Abstract
The adenovirus (Ad) E1b55K and E4orf6 gene products assemble an E3 ubiquitin ligase complex that promotes degradation of cellular proteins. Among the known substrates are p53 and the Mre11-Rad50-Nbs1 (MRN) complex. Since members of the RecQ helicase family function together with MRN in genome maintenance, we investigated whether adenovirus affects RecQ proteins. We show that Bloom helicase (BLM) is degraded during adenovirus type 5 (Ad5) infection. BLM degradation is mediated by E1b55K/E4orf6 but is independent of MRN. We detected BLM localized at discrete foci around viral replication centers. These studies identify BLM as a new substrate for degradation by the adenovirus E1b55K/E4orf6 complex.
Cellular DNA repair proteins are associated with virus infection (reviewed in references 37 and 65). While cellular repair proteins accumulate at viral replication centers (24, 40, 52, 57, 59, 67, 72) and process viral DNA ends (20, 57), viruses employ multiple strategies to manipulate DNA damage signaling and repair pathways (reviewed in references 19 and 65). Infection with adenovirus (Ad) deleted of early region E4 activates the DNA damage response and results in joining of linear viral genomes into concatemers (3, 9, 13, 14, 24, 40, 57). Processing and ligation of viral ends involve the Mre11-Rad50-Nbs1 (MRN) complex and components of the nonhomologous end-joining pathway (3, 9, 57). Adenoviral E4 gene products E4orf3 and E4orf6, respectively, mislocalize and degrade cellular repair proteins to promote viral infection (3, 14, 23, 24, 35, 40, 57, 58). The viral E1b55K and E4orf6 proteins assemble an E3 ubiquitin ligase complex together with cellular proteins Cullin 5 and Elongin B/C (28, 47). Cellular proteins identified as degradation substrates of E1b55K/E4orf6 include the MRN complex (6, 14, 51, 57), p53 (15, 28, 43, 47-50, 55), integrin α3 (22), and DNA ligase IV (3). The E1b55K/E4orf6 ubiquitin ligase activity is suggested to create a cellular environment that promotes efficient virus growth through degradation of these and other targets (4, 5, 7, 68).
Studies of DNA double-strand breaks have identified a large number of factors required for correct recognition, processing, and repair (reviewed in references 30 and 41). The MRN complex senses DNA damage and functions together with nucleases and helicases to process DNA ends and promote repair (36, 41). The human RecQ helicases (RecQ1, WRN, BLM, RecQ4, and RecQ5) are a family of proteins involved in maintaining genome integrity (8, 21, 53). The Bloom helicase (BLM) is implicated in processive resection of DNA breaks (27, 31, 34, 44, 45, 56, 62, 69). Since proteins involved in DNA end processing and repair are deactivated by adenovirus, we investigated whether human RecQ helicases are altered during infection.
E1b55K and E4orf6 induce proteasome-mediated degradation of BLM.
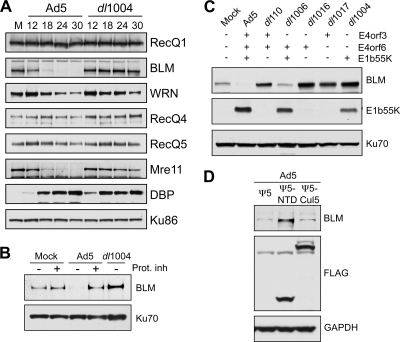
We examined the effect of Ad infection on human RecQ helicases (Fig. 1). Immunoblotting of lysates revealed that levels of BLM decreased significantly in cells infected with wild-type Ad5 virus but not E4-deleted virus (dl1004). The kinetics for BLM decrease mirrored the previously described degradation of Mre11 (Fig. 1A), which is proteasome mediated and E4 dependent (14, 51, 57). Addition of proteasome inhibitors during Ad5 infection confirmed that BLM degradation was proteasome dependent (Fig. 1B). The protein levels of RecQ1, RecQ4, and RecQ5 helicases were not altered during infection. Although levels of WRN decreased, this was not dependent upon proteasome activity and the E4 region (Fig. 1A and data not shown).
FIG. 1.
Adenovirus infection induces proteasome-mediated degradation of BLM. (A) The steady-state levels of cellular RecQ helicases were examined over a time course of adenovirus infection. HeLa cells were either mock infected (M) or infected with wild-type Ad5 (multiplicity of infection [MOI] of 10) or the E4-deleted mutant dl1004 (MOI of 25). Cells were harvested at the indicated hours postinfection (hpi) for analysis by immunoblotting (14). Specific antibodies were used to detect RecQ1 (Santa Cruz), WRN (BD Biosciences), RecQ4 (Cell Signaling), and RecQ5 (gift from P. Janscak) (32). To generate the anti-BLM antibody, rabbits were immunized with a purified recombinant protein consisting of a His-tagged BLM fragment (amino acid residues 1 to 439). The purified anti-BLM antiserum (designated 7099) was tested for specificity by immunoblotting and immunofluorescence (data not shown). The viral DBP (detected with monoclonal antibody B6, from A. Levine) served as a control for infection, and degradation of cellular proteins was confirmed by inclusion of the Mre11 positive control (Genetex). Antibody to Ku86 (Santa Cruz) served as a loading control. Although the levels of WRN were slightly affected, only BLM was reduced in an E4-dependent manner analogous to Mre11 degradation. (B) BLM degradation is proteasome dependent. Cells were infected with Ad5 (MOI of 10) or dl1004 (MOI of 25), and at 12 hpi, proteasome inhibitors (Prot. inh) (10 μM MG132 and 1 μM epoxomicin) were added to the cells for a further 12 h. Degradation of BLM by Ad5 was prevented by the proteasome inhibitors. Ku70 served as a loading control. (C) E1b55K and E4orf6 are required for degradation of BLM during adenovirus infection. BLM levels were compared in HeLa cells infected for 24 h with wild-type Ad5 or mutants lacking genes from the E1 and E4 regions as indicated (2, 10, 11, 26). Compared with results for mock-infected cells, BLM levels were reduced only during infection with viruses that express both E1b55K and E4orf6. E1b55K (detected with monoclonal antibody 2A6, from A. Levine) and Ku70 (antibody from Santa Cruz) served as controls for infection and gel loading, respectively. (D) The Cul5 complex is required for BLM degradation. Cells were infected with Ad5 (MOI of 10), superinfected (MOI of 50) with Ψ5, Ψ5-Cul5, or Ψ5-NTD (68), and harvested at 24 h after the primary infection. In these infections, the FLAG antibody (Sigma) demonstrates expression of Cul5 or NTD-Cul5 from the Ψ5 viruses, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Research Diagnostics Inc.) serves as a loading control.
To identify viral proteins responsible for BLM degradation, we infected HeLa cells with mutant viruses with specific genes deleted (2, 10, 11, 26). BLM levels decreased during infection with a viral mutant that does not express E4orf3 (dl1006) but were unaffected by mutants that lack either E1b55K or E4orf6 (Fig. 1C). The E1b55K/E4orf6 complex is therefore required for BLM degradation. To determine whether BLM degradation was mediated by the Cul5 complex, we utilized a dominant negative version of Cul5, consisting of the N-terminal domain (NTD-Cul5), which prevents substrate ubiquitination (68). BLM degradation was prevented by NTD-Cul5 but was unaffected by empty vector and full-length Cul5 (Fig. 1D). Therefore, BLM undergoes proteasome-mediated degradation by the E1b55K/E4orf6 ubiquitin ligase complex containing Cul5.
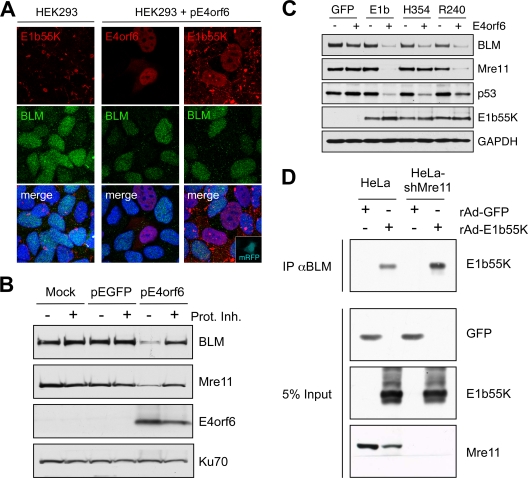
Further experiments examined requirements for BLM degradation (Fig. 2). The E1b55K protein is in an aggresome structure in HEK293 cells (1, 25, 38) and relocalizes to the nucleus upon E4orf6 expression (15). BLM was diffusely nuclear in these cells but was reduced in cells that were transfected to express E4orf6 and have nuclear E1b55K (Fig. 2A). BLM degradation in E4orf6-transfected HEK293 cells was confirmed by immunoblotting and was rescued by proteasome inhibitors (Fig. 2B). BLM was also degraded when E4orf6 was expressed from an adenovirus vector (48) in U2OS cells that stably express E1b55K (14) (Fig. 2C). These data demonstrate that E1b55K and E4orf6 are sufficient for degradation of BLM.
FIG. 2.
E1b55K and E4orf6 are sufficient for degradation of BLM. (A) Immunofluorescence reveals that in HEK293 cells the BLM protein is located throughout the nucleoplasm and E1b55K is in cytoplasmic aggregates (left). When HEK293 cells were transfected with E4orf6 (15), BLM levels were reduced (middle). In cells transfected with plasmids expressing E4orf6 and monomeric red fluorescent protein (mRFP) (at a 9:1 ratio), transfected cells demonstrated nuclear E1b55K, with reduced BLM levels (right). Cells were fixed for staining (14) at 24 h after transfection, and DAPI (4′,6-diamidino-2-phenylindole) staining indicates the location of the nuclei in all merged images. (B) Expression of E4orf6 by transfection of HEK293 cells demonstrated a proteasome-dependent decrease in BLM levels. Cells were harvested at 20 h posttransfection for analysis by immunoblotting with specific antibodies. Degradation was abrogated by proteasome inhibitors (10 μM MG132 and 1 μM epoxomicin). Mre11 served as a control for degradation, and Ku70 served as a loading control. (C) E1b55K and E4orf6 are sufficient for BLM degradation. E4orf6 was expressed by rAd vector transduction (48) of U2OS cells that stably express GFP or E1b55K (14), and protein levels were assessed by immunoblotting with the indicated antibodies. Antibodies to Mre11 (Genetex) and p53 (Calbiochem) served as controls for degradation, and Ku70 served as a loading control. (D) E1b55K coimmunoprecipitates with BLM in the absence of Mre11. HeLa cells or HeLa-shMre11 cells were infected (MOI of 50) with rAd-GFP and rAd-E1b55K (48) for 24 h, and lysates were subjected to immunoprecipitation with the BLM antibody (IP αBLM). Immunoblotting of the precipitated proteins demonstrated that E1b55K could be pulled down by BLM in lysates from both cells.
BLM is a member of the BRCA1-associated genome surveillance complex (BASC) supercomplex of proteins, which interact with the BRCA1 protein (61). Since the MRN complex is also part of the BASC supercomplex (61), it was possible that BLM degradation was an indirect outcome of its association with MRN. We found that BLM is efficiently degraded during Ad5 infection of HeLa cells stably expressing short hairpin RNA (shRNA) against Mre11 (HeLa-shMre11 cells) (60) and in Nijmegen breakage syndrome cells with mutant Nbs1 (16) (data not shown). This suggests that BLM degradation by E1b55K/E4orf6 is independent from interaction with MRN. This was confirmed using U2OS cell lines expressing separation-of-function mutants of E1b55K (14, 51). The H354 insertion mutant degrades p53 but is defective for MRN degradation, while the R240 mutant degrades MRN but not p53. E4orf6 expression in cell lines expressing E1b55K, H354, and R240 degraded BLM (Fig. 2C). Therefore, the region of E1b55K required for Mre11 degradation is different from that required to degrade BLM.
The E1b55K protein provides substrate specificity to the ubiquitin ligase complex through binding to cellular targets (14, 15, 38, 47, 49-51, 54, 66). To examine interactions, we performed immunoprecipitations with BLM-specific antibody on lysates from cells infected with recombinant adenovirus (rAd) expressing green fluorescent protein (GFP) or E1b55K. Immunoblotting demonstrated that E1b55K coimmunoprecipitates BLM (Fig. 2D). BLM could also immunoprecipitate E1b55K in the absence of Mre11 from cell lines stably expressing shMre11 (60). Interaction was also verified in cells transiently transfected with affinity-tagged E1b55K (data not shown). These data demonstrate association of BLM with E1b55K, independently of the interaction between E1b55K and MRN.
BLM localizes to sites of active viral replication.
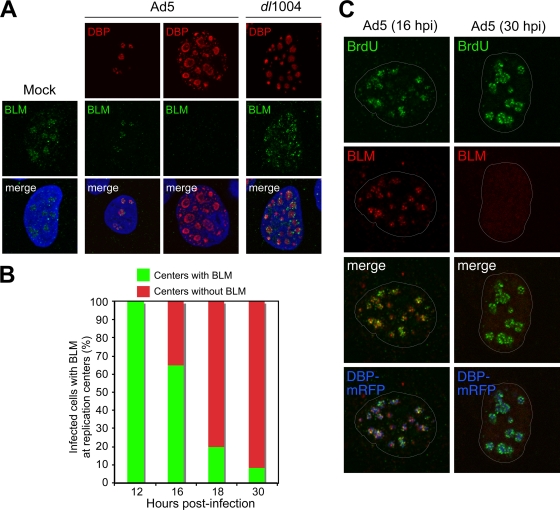
We examined localization of BLM in Ad-infected cells (Fig. 3). The virally encoded single-stranded DNA (ssDNA) binding protein (DBP) marks sites of viral DNA replication in infected nuclei (46). In uninfected cells, BLM was localized throughout the nucleus, in distinct foci, and in the nucleolus, as previously reported (29, 44, 71, 73). In Ad5-infected cells with small DBP centers, representing early-stage infection, BLM localized to foci at DBP centers (Fig. 3A). BLM staining was not detected in Ad5-infected cells with large, spherical-shaped DBP centers, representing late-stage infection (46). Quantitation of staining patterns showed that at early times all viral replication centers displayed colocalizing BLM foci but that this decreased as infection progressed (Fig. 3B). In cells infected with E4-deleted virus, BLM localized at viral replication centers and did not decrease (Fig. 3A). Pulse-labeling with bromodeoxyuridine (BrdU) incorporation reveals sites of active adenoviral DNA replication (46). Immunofluorescence using antibodies to BLM and BrdU revealed partial colocalization in foci at DBP centers (Fig. 3C). This suggests that BLM accumulates near sites of active viral replication during early stages of infection.
FIG. 3.
Accumulation of BLM at sites of viral replication. (A) HeLa cells were infected with wild-type Ad5 (MOI of 10) or E4 mutant dl1004 (MOI of 25). Localization of BLM was examined by immunofluorescence (14) after preextraction to remove soluble protein prior to fixation and compared to that for mock-infected cells (left). In uninfected cells, BLM localized to nucleoli and ND10 structures. In Ad5-infected cells, BLM was either located in discrete foci surrounding early viral replication centers (as detected with an antibody to the viral DBP) or undetectable in cells with large centers (representing late-stage infection). In cells infected with E4-deleted virus dl1004, all infected cells displayed BLM accumulated at viral centers. DAPI staining indicates the location of the nuclei in all merged images. (B) Quantification of infected cells with BLM at viral replication centers over a time course of wild-type Ad5 infection. At each time point, the relative number of cells with BLM in the two patterns demonstrated by the representative images shown in panel A was determined by examining 100 infected cells. (C) BLM at sites of active viral replication. HeLa cells were transfected with DBP-mRFP and, after 8 h, were infected with Ad5 (MOI of 10). At early (16 hpi) and late (30 hpi) stages of infection, cells were pulsed with BrdU for 30 min to label sites of DNA replication (46) and detected with a BrdU-specific antibody (Sigma). In preextracted cells, BLM could be detected adjacent to sites of ongoing viral replication. By 30 hpi, BLM had been degraded.
BLM does not affect levels of DNA accumulation or formation of concatemers.
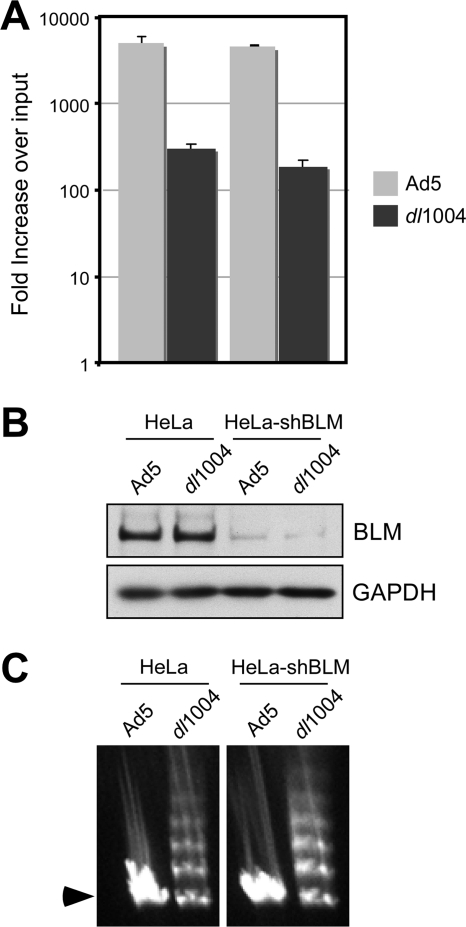
Since BLM is implicated in DNA end processing, we examined whether BLM affected viral DNA replication and formation of viral genome concatemers in E4-defective virus infection (Fig. 4). E4-deleted mutants are defective in accumulation of replicated viral DNA (12, 64), and this is overcome in cells deficient for components of the MRN complex (24, 35, 39). To test whether BLM contributes to inhibition of adenovirus replication, we assessed E4-deleted virus replication in cells deficient for BLM (Fig. 4A). Measurement of virus DNA accumulation by quantitative PCR (qPCR) (35) demonstrated that E4-deleted virus remained defective for replication in HeLa cells stably expressing shRNA against BLM (HeLa-shBLM cells). We also analyzed viral DNA by pulsed-field gel electrophoresis (PFGE) (Fig. 4C). In cells infected with Ad5, the viral genome was detected in a single band representing linear monomers (9, 57, 63). The E4-deleted virus genomes were detected in slower-migrating bands representing concatemers of viral DNA in both HeLa and HeLa-shBLM cells. Together, these results demonstrate that BLM is not responsible for inhibition of E4-deleted Ad replication or concatemerization of viral genomes. These conclusions were confirmed in patient-derived Bloom syndrome cells lacking functional BLM (data not shown).
FIG. 4.
BLM is not responsible for defective viral replication or concatemer formation. (A) Viral replication was assessed in HeLa cells and HeLa cells stably expressing shRNA against BLM (HeLa-shBLM cells). The shRNA sequence 5′-TGCCAATGACCAGGCGATC was inserted into the pSUPER.retro.puro vector (Orbigen), and Phoenix retroviral packaging cells were used to produce retrovirus. HeLa cells were transduced with retrovirus, and the HeLa-shBLM cells were selected and maintained in puromycin. Cells were infected with Ad5 (MOI of 10) or dl1004 (MOI of 3) in triplicate, and DNA was extracted for quantitative PCR at 4 and 30 hpi as previously described (35). Accumulation of viral DNA is represented as fold increase over input viral DNA, as determined at the 4-h time point, and error bars represent standard errors of the means (SEM) from the triplicate samples. (B) Immunoblotting of lysates from infected cells at the 4-h time point confirmed knockdown of BLM levels in HeLa-shBLM cells. GAPDH served as a loading control. (C) Concatemer formation was examined by PFGE analysis of DNA at 48 hpi with Ad5 (MOI of 25) and dl1004 (MOI of 50) as previously described (57). Viral DNA was visualized by staining the gel in ethidium bromide, and the position of the linear viral genome is indicated by an arrowhead. Formation of concatemers by dl1004 was not dependent on BLM.
During Ad infection, the DNA damage signaling and repair machinery is manipulated in multiple ways (65). In this report, we identified BLM as a novel degradation substrate for E1b55K/E4orf6. Despite sequence and structural homology across the human RecQ helicases, BLM is the only member degraded by E1b55K/E4orf6. Although BLM and the MRN complex are both members of the BASC supercomplex (61), we demonstrated that they are degraded independently. There is no obvious homology among the substrates for E1b55K, suggesting that different regions of E1b55K mediate interactions with distinct substrates (51). It will be interesting to determine whether E1b55K/E4orf6 proteins of different Ad serotypes degrade BLM or other RecQ helicases.
Adenovirus inactivation of cellular DNA repair proteins prevents inhibition of viral DNA replication and genome processing (3, 24, 35, 39, 57). In our studies with BLM, we have been unable to assign a functional relevance to degradation. Unlike results with the MRN complex (35, 39), knocking down levels of BLM did not rescue the E4-deleted virus replication defect or prevent concatemerization. Prior to its degradation by E1b55K/E4orf6, BLM was detected adjacent to sites of viral replication. This recruitment may play a positive role in early steps of viral infection. It is possible that replication integrity and resulting progeny genomes are affected by BLM. Redundancy across the RecQ helicase family may functionally substitute in the absence of BLM. BLM suppresses hyperrecombination (17, 18) and functions to resolve recombination intermediates (70). These BLM activities could affect Ad replication by resolving incomplete intermediates or could prevent serotype mixing by suppressing homologous recombination between coinfected viruses. Analysis of the sequences at end-to-end junctions in concatemers has demonstrated heterogeneity in the degree of sequence loss (33, 63), suggesting incomplete replication or processing of viral DNA ends. Given its emerging role in end processing/resection of DNA breaks (27, 42, 45), it is possible that BLM functions with MRN to modify the ends of viral genomes. Comparison of sequences for concatemer junctions formed in the presence and absence of BLM may reveal processing differences. Further understanding of interactions with adenovirus will also provide insights into the role of BLM in cellular functions.
Acknowledgments
We thank A. Berk, P. Branton, P. Concannon, C. Her, P. Janscak, G. Ketner, A. Levine, D. Ornelles, P. van der Vliet, and J. Wilson for generous gifts of reagents. We thank members of the Weitzman lab for discussions and critical reading of the manuscript.
Work in the Weitzman lab was partially supported by a Pioneer Developmental Chair from the Salk Institute and by NIH grant CA097093 (M.D.W.). Work in the Karlseder lab was supported by NIH grant AG025837 (J.K.). N.I.O. was supported in part by a gift from the H. A. & Mary K. Chapman Charitable Trust.
Footnotes
Published ahead of print on 1 December 2010.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackford, A. N., and R. J. Grand. 2009. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J. Virol. 83:4000-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette, P., and P. E. Branton. 2009. Manipulation of the ubiquitin-proteasome pathway by small DNA tumor viruses. Virology 384:317-323. [DOI] [PubMed] [Google Scholar]
- 6.Blanchette, P., et al. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchette, P., et al. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohr, V. A. 2008. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 33:609-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 10.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 11.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bridge, E., S. Medghalchi, S. Ubol, M. Leesong, and G. Ketner. 1993. Adenovirus early region 4 and viral DNA synthesis. Virology 193:794-801. [DOI] [PubMed] [Google Scholar]
- 13.Carson, C. T., et al. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carson, C. T., et al. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerosaletti, K. M., et al. 2000. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15:281-286. [DOI] [PubMed] [Google Scholar]
- 17.Chaganti, R. S., S. Schonberg, and J. German. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 71:4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan, K. L., P. S. North, and I. D. Hickson. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaurushiya, M. S., and M. D. Weitzman. 2009. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8:1166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 80:10346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu, W. K., and I. D. Hickson. 2009. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer 9:644-654. [DOI] [PubMed] [Google Scholar]
- 22.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans, J. D., and P. Hearing. 2003. Distinct roles of the adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans, J. D., and P. Hearing. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleisig, H. B., et al. 2007. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene 26:4797-4805. [DOI] [PubMed] [Google Scholar]
- 26.Ginsberg, H. S., L. L. Moldawer, and G. A. Prince. 1999. Role of the type 5 adenovirus gene encoding the early region 1B 55-kDa protein in pulmonary pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 96:10409-10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gravel, S., J. R. Chapman, C. Magill, and S. P. Jackson. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishov, A. M., et al. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson, S. P., and J. Bartek. 2009. The DNA-damage response in human biology and disease. Nature 461:1071-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, F. B., et al. 2000. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 32.Kanagaraj, R., N. Saydam, P. L. Garcia, L. Zheng, and P. Janscak. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karen, K. A., P. J. Hoey, C. S. Young, and P. Hearing. 2009. Temporal regulation of the Mre11-Rad50-Nbs1 complex during adenovirus infection. J. Virol. 83:4565-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusano, K., M. E. Berres, and W. R. Engels. 1999. Evolution of the RECQ family of helicases: a drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics 151:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakdawala, S. S., et al. 2008. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 82:8362-8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamarche, B. J., N. I. Orazio, and M. D. Weitzman. 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119-126. [DOI] [PubMed] [Google Scholar]
- 38.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew, S. S., and E. Bridge. 2008. Nbs1-dependent binding of Mre11 to adenovirus E4 mutant viral DNA is important for inhibiting DNA replication. Virology 374:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathew, S. S., and E. Bridge. 2007. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365:346-355. [DOI] [PubMed] [Google Scholar]
- 41.Mimitou, E. P., and L. S. Symington. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst.) 8:983-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mimitou, E. P., and L. S. Symington. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. U. S. A. 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neff, N. F., et al. 1999. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol. Biol. Cell 10:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nimonkar, A. V., A. Z. Ozsoy, J. Genschel, P. Modrich, and S. C. Kowalczykowski. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 105:16906-16911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Querido, E., et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Querido, E., et al. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Querido, E., et al. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth, J., et al. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 72:8510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz, R. A., et al. 2008. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 82:9043-9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz, R. A., et al. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma, S., K. M. Doherty, and R. M. Brosh, Jr. 2006. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398:319-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 58.Stracker, T. H., et al. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vo, A. T., et al. 2005. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 6:438-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Y., et al. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 62.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. U. S. A. 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weitzman, M. D., C. E. Lilley, and M. S. Chaurushiya. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64:61-81. [DOI] [PubMed] [Google Scholar]
- 66.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, L., et al. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 70.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 71.Yankiwski, V., R. A. Marciniak, L. Guarente, and N. F. Neff. 2000. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. U. S. A. 97:5214-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, X., et al. 2008. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in simian virus 40-infected primate cells. J. Virol. 82:5316-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong, S., et al. 1999. A role for PML and the nuclear body in genomic stability. Oncogene 18:7941-7947. [DOI] [PubMed] [Google Scholar]