Abstract
Alternative RNA processing mechanisms, including alternative splicing and alternative polyadenylation, are increasingly recognized as important regulators of gene expression. This article will focus on what has recently been described about alternative polyadenylation in development, differentiation, and disease in higher eukaryotes. We will also describe how the evolving global methodologies for examining the cellular transcriptome, both experimental and bioinformatic, are revealing new details about the complex nature of alternative 3′ end formation, as well as interactions with other RNA-mediated and RNA processing mechanisms.
RNA processing has long been known to be involved in regulation of gene expression. Such processing encompasses many modification events, including capping, splicing, polyadenylation, editing, and base modifications. This article will focus on polyadenylation, the process by which messenger RNAs (mRNAs) acquire a poly(A) tail at their 3′ ends, and alternative polyadenylation, the process by which alternative 3′ ends of an mRNA become polyadenylated. It should also be noted that due to space limitations, not all work on this subject could be discussed here; we have highlighted some important areas of current research.
Poly(A) tails are found on the 3′ end of nearly every fully processed eukaryotic mRNA, and influence mRNA stability, translation, and transport (reviewed in 1-3). Polyadenylation is a two-step process (reviewed in 4-7), first involving specific endonucleolytic cleavage at a site determined by binding of polyadenylation factors. The second step involves polymerization of the adenosine tail to ~200 residues in higher eukaryotes and ~70 As in yeast; tail length is quite organism-specific. As has been appreciated more completely in recent years, 3′ end formation is interconnected to other mRNA processing events, mRNA transcription, and transcription termination (8-11).
Almost all eukaryotic polyadenylation signals contain a core upstream element, the consensus sequence AAUAAA (or a variant) ~10-35 nucleotides upstream of the actual site of poly(A) addition (reviewed in 6-7, 12). In addition, sequences ~14-70 nucleotides downstream (core downstream elements) are known to be involved in directing polyadenylation (reviewed in 6-7 and references therein). Auxiliary elements upstream and downstream of the AAUAAA sequence have also been characterized that can enhance polyadenylation efficiency, often mediated by specific binding of additional trans-acting factors (13-22). The basal mammalian polyadenylation and cleavage machinery is comprised of four basal multi-subunit protein factors, which assemble on the RNA before any reaction takes place (reviewed in 6,23). Many additional, auxiliary protein factors have also been identified (reviewed in 7, 23-24). In a tightly coupled, two-step process, cleavage occurs 10-30 residues after the core upstream element, followed by addition of the poly(A) tail by poly(A) polymerase.
The complexity of polyadenylation and interaction with other mRNA processing reactions and transcription indicate that polyadenylation is used to regulate gene expression in a variety of developmental and proliferative “schemes.” Most pre-mRNAs in the cell are not efficiently processed, and therefore, even small changes in the overall processing efficiency of a particular pre-mRNA may have a great effect (see Figures 1, 2, and 3). Frequently, several auxiliary factors bind to the pre-mRNA and either competes and/or cooperates with each other to modulate the mechanisms of cleavage and polyadenylation. Small alterations on the concentrations of any of these factors in a particular physiological state of the cell will affect the outcome of the pre-mRNA 3′ end processing and certainly impacts on gene expression. Alternative processing of mRNAs is another important regulatory level in gene expression control as it is now appreciated that over half of the mRNAs in the human genome are alternatively polyadenylated (12). In addition, the utilization of alternative polyadenylation as a regulator of gene expression is evolutionarily conserved in plants (25). Therefore, an understanding of the mechanisms that control alternative polyadenylation has become of great interest and importance over the past few years.
Figure 1.
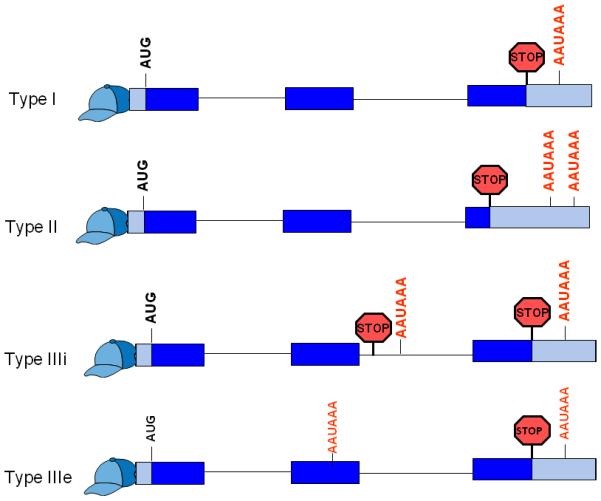
Schematic of polyadenylation events. Light blue boxes, untranslated regions; darker blue boxes, coding regions; lines, introns. mRNAs with Type I polyadenylation only have one polyadenylation signal in the 3′ most exon. mRNAs with Type II polyadenylation have more than one polyadenylation signal in the 3′ most exon. Type III polyadenylation is alternative splicing coupled with alternative polyadenylation; Type IIIi signals have one or more polyadenylation signals in upstream introns; Type IIIe signals have one or more polyadenylation signals in upstream exons.
Figure 2.
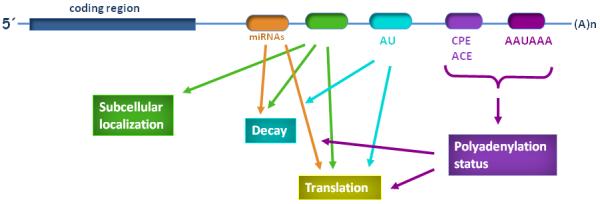
Many RNA cis-acting elements present in the 3′ UTR can affect polyadenylation as well as other RNA processing or RNA mediated events, and these effects are intertwined. Regulation of gene expression by the 3′UTR depends on the presence of the cis-acting regulatory element and may influence nuclear and cytoplasmic polyadenylation (pA signal and cytoplamic polyadenylation element - CPE), the stability of the mRNA produced (for example by AU elements), the subcellular localization of the mRNA, and translation or mRNA decay. Additionally, the presence of microRNA target sites in the 3′UTR may influence gene expression. The presence of the trans-acting factors that bind to these elements or the microRNA that target the mRNA in the cell or tissue, at a particular stage and at the correct concentration, is essential for regulation to occur. Alternative polyadenylation in the 3′UTR generates different mRNA isoforms containing different cis-acting elements. Adapted from Conne et al., 2000 (with permission of the authors).
Figure 3.
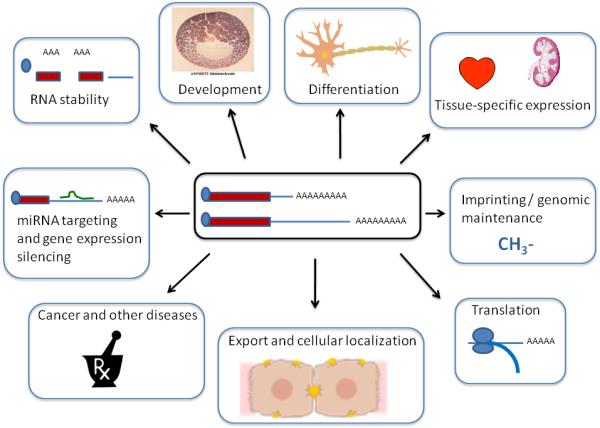
Alternative polyadenylation of mRNA, as shown in the center of the figure, can affect many other cellular or organismal events. See text for details.
Classifications of alternative polyadenylation
We recently described three different types of polyadenylation and alternative polyadenylation in higher eukaryotic mRNAs (26, see also Figure 1). In Type I polyadenylation, only one polyadenylation signal is present in the 3′ UTR, thus resulting in only one mRNA isoform. In Type II alternative polyadenylation, more than one polyadenylation signal is present but are only present in a common terminal exon. In this type of alternative polyadenylation, more than one resulting mRNA is produced, but with no effect on the encoded protein; however, due to possible alteration of mRNA stability/translatability/other downstream effects (Figure 2), there may be changes to the amount of protein produced if alternative polyadenylation signals are chosen. Type III alternative polyadenylation involves alternative polyadenylation signals that are present in upstream introns or exons, thus invoking alternative splicing along with alternative polyadenylation. We have further defined here Type III polyadenylation as Type IIIi (intronic alternative polyadenylation signal) or Type IIIe (exonic alternative polyadenylation signal). These types of alternative polyadenylation may or may not result in different protein products being produced, depending on the stability of the mRNA, the presence of an in-frame stop codon, and the overall translation competence of the mRNA. In this review, we will examine how alternative polyadenylation can modulate or regulate gene expression under various biological circumstances, or be regulated itself.
Alternative polyadenylation during different cellular states or programs
In the past few years, exciting results have further highlighted the crucial role of alternative polyadenylation in the control of global physiological events such as proliferation, differentiation, transformation and development programs. All these biological processes are dependent on the accurate regulation of a number of genes in a very precise temporal manner, which will enable the cells to develop properly. This is particularly important in Type III alternative polyadenylation (26), where different transcript isoforms originated by alternative polyadenylation signal usage will give rise to different proteins with diverse functions in the cell. However, no less important is the global change observed in Type II alternative polyadenylation during these physiological events, which has been adequately illustrated by several recent global analyses.
It has been long established that differences in 3′UTRs can confer different stabilities and/or translation competencies to the corresponding mRNAs. Obviously, longer 3′UTRs contain typically more cis regulatory elements in the pre-mRNA than short 3′UTRs, and these cis elements may be targets of RNA binding proteins involved in mRNA stability, localization and translation (Figure 2). In addition, 3′UTRs are the most common location in mRNAs for microRNA (miRNA) binding sites to be found; such miRNAs are known to regulate mRNA expression through inhibiting mRNA translation or by mRNA cleavage and subsequent degradation. The number of miRNA binding sites, as well as the AU-rich content, is predominantly higher in longer 3′UTRs (27-29). Consequently, the differential utilization of one or the other polyadenylation signal implies that these RNA sequence elements will be differentially used, affecting the fate of the transcript produced and ultimately modulating the expression of the gene.
In transcriptome-wide studies, alternative polyadenylation leading to 3′UTR shortening has been shown to occur during development of spermatocytes (30), in proliferating T cells (29) and in some cancer cell lines (31). Conversely, mRNA 3′UTR lengthening due to alternative polyadenylation has been also identified in ovulated oocytes and zygotes (32), developing mouse embryos (27) and in neurological tissues (33). Additionally, in the generation of pluripotent stem cells from different cell types, the length of the 3′UTR is modified by alternative polyadenylation (34).
For over 30 years it has been known that T lymphocyte activation leads to increases in RNA polyadenylation and protein synthesis, even though the transcription rate remains unchanged (35-36). Only recently, however, it was described that upon T cell activation there is an increase in proximal poly(A) signal usage, thereby leading to shorter 3′UTRs (29). These conclusions were drawn from a genome-wide study using murine T lymphocytes, while more restricted earlier work had already shown that alternative polyadenylation is regulated in some genes in T cells (37, 38). In the Sandberg et al. work, the authors performed a global analysis of alternative polyadenylation using CD4+ T lymphocytes isolated from mice, and activated the lymphocytes with antibodies triggering the TCR/CD3 complex (29). Using RNA prepared from resting and activated T cells they hybridized a microarray and strikingly, 86% of changes in genes with tandem 3′UTRs were congruent with the trend of decrease of the 3′UTR length. Surprisingly, those genes undergoing 3′ exon switching did not show the same effect of 3′UTR shortening upon T cell activation, suggesting that the regulation of this particular mechanism of alternative polyadenylation is splicing-independent. The variation in alternative polyadenylation could not be detected upon 6 hours of activation, but was evident after 48 hours post activation. This suggests that signal-induced alternative poly(A) signal selection is not an immediate event, presumably because it is necessary to transcribe and/or modulate basal and/or auxiliary polyadenylation/cleavage factors. These observations lead to the still unanswered question on how the signals at the membrane surface are transduced to the cleavage/polyadenylation machinery.
Interestingly, the length of the 3′UTRs is consistently shorter in proliferating cell lines than those shown in primary animal tissue, leading to the hypothesis of alternative polyadenylation being correlated with proliferation (29). To understand the impact of 3′UTR shortening in protein expression, different 3′UTRs were fused to a luciferase reporter gene and then luciferase activity was assessed. The results showed that longer 3′UTR conferred a decrease in protein translation. This would indicate that negative post-transcriptional regulators exist downstream of the proximal poly(A) signal. These signals, therefore, only exist in and can only act on the longer isoform that relies on the distal site. This was proven to be correct for Hip2, Huntingtin Interacting Protein 2, a predicted ubiquitin ligase, involved in polyglutamine diseases. A double mutation in miR-21 and miR-155 seed matches (a perfect Watson–Crick match between miRNA and target at miRNA positions 2–7 or 8) contained in the longer 3′UTR restored luciferase expression to the levels obtained with the shorter transcript. This study revealed for the first time that the global mechanism of alternative polyadenylation may be part of a genetic program that can be correlated with the state of cell proliferation, where usage of a proximal poly(A) signal in the 3′UTR may enable the genes to avoid repression induced by miRNAs that target the longer 3′UTR.
Following the finding that alternative polyadenylation is correlated with cellular proliferation and differentiation, it was not surprising that a link was established between alternative polyadenylation and miRNA-mediated repression in cancer cell lines (31). It had been previously reported that oncogenes, for example HMGA2, could be activated via the loss of their miRNAs target sites (39, 40). Additionally, in mantle cell lymphomas, the length of the Cyclin D1 3′UTR was shortened due to mutations that created premature poly(A) signals, leading to an increase in mRNA stability (41). Mayr and Bartel analyzed by Northern blotting the expression of 23 genes containing more than one poly(A) signal in the 3′UTR, comparing cancer cell lines and normal tissues, and they found that most of these genes expressed their shorter isoform in cancer cell lines, due to the utilization of the most proximal poly(A) signal. By comparison with the data produced by Sandberg et al., the authors concluded that cancer cell lines were more prone to produce the shorter 3′UTR isoforms, and importantly, that this effect was linked to the transformation potential of those cells (31). Furthermore, using a luciferase reporter gene fused to the IMP-1 3′UTRs, a shortening of the UTR was shown to result in the loss of miRNA target sites and an increase in protein production. Importantly, a shorter isoform of IMP-1 promoted oncogenic transformation. This work has important implications for the understanding of cancer as it demonstrates that one of the mechanisms of oncogenic transformation is the loss of miRNA sites in the mRNAs of oncogenes. These genes can escape miRNA-mediated repression through 3′UTR shortening due to the pattern of alternative polyadenylation that occurs in cancer cells.
It is worthy to note that loss of miRNA regulation in cancer cell lines accounts for only a quarter to two-thirds of the increase in protein expression levels observed with short 3′UTRs (31). Thus, for the majority of the cell lines there are other regulatory events that must be taken into account. These may be due to the presence of RNA binding proteins, for example, a hypothesis that has not yet been explored. Nevertheless, these findings imply that alternative polyadenylation is a prevalent mechanism in oncogenic transformation and also in physiological T cell proliferation, and may as well be part of a global gene expression program. This concept has been extended to primary tumor samples from a mouse leukemia/lymphoma model (42). It is remarkable that in these tumors, alternative polyadenylation seems to define molecular signatures that can distinguish similar tumor subtypes with more than 70% of accuracy. In addition, certain polyadenylation factors such as CstF-77 were found to be upregulated in the mouse lymphomas (42). These results anticipate the future usage of different groups of genes as biomarkers with diagnostic potential.
All these studies have brought to the spotlight the importance of alternative polyadenylation in modulating gene expression in different physiological conditions. Although some general conclusions can be drawn from these studies, a specific pattern of poly(A) signal usage in the 3′UTR depending on the cellular state should not be definitively established, as alternative polyadenylation is a complex mechanism and depends on a variety of factors. Given the current interest in alternative polyadenylation as a regulatory mechanism in development and disease, many details will be elucidated in the near future.
Alternative polyadenylation and coordination with other RNA processing events
Although polyadenylation and other mRNA processing events were originally thought of as distinct and separate events, it has more recently been appreciated that all RNA processing events are intertwined and inter-connected (reviewed in 8, 10-11). These events include interactions between polyadenylation and splicing, transcription, transcription termination, translation, export, and stability (43, 9, 11). The protein and RNA factors involved in many of these interconnected processes are also now known to be involved in regulating alternative splicing and alternative polyadenylation (21, 44-46). Using high throughput sequencing of RNA isolated by crosslinking-immunoprecipitation (HITS-CLIP) analysis, Darnell and co-workers found such an example in a global study aiming at identifying targets for NOVA, a well-characterized splicing factor. These studies revealed that NOVA could also regulate alternative polyadenylation in the brain (47). Like other auxiliary factors, NOVA can inhibit or activate the usage of poly(A) signals depending on its position in relation to the poly(A) signals; near the poly(A) signal, NOVA seems to inhibit polyadenylation, but when located further away from poly(A) signals it seems to perform a role more in concert with its splicing-promoting nature (47). Moreover, in 9 of 12 genes analyzed containing alternative poly(A) signals in their 3′UTRs, NOVA induced production of mRNAs with longer 3′UTRs by distal poly(A) signal choice (47). As these are areas of active investigation, more interconnections and cross-regulations will likely be revealed in the future.
The impact of genome-wide methodologies on deciphering alternative polyadenylation
The explosion of methodologies that can examine the entire transcriptome of an organism or cell has changed much about how we view and interpret data with regard to alternatively polyadenylated transcripts. Researchers now can use deep sequencing, microarrays, serial analysis of gene expression (SAGE) data, Ref-Seq, RNA-Seq, and chromatin immunoprecipitation (ChIP)-Seq, all combined with ever-evolving powerful bioinformatic algorithms, in order to explore all the transcripts produced in a particular cell, even those that are relatively rare. These global methodologies will undoubtedly continue to evolve and change as they reveal a level of detail that is unprecedented.
With a view to produce a systematic and detailed analysis of alternative polyadenylation-mediated regulation during development, Ji et al. (27) combined SAGE data generated for all stages of mouse developments, with mouse EST and microarray data, to show that before the 8th embryonic day (E8.0) 3′UTRs lengthen rapidly. From this stage and until birth this lengthening continues, although at a slower rate. Finally, after birth and during postnatal development no global changes in 3′UTR can be identified (27). The combination of SAGE and microarray data further allowed the authors to determine that 3′UTR lengthening is associated with an up-regulation in genes involved in morphogenesis and differentiation, and a down-regulation of genes involved in cell proliferation. To experimentally validate their in silico data, the authors used C2C12, a myoblast cell line that can be differentiated into myotubes, and reporter genes, to compare the efficiency of polyadenylation signals in conditions of cell growth and cell differentiation. In myotubes, the efficiency of certain polyadenylation signals was weaker, implying that the effectiveness of the mechanism is diminished, most probably due to low concentrations of some basal cleavage and/or polyadenylation factors. Not surprisingly, the levels of expression of genes coding for proteins of the pre-mRNA 3′ end processing machinery were, in general, lower than other genes during C2C12 differentiation, in particular those coding for the polypeptide components of the cleavage stimulation factor (CstF;27). Notably, this result is in agreement with very early studies on alternative polyadenylation performed more than a decade ago. In what is now considered a hallmark in the study of the significance of alternative polyadenylation in biological processes, Manley and colleagues showed that plasma B cells have lower levels of CstF-64 than that found in pre-B cells, thus affecting the utilization of alternative poly(A) signals in the immunoglobulin genes (48).
While new methods of genome wide transcript measurement such as deep sequencing (ChIP-Seq and RNA-Seq) are becoming widely available, microarray expression analyses are still informative. However, when analyzing microarray expression data, consideration needs to be taken that differences in expression of longer versus shorter 3′UTRs may not be due solely to differences in alternative polyadenylation, but also to differences in transcript stability. The method of cDNA priming to generate the microarrays is of the utmost importance, as those that use random primers will not discriminate between transcripts containing or not the poly(A) tails. Recently, the new rmodel algorithm was developed to analyze microarrays from the transcriptionally silent period of mouse oocyte development (49). Of notice, this new method enables the researchers to distinguish between transcripts that are alternatively polyadenylated from those that have different stabilities using microarray data. Hence, Graber and collaborators have found three classes of transcript changes in oocytes between the germinal vesicle (GV) and metaphase II (MII) transition: complete degradation, deadenylation and cleavage producing stable 5′ fragments (49). Thus alternative polyadenylation signals may contribute to different needs in the egg.
Examination of the transcriptome in a variety of human and mouse tissues using RNA-Seq methods and comparing commonly expressed genes active in every tissue revealed that there are an abundance of such common or “core” genes, making up the majority of the transcripts produced (50). This work also revealed important insights into 3′ end processing of mRNAs that were differentially expressed: mRNAs that were especially expressed in brain had unusually long 3′ UTRs and longer 3′ UTRs were correlated with expression of genes involved in development, morphogenesis and signal transduction. These correlations speak to added complexity of 3′ end formation coupled with regulation of other cellular events.
Alternative polyadenylation in Drosophila
Studies of alternative polyadenylation in Drosophila melanogaster have enjoyed a long history of revealing interesting findings. The Drosophila melanogaster gene polo codes for a highly conserved serine/threonine protein kinase that has a key role in several functions during cell division. Transcription of polo generates two mRNA isoforms that differ in their 3′UTRs due to the alternative usage of two poly (A) signals (51). Interestingly, utilization of the proximal poly(A) signal is dependent on RNA elements localized upstream of the proximal poly(A) signal. These elements are necessary for the assembly of a protein complex containing PTB (polypyrimidine tract binding protein), CstF-64, and hnRNPC (52). Thus, this work suggests that cis-acting regulatory elements and trans-acting protein factors involved in human polyadenylation are also conserved in Drosophila.
The homologue of the human CstF-77 in Drosophila is called suppressor of forked (su(f)) (53-54), a gene that generates three mRNA isoforms by alternative polyadenylation (53). While the two longer transcripts are produced by the use of two poly(A) signals present in the 3′UTR, utilization of a poly(A) signal present in intron 4 results in the synthesis of a truncated transcript that is likely to be degraded by cellular surveillance (55). As the synthesis of this transcript is dependent on Su(f) activity (56) it was suggested that this protein, as part of the Drosophila CstF complex, may regulate its own levels in non-proliferating tissues by favoring the use of the intronic polyadenylation site (57). Interestingly, although there is no extensive sequence similarity between su(f) and hCstF-77, there is a strong conservation of the autoregulatory mechanism originated by alternative polyadenylation that operates in their expression. The human su(f) homologue CstF-77 transcription unit also produces three mRNAs due to alternative polyadenylation; two of these transcripts differ only in the 3′UTR as a result of two poly(A) signals and the smaller mRNA is synthesized by usage of a poly(A) site present in intron 3 (58-59). Levels of the smaller transcript are 5-fold higher in mouse B cells compared to the mitotically active plasma cells (59), which is in agreement with the observations of accumulation of Su(f) in mitotic tissues in Drosophila melanogaster (56). Finally, the S. cerevisiae homologue of CstF-77 (Rna 14) also has preserved the upstream alternative polyadenylation in its mRNA, but not the genomic exon-intron structure (ref).
The Drosophila gene enhancer of rudimentary, e(r), is an example of sex-specific alternative polyadenylation. It uses two poly(A) signals in the 3′UTR: a proximal non-canonical poly(A) signal, UAUAAA, and the distal canonical poly(A) signal AAUAAA (60). Interestingly, the use of each poly(A) signal is sex-specific (60-61). While the smaller mRNA, resulting from the use of the proximal polyadenylation signal, is expressed in both male and female adults, the longer mRNA is specifically expressed in adult females (e(r)-fs). Analysis of e(r)-fs mRNA shows that its expression is controlled by three GU-rich elements present downstream the proximal poly(A) signal and RNA-binding assays showed that the female-specific sex-lethal protein (Sxl) competes with CstF to bind to these elements (61). As a result, in the female germline the binding of CstF is blocked, preventing proximal poly(A) signal utilization and allowing the use of the distal poly(A) signal and synthesis of the e(r)-fs mRNA. As males do not express Sxl, CstF can bind to the GU-rich elements and therefore the proximal poly(A) signal is used, and the smaller non-sex specific e(r) mRNA synthesized (61). Analysis of the expression of the e(r)-fs mRNA in isolated ovaries shows that this transcript is less efficiently translated, suggesting that the 3′UTR present in this transcript is important to repress translation of the e(r)-fs mRNA in the female germline (61). Therefore, alternative polyadenylation of the e(r) represents a mechanism through which regulation of translation can be achieved in a sex-specific manner.
Alternative polyadenylation and effects of chromatin architecture and imprinting
Epigenetic modifications of DNA structure have recently been implicated as having multiple impacts on RNA transcription and processing. Wood and co-workers (62) identified that alternative polyadenylation at three of five possible alternative sites in the imprinted mouse gene H13 are influenced by methylation, primarily at CpG islands not close to the polyadenylation signals (62). This is thought to occur because of three possible mechanisms: A) methylation-sensitive formation of transcription initiation complexes, B) methylation prevents binding of polyadenylation factors and brings about promoter occlusion, or C) both A and B. This novel mechanism of imprinted gene regulation is still not well understood but is under investigation.
Nucleosome composition has also been implicated as a factor in alternative polyadenylation. It was found that in genes with multiple alternative polyadenylation signals, a higher downstream nucleosome affinity was associated with higher polyadenylation signal usage, independently of known motifs that function at the RNA level (63). In addition, the Weissman lab used genomic tiling arrays to examine a fraction of the human genome representing mRNA 3′ ends, and, in addition to discovering that more 3′ mRNA ends exist than have previously been annotated, that there is a correlation between histone modifications at or near the sites of polyadenylation (64). They discovered that lysine 36 methylation is substantially decreased at or near polyadenylation signals. The involvement of chromatin structure in this regulatory framework adds yet another level of complexity to the regulation of gene expression.
Conclusions
Several of the most pertinent questions still to be fully answered are how the cell chooses one polyadenylation signal instead of another and how is this selection regulated? The molecular mechanisms that lead to the usage of a proximal poly(A) signal in detriment of a more distal one are still poorly understood. However, it has been known for more than a decade that the efficiency of polyadenylation can be regulated by either cis or trans acting factors. Variations in the tight regulation of the concentration of these protein factors in the cell may define major changes in the cellular behavior. Alterations of the levels of the basal factor CstF leads to the preferential recognition of weaker polyadenylation signals in B cells (26, 48, 65-66). Interestingly, several cleavage and polyadenylation factors, including CPSF1 and CstF2 are up-regulated in cancer cells (31). Moreover, during spermatogenesis, several cleavage and polyadenylation factors vary during the different developmental stages (30, 67). It remains to be addressed whether these alterations are sufficient to regulate the alternative polyadenylation switch observed in cancer cells and during development.
Not only do changes in the expression of basal cleavage and polyadenylation factors play a role in polyadenylation site switching decisions but auxiliary proteins may be involved. Indeed, the levels of auxiliary protein factors have been shown to be crucial determinants in polyadenylation efficiency and in establishing the pattern of alternative polyadenylation, as in the case of MeCP2 (46), COX-2 (20-21) and pro-thrombin genes (44).
The roles of different alternative splicing-generated isoforms of cleavage/polyadenylation factors, as well the significance of post-translational modifications of these factors, are still largely unexplored. It was recently shown that alternative splicing generates multiple different isoforms of the basal factor CstF-64, that are expressed specifically in the nervous system, suggesting a specific role in this tissue (68). Also, several alternative polyadenylation variants of HuR mRNAs with different stabilities were recently described, and moreover, it was shown that HuR could regulate its own expression (69). Interestingly, HuR is involved in the stabilization of ARE-containing mRNAs (70) and also in polyadenylation (71). Thus, modulation of HuR alternative polyadenylation may potentially control the stabilization and/or polyadenylation efficiency of several RNAs.
Therefore, we conclude that alternative polyadenylation is a particularly complex mechanism: it involves A) basal factors necessary for the process of cleavage and polyadenylation of all pre-mRNAs, B) auxiliary cis and trans-acting factors, that working in cooperation with factors from the transcription and splicing machinery, modulate the usage of some but not all polyadenylation signals, C) gene-, or development-, or disease-specific factors, and also D) chromatin modifications that appear to modulate alternative polyadenylation by altering the kinetics of the RNA polymerase II or accessibility to the polyadenylation signals, for example (see Figures 2 and 3). The combinatorial expression of all these in a particular cell at a particular moment will likely determine the output of alternative polyadenylation, and provide a rich ground for many possible levels of gene expression regulation.
Acknowledgements
The authors wish to thank members of their laboratories for critical reading of the manuscript. The authors also wish to thank the following agencies for funding: NIH 5R21HG005129-02 and 5R03HD054559-02 to CSL, the European Regional Development Fund (FEDER) and the Programa Operacional Ciência, Tecnologia e Inovação 2010 (POCI and POCTI 2010) from the Fundação para a Ciência e Tecnologia (FCT) to AM.
References
- 1.Wickens M, Anderson P, Jackson RJ. Life and death in the cytoplasm: messages from the 3′ end. Curr Op Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson A, Peltz SW. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Ann Rev Biochem. 1996;65:693. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Reviews Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edmonds M. A history of poly(A) sequences: from formation to factors to function. Prog Nucl Acids Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 7.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucl Acids Res. 2010;38:2757. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 9.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 10.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian B, Hu H, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucl Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreira A, Wollerton M, Monks J, Proudfoot NJ. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz CS, Alwine JC. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucl Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end and pre-mRNA processing through a trans-acting factor. Nucl Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′ untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 19.Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2: Nucl Acids Res. 2005;33:2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′ UTR. RNA. 2007;13:1103. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing D, Li QQ. Alternative polyadenylation. Plant Sig Behav. 2009;4:440–442. doi: 10.4161/psb.4.5.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz CS. Alternative polyadenylation: a twist on 3′ end formation. ACS Chemical Biology. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- 27.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–33. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legendre M, Ritchie W, Lopez F, Gautheret D. Differential repression of alternative transcripts: a screen for miRNA targets. PLos Computational Biology. 2006;2:0333–0342. doi: 10.1371/journal.pcbi.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucl Acids Res. 2007;35:234. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coleman MS, Hutton JJ, Bollum FJ. Terminal riboadenylate transferase in human lymphocytes. Nature. 1974;248:407–409. doi: 10.1038/248407a0. [DOI] [PubMed] [Google Scholar]
- 36.Hauser H, Knippers R, Schafer KP. Increased rate for RNA-polyadenylation. An early response in Concanavalin A activated lymphocytes. Exp Cell Res. 1978;111:175–184. doi: 10.1016/0014-4827(78)90247-1. [DOI] [PubMed] [Google Scholar]
- 37.Edwalds-Gilbert G, Veraldi K, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucl Acids Res. 1998;25:2547. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuvpilo S, Zimmer M, Kerstan A, Glöckner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, Schmitt E, Serfling E. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- 39.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:5818. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, Chan WC, Greiner TC, Weisenburger DD, Vose J, Armitage JO, Gascoyne RD, Connors JM, Campo E, Montserrat E, Bosch F, Smeland EB, Kvaloy S, Holte H, Delabie J, Fisher RI, Grogan TM, Miller TP, Wilson WH, Jaffe ES, Staudt LM. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial Control of Signal-Induced Exon Repression by hnRNP L and PSF. Mol Cell Biol. 2007;19:6972. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newnham CM, Hall-Pogar T, Liang S, Tian B, Hu J, Lutz CS. Alternative polyadenylation of MeCP2: influence of cis-acting elements and transacting factors. RNA Biology. 2010 doi: 10.4161/rna.7.3.11564. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 49.Salisbury J, Hutchison KW, Wigglesworth K, Eppig JJ, Graber JH. Probe-level analysis of expression microarrays characterizes isoform-specific degradation during mouse oocyte maturation. PLoS One. 2009;4:e7479. doi: 10.1371/journal.pone.0007479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. Plos Computational Biology. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–6. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 52.Moreira A, Pinto P, Sunkel C, Carmo A. Upstream sequence elements are required for mRNA 3′-end formation of the Drosophila melanogaster cell cycle POLO kinase. FASEB J. 2005;19:A87. [Google Scholar]
- 53.Mitchelson A, Simonelig M, Williams C, O’Hare K. Homology with Saccharomyces cerevisiae RNA14 suggests that phenotypic suppression in Drosophila melanogaster by suppressor of forked occurs at the level of RNA stability. Genes Dev. 1993;7:241. doi: 10.1101/gad.7.2.241. [DOI] [PubMed] [Google Scholar]
- 54.Takagaki Y, Manley JL. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 55.Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 56.Audibert A, Simonelig M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc Natl Acad Sci USA. 1998;95:14302. doi: 10.1073/pnas.95.24.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juge F, Audibert A, Benoit B, Simonelig M. Tissue-specific autoregulation of Drosophila suppressor of forked by alternative poly(A) site utilization leads to accumulation of the suppressor of forked protein in mitotically active cells. RNA. 2000;6:1529. doi: 10.1017/s1355838200001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan Z, Zhang H, Hague LK, Lee JY, Lutz CS, Tian B. An intronic polyadenylation site in human and mouse CstF-77 genes suggests an evolutionarily conserved regulatory mechanism. Gene. 2006;366:325. doi: 10.1016/j.gene.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 60.Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI. Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics. 1994;138:1163. doi: 10.1093/genetics/138.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gawande B, Robida MD, Rahn A, Singh R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006;25:1263. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc’his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008;18:1224. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinicic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze M. Increase in 64 Kilodalton subunit of the cleavage and polyadenylation stimulatory factor during G0 to S phase transition. Proc Natl Acad Sci USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace AM, Denison TL, Attaya EN, MacDonald CC. Developmental distribution of the polyadenylation protein CstF-64 and the variant tauCstF-64 in mouse and rat testis. Biol Reprod. 2004;70:1080. doi: 10.1095/biolreprod.103.022947. [DOI] [PubMed] [Google Scholar]
- 68.Shankarling GS, Coates PW, Dass B, MacDonald CC. A family of splice variants of CstF-64 expressed in vertebrate nervous systems. BMC Mol Biol. 2009;10:22. doi: 10.1186/1471-2199-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-regulation. Nucl Acids Res. 2009;37:3612. doi: 10.1093/nar/gkp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;15:188. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu H, Zhou HL, Hasman RA, Lou H. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem. 2007;282:2203. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 72.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat Med. 2000;6:637. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]


