Abstract
RNA helicase A (RHA) has been shown to promote HIV-1 replication at both the translation and reverse transcription stages. A prerequisite step for reverse transcription involves the annealing of 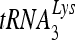 , the primer for reverse transcription, to HIV-1 RNA.
, the primer for reverse transcription, to HIV-1 RNA.  annealing is a multistep process that is initially facilitated by Gag prior to viral protein processing. Herein, we report that RHA promotes this annealing through increasing both the quantity of
annealing is a multistep process that is initially facilitated by Gag prior to viral protein processing. Herein, we report that RHA promotes this annealing through increasing both the quantity of  annealed by Gag and the ability of
annealed by Gag and the ability of 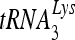 to prime the initiation of reverse transcription. This improved annealing is the result of an altered viral RNA conformation produced by the coordinate action of Gag and RHA. Since RHA has been reported to promote the translation of unspliced viral RNA to Gag protein, our observations suggest that the conformational change in viral RNA induced by RHA and newly produced Gag may help facilitate the switch in viral RNA from a translational mode to one facilitating
to prime the initiation of reverse transcription. This improved annealing is the result of an altered viral RNA conformation produced by the coordinate action of Gag and RHA. Since RHA has been reported to promote the translation of unspliced viral RNA to Gag protein, our observations suggest that the conformational change in viral RNA induced by RHA and newly produced Gag may help facilitate the switch in viral RNA from a translational mode to one facilitating 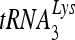 annealing.
annealing.
Upon cellular infection by HIV-1, the viral genomic RNA is reverse transcribed into double-stranded DNA that integrates into the host cell chromosomes as a provirus. Transcription of the proviral DNA results in the production of a full-length, unspliced viral RNA which, upon exit from the nucleus, is initially translated into Gag and GagPol and later encapsidated into the assembling Gag/GagPol viral particles as the viral RNA genome. Associated with the viral genomic RNA is 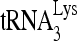 , the primer used for initiating the reverse transcription of minus-strand strong-stop cDNA from viral RNA (32). The 3′-terminal 18 nucleotides (nt) of
, the primer used for initiating the reverse transcription of minus-strand strong-stop cDNA from viral RNA (32). The 3′-terminal 18 nucleotides (nt) of 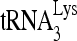 anneal to a complementary 18-nt sequence immediately downstream of the U5 region of HIV-1 RNA termed the primer binding site (PBS) (32). In addition, several other viral sequences upstream of the PBS have been proposed to interact with primer
anneal to a complementary 18-nt sequence immediately downstream of the U5 region of HIV-1 RNA termed the primer binding site (PBS) (32). In addition, several other viral sequences upstream of the PBS have been proposed to interact with primer 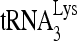 , including an A-rich loop that complements the U-rich anticodon of
, including an A-rich loop that complements the U-rich anticodon of 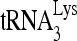 (28, 46, 54) and an 8-nt motif named the primer activation signal (PAS) that complements sequences in the TψC arm of
(28, 46, 54) and an 8-nt motif named the primer activation signal (PAS) that complements sequences in the TψC arm of 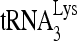 (8).
(8).
The annealing of 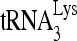 to viral RNA is likely to be a multistep process, initially facilitated in the cytoplasm by the nucleocapsid (NC) domain in Gag (12, 17) and the reverse transcriptase sequence in GagPol (13, 31), prior to the processing of Gag and GagPol that occurs during viral budding.
to viral RNA is likely to be a multistep process, initially facilitated in the cytoplasm by the nucleocapsid (NC) domain in Gag (12, 17) and the reverse transcriptase sequence in GagPol (13, 31), prior to the processing of Gag and GagPol that occurs during viral budding. 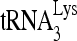 annealing by Gag is, however, not optimal, and evidence suggests that during Gag and GagPol processing, mature nucleocapsid protein (NCp7) can refine annealing to produce a more efficient primer for initiating reverse transcription (22). In this work, we will provide evidence that RNA helicase A (RHA) is also involved in improving
annealing by Gag is, however, not optimal, and evidence suggests that during Gag and GagPol processing, mature nucleocapsid protein (NCp7) can refine annealing to produce a more efficient primer for initiating reverse transcription (22). In this work, we will provide evidence that RNA helicase A (RHA) is also involved in improving 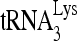 annealing by Gag.
annealing by Gag.
RNA helicases are enzymes that unwind double-stranded stretches of RNA, and they are involved in almost every aspect of RNA metabolism (15). While many viruses have been found to encode RNA helicase or RNA helicase-like proteins to facilitate their replication, retroviruses employ cellular counterparts in their life cycle (30). A number of RNA helicases have been reported to participate in HIV-1 replication (11, 30, 34, 37), including RHA. RHA was found to be involved in multiple steps of HIV-1 replication, including transcription (18), export of unspliced viral RNA from the nucleus (36), translation (9), particle assembly, and reverse transcription (43). In addition to HIV-1, RHA also participates in the replication cycles of other retroviruses (9, 10) and other RNA viruses such as picornaviruses (35) and flaviviruses (23, 29).
Human RHA, also known as DHX9, can unwind double-stranded stretches of both RNA and DNA using ATP hydrolysis as a general energy source (52). RHA is retained mainly in the nucleus, probably by arginine methylation of C-terminal arginine-glycine-glycine (RGG) repeats (44). However, RHA does shuttle back and forth between the nucleus and the cytoplasm (5, 19). RHA possesses two double-stranded RNA binding domains at the N terminus and a classical DEAD box/helicase domain with a conserved ATP binding motif in the center (51).
We have investigated whether RHA helps promote 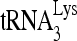 annealing to viral RNA because of a previous report that downregulation of RHA in cells producing HIV-1 resulted in a decrease in new viral cDNA synthesis upon infection of cells with these viruses (43). Herein, we report that RHA and Gag together improve the quantity and quality of
annealing to viral RNA because of a previous report that downregulation of RHA in cells producing HIV-1 resulted in a decrease in new viral cDNA synthesis upon infection of cells with these viruses (43). Herein, we report that RHA and Gag together improve the quantity and quality of 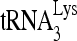 annealing to viral RNA by altering the conformation of the viral RNA. Since RHA has been reported to increase the efficiency of translation of unspliced viral mRNA in a number of retroviruses (9, 36), we propose that the switch from viral RNA translation to its annealing with
annealing to viral RNA by altering the conformation of the viral RNA. Since RHA has been reported to increase the efficiency of translation of unspliced viral mRNA in a number of retroviruses (9, 36), we propose that the switch from viral RNA translation to its annealing with 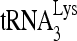 may be regulated by this conformational change.
may be regulated by this conformational change.
MATERIALS AND METHODS
Plasmids.
SVC21.BH10 is a simian virus 40-based vector that contains full-length wild-type HIV-1 proviral DNA (45). BH10.Pr− is SVC21.BH10 containing an inactive viral protease with a D25G point mutation. pTT5-SH5 (a gift of Yves Durocher, Biotechnology Research Institute, Montreal) was modified by creating NotI and AscI restriction sites in the multicloning region by using PCR. hGag is vector containing DNA that is codon-optimized for expressing HIV-1 Gag in mammalian cells (24) and is a gift from Y. Huang and G. Nabel, Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, MD. hGag was amplified by PCR using primers 5′-CGGGCGGCCGCCGGCGCCCGCGCCAGCGTG-3′ and 5′-CGGGGCGCGCCTTATTGTGACGAGGGGTCGCTGCC-3′, digested by NotI and AscI (restriction sites are underlined), inserted into modified pTT5-SH5 to get the plasmid pTT5-SH5-hGag used to purify N-terminally His-tagged HIV-1 Gag protein from 293E cells (16). RHA cDNA was a gift from Chee-Gun Lee, University of Medicine and Dentistry of New Jersey. RHA open reading frame (ORF) was amplified by PCR using primers 5′-CGGGCGGCCGCCGGTGACGTTAAAAATTTTC-3′ (forward) and 5′-CGGGGCGCGCCTCATTAATAGCCGCCACCTCC-3′ (reverse), digested by NotI and AscI (restriction sites are underlined), and inserted into pTT5-SH5 to get the plasmid pTT5-SH5-RHA used to purify N-terminally His-tagged RHA. The K417R mutation was created by using site-directed mutagenesis (Clontech) to construct the plasmid pTT5-SH5-RHAmu that was used to express enzymatically inactive RHA(K417R) in 293E cells.
Purification of RHA and HIV-1 Gag proteins from 293E cells.
HEK 293E cells stably expressing Epstein-Barr virus nuclear antigen-1 were obtained from Yves Durocher (Biotechnology Research Institute, Montreal, Canada), grown in F17 medium (Invitrogen) supplemented by 2 mM l-glutamine and 0.1% Pluronic F-68 (GIBCO), and transfected by using 25-kDa linear polyethylenimine (PEI; pH 7.0) (Polysciences Inc.). We followed the procedures developed by Durocher et al. (42) for expressing proteins in these cells. Briefly, 293E cells were transfected by pTT5-SH5-RHA, pTT5-SH5-RHAmu, or pTT5-SH5-hGag. Forty-eight hours later, transfected cells were collected, washed with ice-cold phosphate-buffered saline, lysed in buffer containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.5% Triton X-100, 10% glycerol, and protease inhibitor cocktail tablets (Roche), pH 7.4, and then centrifuged at 22,000 × g for 30 min at 4°C. Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) was added into cleared supernatant and incubated at 4°C for 2 h to capture His-tagged proteins. Precipitated protein was eluted by 250 mM imidazole solution, characterized by Coomassie brilliant blue R250 staining and Western blot analysis with specific antibodies, dialyzed against 25 mM NaH2PO4 (pH 7.4), 150 mM NaCl, and 10% glycerol, and then stored at −80°C.
Cell culture and transfection.
HEK-293T cells were obtained from the American Type Culture Collection (ATCC), grown in complete Dulbecco's modified Eagle's medium (DMEM), and transfected using Lipofectamine 2000 (Invitrogen).
RNA preparation.
T7 RNA polymerase (T7 MEGAscript kit; Ambion) was used for the in vitro synthesis of the first 386 nt of the 5′ region of HIV-1 RNA, containing sequences from R (+1) through the codon for the 18th amino acid of Gag. DNA templates for RNA synthesis were amplified by PCR from vectors containing the sequences for HIV-1 BH10, with or without deletions of the PBS or the dimerization initiation site (DIS). The forward primer contains the T7 promoter at its 5′-end followed by the sequence complementary to positions +1 to +18 (5′-GCGTTAATACGACTCACTATAGGGAGAGGTCTCTCTGGTTAGACC-3′; T7 promoter is underlined). The reverse primer is complementary to positions +369 to +386 (5′-TTCCCATCGATCTAATTC-3′). PCR products were agarose gel purified and used as templates in the T7 transcription reaction. These PCR products were also mutated by fusion PCR to get the templates for synthesis of mutated HIV-1 RNAs. The primers used in fusion PCR are listed in Table 1.
TABLE 1.
Primers used in fusion PCR to make templates for in vitro synthesis of mutated HIV-1 RNAs
| Primer | Sequence | Mutated HIV-1 RNA |
|---|---|---|
| HIVm1-Forward | 5′-CGACGTCTGCAGCGGGTGTGACTCTGGTAACTA-3′ | HIV mu(nt 101-120) |
| HIVm1-Reverse | 5′-CCGCTGCAGACGTCGACTACTTGAAGCACTCA-3′ | |
| HIVm2-Forward | 5′-CGACGTCTGCAGCGGGAGATCCCTCAGACCCTT-3′ | HIV mu(nt 121-140) |
| HIVm2-Reverse | 5′-CCGCTGCAGACGTCGCACAACAGACGGGCACA-3′ | |
| HIVm3-Forward | 5′-CGACGTCTGCAGCGGAGTCAGTGTGGAAAATC-3′ | HIV mu(nt 141-160) |
| HIVm3-Reverse | 5′-CCGCTGCAGACGTCGATCTCTAGTTACCAGAGT-3′ | |
| HIVm4-Forward | 5′-CGACGTCTGCAGCGGAGCAGTGGCGCCCGAACA-3′ | HIV mu(nt 161-180) |
| HIVm4-Reverse | 5′-CCGCTGCAGACGTCGTGACTAAAAGGGTCTGAG-3′ | |
| HIVm5-Forward | 5′-CGACGTCTGCAGCGGACCAGAGGAGCTCTCTCG-3′ | HIV mu(nt 201-220) |
| HIVm5-Reverse | 5′-CCGCTGCAGACGTCGCAAGTCCCTGTTCGGGCG-3′ | |
| HIVm6-Forward | 5′-CGACGTCTGCAGCGGCGCAGGACTCGGCTTGCT-3′ | HIV mu(nt 221-240) |
| HIVm6-Reverse | 5′-CCGCTGCAGACGTCGTGGTTTCCCTTTCGCTTT-3′ |
Two cDNA oligomers (5′-GCGTAATACGACTCACTATAGGGAGAGC CCGGATAGCTCAGTCGGTAGAGCATCAGACTTTTAATCTGAGGGT CCAGGGTTCAAGTCCCTGTTCGGGCGCCA-3′ and 5′-TGGCGCCCG AACAGGGACTTGAACCCTGGACCCTCAGATTAAAAGTCTGATGCT CTACCGACTGAGCTATCCGGGCTCTCCCTATAGTGAGTCGTATTA CGC-3′) were heat annealed to get the template for synthesis of 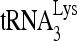 (T7 promoter is underlined). RNA was synthesized using the T7 polymerase. Reactions were stopped by the addition of 2 units of TURBO DNase I and further incubation at 37°C for 15 min. RNA transcripts were purified by Trizol extraction, precipitated with isopropanol, resuspended in 10 mM Tris (pH 7.5) and 100 mM NaCl, and then stored at −80°C.
(T7 promoter is underlined). RNA was synthesized using the T7 polymerase. Reactions were stopped by the addition of 2 units of TURBO DNase I and further incubation at 37°C for 15 min. RNA transcripts were purified by Trizol extraction, precipitated with isopropanol, resuspended in 10 mM Tris (pH 7.5) and 100 mM NaCl, and then stored at −80°C.
Partial duplex RNAs used for helicase assay were prepared as follows: for the 186-nt RNA strand, DNA templates were amplified from luciferase ORF contained in pGL3-Basic (Promega) by PCR using primers 5′-CGCTAATACGACTCACTATAGGGAGAGTCTATTTCGTTCATCCA-3′ (T7 promoter region is underlined, forward) and 5′-TTGCAGGACCACTTCTGCGCTCG-3′ (reverse). For the 34-nt RNA strand, DNA templates were prepared by heat annealing two DNA oligomers, 5′-CGCTAATACGACTCACTATAGGGAGAGAGCCGGTGAGCGTGGGTCTCGCGGTAT-3′ and 5′-ATACCGCGAGACCCACGCTCACCGGCTCTCTCCCTATAGTGAGTCGTATTAGCG-3′ (T7 promoter region is underlined). RNAs were synthesized in vitro from the templates above using T7 polymerase. The shorter RNA fragment was labeled with 32pCp using RNA ligase. The nucleotide sequences of the two strands are as follows: 186-nt strand, 5′-GGGAGAGUCUAUUUCGUUCAUCCAUAGUUGCCUGACUCCCCGUCGUGUAGAUAACUACGAUACGGGAGGGCUUACCAUCUGGCCCCAGUGCUGCAAUGAUACCGCGAGACCCACGCUCACCGGCUCCAGAUUUAUCAGCAAUAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGUGGUCCUGCAA-3′; 34-nt strand, 5′-GGGAGAGAGCCGGUGAGCGUGGGUCUCGCGGUAU-3′ (duplex region is underlined). Samples (10 pmol) of each transcript were mixed and heat annealed, and the resulting partial duplex was stored at −80°C.
32P-labeled RNA was further purified by using Microspin G-25 columns (GE Healthcare) and then stored at −20°C.
RHA helicase assay.
The RNA helicase assay was performed as described previously (27) with minor modifications. Reaction mixtures (20 μl) contained 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 2 units RNasin, 2.5 mM NaH2PO4, 15 mM NaCl, 100 fmol of partial duplex RNA, and, where indicated, 2 mM ATP. RHA was added in reactions as indicated in figure legends. After incubation for 10 min at 37°C, the reactions were stopped by the addition of 5 μl of a solution containing 2% SDS, 10 mM CaCl2, 250 μg/ml proteinase K, 40% glycerol, bromphenol blue, and xylene cyanol. For positive controls, duplex RNA was separated into monomers by heating at 95°C for 5 min, followed by rapid cooling in ice. Samples were electrophoresed on a 10% native polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE). 32P-RNAs were visualized and quantitated using a PhosphorImager (Amersham Pharmacia Biotech).
In vitro annealing assay.
In the in vitro annealing assay, synthetic HIV-1 RNA and 32P-labeled 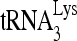 were first incubated at 37°C for 10 min in the presence of 1,000 fmol of bovine serum albumin (BSA) or RHA in a total volume of 20 μl of a buffer containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 2 units RNasin, 2.5 mM NaH2PO4, 15 mM NaCl, and, where indicated, 2mM ATP. The viral RNA/
were first incubated at 37°C for 10 min in the presence of 1,000 fmol of bovine serum albumin (BSA) or RHA in a total volume of 20 μl of a buffer containing 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 2 units RNasin, 2.5 mM NaH2PO4, 15 mM NaCl, and, where indicated, 2mM ATP. The viral RNA/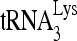 ratio was either 30 fmol viral RNA/100 fmol
ratio was either 30 fmol viral RNA/100 fmol 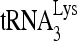 or 100 fmol viral RNA/30 fmol
or 100 fmol viral RNA/30 fmol 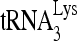 . Different Gag concentrations (up to 40,000 fmol) were then added, and the mixtures were incubated at 25°C for 1 to 45 min as indicated. The reaction was stopped by addition of 5 μl of a solution containing 2% SDS, 10 mM CaCl2, 250 μg/ml proteinase K, 40% glycerol, bromphenol blue, and xylene cyanol. After 20 min incubation, the samples were electrophoresed on a 5% native polyacrylamide gel in 0.5× TBE. 32P-labeled RNAs were visualized and quantitated by using a PhosphorImager.
. Different Gag concentrations (up to 40,000 fmol) were then added, and the mixtures were incubated at 25°C for 1 to 45 min as indicated. The reaction was stopped by addition of 5 μl of a solution containing 2% SDS, 10 mM CaCl2, 250 μg/ml proteinase K, 40% glycerol, bromphenol blue, and xylene cyanol. After 20 min incubation, the samples were electrophoresed on a 5% native polyacrylamide gel in 0.5× TBE. 32P-labeled RNAs were visualized and quantitated by using a PhosphorImager.
DNA oligomers annealed to viral RNA in vitro included the following: oligo 1, 5′-TCTTGCCGTGCGCGCTTC-3′(nt 254 to 271); oligo 2, 5′-GCACCCATCTCTCTCCTT-3′(nt 326 to 343); cPBS, 5′-GTCCCTGTTCGGGCGCCA-3′; DNA-tRNA, 5′-GCCCGGATAGCTCAGTCGGTAGAGCATCAGACTTTTAATCTGAGGGTCCAGGGTTCAAGTCCCTGTTCGGGCGCCA-3′.
Packaging of 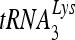 .
.
Virus purification and viral RNA isolation were performed as described previously (47). The relative abundance of 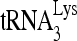 or HIV-1 genomic RNA was determined by dot blot hybridization using 5′-32P-end-labeled 18-mer DNA probe specific for the 3′ end of
or HIV-1 genomic RNA was determined by dot blot hybridization using 5′-32P-end-labeled 18-mer DNA probe specific for the 3′ end of 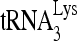 (5′-TGGCGCCCGAACAGGGAC-3′) or 5′-32P-end-labeled 18-mer DNA probe specific for the 5′ end of HIV-1 genomic RNA (5′-AGCCGAGTCCTGCGTCGA-3′). The relative abundance of
(5′-TGGCGCCCGAACAGGGAC-3′) or 5′-32P-end-labeled 18-mer DNA probe specific for the 5′ end of HIV-1 genomic RNA (5′-AGCCGAGTCCTGCGTCGA-3′). The relative abundance of 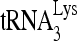 versus HIV-1 genomic RNA was determined by using a PhosphorImager and then normalized with data from the control sample.
versus HIV-1 genomic RNA was determined by using a PhosphorImager and then normalized with data from the control sample.
Northern analysis.
Total cellular RNA of 5 μg was separated in a denaturing 6% polyacrylamide gel, stained with ethidium bromide, visualized under UV light to determine the 5S rRNA level, transferred to hybridization membranes (Perkin-Elmer), and hybridized with a 5′-32P-end-labeled 18-mer DNA specific for the 3′ end of 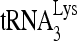 . The specific signals were quantitated using a PhosphorImager.
. The specific signals were quantitated using a PhosphorImager.
Primer extension.
The extension ability of 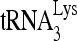 annealed in vivo or in vitro to viral RNA was measured as described previously (22). For 6-nt extension, reactions were carried out in a volume of 20 μl containing 50 mM Tris-HCl (pH 7.8), 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP (0.16 μM), and 0.05 mM ddATP (instead of dATP, thereby terminating the reaction at six bases), 50 ng of HIV-1 reverse transcriptase (RT), and RNase inhibitor (Amersham Pharmacia). For 1-nt extension, reactions were carried out in 20 μl of RT reaction mixture containing only 0.16 μM [α-32P]dCTP. After incubation for 15 min at 37°C, the samples were precipitated with isopropanol and were separated on a denaturing 6% polyacrylamide gel. The relative amounts of extended
annealed in vivo or in vitro to viral RNA was measured as described previously (22). For 6-nt extension, reactions were carried out in a volume of 20 μl containing 50 mM Tris-HCl (pH 7.8), 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP (0.16 μM), and 0.05 mM ddATP (instead of dATP, thereby terminating the reaction at six bases), 50 ng of HIV-1 reverse transcriptase (RT), and RNase inhibitor (Amersham Pharmacia). For 1-nt extension, reactions were carried out in 20 μl of RT reaction mixture containing only 0.16 μM [α-32P]dCTP. After incubation for 15 min at 37°C, the samples were precipitated with isopropanol and were separated on a denaturing 6% polyacrylamide gel. The relative amounts of extended 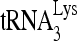 were determined by using a PhosphorImager. The relative amounts of viral RNA in the reactions was also determined by measuring the ability of DNA nt 801 to 830 (5′-TCTAATTCTCCCCCGCTTAATACTGACGCT-3′) annealed at room temperature to the viral RNA to prime a 6-base deoxynucleoside triphosphate (dNTP) extension (5′-CTCGCA-3′).
were determined by using a PhosphorImager. The relative amounts of viral RNA in the reactions was also determined by measuring the ability of DNA nt 801 to 830 (5′-TCTAATTCTCCCCCGCTTAATACTGACGCT-3′) annealed at room temperature to the viral RNA to prime a 6-base deoxynucleoside triphosphate (dNTP) extension (5′-CTCGCA-3′).
siRNA.
Small interfering RNA (siRNA)RHA (5′-GGAAACCAAGCAUAUAGAU-3′, sense; 5′-AUCUAUAUGCUUGGUUUCC-3′, antisense) was designed to target the 3′ untranslated region of RHA mRNA sequence at nt 4116 to 4134 (GenBank NM_001357), purchased from Ambion Inc., and transfected by using Lipofectamine 2000 (Invitrogen).
Virus infectivity assay.
Single-cycle infectivities were determined by challenging 105 TZM-bl indicator cells (48) with viruses corresponding to 5 ng of CAp24 and measuring the induced expression of luciferase activity in cell lysates after 28 h. TZM-bl indicator cells were generated from the stably transfected CD4+ HeLa cell line JC53 by introducing separate integrated copies of the luciferase and β-galactosidase genes under the control of the HIV-1 promoter. Infectivity was detected by the induction of luciferase activity, which was measured using luminometer Lumat LB9507 (EG&G Berthold, Bad Wildbad, Germany).
Western blot analysis.
Western blots were probed with rabbit anti-HIV RT, mouse anti-CAp24 antibody (NIH AIDS Research and Reference Reagent Program), β-actin monclonal antibody (MAb) (Sigma), RHA MAb (M01; Abnova Inc.), or polyhistidine MAb (Sigma) as the primary antibody. Protein bands were detected by enhanced chemiluminescence (ECL; Perkin-Elmer) and quantitated using ImageJ 1.35s public domain software (NIH).
Statistical analysis.
Student's t test was employed in statistical analyses. The lowest level of significance was set at a P value of <0.05.
RESULTS
RHA is required for 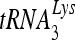 annealing to viral RNA in HIV-1.
annealing to viral RNA in HIV-1.
In transfected cells producing HIV-1, reduction of RHA results in HIV-1 with a reduced ability to synthesize cDNA by reverse transcription (43). Herein, we explore the mechanism responsible for this. RHA in 293T cells producing virus was reduced by siRNARHA, and protein patterns in cell and viral lysates were examined by Western blotting (Fig. 1A). As previously observed (9, 43), the reduction in cellular and viral RHA is not accompanied by a reduction in the viral content of RTp66/51.
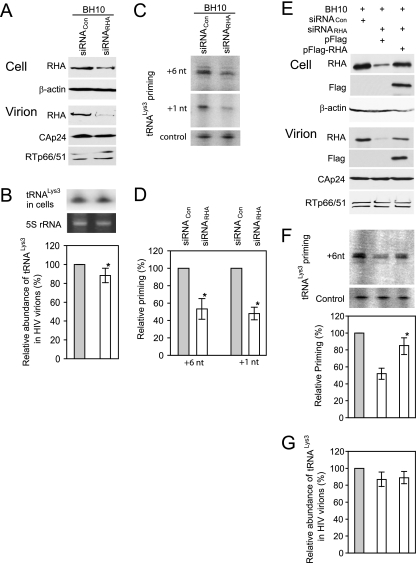
FIG. 1.
RHA is required for 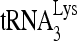 annealing to viral RNA in HIV-1. 293T cells were first transfected with siRNACon or siRNARHA, and 16 h later, they were transfected with DNA coding for HIV-1 (BH10). (A) Western blots of cell lysates probed with antibodies to RHA or β-actin, or viral lysates, containing equal amounts of CAp24, probed with antibodies to RHA, CAp24, or RTp66/p51. (B) Effects of decreased cellular RHA on
annealing to viral RNA in HIV-1. 293T cells were first transfected with siRNACon or siRNARHA, and 16 h later, they were transfected with DNA coding for HIV-1 (BH10). (A) Western blots of cell lysates probed with antibodies to RHA or β-actin, or viral lysates, containing equal amounts of CAp24, probed with antibodies to RHA, CAp24, or RTp66/p51. (B) Effects of decreased cellular RHA on 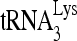 levels in cells and viruses. Upper panel, Northern blot analysis to determine
levels in cells and viruses. Upper panel, Northern blot analysis to determine 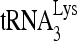 content in cell lysates. Middle panel, ethidium bromide staining of 5S rRNA in the same amount of cell lysate, to confirm that equal amounts of cellular RNA were used. Lower panel,
content in cell lysates. Middle panel, ethidium bromide staining of 5S rRNA in the same amount of cell lysate, to confirm that equal amounts of cellular RNA were used. Lower panel, 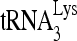 /viral RNA ratio, determined by hybridizing dot blots of total viral RNA with DNA probes specific for
/viral RNA ratio, determined by hybridizing dot blots of total viral RNA with DNA probes specific for 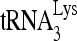 or viral RNA. Average values ± standard errors (SE) of results of quadruplicate experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test). (C)
or viral RNA. Average values ± standard errors (SE) of results of quadruplicate experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test). (C) 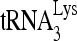 -primed initiation of reverse transcription. Total viral RNA was isolated, and
-primed initiation of reverse transcription. Total viral RNA was isolated, and 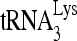 annealed to viral RNA was extended using reverse transcriptase by either 6 nt (upper panel) or 1 nt (middle panel). The extended
annealed to viral RNA was extended using reverse transcriptase by either 6 nt (upper panel) or 1 nt (middle panel). The extended 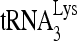 products are resolved by 1D PAGE and detected by autoradiography. The control gel represents the 6-nt extension of a DNA primer annealed to viral RNA downstream of the PBS and is used to show that equal amounts of viral RNA were used in each extension reaction. (D) The values of the 6- or 1-nt-extended
products are resolved by 1D PAGE and detected by autoradiography. The control gel represents the 6-nt extension of a DNA primer annealed to viral RNA downstream of the PBS and is used to show that equal amounts of viral RNA were used in each extension reaction. (D) The values of the 6- or 1-nt-extended 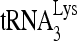 products, quantitated using a PhosphorImager, were normalized to those values obtained with virions produced from cells cotransfected with BH10 and siRNACon and are represented graphically. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test). (E to G). Rescue of the initiation of reverse transcription inhibited by siRNARHA by expression of RHA resistant to siRNARHA. 293T cells were transfected with either siRNARHA or siRNACon, and 16 h later they were cotransfected with DNA coding for HIV-1 (BH10) and either pFlag or pFlag-RHA, an RHA resistant to siRNARHA. (E) Western blots of cellular lysates probed with antibodies to RHA, Flag, and β-actin or of viral lysates containing equal amounts of CAp24, probed with antibodies to RHA, Flag, CAp24, and RTp66/p51. (F)
products, quantitated using a PhosphorImager, were normalized to those values obtained with virions produced from cells cotransfected with BH10 and siRNACon and are represented graphically. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test). (E to G). Rescue of the initiation of reverse transcription inhibited by siRNARHA by expression of RHA resistant to siRNARHA. 293T cells were transfected with either siRNARHA or siRNACon, and 16 h later they were cotransfected with DNA coding for HIV-1 (BH10) and either pFlag or pFlag-RHA, an RHA resistant to siRNARHA. (E) Western blots of cellular lysates probed with antibodies to RHA, Flag, and β-actin or of viral lysates containing equal amounts of CAp24, probed with antibodies to RHA, Flag, CAp24, and RTp66/p51. (F) 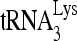 -primed initiation of reverse transcription. Upper panel, 6-nt-extended
-primed initiation of reverse transcription. Upper panel, 6-nt-extended  , resolved by 1D PAGE and detected by autoradiography. The control panel is similar to that described for panel C. In the lower panel, bands in the upper panel were quantitated using a PhosphorImager, and the graphed results are normalized to the value obtained with virions produced from cells cotransfected with BH10 and siRNACon. Average values ± SE of results of three experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test) compared to virions from cells transfected with siRNARHA and pFlag. (G) The
, resolved by 1D PAGE and detected by autoradiography. The control panel is similar to that described for panel C. In the lower panel, bands in the upper panel were quantitated using a PhosphorImager, and the graphed results are normalized to the value obtained with virions produced from cells cotransfected with BH10 and siRNACon. Average values ± SE of results of three experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test) compared to virions from cells transfected with siRNARHA and pFlag. (G) The  /viral RNA ratio in virions was determined as in panel B. Average values ± SE of results of quadruplicate experiments are shown.
/viral RNA ratio in virions was determined as in panel B. Average values ± SE of results of quadruplicate experiments are shown.
Northern blots of cellular RNA containing equivalent amounts of 5S rRNA were probed with DNA specific for  and indicate that reduced cellular RHA does not affect steady-state levels of cellular
and indicate that reduced cellular RHA does not affect steady-state levels of cellular  (Fig. 1B, upper panel). The amounts of
(Fig. 1B, upper panel). The amounts of 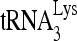 and viral genomic RNA in viruses were determined using dot blots of total viral RNA probed with DNA specific for
and viral genomic RNA in viruses were determined using dot blots of total viral RNA probed with DNA specific for 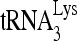 and viral RNA, respectively. The
and viral RNA, respectively. The 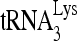 /viral RNA ratio plotted in Fig. 1B, lower panel, indicates that reduced RHA produces a 10% decrease in
/viral RNA ratio plotted in Fig. 1B, lower panel, indicates that reduced RHA produces a 10% decrease in 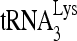 packaging into virions.
packaging into virions.
We next determined the effect of decreasing cell and viral RHA upon the ability of 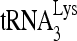 to be extended in an in vitro reverse transcription system, using deproteinized total viral RNA as the source of primer
to be extended in an in vitro reverse transcription system, using deproteinized total viral RNA as the source of primer 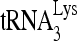 annealed in vivo to viral genomic RNA (Fig. 1C and D). The sequence in which the first six dNTPs are incorporated is 5′-CTGCTA-3′, and we examined
annealed in vivo to viral genomic RNA (Fig. 1C and D). The sequence in which the first six dNTPs are incorporated is 5′-CTGCTA-3′, and we examined 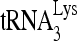 extended either by these six nucleotides (nt), by replacing dATP with ddATP, or by only the first nucleotide, dCTP. Figure 1C shows the one-dimensional (1D) PAGE patterns of both 6- and 1-nt extension products using RNA from viruses produced from cells with normal or reduced RHA content. Equal amounts of viral genomic RNA, determined by dot blot hybridization, were used in these reactions, and this was further tested for with a control (Fig. 1, “control”) that involves annealing a DNA primer complementary to viral RNA sequences downstream of the PBS and extending it by 6 nt (Materials and Methods). The results shown in Fig. 1C were quantitated by using a PhosphorImager, normalized to results with siRNACon, and shown graphically in Fig. 1D. Decreasing endogenous RHA results in about 40% and 50% reductions in 6-nt and 1-nt extensions, respectively, indicating that the initiation, rather than elongation, of reverse transcription was affected. The decreases in
extended either by these six nucleotides (nt), by replacing dATP with ddATP, or by only the first nucleotide, dCTP. Figure 1C shows the one-dimensional (1D) PAGE patterns of both 6- and 1-nt extension products using RNA from viruses produced from cells with normal or reduced RHA content. Equal amounts of viral genomic RNA, determined by dot blot hybridization, were used in these reactions, and this was further tested for with a control (Fig. 1, “control”) that involves annealing a DNA primer complementary to viral RNA sequences downstream of the PBS and extending it by 6 nt (Materials and Methods). The results shown in Fig. 1C were quantitated by using a PhosphorImager, normalized to results with siRNACon, and shown graphically in Fig. 1D. Decreasing endogenous RHA results in about 40% and 50% reductions in 6-nt and 1-nt extensions, respectively, indicating that the initiation, rather than elongation, of reverse transcription was affected. The decreases in 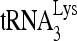 packaging shown in Fig. 1B are small compared to the decreases seen in 6- or 1-nt extension of
packaging shown in Fig. 1B are small compared to the decreases seen in 6- or 1-nt extension of 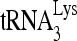 , suggesting that factors other than reduced
, suggesting that factors other than reduced 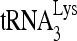 packaging are responsible for the decrease in
packaging are responsible for the decrease in 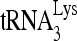 priming. These changes in the initiation of reverse transcription seen here correlate well with reported decreases in both viral infectivity and synthesis of minus-strand strong-stop cDNA that are seen upon new infection by viruses produced from cells with reduced RHA content (43).
priming. These changes in the initiation of reverse transcription seen here correlate well with reported decreases in both viral infectivity and synthesis of minus-strand strong-stop cDNA that are seen upon new infection by viruses produced from cells with reduced RHA content (43).
A well-recognized pitfall of siRNA experiments is possible off-target effects. We therefore constructed an exogenous Flag-tagged RHA gene (pFlag-RHA) whose mRNA product lacks siRNARHA target sequences, so as to determine the effect of increasing expression of RHA upon 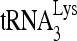 priming in the presence of highly reduced endogenous RHA. 293T cells were first transfected with siRNA and, 16 h later, were cotransfected with DNA coding for HIV-1 (BH10) and either a vector coding only for the Flag tag (pFlag) or a vector coding also for Flag-RHA. Western blot analysis of cell and viral lysates indicates that the strong reduction in cellular or viral RHA caused by siRNARHA is partially rescued by the expression of exogenous RHA (Fig. 1E). The viral content of RTp66/51, relative to CAp24, remains unaffected. The ability of exogenous RHA to rescue reduced
priming in the presence of highly reduced endogenous RHA. 293T cells were first transfected with siRNA and, 16 h later, were cotransfected with DNA coding for HIV-1 (BH10) and either a vector coding only for the Flag tag (pFlag) or a vector coding also for Flag-RHA. Western blot analysis of cell and viral lysates indicates that the strong reduction in cellular or viral RHA caused by siRNARHA is partially rescued by the expression of exogenous RHA (Fig. 1E). The viral content of RTp66/51, relative to CAp24, remains unaffected. The ability of exogenous RHA to rescue reduced 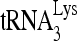 priming (6-nt extension) by siRNARHA is shown in Fig. 1F. The
priming (6-nt extension) by siRNARHA is shown in Fig. 1F. The 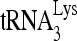 priming was decreased only slightly by siRNARHA in the presence of exogenous RHA. The partial rescue of
priming was decreased only slightly by siRNARHA in the presence of exogenous RHA. The partial rescue of 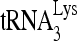 priming of reverse transcription most likely reflects the partial rescue of viral RHA by expression of exogenous RHA. However, expression of exogenous RHA did not significantly change the packaging of
priming of reverse transcription most likely reflects the partial rescue of viral RHA by expression of exogenous RHA. However, expression of exogenous RHA did not significantly change the packaging of 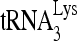 in HIV-1 virions (Fig. 1G).
in HIV-1 virions (Fig. 1G).
RHA is required for 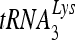 annealing to viral RNA in protease-negative HIV-1.
annealing to viral RNA in protease-negative HIV-1.
The selective concentration and annealing of 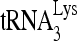 that occurs in wild-type HIV-1 also occurs in protease-negative virions. As Gag-facilitated annealing may represent the first step in
that occurs in wild-type HIV-1 also occurs in protease-negative virions. As Gag-facilitated annealing may represent the first step in 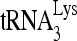 annealing to viral RNA, we determined whether RHA contributes to the annealing of
annealing to viral RNA, we determined whether RHA contributes to the annealing of 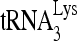 by Gag in vivo by investigating the effect of decreased RHA upon 6-nt extension of
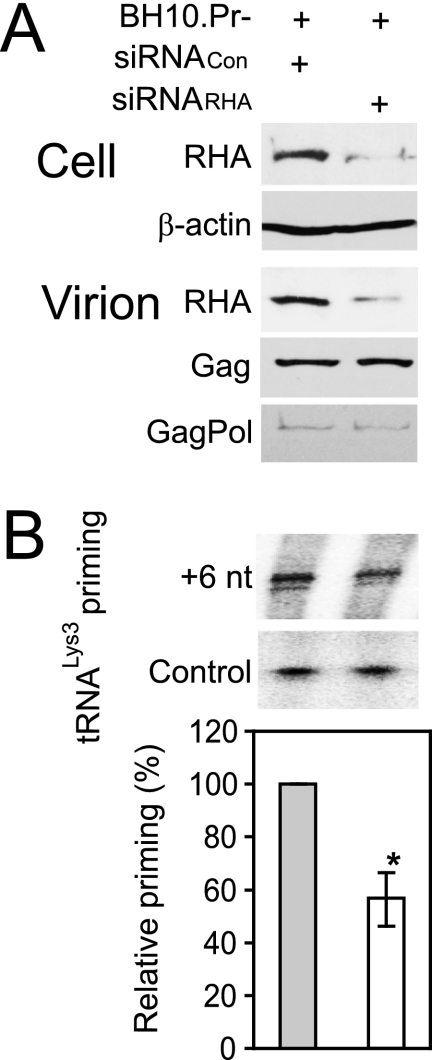
by Gag in vivo by investigating the effect of decreased RHA upon 6-nt extension of 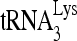 in protease-negative (Pr−) virions that are unable to process Gag or GagPol. Western blot analysis shows a reduction in both the cellular expression and viral incorporation of RHA in the presence of siRNARHA (Fig. 2A). We further found that the decreases in cellular and viral RHA are correlated with a significant decrease in the ability of
in protease-negative (Pr−) virions that are unable to process Gag or GagPol. Western blot analysis shows a reduction in both the cellular expression and viral incorporation of RHA in the presence of siRNARHA (Fig. 2A). We further found that the decreases in cellular and viral RHA are correlated with a significant decrease in the ability of 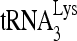 to be extended (Fig. 2B), suggesting that the effect of RHA upon
to be extended (Fig. 2B), suggesting that the effect of RHA upon 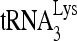 -primed initiation of reverse transcription in wild-type viruses may occur prior to the processing of Gag by viral protease. Thus, the influence of RHA upon Gag-facilitated
-primed initiation of reverse transcription in wild-type viruses may occur prior to the processing of Gag by viral protease. Thus, the influence of RHA upon Gag-facilitated 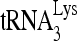 annealing is the focus of this report.
annealing is the focus of this report.
FIG. 2.
RHA is required for  annealing to viral RNA in protease-negative HIV-1. 293T cells were first transfected with siRNACon or siRNARHA and then transfected 16 h later with DNA coding for protease-negative HIV-1 (BH10.Pr−). (A) Western blots of cellular and viral lysates, containing equal amounts of β-actin or Gag, respectively, were probed with antibodies to RHA and β-actin (Cell) or, for viruses, with antibodies to RHA, CAp24 (Gag), or RTp66/p51 (GagPol). (B)
annealing to viral RNA in protease-negative HIV-1. 293T cells were first transfected with siRNACon or siRNARHA and then transfected 16 h later with DNA coding for protease-negative HIV-1 (BH10.Pr−). (A) Western blots of cellular and viral lysates, containing equal amounts of β-actin or Gag, respectively, were probed with antibodies to RHA and β-actin (Cell) or, for viruses, with antibodies to RHA, CAp24 (Gag), or RTp66/p51 (GagPol). (B)  -primed initiation of reverse transcription. Upper panel, 6-nt-extended
-primed initiation of reverse transcription. Upper panel, 6-nt-extended  , resolved by 1D PAGE and detected by autoradiography. The control panel represents the 6-nt extension of a DNA primer annealed to viral RNA downstream of the PBS and shows that equivalent amounts of viral RNA were used in each extension reaction. Lower panel, the 6-nt-extended
, resolved by 1D PAGE and detected by autoradiography. The control panel represents the 6-nt extension of a DNA primer annealed to viral RNA downstream of the PBS and shows that equivalent amounts of viral RNA were used in each extension reaction. Lower panel, the 6-nt-extended  products were quantitated using a PhosphorImager, and the graphed results shown in the lower panel are normalized to the value obtained with virions produced from cells cotransfected with BH10. Pr- and siRNACon. Average values ± standard errors (SE) of results of three experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test).
products were quantitated using a PhosphorImager, and the graphed results shown in the lower panel are normalized to the value obtained with virions produced from cells cotransfected with BH10. Pr- and siRNACon. Average values ± standard errors (SE) of results of three experiments are shown. Asterisk indicates a P value of <0.05 (Student's t test).
RHA promotes 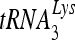 annealing to viral RNA in vitro.
annealing to viral RNA in vitro.
To confirm that RHA has a regulatory role in the annealing of 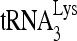 , we investigated the effect of RHA upon the annealing of 32P-
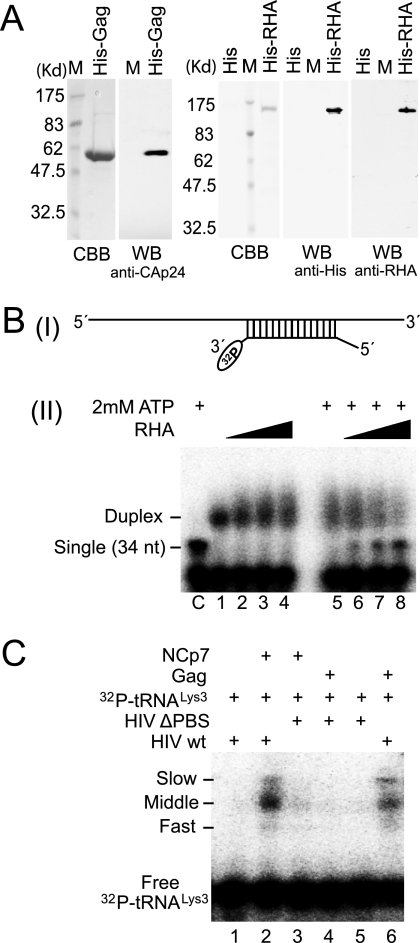
, we investigated the effect of RHA upon the annealing of 32P-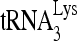 to viral RNA in vitro. Both Gag and RHA were purified from 293E cells, as shown by Western blot analysis (Fig. 3A). The presence of helicase activity in the purified RHA was determined by measuring its ability to unwind an RNA duplex composed of two strands: a longer strand of 186 nt and a shorter, complementary strand of 34 nt, which was 3′-end labeled with 32pCp. Unwinding this duplex will result in a labeled band with an increased electrophoretic mobility. The purified RHA possesses the ability to unwind the RNA duplex in an ATP-dependent manner (Fig. 3B).
to viral RNA in vitro. Both Gag and RHA were purified from 293E cells, as shown by Western blot analysis (Fig. 3A). The presence of helicase activity in the purified RHA was determined by measuring its ability to unwind an RNA duplex composed of two strands: a longer strand of 186 nt and a shorter, complementary strand of 34 nt, which was 3′-end labeled with 32pCp. Unwinding this duplex will result in a labeled band with an increased electrophoretic mobility. The purified RHA possesses the ability to unwind the RNA duplex in an ATP-dependent manner (Fig. 3B).
FIG. 3.
Protein purification and activity determination assay in vitro. (A) Purification of full-length Gag and enzymatically active RHA. WB, Western blots of full-length, His-tagged Gag or RHA purified from 293E cells, and resolved by 1D SDS-PAGE. Blots are probed with anti-CAp24 or with antibodies to either His or RHA. CBB, proteins in SDS-PAGE, stained with Coomassie brilliant blue R250. M, protein size markers (BioRad). (B) Helicase activity assay for purified RHA. Upper panel (I), schematic of the partial duplex RNA used in helicase activity assay. Lower panel (II), unwinding of this RNA duplex will result in a fast electrophoretic mobility of the 34-nt-labeled RNA strand. Boiled duplex RNA was included as a single control (lane C). The increasing amounts of RHA added in reactions were 0, 500, 1,000, and 2,000 fmol. RHA in the absence of ATP did not show unwinding activity. (C) Full-length Gag purified from 293E cells has 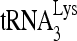 annealing activity. 32P-
annealing activity. 32P-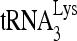 (100 fmol) was annealed to 30 fmol synthetic viral RNA, using either 30 pmol synthetic NCp7 or 20 pmol full-length Gag. The annealing reaction mixture was incubated at 25°C for 45 min, and free
(100 fmol) was annealed to 30 fmol synthetic viral RNA, using either 30 pmol synthetic NCp7 or 20 pmol full-length Gag. The annealing reaction mixture was incubated at 25°C for 45 min, and free 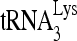 was resolved from
was resolved from 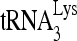 annealed to viral RNA by electrophoresis in 5% native polyacrylamide gel and was visualized by using a PhosphorImager.
annealed to viral RNA by electrophoresis in 5% native polyacrylamide gel and was visualized by using a PhosphorImager.
To study the effect of RHA upon 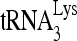 annealing in vitro, full-length synthetic
annealing in vitro, full-length synthetic 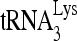 was 3′-end labeled with 32pCp and annealed to a 386-nt synthetic HIV-1 RNA, containing sequences from R through the codon for the 18th amino acid of Gag. The in vitro annealing of 32P-
was 3′-end labeled with 32pCp and annealed to a 386-nt synthetic HIV-1 RNA, containing sequences from R through the codon for the 18th amino acid of Gag. The in vitro annealing of 32P-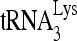 to viral RNA by either Gag or NCp7 was resolved in native 1D PAGE (Fig. 3C). When annealing is facilitated with either NCp7 or Gag, three
to viral RNA by either Gag or NCp7 was resolved in native 1D PAGE (Fig. 3C). When annealing is facilitated with either NCp7 or Gag, three 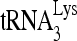 -viral RNA binary complex bands of different electrophoretic mobilities (slow, middle, and fast) were detected, with the middle band being the dominant species. These bands represent 32P-
-viral RNA binary complex bands of different electrophoretic mobilities (slow, middle, and fast) were detected, with the middle band being the dominant species. These bands represent 32P-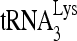 bound to the PBS region of the viral RNA, since no bands representing annealing are seen when the PBS was deleted from viral RNA (HIVΔPBS).
bound to the PBS region of the viral RNA, since no bands representing annealing are seen when the PBS was deleted from viral RNA (HIVΔPBS).
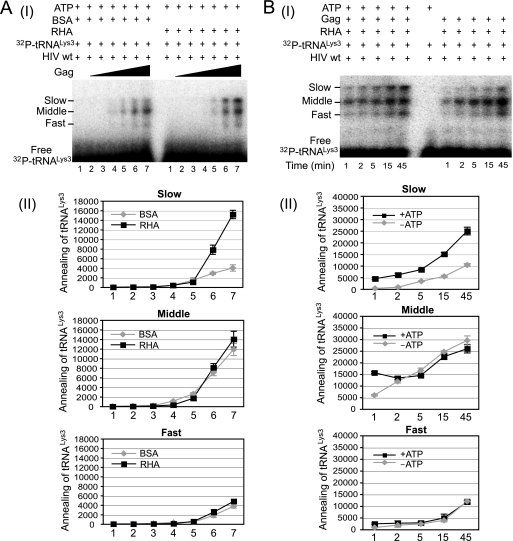
In Fig. 4A, part I, we examined the effect of adding RHA to the annealing reaction facilitated by increasing concentrations of Gag. All reaction mixtures contain the ATP required for RHA activity and include either BSA or RHA. When RHA is absent, there is an increase in 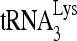 -annealed complex with increasing concentrations of Gag. However, the presence of RHA results in an increasing amount of the slow-migrating band. Quantitation of these bands is presented graphically in Fig. 4A, part II. The presence of RHA specifically increases the slow-migrating band almost 4-fold, without significantly affecting the amount of annealed complex represented by the middle and fast-migrating bands.
-annealed complex with increasing concentrations of Gag. However, the presence of RHA results in an increasing amount of the slow-migrating band. Quantitation of these bands is presented graphically in Fig. 4A, part II. The presence of RHA specifically increases the slow-migrating band almost 4-fold, without significantly affecting the amount of annealed complex represented by the middle and fast-migrating bands.
FIG. 4.
RHA enhances the formation of a slow mobility 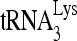 /viral RNA binary complex. (A) RHA promotes
/viral RNA binary complex. (A) RHA promotes 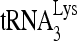 annealing. Upper panel (I), 100 fmol 32P-
annealing. Upper panel (I), 100 fmol 32P-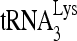 was annealed to 30 fmol synthetic viral RNA, using increasing concentrations of Gag, and in the additional presence of 1 pmol of either RHA or BSA. Femtomoles of Gag added were, from lane 1 to 7, 0, 50, 200, 500, 1,000, 5,000, and 20,000. Lower panel (II), the slow-, middle, and fast-migrating annealing complexes were quantitated using a PhosphorImager. Relative phosphorImager signals represent the annealing of
was annealed to 30 fmol synthetic viral RNA, using increasing concentrations of Gag, and in the additional presence of 1 pmol of either RHA or BSA. Femtomoles of Gag added were, from lane 1 to 7, 0, 50, 200, 500, 1,000, 5,000, and 20,000. Lower panel (II), the slow-, middle, and fast-migrating annealing complexes were quantitated using a PhosphorImager. Relative phosphorImager signals represent the annealing of 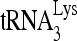 . Average values ± standard errors (SE) of results of three experiments are shown. (B) Time course of annealing, showing an ATP requirement for RHA-induced formation of the slow-migrating annealing complex. Upper panel (I), in the presence or absence of 2 mM ATP, 100 fmol of 32P-
. Average values ± standard errors (SE) of results of three experiments are shown. (B) Time course of annealing, showing an ATP requirement for RHA-induced formation of the slow-migrating annealing complex. Upper panel (I), in the presence or absence of 2 mM ATP, 100 fmol of 32P-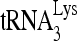 was annealed by 20 pmol Gag to 30 fmol of viral RNA in the presence of 1 pmol RHA. Annealing reaction mixtures were incubated from 1 to 45 min at 25°C. A reaction without RHA and Gag was used as a control (middle lane). Lower panel (II), the slow-, middle, and fast-migrating bands in part I were quantitated by using a PhosphorImager. Relative average values ± SE of results of three experiments are shown.
was annealed by 20 pmol Gag to 30 fmol of viral RNA in the presence of 1 pmol RHA. Annealing reaction mixtures were incubated from 1 to 45 min at 25°C. A reaction without RHA and Gag was used as a control (middle lane). Lower panel (II), the slow-, middle, and fast-migrating bands in part I were quantitated by using a PhosphorImager. Relative average values ± SE of results of three experiments are shown.
RHA uses ATP as a general energy source for its unwinding activity of double-stranded RNA (52). Thus, the influence of ATP on the action of RHA upon 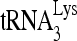 annealing was examined in two ways. In Fig. 4B, part I, the RHA-induced increase in the slow-migrating band does occur in the absence of ATP, but its appearance is speeded up when ATP is present, indicating that the spontaneous appearance of the slow-migrating band is accelerated by RHA helicase activity. Quantitation of these bands is presented graphically in Fig. 4B, part II, and indicates that the helicase activity of RHA is critical to the rate of formation of the slow-migrating band.
annealing was examined in two ways. In Fig. 4B, part I, the RHA-induced increase in the slow-migrating band does occur in the absence of ATP, but its appearance is speeded up when ATP is present, indicating that the spontaneous appearance of the slow-migrating band is accelerated by RHA helicase activity. Quantitation of these bands is presented graphically in Fig. 4B, part II, and indicates that the helicase activity of RHA is critical to the rate of formation of the slow-migrating band.
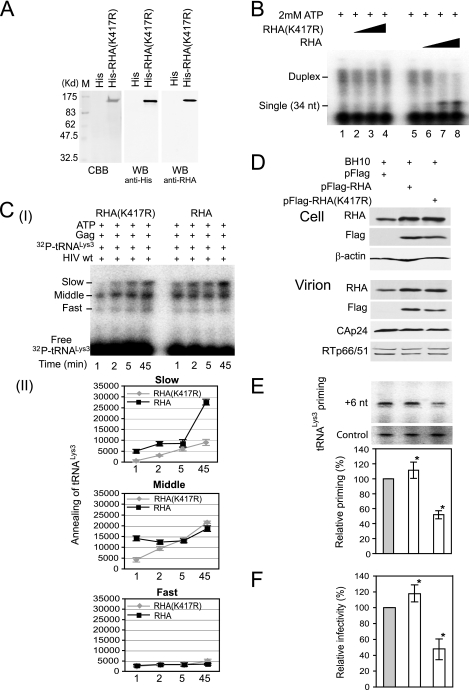
Alternatively, we tested the ability of a mutant RHA unable to utilize ATP to facilitate formation of the slow-migrating band. RHA containing the K417R mutation has a reduced ability to bind ATP (4, 39). This mutant RHA was purified from 293E cells (Fig. 5A) and shows a reduced ability to both unwind double-stranded RNA (Fig. 5B) and stimulate formation of the slow-migrating 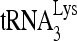 -viral RNA binary complex (Fig. 5C). The effect of wild-type or mutant RHA upon
-viral RNA binary complex (Fig. 5C). The effect of wild-type or mutant RHA upon 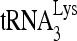 annealing is also reflected in vivo (Fig. 5D to F). Overexpression of wild-type or mutant RHA increases both cellular and viral content of either RHA (Fig. 5D). However, in contrast to the overexpression of exogenous wild-type RHA, which increases slightly the initiation of reverse transcription (6-nt extension), overexpression of RHA(K417R) decreased it by 50% (Fig. 5E), presumably because RHA(K417R) acts as a dominant negative regulator by competing with endogenous wild-type RHA. These changes in the ability of virions to initiate reverse transcription are correlated with similar changes in the infectivity of the viruses (Fig. 5F).
annealing is also reflected in vivo (Fig. 5D to F). Overexpression of wild-type or mutant RHA increases both cellular and viral content of either RHA (Fig. 5D). However, in contrast to the overexpression of exogenous wild-type RHA, which increases slightly the initiation of reverse transcription (6-nt extension), overexpression of RHA(K417R) decreased it by 50% (Fig. 5E), presumably because RHA(K417R) acts as a dominant negative regulator by competing with endogenous wild-type RHA. These changes in the ability of virions to initiate reverse transcription are correlated with similar changes in the infectivity of the viruses (Fig. 5F).
FIG. 5.
The K417R mutation reduces the ability of RHA to promote new annealing of 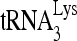 in vitro and in vivo. (A) Purification and characterization of enzymatically inactive RHA(K417R). WB, Western blots of RHA(K417R) purified from 293E cells and resolved by 1D SDS-PAGE. Blots are probed with antibodies to either His or RHA. CBB, proteins in SDS-PAGE are stained with Coomassie brilliant blue R250. M, protein size markers (BioRad). (B) Helicase activity assay for purified RHA(K417R), as described in the legend to Fig. 3. The increasing amounts of RHA added in reactions were 0, 500, 1,000, and 2,000 fmol. RHA(K417R) in the presence of ATP did not show unwinding activity. (C) Effect of RHA(K417R) upon
in vitro and in vivo. (A) Purification and characterization of enzymatically inactive RHA(K417R). WB, Western blots of RHA(K417R) purified from 293E cells and resolved by 1D SDS-PAGE. Blots are probed with antibodies to either His or RHA. CBB, proteins in SDS-PAGE are stained with Coomassie brilliant blue R250. M, protein size markers (BioRad). (B) Helicase activity assay for purified RHA(K417R), as described in the legend to Fig. 3. The increasing amounts of RHA added in reactions were 0, 500, 1,000, and 2,000 fmol. RHA(K417R) in the presence of ATP did not show unwinding activity. (C) Effect of RHA(K417R) upon 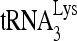 annealing. Upper panel (I), 100 fmol of 32P-
annealing. Upper panel (I), 100 fmol of 32P-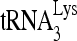 was annealed by 20 pmol Gag to 30 fmol of viral RNA in the presence of 1 pmol RHA or RHA(K417R) and 2 mM ATP. Annealing reaction mixtures were incubated from 1 to 45 min at 25°C. Lower panel (II), the slow-, middle, and fast-migrating bands in part I were quantitated by using a PhosphorImager. Relative phosphorImager signals represent the annealing of
was annealed by 20 pmol Gag to 30 fmol of viral RNA in the presence of 1 pmol RHA or RHA(K417R) and 2 mM ATP. Annealing reaction mixtures were incubated from 1 to 45 min at 25°C. Lower panel (II), the slow-, middle, and fast-migrating bands in part I were quantitated by using a PhosphorImager. Relative phosphorImager signals represent the annealing of 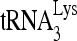 . Average values ± standard errors (SE) of results of three experiments are shown. (D to F) 293T cells were cotransfected with DNA coding for HIV-1 (BH10) and either pFlag, pFlag-RHA, or pFlag-RHA(K417R). (D) Western blots of cellular lysates probed with antibodies to RHA, Flag, and β-actin or of viral lysates containing equal amounts of CAp24, probed with antibodies to RHA, Flag, CAp24, and RTp66/p51. (E)
. Average values ± standard errors (SE) of results of three experiments are shown. (D to F) 293T cells were cotransfected with DNA coding for HIV-1 (BH10) and either pFlag, pFlag-RHA, or pFlag-RHA(K417R). (D) Western blots of cellular lysates probed with antibodies to RHA, Flag, and β-actin or of viral lysates containing equal amounts of CAp24, probed with antibodies to RHA, Flag, CAp24, and RTp66/p51. (E) 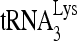 -primed initiation of reverse transcription. Upper panel, 6-nt-extended
-primed initiation of reverse transcription. Upper panel, 6-nt-extended 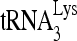 , resolved by 1D PAGE and detected by autoradiography. The control panel shows that equal amounts of viral RNA were used in each extension reaction, as described in the legend in Fig. 2B. Bands in the upper panel were quantitated using a PhosphorImager, and the graphed results are normalized to the value obtained with virions produced from cells cotransfected with BH10 and pFlag. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to the control. (F) Single-round viral infectivity was measured using TMZ-bl cells. The results were normalized to virions produced from cells cotransfected with BH10 and pFlag. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to the control.
, resolved by 1D PAGE and detected by autoradiography. The control panel shows that equal amounts of viral RNA were used in each extension reaction, as described in the legend in Fig. 2B. Bands in the upper panel were quantitated using a PhosphorImager, and the graphed results are normalized to the value obtained with virions produced from cells cotransfected with BH10 and pFlag. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to the control. (F) Single-round viral infectivity was measured using TMZ-bl cells. The results were normalized to virions produced from cells cotransfected with BH10 and pFlag. Average values ± SE of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to the control.
Differences in the electrophoretic mobilities of  annealing complexes reflect different viral RNA conformations.
annealing complexes reflect different viral RNA conformations.
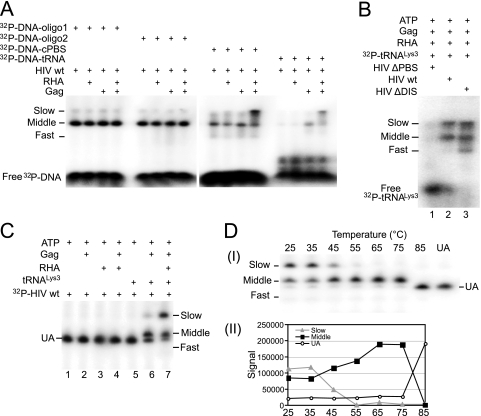
There is only one PBS per viral RNA molecule, and the absence of 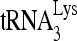 annealing to viral RNA in which the PBS has been deleted (Fig. 4C) indicates that the decreased mobility of the slow-migrating band is not due to an increase in the number of
annealing to viral RNA in which the PBS has been deleted (Fig. 4C) indicates that the decreased mobility of the slow-migrating band is not due to an increase in the number of  molecules bound per genomic RNA. To further understand how the slow-migrating annealing complex is formed, we examined the effect of replacing 32P-
molecules bound per genomic RNA. To further understand how the slow-migrating annealing complex is formed, we examined the effect of replacing 32P- in the annealing reaction with a labeled 18-nt DNA complementary to either the PBS or to the regions in the viral RNA downstream of the PBS (Fig. 6A). Unlike
in the annealing reaction with a labeled 18-nt DNA complementary to either the PBS or to the regions in the viral RNA downstream of the PBS (Fig. 6A). Unlike  , annealing of short DNA oligomers to the viral RNA occurs independently of Gag. Annealed oligomers complementary to nt 254 to 271 (oligo 1) or 326 to 343 (oligo 2) show a strong hybridization complex migrating in gels with middle band mobility, which is not affected by the presence or absence of RHA and/or Gag, while the oligomer annealed to the PBS (nt 182 to 200) shows a weaker middle band annealing and a strong mobility shift to the slow-migrating band when RHA and Gag are present together. Thus, annealing to the PBS seems to be required for generating the RHA-induced slow-migrating complex, whose formation is independent of the molecular weight of the annealed component. This was further verified by the annealing of a longer DNA oligomer (32P-DNA-tRNA) representing the full-length
, annealing of short DNA oligomers to the viral RNA occurs independently of Gag. Annealed oligomers complementary to nt 254 to 271 (oligo 1) or 326 to 343 (oligo 2) show a strong hybridization complex migrating in gels with middle band mobility, which is not affected by the presence or absence of RHA and/or Gag, while the oligomer annealed to the PBS (nt 182 to 200) shows a weaker middle band annealing and a strong mobility shift to the slow-migrating band when RHA and Gag are present together. Thus, annealing to the PBS seems to be required for generating the RHA-induced slow-migrating complex, whose formation is independent of the molecular weight of the annealed component. This was further verified by the annealing of a longer DNA oligomer (32P-DNA-tRNA) representing the full-length  sequence (76 nt) to the viral RNA. This annealing is Gag dependent, but in spite of the difference in size (76 nt versus 18 nt), the middle and slow bands migrate with the same mobility as when an 18-mer DNA is annealed, suggesting that the change in mobility is primarily due to a change in viral RNA conformation.
sequence (76 nt) to the viral RNA. This annealing is Gag dependent, but in spite of the difference in size (76 nt versus 18 nt), the middle and slow bands migrate with the same mobility as when an 18-mer DNA is annealed, suggesting that the change in mobility is primarily due to a change in viral RNA conformation.
FIG. 6.
Factors influencing the electrophoretic mobility of viral RNA. (A) Influence of RHA upon the annealing of 32P-DNA oligomers to viral RNA in vitro. 32P-DNA oligomers (100 fmol) were annealed to 30 fmol synthetic viral RNA in the presence or absence of 20 pmol Gag and 1 pmol RHA. Annealing reaction mixtures were incubated at 25°C for 45 min. Free and bound DNA oligomers were resolved by electrophoresis in a 5% native polyacrylamide gel and visualized by using a PhosphorImager. (B) 1D PAGE showing the influence of deletions of the PBS or the DIS upon the electrophoretic mobility of annealed products. Annealing was carried out at 25°C for 45 min, and the annealing conditions contained 30 fmol 32P-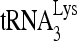 , 100 fmol of wild-type or mutant viral RNA, 20 pmol Gag, 2 mM ATP, and 1 pmol RHA. (C) 1D PAGE showing the influence of RHA, Gag, and
, 100 fmol of wild-type or mutant viral RNA, 20 pmol Gag, 2 mM ATP, and 1 pmol RHA. (C) 1D PAGE showing the influence of RHA, Gag, and 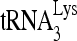 upon the electrophoretic mobility of annealed and unannealed viral RNA. Annealing was carried out at 25°C for 45 min, and the complete annealing conditions (lane 7) contained 2 mM ATP, 100 fmol
upon the electrophoretic mobility of annealed and unannealed viral RNA. Annealing was carried out at 25°C for 45 min, and the complete annealing conditions (lane 7) contained 2 mM ATP, 100 fmol 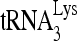 , 20 pmol Gag, 1 pmol RHA, and 30 fmol of 32P-viral RNA. (D) Aliquots from the sample analyzed in panel C, lane 7, were each incubated for 2 min at increasing temperatures, quick-cooled on ice, and resolved by 1D PAGE (part I). Unannealed (UA) viral RNA and the slow- and middle migrating annealed complexes were quantitated using a PhosphorImager, and the results were graphed (part II).
, 20 pmol Gag, 1 pmol RHA, and 30 fmol of 32P-viral RNA. (D) Aliquots from the sample analyzed in panel C, lane 7, were each incubated for 2 min at increasing temperatures, quick-cooled on ice, and resolved by 1D PAGE (part I). Unannealed (UA) viral RNA and the slow- and middle migrating annealed complexes were quantitated using a PhosphorImager, and the results were graphed (part II).
Figure 6A also shows that a small amount of slow-migrating annealing complex appears to be present independently of either Gag or annealing that is specifically to the PBS and probably represents a small fraction of the slow-migrating viral RNA conformer that is in equilibrium with the main viral RNA conformer. However, driving the equilibrium toward slow-migrating conformer requires not only Gag but RHA, and a specific annealing to the PBS appears to be required for stabilization.
In Fig. 6B, we have examined whether the RHA-induced mobility shift from the middle to the slow-migrating band could be due to a shift from the monomeric to the dimeric form of the viral RNA. 32P-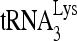 was annealed in vitro to either wild-type viral RNA (HIV wt, lane 2), viral RNA with a deleted PBS (HIVΔPBS, lane 1), or viral DNA with a deleted DIS (HIVΔDIS, lane 3). Although removal of the DIS prevents formation of the dimeric viral RNA (14), this mutation does not prevent formation of the slow-migrating hybridization complex. The fast-migrating band is more clearly seen in lane 3. It may represent another conformation of the
was annealed in vitro to either wild-type viral RNA (HIV wt, lane 2), viral RNA with a deleted PBS (HIVΔPBS, lane 1), or viral DNA with a deleted DIS (HIVΔDIS, lane 3). Although removal of the DIS prevents formation of the dimeric viral RNA (14), this mutation does not prevent formation of the slow-migrating hybridization complex. The fast-migrating band is more clearly seen in lane 3. It may represent another conformation of the 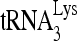 -viral RNA binary complex whose relative amount has been increased as a result of the ΔDIS mutation. In the previous in vitro annealing experiments described in this work, we used a 3-molar excess of 32P-
-viral RNA binary complex whose relative amount has been increased as a result of the ΔDIS mutation. In the previous in vitro annealing experiments described in this work, we used a 3-molar excess of 32P-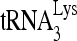 to viral RNA, but to see if the
to viral RNA, but to see if the 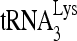 /viral RNA ratio had any effect upon the results, the experiments shown in Fig. 6B were done with a 3-molar excess of viral RNA to
/viral RNA ratio had any effect upon the results, the experiments shown in Fig. 6B were done with a 3-molar excess of viral RNA to 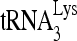 , thus accounting for the complete annealing of
, thus accounting for the complete annealing of 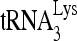 in these experiments.
in these experiments.
Further evidence that RHA is altering the viral RNA conformation is shown by exposure of 3′-32P-viral RNA to RHA in the presence or absence of unlabeled 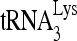 (Fig. 6C). Annealing conditions were similar to those used in Fig. 4 and 5, with an incubation time of 45 min. The first 5 lanes show that, whatever combination of ATP, Gag, or RHA the viral RNA is exposed to, in the absence of
(Fig. 6C). Annealing conditions were similar to those used in Fig. 4 and 5, with an incubation time of 45 min. The first 5 lanes show that, whatever combination of ATP, Gag, or RHA the viral RNA is exposed to, in the absence of 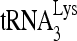 , there is no detectable change in the electrophoretic mobility of the unannealed (UA) viral RNA. The 6th lanes show the small decrease in electrophoretic mobility of viral RNA when
, there is no detectable change in the electrophoretic mobility of the unannealed (UA) viral RNA. The 6th lanes show the small decrease in electrophoretic mobility of viral RNA when 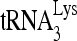 is annealed to the viral RNA in the absence of RHA, with a corresponding diminishment of unannealed viral RNA. As the experimental conditions in lanes 2 and 6 are identical except for the addition of
is annealed to the viral RNA in the absence of RHA, with a corresponding diminishment of unannealed viral RNA. As the experimental conditions in lanes 2 and 6 are identical except for the addition of 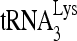 to lane 6, the small reduction in electrophoretic mobility is likely to be due to a change in molecular weight of the annealed complex. As described for Fig. 6A, the small amount of slow-migrating complex generated in the absence of RHA suggests that this conformation of viral RNA is a minor component in the total viral RNA without exposure to RHA, but that can only be seen when stabilized through annealing. However, the switch to the slow mobility complex is greatly enhanced when RHA is also present (lane 7) and is accompanied by a significant decrease in UA viral RNA.
to lane 6, the small reduction in electrophoretic mobility is likely to be due to a change in molecular weight of the annealed complex. As described for Fig. 6A, the small amount of slow-migrating complex generated in the absence of RHA suggests that this conformation of viral RNA is a minor component in the total viral RNA without exposure to RHA, but that can only be seen when stabilized through annealing. However, the switch to the slow mobility complex is greatly enhanced when RHA is also present (lane 7) and is accompanied by a significant decrease in UA viral RNA.
Further confirmation of the different viral RNA conformations present in the slow- and middle migrating annealing complexes is shown in Fig. 6D, where the sample analyzed electrophoretically in Fig. 6C, lane 7, was subjected to increases in temperature. The early melting (Tm = 45°C) of the slow-migrating annealing complex was correlated with an increase in the amount of middle migrating annealing complex, without any loss in annealed 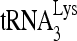 . At a Tm of 85°C, the rapid loss of the middle migrating annealing complex is associated with a similar increase in UA viral RNA, suggesting the loss of annealed
. At a Tm of 85°C, the rapid loss of the middle migrating annealing complex is associated with a similar increase in UA viral RNA, suggesting the loss of annealed 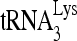 from viral RNA at these temperatures.
from viral RNA at these temperatures.
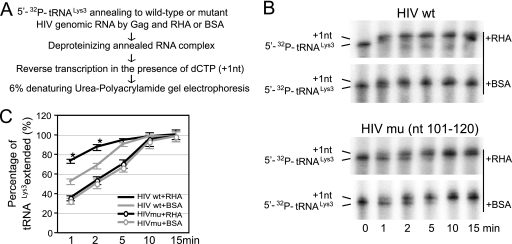
Viral RNA sequences critical to the formation of the slow-migrating annealed complex.
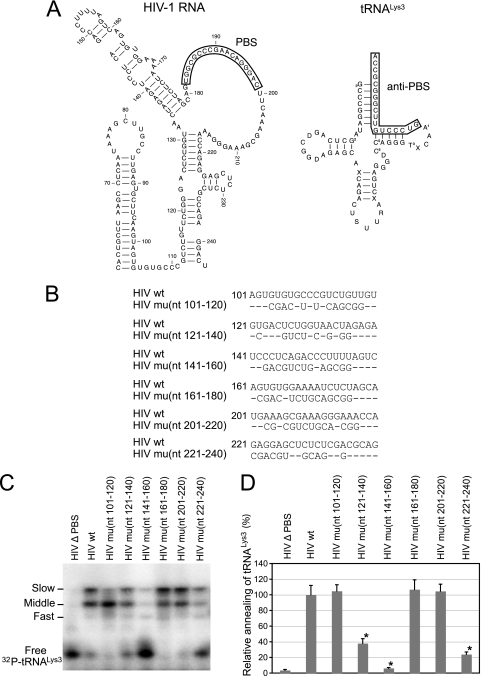
To determine what sequences of the viral RNA contribute to the formation of the slow-migrating 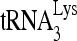 -viral RNA binary complex, viral RNA sequences between nt 101 and 221 were mutated by substitution with the same random 20-nt RNA sequence (5′-AGUCGACGUCUGCAGCGGGU-3′) (Fig. 7B). Figure 7A shows one of the predicted secondary structures of both the U5/leader sequence in HIV-1 and of
-viral RNA binary complex, viral RNA sequences between nt 101 and 221 were mutated by substitution with the same random 20-nt RNA sequence (5′-AGUCGACGUCUGCAGCGGGU-3′) (Fig. 7B). Figure 7A shows one of the predicted secondary structures of both the U5/leader sequence in HIV-1 and of 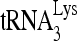 , from which the location of these substitution mutations can be ascertained. Figure 7C shows the 1D PAGE pattern of 32P-
, from which the location of these substitution mutations can be ascertained. Figure 7C shows the 1D PAGE pattern of 32P-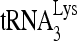 annealed by Gag to an excess of either wild-type or mutant viral RNA in the presence of RHA and ATP. Figure 7D compares the overall annealing of
annealed by Gag to an excess of either wild-type or mutant viral RNA in the presence of RHA and ATP. Figure 7D compares the overall annealing of 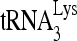 between wild-type and mutant viral RNAs. Aside from the PBS, mutation of sequences between nt 121 and 140, 141 and 160, and 221 and 240 dramatically reduced the annealing of
between wild-type and mutant viral RNAs. Aside from the PBS, mutation of sequences between nt 121 and 140, 141 and 160, and 221 and 240 dramatically reduced the annealing of 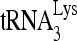 to viral RNA. In contrast, mutation of other sequences did not change the overall annealing of
to viral RNA. In contrast, mutation of other sequences did not change the overall annealing of 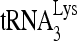 to viral RNA but affected the switch between the middle and slow-migrating conformer of the
to viral RNA but affected the switch between the middle and slow-migrating conformer of the 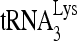 -viral RNA binary complex. Most notably, mutation of sequences between nt 101 and nt 120 completely blocked the formation of slow-migrating complex, indicating that these sequences are critical to the RHA-induced conformation change.
-viral RNA binary complex. Most notably, mutation of sequences between nt 101 and nt 120 completely blocked the formation of slow-migrating complex, indicating that these sequences are critical to the RHA-induced conformation change.
FIG. 7.
RNA sequences contributing to the RHA-induced change in conformation of 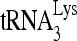 -viral RNA binary complex. 32P-
-viral RNA binary complex. 32P-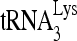 (30 fmol) was annealed in the presence of 20 pmol of Gag and 1 pmol RHA to 100 fmol of wild-type (wt) viral RNA or mutant (mu) viral RNA containing 20-nt substitution mutations. (A) Secondary structure of PBS stem-loop region of HIV-1 RNA or
(30 fmol) was annealed in the presence of 20 pmol of Gag and 1 pmol RHA to 100 fmol of wild-type (wt) viral RNA or mutant (mu) viral RNA containing 20-nt substitution mutations. (A) Secondary structure of PBS stem-loop region of HIV-1 RNA or 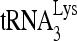 . (B) Mutated RNA sequences. (C) After deproteinization, annealed complexes were resolved using 1D PAGE. (D) Quantitation of total annealing of
. (B) Mutated RNA sequences. (C) After deproteinization, annealed complexes were resolved using 1D PAGE. (D) Quantitation of total annealing of 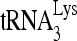 to mutant viral RNA observed in panel C, relative to annealing to wild-type HIV-1 RNA. Average values ± standard errors (SE) of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to HIV wt.
to mutant viral RNA observed in panel C, relative to annealing to wild-type HIV-1 RNA. Average values ± standard errors (SE) of results of three experiments are shown. Asterisks indicate a P value of <0.05 (Student's t test) compared to HIV wt.
 -viral RNA binary complexes enriched in the RHA-induced slow-migrating conformation display a higher efficiency of reverse transcription initiation.
-viral RNA binary complexes enriched in the RHA-induced slow-migrating conformation display a higher efficiency of reverse transcription initiation.
We have examined the ability of 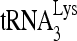 to initiate reverse transcription in annealing complex mixtures that do and do not contain the slow-migrating
to initiate reverse transcription in annealing complex mixtures that do and do not contain the slow-migrating 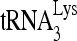 -viral RNA binary complex. The
-viral RNA binary complex. The 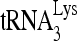 was 5′-end labeled with [γ-32P]ATP, annealed by Gag to an excess of either wild-type or mutant (nt 101 to 120) viral RNA in the presence or absence of RHA. The RNA was then deproteinized and extended 1 nt in an in vitro reverse transcription reaction mixture containing dCTP (Fig. 8A). The extended and unextended 32P-
was 5′-end labeled with [γ-32P]ATP, annealed by Gag to an excess of either wild-type or mutant (nt 101 to 120) viral RNA in the presence or absence of RHA. The RNA was then deproteinized and extended 1 nt in an in vitro reverse transcription reaction mixture containing dCTP (Fig. 8A). The extended and unextended 32P-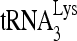 primers were resolved on denaturing 6% polyacrylamide gels (Fig. 8B), and the percentage of the 32P-
primers were resolved on denaturing 6% polyacrylamide gels (Fig. 8B), and the percentage of the 32P- 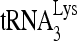 that was extended is graphed (Fig. 8C). After dCTP incorporation for 1 min, it is clear that 32P-
that was extended is graphed (Fig. 8C). After dCTP incorporation for 1 min, it is clear that 32P- 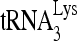 annealed to wild-type viral RNA in the presence of RHA is more efficient at incorporating dCTP than that annealed in the absence of RHA. Efficiency of dCTP incorporation is lowest for the 32P-
annealed to wild-type viral RNA in the presence of RHA is more efficient at incorporating dCTP than that annealed in the absence of RHA. Efficiency of dCTP incorporation is lowest for the 32P-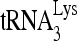 annealed to the viral RNA containing the mutation (nt 101 to 120) and is not affected by the presence or absence of RHA. The highest rate of dCTP incorporation may primarily reflect the efficient initiation of reverse transcription from the slow-migrating
annealed to the viral RNA containing the mutation (nt 101 to 120) and is not affected by the presence or absence of RHA. The highest rate of dCTP incorporation may primarily reflect the efficient initiation of reverse transcription from the slow-migrating 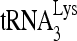 -viral RNA binary complex.
-viral RNA binary complex.
FIG. 8.
Initiation of reverse transcription from 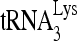 -viral RNA complexes lacking or enriched in the slow-migrating conformation. 5′-32P-
-viral RNA complexes lacking or enriched in the slow-migrating conformation. 5′-32P-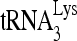 (30 fmol) were annealed in the presence of 40 pmol Gag and 1 pmol RHA or BSA to 100 fmol wild-type viral RNA. The annealed RNA complex was deproteinized by using proteinase K/phenol-chloroform extraction, precipitated by isopropanol, resuspended in 10 mM Tris, pH 7.5, extended for various lengths of time by 1 nt (dCTP) using HIV-1 reverse transcriptase, denatured by boiling, and then resolved by 1D PAGE. (A) Diagram of experimental procedure. (B) Time course of reverse transcription after
(30 fmol) were annealed in the presence of 40 pmol Gag and 1 pmol RHA or BSA to 100 fmol wild-type viral RNA. The annealed RNA complex was deproteinized by using proteinase K/phenol-chloroform extraction, precipitated by isopropanol, resuspended in 10 mM Tris, pH 7.5, extended for various lengths of time by 1 nt (dCTP) using HIV-1 reverse transcriptase, denatured by boiling, and then resolved by 1D PAGE. (A) Diagram of experimental procedure. (B) Time course of reverse transcription after 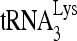 annealing to viral RNA. Lane 0, annealed
annealing to viral RNA. Lane 0, annealed 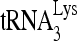 that was not extended by 1 nt in the absence of reverse transcriptase was loaded in the gel to represent the total annealed
that was not extended by 1 nt in the absence of reverse transcriptase was loaded in the gel to represent the total annealed 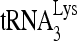 used in each reaction. (C) Quantitation of extended
used in each reaction. (C) Quantitation of extended 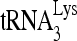 (+1 nt) by using a PhosphorImager was normalized with total annealed
(+1 nt) by using a PhosphorImager was normalized with total annealed 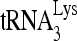 (lane 0) and presented as a percentage. Average values ± standard errors (SE) of results of three experiments are shown. Asterisks indicate P < 0.05 (Student's t test) compared to the +BSA control.
(lane 0) and presented as a percentage. Average values ± standard errors (SE) of results of three experiments are shown. Asterisks indicate P < 0.05 (Student's t test) compared to the +BSA control.
DISCUSSION
Even though RHA has been shown to be incorporated into virions (43), we believe that RHA promotes the annealing of 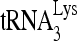 to viral RNA in the cytoplasm, prior to viral RNA incorporation into the virus. The annealing of
to viral RNA in the cytoplasm, prior to viral RNA incorporation into the virus. The annealing of 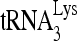 by Gag, either in vitro or in protease-negative viruses, suggests that the selective incorporation of
by Gag, either in vitro or in protease-negative viruses, suggests that the selective incorporation of 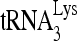 into mature virions may reflect an earlier concentration of
into mature virions may reflect an earlier concentration of 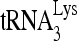 at a cellular site of early viral assembly, where
at a cellular site of early viral assembly, where 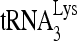 annealing to viral RNA also occurs, prior to viral budding and processing of Gag and GagPol (22). The evidence for the formation of a
annealing to viral RNA also occurs, prior to viral budding and processing of Gag and GagPol (22). The evidence for the formation of a 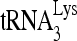 annealing complex in the cytoplasm has been summarized elsewhere (33), and direct evidence for the annealing of
annealing complex in the cytoplasm has been summarized elsewhere (33), and direct evidence for the annealing of 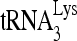 to HIV-1 RNA in infected cells was recently reported (50).
to HIV-1 RNA in infected cells was recently reported (50).
The reduced infectivity of HIV-1 produced from cells with reduced RHA (9, 43) is correlated with the reduced activity of reverse transcription in vivo (43). In this report, we demonstrate that this reduction in reverse transcription results from an inhibition of 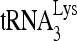 annealing to viral RNA, and we show that enzymatically active RHA, in cooperation with Gag, increases the in vitro annealing of
annealing to viral RNA, and we show that enzymatically active RHA, in cooperation with Gag, increases the in vitro annealing of 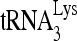 by Gag. Previous reports have speculated on the role of RHA in rearranging chromatin (20) or ribonucleoprotein (9) but without direct evidence. Here, we observed an RHA-induced change in viral RNA conformation that is associated with the increase in Gag-facilitated
by Gag. Previous reports have speculated on the role of RHA in rearranging chromatin (20) or ribonucleoprotein (9) but without direct evidence. Here, we observed an RHA-induced change in viral RNA conformation that is associated with the increase in Gag-facilitated 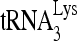 annealing. While reduction in RHA also produces a much smaller change in the incorporation of
annealing. While reduction in RHA also produces a much smaller change in the incorporation of 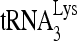 into virions (∼10% decrease; Fig. 1B), this probably represents an off-target effect of siRNARHA, other than the change in viral RNA conformation reported here, since
into virions (∼10% decrease; Fig. 1B), this probably represents an off-target effect of siRNARHA, other than the change in viral RNA conformation reported here, since 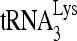 incorporation into HIV-1 occurs independently of viral RNA packaging (38).
incorporation into HIV-1 occurs independently of viral RNA packaging (38).
We show using truncated viral RNA that a 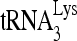 -viral RNA binary complex formed in vitro can display different conformations upon exposure to both Gag and RHA that are distinguished by their electrophoretic mobilities in native polyacrylamide gels (Fig. 3 and 4). The RHA-induced alteration in conformation is dependent upon forming a PBS duplex but not necessarily with
-viral RNA binary complex formed in vitro can display different conformations upon exposure to both Gag and RHA that are distinguished by their electrophoretic mobilities in native polyacrylamide gels (Fig. 3 and 4). The RHA-induced alteration in conformation is dependent upon forming a PBS duplex but not necessarily with 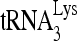 . Thus, this conformation can be achieved if the PBS is annealed with either
. Thus, this conformation can be achieved if the PBS is annealed with either 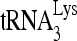 (which requires Gag for annealing) or an 18-nt DNA oligomer complementary to the PBS (which anneals independently of Gag). If viral RNA is annealed in the presence of Gag and RHA with an 18-nt DNA oligomer complementary to some other region in the viral RNA genome, no conformational change is seen (Fig. 6), indicating that the altered electrophoretic mobility of viral RNA is not due to a change in molecular weight but to alterations in the intramolecular interactions of viral RNA.
(which requires Gag for annealing) or an 18-nt DNA oligomer complementary to the PBS (which anneals independently of Gag). If viral RNA is annealed in the presence of Gag and RHA with an 18-nt DNA oligomer complementary to some other region in the viral RNA genome, no conformational change is seen (Fig. 6), indicating that the altered electrophoretic mobility of viral RNA is not due to a change in molecular weight but to alterations in the intramolecular interactions of viral RNA.
The change in viral RNA conformation enhanced by RHA was also confirmed by studying the thermal stability of the slow- and middle migrating complexes containing 32P-viral RNA. As shown in Fig. 6D, increasing temperature causes a shift from slow-migrating conformation to middle migrating conformation, without any loss of the annealed 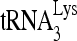 . Only at higher temperatures does the middle migrating band disappear, accompanied by a similar increase in unannealed viral RNA. The data show that the slow-migrating annealing complex conformation is less stable than the middle migrating annealing complex conformation, which may be associated with the fact that the formation of the slow-migrating conformation requires energy input, i.e., ATP-dependent RNA helicase activity. The coordinate action of RHA plus Gag results in a doubling of
. Only at higher temperatures does the middle migrating band disappear, accompanied by a similar increase in unannealed viral RNA. The data show that the slow-migrating annealing complex conformation is less stable than the middle migrating annealing complex conformation, which may be associated with the fact that the formation of the slow-migrating conformation requires energy input, i.e., ATP-dependent RNA helicase activity. The coordinate action of RHA plus Gag results in a doubling of 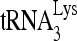 annealing over that achieved by Gag alone over the same time period (Fig. 4B, part II, and Fig. 5C, part II, top panels). If slow- and middle migrating annealing complex conformations had the same rate of annealing, the proportion of each in solution should not affect this rate. Therefore, annealing complex with the slow-migrating conformation must have a better rate of annealing than that with middle migrating conformation.
annealing over that achieved by Gag alone over the same time period (Fig. 4B, part II, and Fig. 5C, part II, top panels). If slow- and middle migrating annealing complex conformations had the same rate of annealing, the proportion of each in solution should not affect this rate. Therefore, annealing complex with the slow-migrating conformation must have a better rate of annealing than that with middle migrating conformation.
The data presented in this work are consistent with the following annealing scenario. The majority of the 5′ region of viral RNA studied here folds into the lowest energy state, vRNA, with a small fraction of the population existing with the slow-migrating conformation, vRNA*. In the presence of Gag, 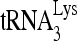 will anneal to both forms, with the major annealed form containing the vRNA conformation. The presence of RHA increases formation of unannealed vRNA*, whose conformation will be stabilized by the Gag-facilitated annealing with
will anneal to both forms, with the major annealed form containing the vRNA conformation. The presence of RHA increases formation of unannealed vRNA*, whose conformation will be stabilized by the Gag-facilitated annealing with 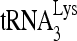 . Both middle and slow-migrating conformations compete for unannealed viral RNA, and the ratio of slow-/middle migrating annealing complexes will depend upon the vRNA*/vRNA of the unannealed viral RNA, which is regulated by RHA. The relative stability in the amount of slow-migrating annealing complex in the presence or absence of RHA indicates that RHA's effect is primarily on the unannealed vRNA.
. Both middle and slow-migrating conformations compete for unannealed viral RNA, and the ratio of slow-/middle migrating annealing complexes will depend upon the vRNA*/vRNA of the unannealed viral RNA, which is regulated by RHA. The relative stability in the amount of slow-migrating annealing complex in the presence or absence of RHA indicates that RHA's effect is primarily on the unannealed vRNA.
The nature of the conformation change to the slow migrating complex is not yet known. In most RNA models, the PBS is part of a single-stranded loop (1, 49), and since the change in viral RNA conformation is seen only when the PBS is annealed with complementary sequences, an initial annealing to the PBS may be required to produce a conformation that allows other regions in viral RNA and 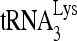 to hybridize with each other to form a
to hybridize with each other to form a 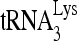 -viral RNA binary complex that shows increases in both the rate of
-viral RNA binary complex that shows increases in both the rate of 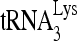 annealing and in the initiation of reverse transcription initiation (Fig. 8).
annealing and in the initiation of reverse transcription initiation (Fig. 8).
We have made a series of substitution mutations in the U5 region upstream of the PBS and in the leader sequence downstream of the PBS in order to determine what sequences might be involved in the RHA-induced conformational switch (Fig. 7). The most informative of these is the mutation replacing nt 101 to 120. These nucleotides, while not affecting 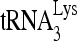 annealing, are critical for obtaining the RHA-induced conformational switch of the
annealing, are critical for obtaining the RHA-induced conformational switch of the 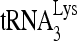 -viral RNA binary complex. It is not yet clear if these sequences contain a binding site for Gag/RHA or if the effect of the mutation is primarily conformational. This substitution mutation results in a significant decrease in the ability to initiate reverse transcription (Fig. 8).
-viral RNA binary complex. It is not yet clear if these sequences contain a binding site for Gag/RHA or if the effect of the mutation is primarily conformational. This substitution mutation results in a significant decrease in the ability to initiate reverse transcription (Fig. 8).
Substitution mutations for nt 161 to 180 and 201 to 220 have little effect upon either annealing or the RHA-induced conformational switch. There is no published evidence that either nt 161 to 180 or nt 201 to 220 have an effect upon the amount of 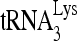 annealed. However, nt 161 to 180 do contain an A-rich loop that appears to interact with the 4 U's of the
annealed. However, nt 161 to 180 do contain an A-rich loop that appears to interact with the 4 U's of the 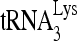 anticodon loop, as determined by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (49). However, earlier chemical probing and enzymatic footprinting had suggested that this interaction might be more important for the initiation of reverse transcription than for
anticodon loop, as determined by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (49). However, earlier chemical probing and enzymatic footprinting had suggested that this interaction might be more important for the initiation of reverse transcription than for 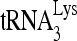 annealing (6, 28, 53). Furthermore, a biological role for the interaction between the A-rich motif and the anticodon of
annealing (6, 28, 53). Furthermore, a biological role for the interaction between the A-rich motif and the anticodon of 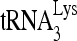 remains controversial, since many strains of HIV-1 do not contain this A-rich motif (21).
remains controversial, since many strains of HIV-1 do not contain this A-rich motif (21).
The effect of substitution mutations in nt 121 to 140, 141 to 160, and 221 to 240 is to inhibit overall annealing. nt 121 to 140 were not found to interact with 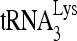 using SHAPE (49); nevertheless, this mutation significantly reduces the Gag-facilitated
using SHAPE (49); nevertheless, this mutation significantly reduces the Gag-facilitated 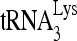 annealing in vitro. This region contains the PAS motif (nt 123 to 130), a sequence that is complementary to sequences in the TψC arm of
annealing in vitro. This region contains the PAS motif (nt 123 to 130), a sequence that is complementary to sequences in the TψC arm of 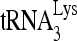 (7) and that has been reported to influence the initiation of reverse transcription but without affecting the amount of
(7) and that has been reported to influence the initiation of reverse transcription but without affecting the amount of 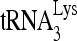 annealed (7, 40). However, annealing conditions used for those reports were different from those used for this report, including the absence of Gag and RHA in the annealing conditions. The large decrease in annealing for mutation in nt 141 to 160 may be due to nt 142 to 147 in this region, which have been identified by SHAPE to interact with
annealed (7, 40). However, annealing conditions used for those reports were different from those used for this report, including the absence of Gag and RHA in the annealing conditions. The large decrease in annealing for mutation in nt 141 to 160 may be due to nt 142 to 147 in this region, which have been identified by SHAPE to interact with 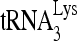 (49). While nt 221 to 240 have not been reported to interact with
(49). While nt 221 to 240 have not been reported to interact with 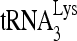 , the mutation of this region dramatically reduced the Gag-facilitated annealing of
, the mutation of this region dramatically reduced the Gag-facilitated annealing of 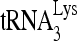 . This could be due to a conformational change in viral RNA that prevents access of
. This could be due to a conformational change in viral RNA that prevents access of 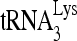 to the viral RNA, which the combined activity of Gag and RHA is unable to resolve.
to the viral RNA, which the combined activity of Gag and RHA is unable to resolve.
RHA has been reported to increase translational efficiency of Gag mRNA by binding to a structural element (the posttranscriptional control element, or PCE) in the RU5 region of viral RNA from several retroviruses, including HIV-1 (10, 25). Since regions in U5 RNA discussed above are believed to be involved in 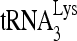 annealing, the possible binding of both
annealing, the possible binding of both 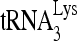 and RHA to similar regions in U5 may represent mutually exclusive events. Thus, RHA appears to be able to facilitate what are probably two mutually exclusive events in the viral life cycle: the translation of Gag mRNA and, in association with Gag protein, the annealing of
and RHA to similar regions in U5 may represent mutually exclusive events. Thus, RHA appears to be able to facilitate what are probably two mutually exclusive events in the viral life cycle: the translation of Gag mRNA and, in association with Gag protein, the annealing of 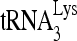 to this mRNA. Thus, when full-length viral RNA first exits the nucleus, it is translated into the major viral proteins Gag and GagPol, but later in the HIV-1 life cycle, translation will be diminished as viral RNA and its associated
to this mRNA. Thus, when full-length viral RNA first exits the nucleus, it is translated into the major viral proteins Gag and GagPol, but later in the HIV-1 life cycle, translation will be diminished as viral RNA and its associated 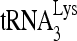 are incorporated into the virus. In fact, a feedback mechanism whereby Gag at high concentration inhibits the translation of its own mRNA has been reported (3), and part of this feedback mechanism may involve the coordinate action of Gag and RHA to alter viral RNA conformation.
are incorporated into the virus. In fact, a feedback mechanism whereby Gag at high concentration inhibits the translation of its own mRNA has been reported (3), and part of this feedback mechanism may involve the coordinate action of Gag and RHA to alter viral RNA conformation.
Two major alternate conformations of the RU5/leader region in HIV-1 RNA found in vitro, showing different electrophoretic mobilities in native gels, have been reported (26). The 5′ leader sequence of HIV-1 RNA was predicted by computer analysis to fold into two major conformations: the LDI (long-distance interaction) form, in which there is a long-distance interaction between the poly(A) and the DIS regions, and the BMH (branched multiple hairpins) form, in which the DIS region base pairing with the poly(A) region is disrupted and it forms a more interactive hairpin (26). The RNA packaging sequence, Ψ, is also presented differently in these two different conformations. BMH RNA has a slower electrophoretic mobility in native gels than LDI RNA, and the LDI-BMH equilibrium can be driven toward the BMH conformation either by mutations or through the addition of mature nucleocapsid (NCp7) (26). Compared to LDI RNA, BMH RNA was shown to more readily dimerize in vitro and be packaged in vivo (41). While a shift in the LDI-BMH equilibrium may facilitate the switch from viral RNA translation to packaging, mutations favoring the formation of the BMH conformer were not found to alter the ability of the viral RNA to be translated (2) or its ability to be reverse transcribed (40). Since RHA and Gag do affect translation and reverse transcription but not dimerization or packaging of viral RNA (9, 43), additional changes in viral RNA not seen in the BMH conformation may be required. Such alterations would require both the energy input of helicase activity and the stabilization by interacting molecules such as 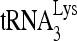 , both of which need to be taken into account in predicting the viral RNA conformation.
, both of which need to be taken into account in predicting the viral RNA conformation.
Acknowledgments
We thank Xia Zhao (Jewish General Hospital, Montreal) for cloning the K417R RHA and siRNARHA, Yves Durocher (Biotechnology Research Institute, Montreal) for plasmid pTT5-SH5 and 293E cells and for technical advice on protein purification using this system, and Chee-Gun Lee (University of Medicine and Dentistry of New Jersey) for RHA cDNA.
This work was supported by a grant from the Canadian Institutes of Health.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1.Abbink, T. E., and B. Berkhout. 2008. HIV-1 reverse transcription initiation: a potential target for novel antivirals? Virus Res. 134:4-18. [DOI] [PubMed] [Google Scholar]
- 2.Abbink, T. E., M. Ooms, P. C. Haasnoot, and B. Berkhout. 2005. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 44:9058-9066. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, E. C., and A. M. Lever. 2006. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 80:10478-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aratani, S., et al. 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol. 21:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aratani, S., et al. 2006. The nuclear import of RNA helicase A is mediated by importin-alpha3. Biochem. Biophys. Res. Commun. 340:125-133. [DOI] [PubMed] [Google Scholar]
- 6.Arts, E. J., et al. 1996. Initiation of (−) strand DNA synthesis from tRNA(3Lys) on lentiviral RNAs: implications of specific HIV-1 RNA-tRNA(3Lys) interactions inhibiting primer utilization by retroviral reverse transcriptases. Proc. Natl. Acad. Sci. U. S. A. 93:10063-10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beerens, N., and B. Berkhout. 2002. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 76:2329-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerens, N., F. Groot, and B. Berkhout. 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 276:31247-31256. [DOI] [PubMed] [Google Scholar]
- 9.Bolinger, C., A. Sharma, D. Singh, L. Yu, and K. Boris-Lawrie. 2010. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 38:1686-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolinger, C., et al. 2007. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 35:2629-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman, F. D., et al. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
12.Cen, S., et al. 1999. The role of Pr55gag in the annealing of
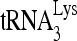 to human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
to human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar] - 13.Cen, S., M. Niu, and L. Kleiman. 2004. The connection domain in reverse transcriptase facilitates the in vivo annealing of tRNALys3 to HIV-1 genomic RNA. Retrovirology 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordin, O., J. Banroques, N. K. Tanner, and P. Linder. 2006. The DEAD-box protein family of RNA helicases. Gene 367:17-37. [DOI] [PubMed] [Google Scholar]
- 16.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, Y. X., et al. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii, R., et al. 2001. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 276:5445-5451. [DOI] [PubMed] [Google Scholar]
- 19.Fujita, H., et al. 2005. Relevance of nuclear localization and functions of RNA helicase A. Int. J. Mol. Med. 15:555-560. [PubMed] [Google Scholar]
- 20.Fuller-Pace, F. V. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldschmidt, V., et al. 2004. Structural variability of the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 279:35923-35931. [DOI] [PubMed] [Google Scholar]
-
22.Guo, F., J. Saadatmand, M. Niu, and L. Kleiman. 2009. Roles of Gag and NCp7 in facilitating
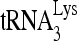 annealing to viral RNA in human immunodeficiency virus type 1. J. Virol. 83:8099-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
annealing to viral RNA in human immunodeficiency virus type 1. J. Virol. 83:8099-8107. [DOI] [PMC free article] [PubMed] [Google Scholar] - 23.He, Q. S., et al. 2008. Comparisons of RNAi approaches for validation of human RNA helicase A as an essential factor in hepatitis C virus replication. J. Virol. Methods 154:216-219. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Y., W. P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hull, S., and K. Boris-Lawrie. 2002. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 76:10211-10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iost, I., M. Dreyfus, and P. Linder. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274:17677-17683. [DOI] [PubMed] [Google Scholar]
- 28.Isel, C., et al. 1999. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 18:1038-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isken, O., et al. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 13:1675-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeang, K. T., and V. Yedavalli. 2006. Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 34:4198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khorchid, A., H. Javanbakht, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 2000. Sequences within Pr160gag-pol affecting the selective packaging of tRNALys into HIV-1. J. Mol. Biol. 299:17-26. [DOI] [PubMed] [Google Scholar]
- 32.Kleiman, L., R. Halwani, and H. Javanbakht. 2004. The selective packaging and annealing of primer tRNALys3 in HIV-1. Curr. HIV Res. 2:163-175. [DOI] [PubMed] [Google Scholar]
- 33.Kleiman, L., C. P. Jones, and K. Musier-Forsyth. 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 584:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konig, R., et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence, P., and E. Rieder. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 83:11356-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J., et al. 1999. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. U. S. A. 96:709-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, J., et al. 2008. The requirement of the DEAD-box protein DDX24 for the packaging of human immunodeficiency virus type 1 RNA. Virology 375:253-264. [DOI] [PubMed] [Google Scholar]
- 38.Mak, J., et al. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakajima, T., et al. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107-1112. [DOI] [PubMed] [Google Scholar]
- 40.Ooms, M., D. Cupac, T. E. Abbink, H. Huthoff, and B. Berkhout. 2007. The availability of the primer activation signal (PAS) affects the efficiency of HIV-1 reverse transcription initiation. Nucleic Acids Res. 35:1649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ooms, M., H. Huthoff, R. Russell, C. Liang, and B. Berkhout. 2004. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham, P. L., A. Kamen, and Y. Durocher. 2006. Large-scale transfection of mammalian cells for the fast production of recombinant protein. Mol. Biotechnol. 34:225-237. [DOI] [PubMed] [Google Scholar]
- 43.Roy, B. B., et al. 2006. Association of RNA helicase A with human immunodeficiency virus type 1 particles. J. Biol. Chem. 281:12625-12635. [DOI] [PubMed] [Google Scholar]
- 44.Smith, W. A., B. T. Schurter, F. Wong-Staal, and M. David. 2004. Arginine methylation of RNA helicase A determines its subcellular localization. J. Biol. Chem. 279:22795-22798. [DOI] [PubMed] [Google Scholar]
- 45.Terwilliger, E. F., E. A. Cohen, Y. C. Lu, J. G. Sodroski, and W. A. Haseltine. 1989. Functional role of human immunodeficiency virus type 1 vpu. Proc. Natl. Acad. Sci. U. S. A. 86:5163-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watts, J. M., et al. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
47.Wei, M., S. Cen, M. Niu, F. Guo, and L. Kleiman. 2005. Defective replication in human immunodeficiency virus type 1 when non-
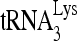 primers are used for reverse transcription. J. Virol. 79:9081-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
primers are used for reverse transcription. J. Virol. 79:9081-9087. [DOI] [PMC free article] [PubMed] [Google Scholar] - 48.Wei, X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson, K. A., et al. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeung, M. L., et al. 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37:6575-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, S., and F. Grosse. 1997. Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem. 272:11487-11494. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, S., and F. Grosse. 2010. Molecular characterization of nuclear DNA helicase II (RNA helicase A). Methods Mol. Biol. 587:291-302. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Z., S. M. Kang, Y. Li, and C. D. Morrow. 1998. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA 4:394-406. [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Z., S. M. Kang, and C. D. Morrow. 1998. Genetic evidence of the interaction between tRNA(Lys,3) and U5 facilitating efficient initiation of reverse transcription by human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 14:979-988. [DOI] [PubMed] [Google Scholar]