Abstract
Maturation of nascent virions, a key step in retroviral replication, involves cleavage of the Gag polyprotein by the viral protease into its matrix (MA), capsid (CA), and nucleocapsid (NC) components and their subsequent reorganization. Bevirimat (BVM) defines a new class of antiviral drugs termed maturation inhibitors. BVM acts by blocking the final cleavage event in Gag processing, the separation of CA from its C-terminal spacer peptide 1 (SP1). Prior evidence suggests that BVM binds to Gag assembled in immature virions, preventing the protease from accessing the CA-SP1 cleavage site. To investigate this hypothesis, we used cryo-electron tomography to examine the structures of (noninfectious) HIV-1 viral particles isolated from BVM-treated cells. We find that these particles contain an incomplete shell of density underlying the viral envelope, with a hexagonal honeycomb structure similar to the Gag lattice of immature HIV but lacking the innermost, NC-related, layer. We conclude that the shell represents a remnant of the immature Gag lattice that has been processed, except at the CA-SP1 sites, but has remained largely intact. We also compared BVM-treated particles with virions formed by the mutant CA5, in which cleavage between CA and SP1 is also blocked. Here, we find a thinner CA-related shell with no visible evidence of honeycomb organization, indicative of an altered conformation and further suggesting that binding of BVM stabilizes the immature lattice. In both cases, the observed failure to assemble mature capsids correlates with the loss of infectivity.
HIV assembly is driven by polymerization of the Gag polyprotein at the host cell's plasma membrane, budding into an enveloped spherical particle called the immature virion (3, 21). Gag has matrix (MA), capsid (CA), and nucleocapsid (NC) domains as well as several shorter segments: SP1 (spacer peptide 1), SP2, and p6 (Fig. 1 A). MA is responsible for Gag-membrane interactions, CA contains most determinants of Gag-Gag interactions, and NC binds the viral RNA (vRNA). To become infectious, immature virions must undergo maturation (3). During maturation, Gag is cleaved by the viral protease (PR), after which MA, CA, and NC become the principal protein constituents of the mature virion (3, 47). MA remains membrane associated, CA reassembles de novo into a capsid, and NC engages the viral RNA. The five cleavage events per Gag subunit proceed in a defined order (Fig. 1A), with the final event separating CA from SP1. Impeding or otherwise perturbing the cleavage program results in incomplete or improper maturation, severely reducing the infectivity of the resulting viral particles (3, 48).
FIG. 1.
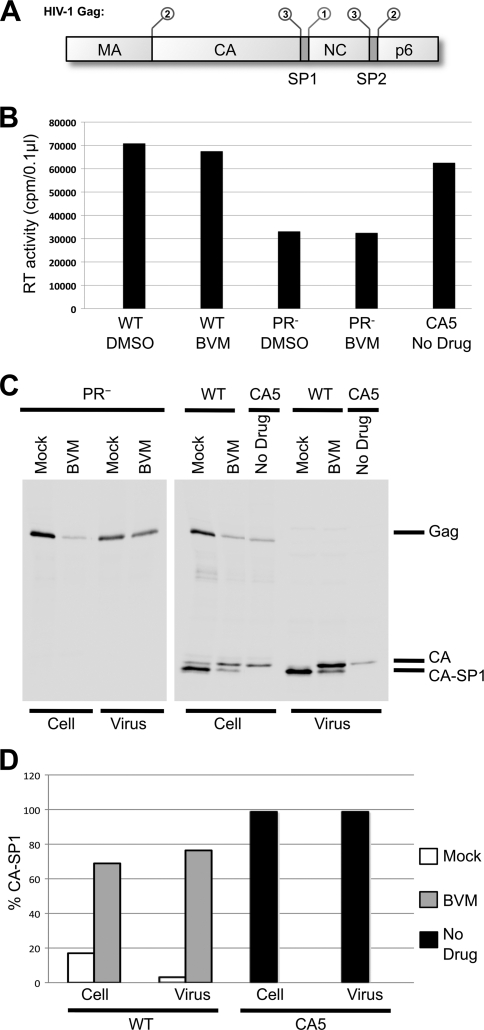
(A) Domain organization and processing program of HIV-1 Gag. The functional domains MA, CA, NC, and p6, plus spacer peptides SP1 and SP2, are indicated. The SP1-NC cleavage occurs first (1), followed by separation of MA from CA and NC-SP2 from p6 (2). The final cleavage events separate NC from SP2 and CA from SP1 (3). (B) Virus particle production was measured by monitoring RT activity in the concentrated virus pellets. RT activity is 2-fold lower in PR− samples, but sufficient quantities of particles to perform cryo-ET were obtained for all samples. (C and D) Detection of virus-associated proteins. HeLa cells transfected with WT pNL4-3 or derivatives and cultured in either the absence of drug or the presence of 4 μg/ml BVM or the equivalent concentration of DMSO were metabolically labeled for 3.5 h with [35S]Met-Cys, and released virions were pelleted by ultracentrifugation. Virus lysates were immunoprecipitated with anti-HIV-Ig, and processing of CA-SP1 to CA was analyzed by SDS-PAGE and fluorography (C) followed by phosphorimager analysis to quantify the percentage of CA-SP1 relative to total CA-SP1 plus CA (D).
Retroviral capsids are polymorphic; in the case of HIV, the predominant species has a conical morphology (9, 10, 13). The CA subunit has an N-terminal domain (NTD) and a C-terminal domain (CTD) connected by a flexible linker. The structures of the NTD and CTD are conserved among different retroviruses, despite little sequence similarity (15, 28, 34, 35, 41, 42, 45). Closed capsids may be described as “fullerene” shells formed from 12 CA pentamers and a variable (and much larger) number of CA hexamers (31). The hexamers have been visualized in cryo-electron microscopy (cryo-EM) studies of CA tubes and sheet-like assemblies (29, 39), and pentamers have been observed in a cryo-EM study of icosahedral capsids of Rous sarcoma virus (RSV) CA (17). Thus, a basic hexagonal lattice with a repeat of ∼9.5 nm is folded into a variety of irregular polyhedral forms, depending on how the pentamers are distributed (9, 10, 13, 14, 31).
The organization of the immature lattice is less well understood. The MA domain contacts the viral membrane, and the rest of the Gag molecule extends radially inwards (49). Cryo-EM and cryo-electron tomography (cryo-ET) have revealed that the Gag shell has ordered patches of a honeycomb-like lattice (11, 52). This lattice is most evident at the radial level assigned to the CA-SP1 portion of Gag. No evidence has yet been presented for pentamers in the immature Gag shell, whose curvature has alternatively been attributed to gaps or irregularities in the lattice (11).
The small molecule 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid (DSB), also known as bevirimat (BVM), PA-457, or MPC-4326, is the first of a new class of anti-HIV drugs termed maturation inhibitors. BVM potently inhibits HIV-1 replication by blocking a late step of the Gag cleavage pathway, preventing scission at the CA-SP1 junction (4, 6, 8, 37, 56). As a result, aberrant noninfectious virions are produced (6, 37, 48). Imaged by conventional thin-sectioning EM, these particles lack the typical mature conical core but contain acentric, roughly spherical, masses and a partial submembranous shell, assumed to be Gag related (37, 48). At higher concentrations, BVM may inhibit Gag assembly and release (20).
The mechanism of action of BVM is still being characterized. Unlike protease inhibitors that block the PR active site, thus preventing all Gag cleavages (51), BVM targets only one such event, that at the CA-SP1 junction. In support of this view, other lentiviruses with different sequences at their CA-SP1 regions are insensitive to BVM treatment (54). Additionally, it has been demonstrated that BVM is incorporated into assembling virus particles in a Gag-dependent manner (55). Furthermore, nearly all reported BVM resistance mutations have mapped to the CA-SP1 junction or within SP1 (2, 37, 38, 53, 56), and the resistance mutants that have been tested incorporate less BVM into particles (53, 55). Finally, BVM is able to prevent cleavage of CA from SP1 only in Gag assembled into particles, not in free Gag in solution (37, 46). These data have led to a model whereby the BVM binding site is formed on Gag assembly, at or nearby the CA-SP1 cleavage site, and binding of BVM blocks PR from accessing that site.
In this study, we sought to investigate the three-dimensional (3D) structure of BVM-treated viral particles. Using cryo-ET, we analyzed samples of BVM-treated HIV-1 and compared them to wild-type (WT) HIV-1, immature HIV-1, and a Gag cleavage mutant that blocks the CA-SP1 site (CA5) (48). The results bear on the relationship between proteolytic cleavage and structural reorganization in HIV maturation and further illuminate the mode of action of BVM.
MATERIALS AND METHODS
Cell culture, plasmids, and BVM.
HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% (vol/vol) fetal bovine serum (FBS), l-glutamine (2 mM), penicillin, and streptomycin. BVM was prepared in dimethyl sulfoxide (DMSO) as described previously (27). DMEM containing BVM at the concentrations indicated below and in the figure legends or an equivalent amount of DMSO was prepared immediately before use and vortexed. Plasmid DNA was purified with a plasmid purification Maxiprep Kit (Qiagen), adjusted to 1 μg/μl, and verified by sequencing. The following plasmids were used to generate virus particles: wild-type HIV-1 molecular clone pNL4-3 (1), pNL4-3 PR− with a D25N substitution to inactivate PR (PR−) (33), and pNL4-3 CA5 (48) (a kind gift of H.-G. Kräusslich).
Virus particle production.
Virions were produced from batches of 2.4 × 107 HeLa cells transfected with Lipofectamine 2000 (Invitrogen). Transfected cells were incubated overnight in medium containing either no drug, 2 μg/ml BVM, or the equivalent volume of DMSO. Cells incubated with BVM or DMSO were then gently washed and incubated for a further 3.5 h with fresh medium containing either 4 μg/ml BVM or the equivalent volume of DMSO. Virions produced during the 3.5-h incubation or following the non-drug-treated overnight incubation were harvested from virus-containing supernatant. The supernatants were clarified by a low-speed centrifugal spin and filtered using a low-molecular-weight (MW) protein-binding 0.4-μm-pore-size filter disc. The virus particles were pelleted by ultracentrifugation at 28,000 rpm for 2 h at 4°C using a Sorvall AH-629 rotor. Excess medium was removed from the pellet before resuspension in 10 μl of ice-cold phosphate-buffered saline (PBS). The virus-containing suspension was centrifuged at 5,000 rpm for 2 min at 4°C to remove debris, and the sample was fixed to inactivate the virus by the addition of an equal volume of freshly prepared 4% (vol/vol) EM-grade paraformaldehyde (Electron Microscopy Sciences).
Verification of virus particle production and BVM activity.
Virus particle production was monitored at specific stages of the procedure. Reverse transcriptase (RT) activity in culture supernatant and in the final virus-containing suspension was monitored by a previously described RT assay (26). Virus-producing cells were fixed and embedded for thin-sectioning EM as described previously (26). In parallel, BVM activity was monitored by metabolic labeling of transfected HeLa cells, preparation of cell and virus lysates, and immunoprecipitation with pooled immunoglobulin from HIV-1-infected patients (HIV-Ig) obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. This biochemical analysis has been described in detail previously (2, 25, 50); however, the BVM concentrations and incubation times were consistent with the virus production procedure used in this study (see above). The pNL4-3 PR− samples were denatured prior to immunoprecipitation to facilitate disruption of assembled Gag complexes, in which epitope masking can otherwise occur (43). The radiolabeled immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and exposed to X-ray film and a phosphorimager plate, and the bands were quantitated by means of Quantity One software (Bio-Rad).
Cryo-electron tomography.
Cryo-ET analysis was performed essentially as described previously (14). In brief, purified virus was mixed with a suspension of 10-nm colloidal gold particles (Ted Pella) to serve as fiducial markers. Drops of 3.75 μl were applied to Quantifoil R2/2 holey carbon grids (Quantifoil Micro Tools) before the grids were blotted and plunge-frozen in liquid ethane with a Vitrobot (FEI). Vitrified specimen-bearing grids were transferred to a cryogenic specimen holder (type 626; Gatan) for data acquisition. Single-axis tilt series were recorded on a Tecnai-12 transmission electron microscope ([TEM] FEI) operating at 120 keV (30). The microscope was equipped with a GIF energy filter (Gatan) operating in zero-loss mode with an energy slit width of 20 eV. Projection images were acquired on a 2,048- by 2,048-pixel charge-coupled-device (CCD) camera using the program SerialEM (40) for automated data collection. For each tilt series, images were recorded in 2° increments over an average angular range of −66° to +66°, at an effective magnification of ×38,500 (7.8-Å pixel size). Electron doses per image ranged between 0.5 and 1.0 e−/Å2, for a cumulative dose of 35 to 70 e−/Å2 per tilt series. Image defocus was set at −4 μm, corresponding to a first contrast transfer function zero at (3.7 nm)−1.
Tomogram reconstruction and image processing.
Unless otherwise indicated, all image analysis and reconstruction operations were carried out using tools in the Bsoft package (32). Tilt series projections were aligned with btrack, using the 10-nm gold particles as fiducial markers. Tomograms were calculated in reciprocal space with btomrec to a resolution cutoff of 30 Å, as described previously (32). Briefly, the 3D Fourier transform of each sample volume was assembled by packing the two-dimensional (2D) transforms of the aligned projections according to the central section theorem (24). The resulting 3D transform was then back-transformed to produce the density map. Tomograms were denoised using 50 iterations of nonlinear anisotropic diffusion filtering (23). Subvolumes containing individual virus particles were extracted from both raw and denoised tomograms for further analysis. Tomogram resolution was assessed in terms of the NLOO-2D criterion (16) applied to three virions per tomogram (threshold, 0.3). Resolution of the averaged lattice structures was assessed in terms of the Fourier shell correlation coefficient, threshold 0.3. Denoised particles were segmented using modeling tools available in Bsoft and visualized using the UCSF Chimera package (44).
Lattice analysis.
Selected particles were initially modeled as spherical shells of points, fitted to the corresponding density map. Subtomograms were extracted around model points associated with protein shell density, rotated according to their modeled position, and averaged. This unaligned average served as the starting template for further rounds of subtomogram alignment and averaging. Subtomogram orientation determination and alignment were carried out with the program bfind, taking into account the missing wedge of information (22, 32). The local 6-fold symmetry of the Gag/CA lattice was applied in intermediate steps, and lattice models were generated independently for each particle analyzed and then combined. For each sample condition analyzed, subvolumes from particles with clearly visible lattice features were pooled and then subjected to further rounds of alignment and averaging to generate the final lattice model. Radial density profiles were calculated from the final averages. Fitting, comparison, and visualization of the final lattice averages were carried out using the UCSF Chimera package (44).
RESULTS
Generation of virus particles in which CA-SP1 cleavage is blocked.
Virus particles were produced by two approaches. In the first, they were isolated from HeLa cells grown in the presence of BVM that were transfected with the wild-type pNL4-3 clone. In the second, they were generated using the CA5 derivative of pNL4-3, which has two point mutations that inhibit cleavage, one at the CA-SP1 processing site and one at the cryptic cleavage site following SP1 residue 4 (48). To allow comparison with immature virions, we also produced particles whose maturation was blocked by an inactivating mutation in the PR active site (33) in cells grown both with and without BVM.
Particle production was monitored by measurement of RT activity in the culture supernatant and in the pelleted virion fraction. This activity was essentially unaffected by BVM treatment but reduced by ∼50% for the protease-defective mutant (Fig. 1B). BVM activity was verified by detecting cell- and virus-associated proteins (Fig. 1C) and by a quantitative biochemical assay for CA-SP1 processing (2) (Fig. 1D). Levels of the unprocessed CA-SP1 polypeptide in both BVM-treated and CA5 virus particles were as expected from previous studies (2, 5, 7, 19), with a CA-SP1/CA ratio of ∼75%/25% for BVM-treated preparations compared to ∼99%/1% for CA5 preparations. Detection of viral proteins also confirmed that pNL4-3 PR− particles were arrested in an immature state as they contained only uncleaved Gag. Thin-section EM (see Fig. S1 in the supplemental material) confirmed that the various particles generated had the expected morphologies (37, 48).
Cryo-electron tomography of BVM-treated virions.
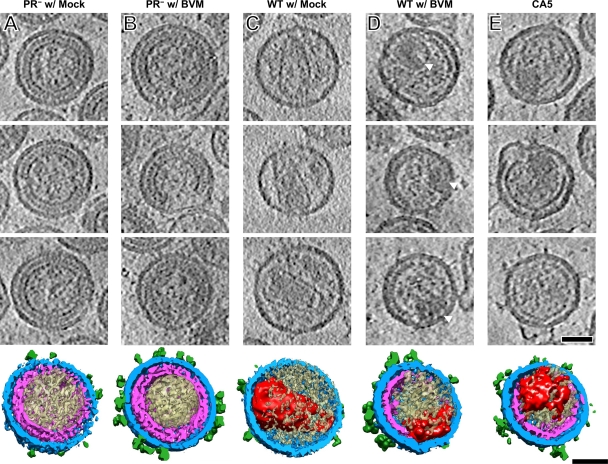
Cryo-ET was performed on five samples: wild-type virions with mock treatment (DMSO), wild-type with 4 μg/ml BVM, immature (PR-deficient) virions with mock treatment, immature (PR-deficient) virions with 4 μg/ml BVM, and the CA5 mutant. At least four tilt series were collected and reconstructed for each sample. The in-plane resolutions achieved were 5 to 6 nm. Individual particles were extracted from the tomograms for further analysis. In all samples, virus particles appeared roughly spherical and exhibited similar size distributions (Table 1). The average particle diameter was ∼125 nm for all samples, with standard deviations (SD) ranging from 9 to 15 nm. Thus, neither BVM treatment nor the CA5 mutation had any significant effect on particle size. However, differences in morphology were observed between immature, mature, and cleavage-blocked particles (Fig. 2).
TABLE 1.
Analysis of virion size
| Virus | Treatment | Particle size (nm) |
No. of particles | |
|---|---|---|---|---|
| Avg diam | SD | |||
| WT | Mock | 127 | 15 | 60 |
| BVM | 124 | 12 | 122 | |
| PR− | Mock | 123 | 14 | 109 |
| BVM | 123 | 9 | 98 | |
| CA5 | 126 | 14 | 114 | |
FIG. 2.
Cryo-electron tomography of five HIV-1-related particles: pNL4-3 PR− (immature) with mock treatment (A), pNL4-3 PR− treated with 4 μg/ml BVM (B), pNL4-3 WT with mock treatment (C), pNL4-3 WT treated with 4 μg/ml BVM (D), and pNL4-3 CA5 (E). The three upper panels in each column show central sections, 0.78 nm thick, through three representative particles from that sample. Below, a segmented surface rendering of the particle in the bottom section is shown. The glycoprotein spikes are shown in green, the membrane plus MA layer is in blue, Gag-related shells are in magenta, core structures are in red, and other internal density is shown in beige. Scale bar, 50 nm.
Immature particles (Fig. 2A) were seen to have a multilayered shell of density corresponding to the Gag lattice (11, 52), underlying the viral envelope. Only 3% of particles had no visible discontinuities in their lattices. The majority of particles, ∼65%, had lattices covering at least 70% of the spherical surface while the remainder had less extensive lattice coverage. These results are similar to findings reported in a recent study of immature HIV-1 virions (18). BVM-treated immature particles were indistinguishable from untreated immature particles (compare Fig. 2A and B). Thus, at the concentrations used, the drug does not affect the assembly of immature virions. Wild-type virions exhibited the typical mature morphology (9, 13), with most containing a prominent conical core and none containing any trace of the immature Gag shell (Fig. 2C). In contrast, BVM-treated wild-type particles (Fig. 2D) were found to contain a partial spherical shell under the viral membrane. These shells varied considerably in extent, and they were less complete than the shells of immature virions, generally not exceeding ∼50% coverage. A minor fraction of particles, ∼18%, had no visible shell. In addition, BVM-treated particles were often observed to contain roundish acentric masses, 30 to 40 nm across (Fig. 2D, white arrowheads). CA5 particles appeared similar in morphology to BVM-treated WT virions (Fig. 2E). On closer inspection, we observed that their shells are significantly thinner than those of BVM-treated WT virions (see below). CA5 virions also frequently contained acentric masses similar to those seen in BVM-treated virions.
Gag-related lattice structures.
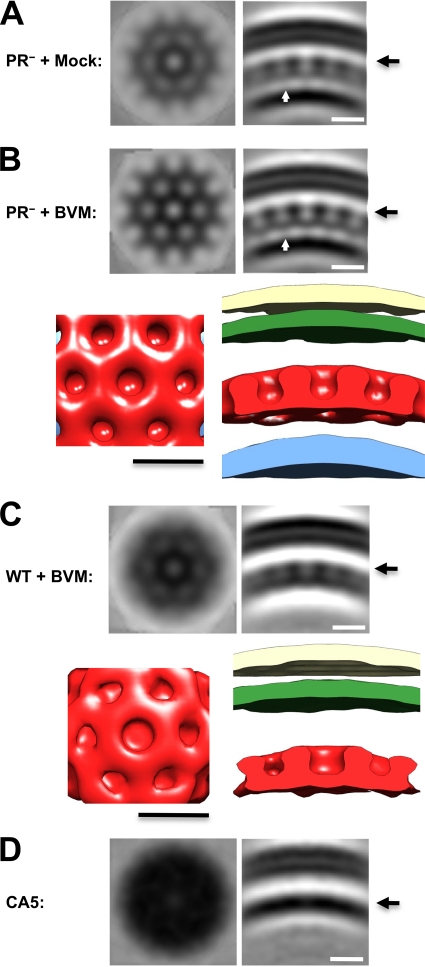
The immature Gag lattice is a continuous curved hexagonal array with a distinctive multilamellar organization (11, 52) (Fig. 2A and B). Strikingly, although proteolytic processing of Gag is almost complete in wild-type BVM-treated particles, most of them contain a shell similar to that of the immature particle. To compare the immature and wild-type BVM-treated structures more closely, we enhanced them by subvolume averaging, thus obtaining density maps with ∼4-nm resolution (Fig. 3 A, B, and C). Individual particles with clearly defined protein shells were selected from whole tomograms for lattice analysis. In the case of samples blocked for CA-SP1 cleavage, particles with more extensive shells were taken. The final averages each combined data from 5 to 15 viral particles. This analysis revealed a similar structure for immature particles, regardless of whether they were assembled in the presence of BVM (Fig. 3A and B). In the following analysis, immature particles assembled in the presence of BVM were used.
FIG. 3.
Structural analysis of immature HIV-1 virions, BVM-treated virions, and CA5 virions. In each case, subtomogram averaging was performed to enhance the structures present in the outer density layers. (A and B) Immature virions from the protease-defective clone pNL4-3 PR−. The samples were produced in the presence of DMSO (mock treatment; A) or BVM (B). White arrows indicate the putative helical bundles formed by SP1 connecting CA and NC/RNA densities. (C) Maturation-inhibited virions produced by WT pNL4-3 produced in the presence of 4 μg/ml BVM. (D) Virions from the mutant clone pNL4-3 CA5 produced in the absence of BVM. In the upper portions of panels A to C, two views of the averaged density map are shown: at left is an in-plane section, 0.78 nm thick, at the level marked with a long arrowhead in the CA density layer; at right is a radial section of similar thickness. In the lower portions of panels B and C are isodensity surface renderings of the respective structures. The viral membrane is shown in beige, the MA layer is in green, the CA layer is in red, and the NC/RNA layer, when visualized, is in blue. CA5 particles display no evidence of a lattice in the averaged in-plane section, from which we conclude that if such a lattice is present, the contrast and/or resolution of this analysis is not sufficient to detect it. Scale bar, 10 nm.
The principal in-plane feature of both the immature and the BVM-treated viral particles is the honeycomb lattice in the layer occupied by CA (Fig. 3A to C). Direct superposition of the respective lattices confirms their close mutual resemblance (Fig. 4). However, when they are aligned on the viral membrane, an inward radial shift by ∼2 nm of the CA layer in the BVM-treated particles is evident (compare Fig. 3B and C and 4A).
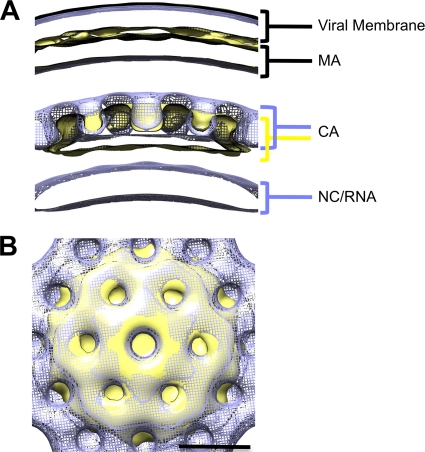
FIG. 4.
Comparison of immature and BVM-treated lattices. Average density maps from PR− virus treated with 4 μg/ml BVM (blue; mesh) and WT virus treated with 4 μg/ml BVM (beige; surface) were aligned and compared. (A) Radial view of density maps aligned with viral membrane present. Density layers are labeled with the corresponding viral structures. The membrane and MA layers are essentially superposable, whereas the CA density layer has shifted inwards in the BVM-treated particle following PR cleavage. (B) Top view of CA lattice. The aligned hexagonal lattices are shown superimposed in this view. Scale bar, 10 nm.
Underlying the honeycomb lattice of immature particles and connecting it to the NC layer are relatively faint but nevertheless significant densities (Fig. 3A and B, white arrows). These densities were also visualized in a recent cryo-ET study of immature virions in a strain inactivated by mutagenesis in which they were proposed to represent hexameric bundles of SP1 α-helices (52). These densities are not visible on the WT BVM-treated virions (Fig. 3C) although it is not yet clear whether this results from the slightly lower resolution of this averaged subtomogram or represents a genuine structural change following separation from the NC domains.
The averaged radial density profiles of the two PR− particles (mock and BVM-treated) confirm that BVM treatment has little, if any, effect on immature particle organization (compare Fig. 5 A and B). The effect of BVM on WT particles is shown in Fig. 5C. In all cases, the outermost layer is resolved into two peaks of approximately equal densities 5 nm apart. We interpret the outer peak as the lipid bilayer of the viral membrane and the inner peak as the membrane-lining layer of MA domains. We do not favor the alternative interpretation that the outer peak might represent the outer leaflet of the bilayer with the inner peak as the inner leaflet plus MA because we do not observe a double peak in tomograms of membrane vesicles (unpublished results). Moreover, we would not expect the two layers to be of nearly equal densities if one were to correspond to a single membrane leaflet and the other to a single membrane leaflet coated with MA protein.
FIG. 5.
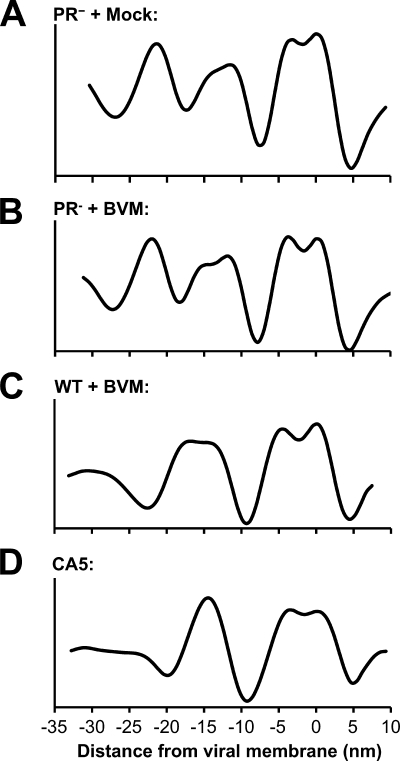
Comparison of radial density profiles of immature virions, BVM-treated virions, and CA5 virions. Radial density profiles were calculated from subtomogram averages of immature virions produced in the presence of DMSO (A), immature virions produced in the presence of BVM (B), WT virions produced in the presence of BVM (C), and CA5 particles produced in the absence of BVM (D). Profiles have been aligned based on the position of the viral membrane.
Moving inwards, the next density peaks in Fig. 5A, B, and C are also about 10 nm thick and also appear as doublets, which we attribute to the two CA domains, NTD (outer peak) and CTD (inner peak) (49). The inward shift of the CA layer in BVM-treated particles is apparent in Fig. 5C. The final peak in the density profile of the immature virions (Fig. 5A and B), ∼25 nm in from the membrane, is presumably contributed by NC plus vRNA (49) and has no counterpart in the BVM-treated virions (Fig. 5C).
As in previous tomographic studies of immature HIV-1 particles (11, 52), we do not resolve subunits in the MA layer, which appears as a sheet of continuous density (Fig. 3). The CA layer presents as a honeycomb-like lattice with a center-to-center unit cell spacing of 8 nm (Fig. 3A to C and 4B). On its underside, we observe faint rod-like densities connecting the CA and NC/RNA layers (Fig. 3A and B, white arrow), which have been proposed to represent bundles of six SP1 α-helices (52). No such features are apparent on the BVM-treated lattice (Fig. 3C), which may indicate that the SP1 domains are less ordered or that they may have folded back onto the CA-CTD layer after the connection to NC was severed.
Comparison of BVM-treated and CA5 mutant virions.
The CA5 mutant has two substitutions, L364I and M368I, which block cleavage at the CA-SP1 site, resulting in the assembly of aberrant particles that fail to mature and are noninfectious (48) (Fig. 1C and D). Because this phenotype is similar to that resulting from BVM treatment, we performed cryo-ET on CA5 particles to provide a basis for comparison. In terms of overall morphology, CA5 particles and BVM-treated WT particles are quite similar, with both having a shell beneath the viral membrane and often containing an irregular acentric mass (compare Fig. 2A and D; see also Fig. S1 in the supplemental material). The radial density profile reveals that the outer double peak (taken to represent the membrane plus MA layer [see above]) is essentially the same as in the BVM-treated particles, and the CA layer is similarly shifted toward the particle center. However, the CA layer is thinner, and, in the radial profile (Fig. 5D), it rises to a higher density than that of the outer double peak, whereas the relative heights of these peaks are reversed in BVM-treated and immature virions (Fig. 5A to C). On attempting to probe the substructure of the CA5-CA layer by subvolume averaging, we could not detect a honeycomb lattice or, indeed, any other kind of lattice (Fig. 3D). It follows that this layer contains CA in a conformation that differs from that adopted in immature WT or BVM-treated virions. It also appears to be in a different conformation from that of the mature CA shell, being thicker and more textured (compare Fig. 2C and E).
Core-like structures.
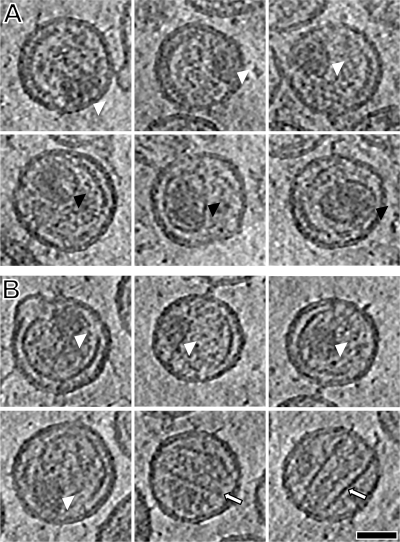
The majority of cleavage-inhibited particles were observed to contain acentric masses. Galleries of BVM-treated and CA5 particles are presented in Fig. 6. In the BVM-treated particles, two types of acentric masses were observed. The first is a compact region of higher density that lacks a clear border structure (Fig. 6A, white arrowheads). The second has a defined shell surrounding the dense contents (Fig. 6A, black arrowheads). In thickness and density, this shell resembles the capsids of authentic mature cores (compare Fig. 2C). However, these structures are smaller (30 to 40 nm in diameter) than native mature cores and differently shaped.
FIG. 6.
Gallery of core structures in BVM-treated and CA5 particles: pNL4-3 WT with BVM treatment (A) and pNL4-3 CA5 (B). Central sections, 0.78 nm thick, through eight representative particles are shown for each sample. Core-like features are indicated by arrowheads: white, small, electron-dense material with no shell; black, defined shell containing electron-dense material; outlined arrows, incomplete tubes and cones. Scale bar, 50 nm.
CA5 particles were also often observed to contain acentric masses, similar to the borderless ones observed after BVM treatment (Fig. 6B, white arrowheads). Less commonly (in ∼10% of cases), internal structures resembling incomplete mature cores were observed (Fig. 6B, outlined arrows). These structures appear as open-ended tubes or cones. Condensed material was usually found adjacent to, rather than inside, these shells. It is noteworthy that virions containing incomplete tubes/cones were not observed to contain the peripheral curved shell commonly seen in CA5 particles, suggesting that these structures may be mutually exclusive, and may represent alternative assemblies derived from the same pool of CA protein.
DISCUSSION
The present observations bear on the mode of action of BVM in inactivating HIV-1. We infer that as immature virions assemble, the drug binds to Gag, presumably to the CA-CTD/SP1 junction; this interaction is apparently innocuous as it does not appreciably affect the assembly of immature virions. However, the drug must be incorporated in order to inhibit maturation and suppress infectivity. We observed two distinctive structures within BVM-treated virions. One is closely related to the Gag shell of immature virions; the other is the acentric mass.
The shell found in BVM-treated virions lacks the inner layer of density present on native (unprocessed) Gag shells that is associated with NC and bound vRNA, and the CA-related layer of density is shifted slightly inwards. These shells are less extensive and more variable in size than the Gag shells in immature virions. However, they have essentially the same lattice structure as the uncleaved Gag shell of immature virions although it is somewhat more sharply defined in the latter case (compare Fig. 3B and C). The latter distinction may reflect better ordering in the immature Gag shells and/or a slight conformational difference between the respective lattices. Notwithstanding, we conclude that although four of the five maturational cleavages (all except at the CA-SP1 site) have taken place, it is only at the SP1-NC site that the cleavage products have fully separated. In fact, about 20% of CA-SP1 is cleaved despite the presence of the drug (Fig. 1C), and it is plausible that the smaller size of the BVM-treated shells reflects erosion by the shedding of some CA-SP1 or CA subunits. Although the Gag cleavage fragments MA and CA are physically quite far from the CTD-SP1 junction where BVM binds, they remain essentially in place, albeit with a slightly shifted CA layer. We infer that BVM binding exercises an allosteric stabilizing effect on the processed Gag shell, in addition to sterically blocking cleavage at the CTD-SP1 site. An apparent stabilizing effect of BVM on CA structures isolated from virions has been measured previously biochemically (56) and supports the proposition that BVM may function beyond blocking CA-SP1 cleavage.
During normal maturation, CA subunits released from the Gag shell assemble into a capsid containing the NC, vRNA, and the replicative enzymes RT and integrase (3, 47) while also leaving a sizeable pool of unassembled CA subunits (12, 36). In BVM-treated virions, the supply of free CA subunits is greatly reduced as most of them remain anchored in the residual shell. However, NC subunits are quantitatively released, and it is plausible that the acentric masses are ribonucleoprotein complexes mainly comprising NC and vRNA. This interpretation is consistent with their propensity to stain positively in conventional thin sections (2, 37; see also Fig. S1 in the supplemental material) and the compaction that is implied from their enhanced density in cryo-tomograms (Fig. 6A).
Some acentric masses have surface shells of about the same thickness as capsids (Fig. 6). We suggest that these shells may form in virions in which CA/CA-SP1 molecules have been released from the Gag-like shell in sufficient quantity to support assembly of a small capsid. The reduced size and altered shape of these putative minicapsids relative to the wild-type conical capsids may represent an effect of appended SP1 on the self-assembly properties of CA or an effect of the continuing presence of BVM, or both.
Similar but not identical departures from normal maturation occur in CA5 particles. Here, we also observed acentric masses without surface shells that we would assign to NC plus vRNA (as above). In addition, we observed a different set of what we take to be aberrant CA assemblies, including open-ended tubes and cones. Their most distinctive feature was an incomplete protein shell tracking the virion envelope (Fig. 2E). This shell is thinner than the CA layer in the BVM-treated particles and does not display its honeycomb lattice structure. On the other hand, it is thicker than that of the mature capsid (compare Fig. 2C and E). We surmise that this represents an imperfectly ordered hexagonal lattice whose periodicity is not expressed with high enough contrast to be discerned in our tomograms (see Fig. 6 of reference 14) and which represents a conformation different from both the immature Gag lattice and the mature CA lattice. It is unclear whether these shells represent conformationally transformed derivatives of the original Gag shells (postprocessing) or de novo assembly products.
The mechanism underlying the formation of the observed incomplete tubes and cones in CA5 particles is also a source of uncertainty though information from other viruses may prove useful by analogy. Quite a high incidence of tubular capsids (∼30%) was observed in cryo-tomograms of wild-type RSV virions (14). As there is no ready way to map a spherical shell on to a tube, it appears likely that incomplete tubes and cones represent de novo assembly products resulting from an altered assembly nucleation event.
It is important to note that some of the differences observed between CA5 and BVM-treated WT virions could be due not only to the presence or absence of BVM but also to the fact that CA5 virions contain ∼100% CA-SP1 whereas BVM-treated WT particles contain a small amount (∼20%) of mature, processed CA protein. In any event, these results provide further evidence in support of the contention that a correctly formed and packaged core is essential for a virion to be infectious. In the presence of BVM, stabilization of the immature CA lattice and prevention of mature capsid formation effectively suppress particle infectivity. The data obtained here with both CA5 and BVM-treated WT virions indicate that CA must be cleaved at both ends for proper maturation to occur.
Taken together, the present results suggest that BVM may inhibit virus infectivity not only by blocking CA-SP1 processing but also by stabilizing the immature lattice (see above). Alternatively, it is possible that the difference seen between BVM-treated WT particles and CA5 particles in terms of lattice formation is due in part to the presence of some mature CA in the BVM-treated WT particles but not in the CA5 particles. This hypothesis could be tested by producing virions with an 80%/20% CA-SP1/CA ratio (19) and analyzing them by cryo-ET.
Supplementary Material
Acknowledgments
We thank Dennis Winkler for support with resources for cryo-electron tomography and guidance in their use, F. Soheilian and K. Nagashima for assistance with thin-section EM, Sherimay Ablan for expert technical assistance with protein immunoprecipitation, and H.-G. Kräusslich for providing the CA5 mutant.
This work was supported by the Intramural Research Programs of NIAMS and NCI and by the NIH Intramural AIDS Targeted Antiviral Program.
Footnotes
Published ahead of print on 24 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adachi, A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., et al. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J. Virol. 80:10957-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson, C. S., and E. O. Freed. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv. Pharmacol. 55:347-387. [DOI] [PubMed] [Google Scholar]
- 4.Adamson, C. S., and E. O. Freed. 2008. Recent progress in antiretrovirals—lessons from resistance. Drug Discov. Today 13:424-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson, C. S., M. Sakalian, K. Salzwedel, and E. O. Freed. 2010. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson, C. S., K. Salzwedel, and E. O. Freed. 2009. Virus maturation as a new HIV-1 therapeutic target. Expert Opin. Ther. Targets 13:895-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamson, C. S., K. Waki, S. D. Ablan, K. Salzwedel, and E. O. Freed. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457). J. Virol. 83:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aiken, C., and C. H. Chen. 2005. Betulinic acid derivatives as HIV-1 antivirals. Trends Mol. Med. 11:31-36. [DOI] [PubMed] [Google Scholar]
- 9.Benjamin, J., B. K. Ganser-Pornillos, W. F. Tivol, W. I. Sundquist, and G. J. Jensen. 2005. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J. Mol. Biol. 346:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs, J. A., et al. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15-20. [DOI] [PubMed] [Google Scholar]
- 11.Briggs, J. A., et al. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090-11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs, J. A., et al. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. [DOI] [PubMed] [Google Scholar]
- 13.Briggs, J. A., T. Wilk, R. Welker, H. G. Kräusslich, and S. D. Fuller. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butan, C., D. C. Winkler, J. B. Heymann, R. C. Craven, and A. C. Steven. 2008. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J. Mol. Biol. 376:1168-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 16.Cardone, G., K. Grünewald, and A. C. Steven. 2005. A resolution criterion for electron tomography based on cross-validation. J. Struct. Biol. 151:117-129. [DOI] [PubMed] [Google Scholar]
- 17.Cardone, G., J. G. Purdy, N. Cheng, R. C. Craven, and A. C. Steven. 2009. Visualization of a missing link in retrovirus capsid assembly. Nature 457:694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson, L. A., et al. 2008. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe 4:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Checkley, M. A., B. G. Luttge, F. Soheilian, K. Nagashima, and E. O. Freed. 2010. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dafonseca, S., et al. 2008. The inhibition of assembly of HIV-1 virus-like particles by 3-O-(3′,3′-dimethylsuccinyl) betulinic acid (DSB) is counteracted by Vif and requires its zinc-binding domain. Virol. J. 5:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demirov, D. G., and E. O. Freed. 2004. Retrovirus budding. Virus Res. 106:87-102. [DOI] [PubMed] [Google Scholar]
- 22.Frangakis, A. S., et al. 2002. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc. Natl. Acad. Sci. U. S. A. 99:14153-14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frangakis, A. S., and R. Hegerl. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239-250. [DOI] [PubMed] [Google Scholar]
- 24.Frank, J. 2006. Electron tomography, 2nd ed. Springer, New York, NY.
- 25.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujioka, T., et al. 1994. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J. Nat. Prod. 57:243-247. [DOI] [PubMed] [Google Scholar]
- 28.Gamble, T. R., et al. 1997. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 29.Ganser-Pornillos, B. K., A. Cheng, and M. Yeager. 2007. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell 131:70-79. [DOI] [PubMed] [Google Scholar]
- 30.Harris, A., et al. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103:19123-19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heymann, J. B., C. Butan, D. C. Winkler, R. C. Craven, and A. C. Steven. 2008. Irregular and semi-regular polyhedral models for Rous sarcoma virus cores. Comput. Math. Methods Med. 9:197-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heymann, J. B., G. Cardone, D. C. Winkler, and A. C. Steven. 2008. Computational resources for cryo-electron tomography in Bsoft. J. Struct. Biol. 161:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, Z., L. Jin, D. L. Peterson, and C. L. Lawson. 1999. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286:83-93. [DOI] [PubMed] [Google Scholar]
- 35.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 36.Lanman, J., et al. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11:676-677. [DOI] [PubMed] [Google Scholar]
- 37.Li, F., et al. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, F., et al. 2006. Determinants of activity of the HIV-1 maturation inhibitor PA-457. Virology 356:217-224. [DOI] [PubMed] [Google Scholar]
- 39.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 40.Mastronarde, D. N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36-51. [DOI] [PubMed] [Google Scholar]
- 41.Mortuza, G. B., et al. 2008. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, Gag assembly and core formation. J. Mol. Biol. 376:1493-1508. [DOI] [PubMed] [Google Scholar]
- 42.Mortuza, G. B., et al. 2004. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 431:481-485. [DOI] [PubMed] [Google Scholar]
- 43.Ono, A., A. A. Waheed, A. Joshi, and E. O. Freed. 2005. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131-14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen, E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 45.Pornillos, O., et al. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakalian, M., et al. 2006. 3-O-(3′,3′-Dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 Gag precursor assembled in vitro. J. Virol. 80:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 48.Wiegers, K., et al. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilk, T., et al. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U. S. A. 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wlodawer, A., and J. Vondrasek. 1998. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 27:249-284. [DOI] [PubMed] [Google Scholar]
- 52.Wright, E. R., et al. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, J., C. H. Chen, and C. Aiken. 2006. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. J. Virol. 80:12095-12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, J., C. H. Chen, and C. Aiken. 2004. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid. Retrovirology 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, J., L. Huang, D. L. Hachey, C. H. Chen, and C. Aiken. 2005. Inhibition of HIV-1 maturation via drug association with the viral Gag protein in immature HIV-1 particles. J. Biol. Chem. 280:42149-42155. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, J., et al. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.