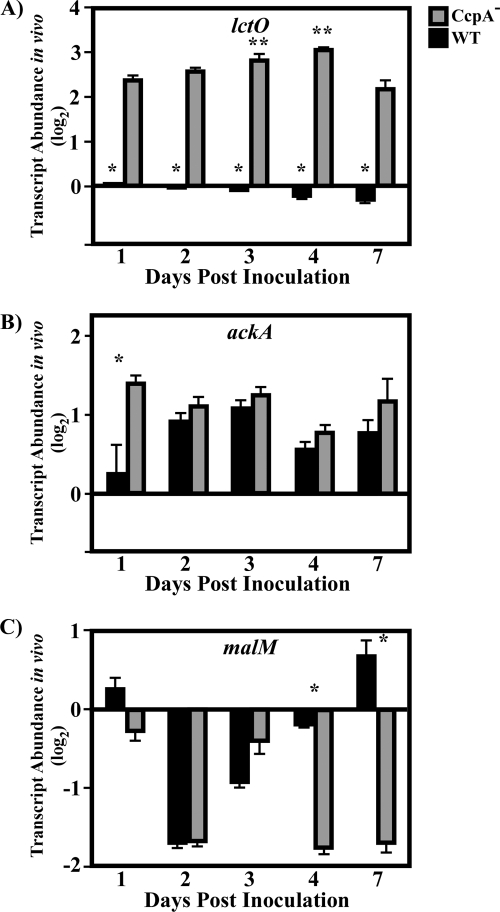
Comparison of the expression of candidate genes between wild-type and mutant strains over the course of infection was used to assess the contributions of individual regulators to transcriptome dynamics in vivo. For this analysis, real-time RT-PCR was used to quantitate the expression of candidate genes in lesions at days 1, 2, 3, 4, and 7 postinfection relative to the expression of a given gene at day 1 postinfection. The first set of genes examined comprised those regulated by glucose and/or CcpA (lctO, ackA, and malM [Table 2; Fig. 2]). For lctO, regulated in vitro by glucose and CcpA, expression in the wild type stayed relatively constant over all 7 days compared with expression at day 1 postinfection (Fig. 4A). In contrast, lctO expression was significantly derepressed in the CcpA− mutant relative to that in the wild type at each time point analyzed, with the highest levels corresponding to the time when lesions reached their maximal areas of involvement (days 3 to 4 postinfection [Fig. 4A]). A different pattern was observed for ackA, regulated in vitro by CcpA but not by glucose. The level of the ackA transcript was significantly higher in the CcpA− mutant only at day 1 postinfection; it was not significantly different on any subsequent day (Fig. 4B). A third pattern of expression was observed for malM, regulated in vitro by glucose but not by CcpA. Here, levels in the wild-type strain were decreased by day 2 postinfection and then increased over the next several days (Fig. 4C). This pattern was not significantly altered in the CcpA− mutant over days 1 to 3 postinfection, but levels were significantly decreased in the mutant at the later time points (days 4 and 7 [Fig. 4C]). This indicates a role for CcpA in the regulation of this gene in more-mature lesions that differs from that observed both earlier in infection and in vitro. Taken together, these data establish that despite an overlap of regulator and signal, these genes have distinctly different patterns of regulation with respect to time when observed in vivo.
FIG. 4.
Analysis of transcript abundance over time for CcpA- and glucose-repressed genes. After the sizes of the lesions were documented (Fig. 4), mice were sacrificed, and total RNA was purified from lesions. The RNA was then reverse transcribed and subjected to RT-PCR analysis using the primers listed in Table S1 in the supplemental material. Data are presented as the ratio of the value for the indicated gene from the CcpA− (CKB206) or wild-type (WT) (HSC5) strain at the indicated time point to the value for the WT strain on day 1 postinfection. Differences between mean values were tested for significance using Student's t test. The data presented are means and standard errors of the means from two independent experiments in which three replicates per experimental group were analyzed in triplicate. (A) Asterisks indicate significant differences (*, P < 0.0001; **, P < 0.01) from the transcript abundance for the CcpA− strain on other days. (B) *, P < 0.0008. (C) *, P < 0.0001.
Analysis of genes activated by CcpA reveals a switch in regulatory activity in vivo.
Although the majority of catabolite-sensitive genes are repressed by CcpA and/or glucose in the medium, several genes are activated by CcpA in vitro (Fig. 1; Table 1). To test the in vivo dynamics of genes activated by CcpA, the abundances of the dexS and xpt transcripts were analyzed. Both genes were activated by CcpA in vitro, though dexS was repressed and xpt activated by glucose (Table 2; Fig. 2). The xpt and dexS transcripts remained relatively constant in wild-type bacterial lesions over the period of observation until day 7, at which time the abundance of the xpt transcript increased 7- to 8-fold (Fig. 5A and B). Although dexS and xpt are activated by CcpA in vitro, the pattern for dexS and xpt regulation in wild-type bacteria in vivo closely resembled the pattern previously observed for the glucose- and CcpA-repressed gene lctO. In contrast to regulatory patterns in wild-type bacteria and the effects of CcpA in vitro, both the xpt and dexS transcripts were significantly derepressed in CcpA− mutant lesions during the middle of the period of observation; this pattern was shared between these two genes on days 2 to 4 postinoculation (Fig. 5A and B); in addition, the dexS gene was also derepressed at the earliest time point, day 1 postinoculation (Fig. 5A). Thus, in contrast to the regulation of dexS and xpt in vitro, inactivation of CcpA causes a derepression of transcript abundance in vivo. The temporal pattern of derepression of the dexS and xpt genes in the CcpA− mutant lesions overlaps with the pattern for the lctO gene on days 2 to 4, indicating that genes affected by CcpA and glucose are misregulated at similar times and that patterns of misregulation are not dependent on the regulatory outcomes of CcpA observed in vitro.
FIG. 5.
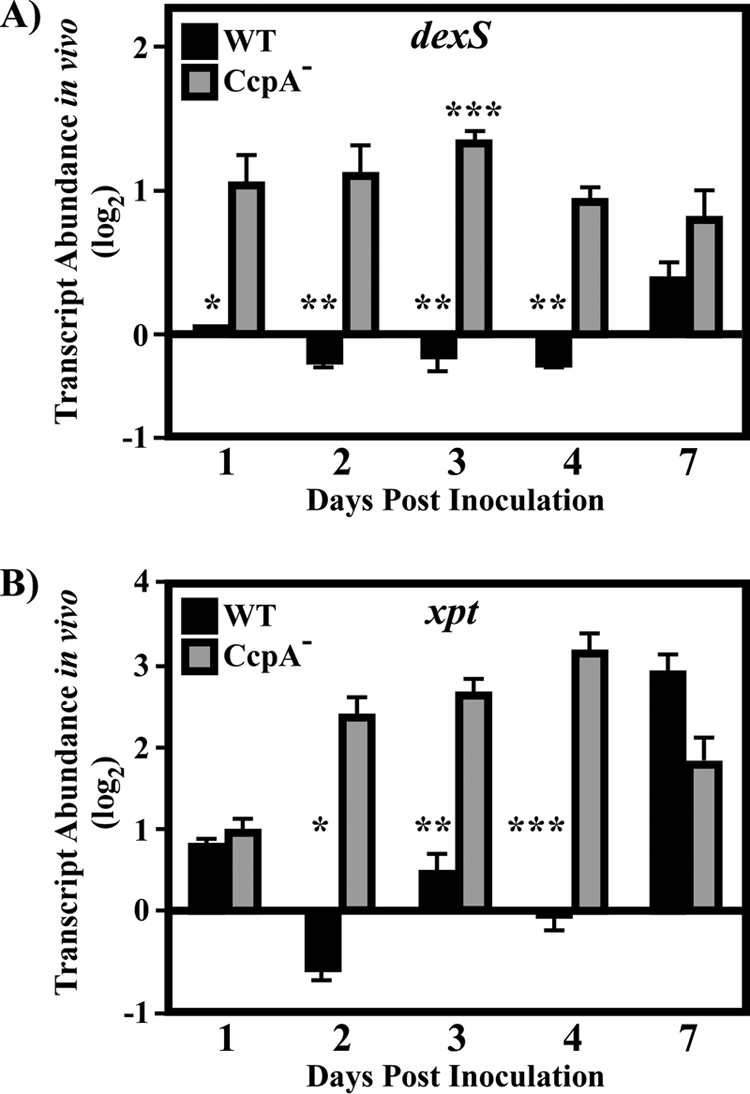
Analysis of transcript abundance over time for CcpA-activated genes. Samples were treated as explained in the legend to Fig. 4. Data are presented as the ratio of the value for the indicated gene from the CcpA− (CKB206) or wild-type (WT) (HSC5) strain at the indicated time point to that for the WT strain on day 1 postinfection. Differences between mean values were tested for significance using Student's t test. The data presented are means and standard errors of the means from two independent experiments in which three replicates per experimental group were analyzed in triplicate. (A) Asterisks indicate significant differences (*, P = 0.0001; **, P < 0.0001; ***, P < 0.05) between the CcpA− strain on day 3 and the CcpA− strain on all other days. (B) *, P = 0.0003; **, P = 0.0006; ***, P = 0.001.
Expression pattern of a LacD.1-regulated gene.
The abundance of the SPy_1507 transcript, which was derepressed in a LacD.1− mutant in vitro (Table 2; Fig. 2), was examined for comparison to patterns observed for glucose- and CcpA-regulated genes. In contrast to these, the SPy_1507 transcript exhibited a more dynamic pattern, with a peak of repression occurring on day 2 postinoculation and tapering off over time (Fig. 6). In addition, the pattern of SPy_1507 expression in the LacD.1− mutant was essentially similar to that observed in the wild-type strain (Fig. 6). A statistically significant 2-fold difference in transcript abundance was observed on day 1 postinfection, indicating that the influence of LacD.1 on SPy_1507 transcription may be strongest at time points early in this observation period and may then wane as the lesions mature. This was in contrast to CcpA- and glucose-regulated genes, such as lctO, dexS, and xpt, which were misregulated at times corresponding to more-mature lesions.
FIG. 6.
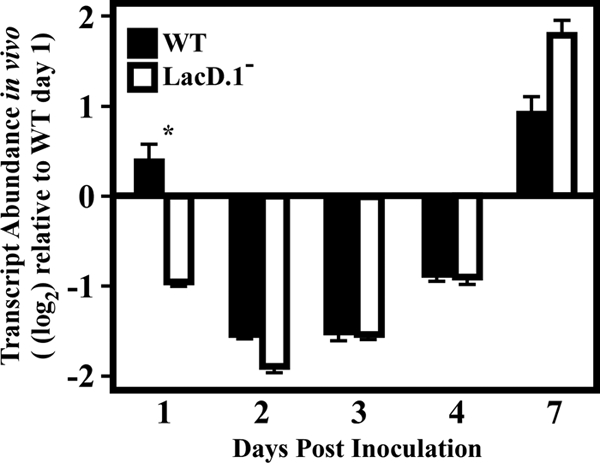
Analysis of the LacD.1-regulated gene Spy_1507 over time. Samples were treated as explained in the legend to Fig. 4. Data are presented as the ratio of the value for the indicated gene from the LacD.1− (JL141) or wild-type (WT) (HSC5) strain at the indicated time point to that for the WT strain on day 1 postinfection. Differences between mean values were tested for significance using Student's t test. The data presented are means and standard errors of the means from two independent experiments in which three replicates per experimental group were analyzed in triplicate. *, P < 0.0001.
Genes coregulated by CcpA and LacD.1 show distinct patterns of regulation.
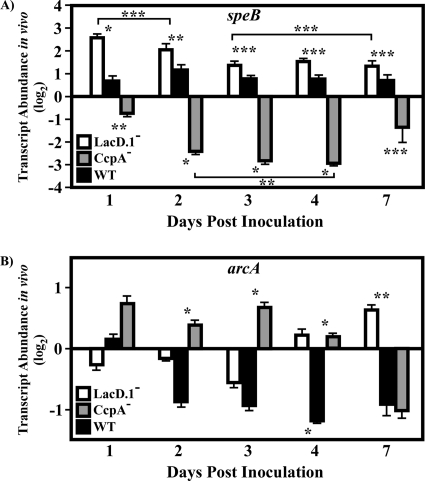
The dynamics of speB and arcA transcripts were next studied in order to better understand the effects of CcpA and LacD.1 on the transcript abundance of coregulated genes. Under in vitro conditions, speB is activated by CcpA and repressed by LacD.1 (22, 28, 38, 39). In contrast, like several genes in vitro, the arc operon (represented here by the arcA transcript) is repressed by CcpA and activated by LacD.1. The speB transcript was upregulated slightly on day 2 postinfection; however, in agreement with previous findings, its abundance was relatively constant over the observation period (Fig. 7A) (26). The arcA transcript dropped 2-fold in abundance on day 2 postinfection and then remained at this level throughout the observation period (Fig. 7B). In CcpA− mutant bacteria, speB transcript abundance differed slightly from that of the wild type on days 1 and 7 postinfection; however, the greatest misregulation (8- to 10-fold) was observed on days 2 to 4 postinfection (Fig. 7A). As with speB, the abundance of the arcA transcript in CcpA− mutant bacteria was significantly different from that in wild-type bacteria on days 2 to 4 postinfection, ranging from 2- to 3-fold higher over this period (Fig. 7B). In contrast to the patterns of transcript misregulation observed for the speB and arcA genes in CcpA− mutant bacteria, the speB transcript abundance in LacD.1− mutant bacteria was significantly altered (5-fold more abundant) from that in the wild type on day 1 postinfection; by day 3 postinfection, the difference from levels in wild-type bacteria dropped to less than 2-fold (Fig. 7A). The arcA transcript was significantly higher in abundance (2-fold) in LacD.1− mutant bacteria on day 4 postinfection, and the difference increased to 3-fold on day 7 postinfection (Fig. 7B). Thus, for these two CcpA- and LacD.1-coregulated genes, the patterns of misregulation are distinct; inactivation of CcpA correlates with peaks of misregulation of these target genes on days 2 to 4 postinfection, while LacD.1 inactivation correlates with peaks of misregulation of target genes either at early (speB) or at late (arcA) time points during the observed period of infection.
FIG. 7.
Analysis of CcpA- and LacD.1-coregulated genes over time. Samples were treated as explained in the legend to Fig. 4. Data are presented as the ratio of the value for the indicated gene from the CcpA− (CKB206), LacD.1− (JL141), or wild-type (WT) (HSC5) strain at the indicated time point to that for the WT strain on day 1 postinfection. Differences between mean values were tested for significance using Student's t test. The data presented are means and standard errors of the means from two independent experiments in which three replicates per experimental group were analyzed in triplicate. (A) *, P < 0.0001; **, P < 0.001; ***, P < 0.05. Brackets indicate groups that are significantly different from each other. (B) *, P < 0.0001; **, P = 0.0116.
DISCUSSION
Extensive profiling of the S. pyogenes transcriptome within infected tissues and host secretions has shown that these niches tend to alter the expression of the same subsets of genes that are known to be sensitive to carbon catabolite regulation in vitro (12, 18, 37, 46). While these data suggest that carbohydrate substrates constitute an important signal used for compartment-specific modulation of virulence genes during infection, the full extent of the transcriptome regulated in response to carbohydrate and the specific contributions of known regulators of carbon catabolite repression to this subset were not known. This question was of particular importance because S. pyogenes possesses multiple carbon catabolite regulatory pathways, including those controlled by CcpA and LacD.1, which may regulate overlapping sets of genes (22, 28). How these pathways are integrated to control transcriptome behavior and the expression of virulence genes was not understood. In this study, we have investigated the respective roles of CcpA and LacD.1 in the regulation of the transcriptome in response to glucose, and we have used time-resolved analyses of transcript abundance from infected tissue to show that CcpA and LacD.1 have discrete patterns of regulation of catabolite-sensitive genes in vivo.
Taking into account the distinct temporal patterns of regulation demonstrated by these two regulators may help to reconcile their roles during infection, particularly for those genes on which they appear to have antagonistic effects. Previously it has been shown that inactivation of CcpA, but not of LacD.1, has significant effects on overall virulence in models of both localized and systemic disease (22, 25, 38). This alteration in the severity of disease with the CcpA mutant correlates with the misregulation of virulence genes, including the speB gene, in diseased tissues (22, 39). The observation that LacD.1 and CcpA often coregulate catabolite-responsive genes and have opposite effects on transcription may relate to the observation that CcpA-dependent regulation of glucose-sensitive genes is readily observable in exponential phase, while LacD.1-dependent regulation is more detectable in stationary phase. The utilization of CcpA- and LacD.1-dependent regulation at different times or in alternate environments would explain why two opposing catabolite-responsive systems exist. This model, where CcpA and LacD.1 have distinct temporal roles in regulating virulence genes, is supported by the time-resolved analyses of transcription during soft tissue infection, which demonstrated that despite shared in vitro signals, LacD.1- and CcpA-coregulated genes are misregulated by the loss of CcpA or LacD.1 at different times over the course of infection. The loss of CcpA signaling leads to the misregulation of target genes at times corresponding to the time of maximum lesion size in the wild-type strain, whereas LacD.1 influences transcript abundance either early or late in the observation period. The ability of the LacD.1 mutant but not the CcpA mutant to regulate target genes similarly to wild-type bacteria at times corresponding to the full development of lesions may explain why the CcpA but not the LacD.1 mutant is attenuated in this virulence model. Little is known about the alterations in host tissues that coincide with lesion development and resolution; therefore, it is possible that the temporal pattern of expression observed for the LacD.1 and CcpA mutants is in part due to the influence of changing environmental signals that modify their regulatory activity, in addition to the central glucose signal. This hypothesis predicts that the influence of LacD.1 and CcpA on virulence depends both on environmental signals, for which time is a proxy within this model of soft tissue infection, and (or perhaps alternatively) on tissue-specific signals that differ between sites of infection. Only due to the influence of time- and/or tissue specific signals is the determination made whether the effects of CcpA or LacD.1 are necessary. This ability to compartmentalize multiple pathways for carbon catabolite repression that have antagonistic effects also underscores the importance of catabolite signals and the fine-tuned control of catabolite regulation in S. pyogenes pathogenesis.
This study has also shown that catabolite regulation in general, and regulation by CcpA in particular, is quite complex. Supporting the former conclusion is the fact that while there was extensive overlap between the subset of genes regulated by CcpA and that regulated by glucose, a number of genes were regulated by glucose but not by CcpA or LacD.1. This suggests that additional catabolite regulators are active under these conditions. The observation that the global regulator of virulence Mga may interact with the upstream elements of the CcpA pathway (1, 20) implicates Mga as an additional contributor to catabolite regulation. The latter suggestion is supported by the observation that a significant number of genes are regulated by CcpA in a glucose-independent manner. In addition, several genes activated by CcpA in vitro (dexS and xpt) are derepressed in CcpA− mutant bacteria during infection, indicating that in different environments, CcpA can either repress or activate a target gene. Structural studies of the mechanism by which CcpA represses transcription have revealed a novel mechanism of signal transduction linking glucose availability to CcpA activity. In this model, the Hpr protein (a phosphocarrier protein involved in high-affinity carbohydrate uptake systems) becomes phosphorylated on a regulatory serine by a kinase activated by high concentrations of fructose 1,6-bisphosphate (indicative of high carbohydrate flux through glycolysis) and then functions as a corepressor by binding to CcpA (15, 47). This Hpr-Ser-P molecule binds to the N subdomain of the periplasmic binding protein (PBP)-like domain of CcpA, both stabilizing the formation of a CcpA dimer and increasing the affinity of the DNA binding domain of CcpA for its target DNA sequence, the catabolite-responsive element (CRE) (35). However, many studies using purified apo-CcpA have shown that CcpA can bind CRE sites in the absence of Hpr-Ser-P, indicating that the effect of Hpr-Ser-P on CcpA may involve more than influencing its affinity for DNA (3, 23, 34). This suggests that CcpA may bind to CRE sites in vivo independently of the Hpr-Ser-P signal or that repression by CcpA may require a conformational change in CcpA when it is prebound to DNA that converts it into a repression-competent form upon interaction with its corepressor.
The ability of CcpA to promote different regulatory outcomes, leading to activation or repression, suggests that additional modulating signals and/or regulatory pathways interact with CcpA in the regulation of specific target genes. In addition, the data presented in this report show that at a subset of CcpA-regulated promoters, CcpA can either repress or activate transcription depending on the environment. In this regard, while much is known structurally about the influence of Hpr-Ser-P on how CcpA binds DNA, little is known about the biochemical mechanism by which CcpA influences transcription. Binding to the CRE site is important; however, not all predicted CRE sites are active, suggesting that the position of the CRE site relative to the promoter is important for regulatory outcome (48). Those CRE sites that overlap the promoter are generally thought to lead to repression by CcpA by interfering with the binding of RNA polymerase. However, there are examples of CRE sites far downstream of promoters leading to repression of the target gene (24). In contrast, those sites located upstream of the −35 promoter sequence are generally involved in activation (48). Recognition of these sites may also require CcpA to interact with Hpr-Ser-P, although CcpA may be in a different conformational state (42). In this regard, activation of the speB gene by CcpA presents an interesting example. Analysis in vitro has suggested that CcpA may bind to a predicted CRE site located upstream of the speB promoter or to a site within the speB open reading frame (22, 39). However, CcpA activation of speB is growth phase independent, yet high levels of Hpr-Ser-P are expected to be present only in the exponential phase of growth, when extracellular glucose levels are high (22). Thus, further analyses of how CcpA may interact with RNA polymerase at the speB promoter and how this may be modulated by additional factors, including Hpr-Ser-P and small organic molecules predicted to bind the PBP domain of CcpA, will be required for a full understanding of how carbohydrate modulates S. pyogenes virulence.
The data from this study also show that LacD.1, like CcpA, can change its regulatory mode based on the environment. For example, disruption of LacD.1 reduced the abundance of the arcA transcript in vivo but resulted in increased abundance in infected tissue. The regulatory activity of LacD.1 functions independently of its aldolase activity but apparently does require the ability to bind dihydroxyacetone phosphate and/or glyceraldehyde-3-phosphate (28). This has given rise to the suggestion that LacD.1 may sense the flow of carbon through glycolysis, although some LacD.1-regulated genes require additional signals (28). It is not well understood how LacD.1 senses these signals or by what mechanism LacD.1 influences the transcription of target genes, although LacD.1 may interact directly with DNA-binding proteins such as RopB (28). This regulator controls the transcription of a number of LacD.1-regulated genes, including speB (29) and the arc operon (11, 32). Since not all LacD.1-regulated genes appear to require RopB, and since it has been observed that LacD.1 can both activate and repress target genes, it seems likely that LacD.1 may interact with other transcription regulators to control the organization of CcpA and LacD.1 pathways in order to prevent their concurrent activity at coregulated genes.
This study has contributed to an emerging literature that implicates carbohydrate signals as important in regulating virulence genes in group A streptococci, as evidenced by the presence of several catabolite-sensitive regulators and the large numbers of genes they control. The observation that ablation of CcpA signaling attenuates virulence in a murine subcutaneous infection model demonstrates that carbon catabolite regulation plays an important role in the adaptation of S. pyogenes to the host milieu. The loss of LacD.1 signaling had a lesser impact on virulence; however, it is possible that this may represent a limitation of the model, given the diverse host tissues that can be infected by S. pyogenes (13). Despite these insights, a number of important questions remain unanswered. These include how the relationship between catabolite regulation and spatiotemporal regulation is controlled and the impact of the latter on virulence. It is also important to understand how LacD.1 and CcpA became adapted to regulate virulence. Further study of these questions will lead to a better understanding of S. pyogenes pathogenesis and of the mechanism by which the ability of S. pyogenes to fine-tune responses to carbohydrate substrates enhances its ability to cause disease in different tissues.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service Grant AI070759 from the National Institutes of Health.
Editor: A. Camilli
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Almengor, A. C., T. L. Kinkel, S. J. Day, and K. S. McIver. 2007. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A streptococcus. J. Bacteriol. 189:8405-8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aung-Hilbrich, L. M., G. Seidel, A. Wagner, and W. Hillen. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319:77-85. [DOI] [PubMed] [Google Scholar]
- 4.Behari, J., and P. Youngman. 1998. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J. Bacteriol. 180:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Brenot, A., K. Y. King, and M. G. Caparon. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55:221-234. [DOI] [PubMed] [Google Scholar]
- 7.Brenot, A., K. Y. King, B. Janowiak, O. Griffith, and M. G. Caparon. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 72:408-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenot, A., B. F. Weston, and M. G. Caparon. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 63:1185-1196. [DOI] [PubMed] [Google Scholar]
- 9.Bunce, C., L. Wheeler, G. Reed, J. Musser, and N. Barg. 1992. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 60:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., et al. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U. S. A. 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, Y. 2009. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73:245-259. [DOI] [PubMed] [Google Scholar]
- 16.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 17.Glantz, S. A. 2005. Primer of biostatistics, 6th ed. McGraw-Hill Professional, New York, NY.
- 18.Graham, M. R., et al. 2005. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 166:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanski, E., P. A. Horwitz, and M. G. Caparon. 1992. Expression of protein F, the fibronectin-binding protein of Streptococcus pyogenes JRS4, in heterologous streptococcal and enterococcal strains promotes their adherence to respiratory epithelial cells. Infect. Immun. 60:5119-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hondorp, E. R., and K. S. McIver. 2007. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 66:1056-1065. [DOI] [PubMed] [Google Scholar]
- 21.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kietzman, C. C., and M. G. Caparon. 2010. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. H., Z. T. Guvener, J. Y. Cho, K. C. Chung, and G. H. Chambliss. 1995. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J. Bacteriol. 177:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. H., Y. K. Yang, and G. H. Chambliss. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56:155-162. [DOI] [PubMed] [Google Scholar]
- 25.Kinkel, T. L., and K. S. McIver. 2008. CcpA-mediated repression of streptolysin S expression and virulence in the group A streptococcus. Infect. Immun. 76:3451-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughman, J. A., and M. Caparon. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loughman, J. A., and M. G. Caparon. 2007. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 64:269-280. [DOI] [PubMed] [Google Scholar]
- 28.Loughman, J. A., and M. G. Caparon. 2006. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 25:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 31.Mertins, S., et al. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189:473-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:122-130. [PubMed] [Google Scholar]
- 34.Ramseier, T. M., J. Reizer, E. Kuster, W. Hillen, and M. H. Saier. 1995. In vitro binding of the CcpA protein of Bacillus megaterium to cis-acting catabolite responsive elements (CREs) of gram-positive bacteria. FEMS Microbiol. Lett. 129:207-213. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher, M. A., et al. 2004. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell 118:731-741. [DOI] [PubMed] [Google Scholar]
- 36.Seidl, K., et al. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelburne, S. A., III, et al. 2005. Growth characteristics of and virulence factor production by group A streptococcus during cultivation in human saliva. Infect. Immun. 73:4723-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shelburne, S. A., III, et al. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 105:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shelburne, S. A., et al. 2010. A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog. 6:e1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203-207. [DOI] [PubMed] [Google Scholar]
- 41.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917-927. [DOI] [PubMed] [Google Scholar]
- 42.Sprehe, M., G. Seidel, M. Diel, and W. Hillen. 2007. CcpA mutants with differential activities in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 12:96-105. [DOI] [PubMed] [Google Scholar]
- 43.Tatusov, R. L., et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 45.Varga, J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186:5221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Virtaneva, K., et al. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. U. S. A. 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. P. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.