Abstract
Intron removal from tRNA precursors involves cleavage by a tRNA splicing endonuclease to yield tRNA 3′-halves beginning with a 5′-hydroxyl, and 5′-halves ending in a 2′,3′-cyclic phosphate. A tRNA ligase then incorporates this phosphate into the internucleotide bond that joins the two halves. Although this 3′-P RNA splicing ligase activity was detected almost three decades ago in extracts from animal and later archaeal cells, the protein responsible was not yet identified. Here we report the purification of this ligase from Methanopyrus kandleri cells, and its assignment to the still uncharacterized RtcB protein family. Studies with recombinant Pyrobaculum aerophilum RtcB showed that the enzyme is able to join spliced tRNA halves to mature-sized tRNAs where the joining phosphodiester linkage contains the phosphate originally present in the 2′,3′-cyclic phosphate. The data confirm RtcB as the archaeal RNA 3′-P ligase. Structural genomics efforts previously yielded a crystal structure of the Pyrococcus horikoshii RtcB protein containing a new protein fold and a conserved putative Zn2+ binding cleft. This structure guided our mutational analysis of the P. aerophilum enzyme. Mutations of highly conserved residues in the cleft (C100A, H205A, H236A) rendered the enzyme inactive suggesting these residues to be part of the active site of the P. aerophilum ligase. There is no significant sequence similarity between the active sites of P. aerophilum ligase and that of T4 RNA ligase, nor ligases from plants and fungi. RtcB sequence conservation in archaea and in eukaryotes implicates eukaryotic RtcB as the long-sought animal 3′-P RNA ligase.
Keywords: ligation, tRNA biosynthesis, RNA processing
Eukaryotes and archaea contain a number of intron-containing tRNA genes. After their transcription the intron is cleaved from the precursor tRNA by the splicing endonuclease. The resulting tRNA halves are then joined by a tRNA ligase to form mature-sized tRNA (1). The cleavage by the tRNA splicing endonuclease leaves the 5′ exon with a 2′,3′-cyclic phosphate terminus and the 3′ exon with a 5′-hydroxyl group (2). The well-known multifunctional yeast tRNA ligase, a class I 5′-P RNA ligase (RNL) (3), is unable to directly join these ends together (4). Instead, the class I 5′-P RNL uses its 2′,3′-cyclic-3′-phophodiesterase and 5′-RNA polynucleotide kinase activities to yield a 2′-phosphate-3′-hydroxyl and a 5′-phosphate, which the yeast enzyme then joins via formation of a 2′-phosphate-3′,5′-phosphodiester bond in an ATP-dependent reaction. In plants, mature tRNAs are formed in a similar manner by the class II 5′-P RNL (5, 6).
Although the 5′-P ligation pathway is known to exist in animals, almost 30 years ago a different RNA ligase activity was discovered in HeLa cell extracts (7). This 3′-P RNL ligates tRNA 3′-halves beginning with a 5′-hydroxyl, and 5′-halves ending in a 2′,3′-cyclic phosphate by incorporating this phosphate into the internucleotide bond that joins the two halves. Attempts to purify this activity to homogeneity were unsuccessful (8). A similar 3′-P RNL activity was later detected in extracts from the archaea Desulfurococcus mobilis and Haloferax volcanii (9–11).
Here we describe the purification of the 3′-P RNL activity from extracts of Methanopyrus kandleri and identify the enzyme as archaeal RtcB. Recombinant RtcB from Pyrobaculum aerophilum is dependent on Zn2+ for activity, and mutations of conserved residues in a putative Zn2+ binding cleft (12) yielded inactive ligase enzymes. Phylogenetic analysis shows RtcB is present in all three domains of life.
Results
Identification of the Archaeal tRNA Splicing Ligase.
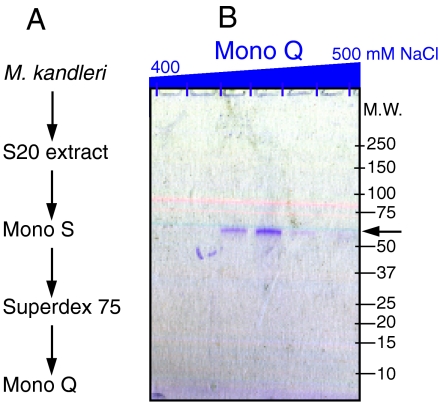
To identify the 3′-P RNL, the activity was purified from M. kandleri extracts in three chromatographic steps (see Materials and Methods). SDS-PAGE analysis of the final active fractions by both silver staining and Coomassie colloidal blue staining revealed only a single protein of about 55 kDa (Fig. 1). This protein (300 ng) was excised from the gel for tryptic digestion and identified by liquid chromatography coupled to tandem mass-spectrometry analysis (13). The mass of 30 tryptic fragments identified the protein as MK1682, which is annotated as an 988-aa long intein-containing precursor of a protein of the RtcB family. Coverage of the RtcB extein (506 aa long, 56-kDa predicted molecular weight) was 51% (Fig. S1). The M. kandleri RtcB appears to be a monomer in solution as revealed by gel filtration (Fig. S2).
Fig. 1.
Isolation of M. kandleri RNA splicing ligase. (A) Purification scheme. RNA ligase activity was purified from soluble protein extract (S20) by three consecutive steps. (B) Purified fractions from the Mono Q column. Proteins bound to the Mono Q HR5/5 column were eluted in a 15-mL linear gradient from 0–500 mM NaCl and collected in 20 fractions. Aliquots of the Mono Q elution fractions in the range from 400–500 mM NaCl were analyzed on a 4–20% gradient polyacrylamide/0.1% SDS gel and stained with Coomassie colloidal blue (Pierce Imperial Stain). The arrow indicates the putative RNA ligase protein.
To verify RtcB as the archaeal 3′-P RNL, we attempted to prepare active recombinant RtcB from a number of archaea; we settled on the P. aerophilum RtcB protein because it had the best activity. In order to facilitate folding of the archaeal protein during overexpression in Escherichia coli and subsequent purification, we cloned the P. aerophilum rtcB ORF as a maltose binding protein (MBP) fusion into the pBAD Myc-His A vector. The fusion protein eluted from the amylose resin with 10 mM maltose and was then directly tested for tRNA ligase activity (Fig. 2).
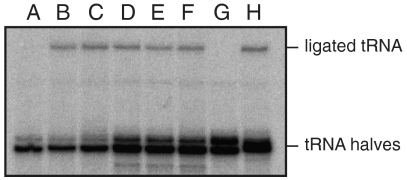
Fig. 2.
RNA ligase-activity of the recombinant Pyrobaculum RtcB protein. Suitable tRNA splicing intermediates (see Materials and Methods) were incubated with the Pyrobaculum RtcB—MBP fusion protein. Lane A: no ATP or GTP added. Lane B: no ATP or GTP but with a heavy metal mix (39). Lanes C–F: the enzyme preparation was preincubated in 0.5 mM ZnCl2 for 15 min on ice before the enzyme was added to the RNA ligation mixture. Lane C: no ATP or GTP added; lane D: with 0.5 mM ATP; lane E: with 0.5 mM GTP; lane F: with 0.5 mM ATP and 0.5 mM GTP. Lane G: no MBP-RtcB enzyme added. Lane H: the positive control—addition of T4 polynucleotide kinase with 3′-phosphatase (PNKp) and T4 RNA ligase 1. Lanes D–H include NTPs that serve as coprecipitant during the ethanol precipitation of the phenol/chloroform extracted RNA ligation mixtures. Hence, more ribonucleic acids are precipitated as indicated by the presence of more tRNA halves in lanes D–H in comparison to lanes A–C.
Suitable tRNA halves bearing a 2′,3′-cyclic phosphate and a 5′-hydroxyl were generated by cleavage of the intron-containing pre-tRNA with the Methanocaldococcus jannaschii tRNA splicing endonuclease (for details, see Materials and Methods). RNA ligase activity is indicated by the formation of ligated tRNA that migrates slower in a 12.5% polyacrylamide/8 M urea gel—as shown by a control ligation reaction with the combined action of the T4 polynucleotide kinase/3′-phosphatase and T4 RNA ligase 1 (Fig. 2, lane H). Incubation of the tRNA halves with recombinant RtcB without any heavy metal mix and without ATP or GTP did not stimulate the formation of ligated tRNA (lane A). However, upon addition of a mixture of heavy metal ions, RtcB was able to ligate the tRNA halves and circularize the linear intron (lane B), suggesting a requirement for a heavy metal ion. We immediately tried Zn2+ (lane C–F), as a Zn2+-binding site was suggested (12) in the Pyrococcus horikoshii RtcB crystal structure (14). Addition of Zn2+ successfully substituted for the heavy metal mix (lane C). Hence, the recombinant RtcB preparations were incubated on ice with 0.5 mM ZnCl2 for 15 min before enzyme addition to the RNA ligation mixture. Addition of ATP (lane D), GTP (lane E), or ATP and GTP (F) did not further stimulate the overall ligation rate observed in lanes B and C.
These data show that Zn2+, but not ATP or GTP, is required for activity of the P. aerophilum RtcB tRNA ligase.
RtcB-Catalyzed Ligation Incorporates the Phosphate of the Cyclic Phosphate into the Phosphodiester Bond.
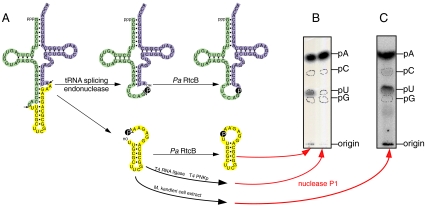
Archaea possess two other RNA ligases besides the 3′-P RtcB RNL: (i) a GTP-dependent 2′,5′-RNA ligase that incorporates the cyclic phosphate into a 2′,5′-phosphodiester bond (15) and (ii) a homolog of T4 RNA ligase that joins a 3′-hydroxyl to a 5′-phosphate in a ATP-dependent manner (16). What differentiates the archaeal tRNA splicing ligase from the other two ligase activities is the incorporation of the 2′,3′-cyclic phosphate into the newly formed 3′,5′-phosphodiester bond. We assayed this type of incorporation by a nearest neighbor analysis (11). Therefore we synthesized by an A-labeled pre-tRNA by adding [α-32P]ATP during transcription and then cleaved the pre-RNA with the M. jannaschii tRNA splicing endonuclease leading to tRNA halves and a linear intron that contains a [32P]labeled 2′,3′-cyclic phosphate terminus (Fig. 3). RNA labeled by the incorporation of [α-32P]ATP during transcription yields upon nuclease P1 hydrolysis [32P]AMP. Based on the mechanism of the three different archaeal tRNA ligases, there are three different endpoints for the labeled 2′,3′-cyclic phosphate after nuclease P1 degradation of the circular intron. (i) The 2′,5′-RNA ligation pathway results in a 2′,5′-phosphodiester bond that is not cleavable by nuclease P1 and a dinucleotide would be visible. (ii) The T4-like RNA ligation pathway removes the cyclic phosphate, and the junction phosphate is provided by the γ-phosphate of ATP. Hence, there would no other label besides [32P]AMP in the nuclease P1 hydrolysate. In the case of the 3′-P RNA ligation pathway, the labeled cyclic phosphate is incorporated into a new 3′,5′-phosphodiester bond, i.e., …p*Ap*Up… for the circular intron. Nuclease P1 hydrolysis of the circular intron would result in [32P]UMP besides [32P]AMP in a ratio of 1∶4.
Fig. 3.
Analysis of the tRNA ligation products. (A) tRNA halves from [α-32P]ATP labeled pre-tRNA was used as substrates for the in vitro RNA ligation assay resulting in ligated tRNA and circular intron. (B) The circular intron formed by the action of the M. kandleri extract, P aerophilum RtcB, or T4 polynucleotide kinase + T4 RNA ligase was recovered from a 12% polyacrylamide/8 M urea gel by passive elution and used for nuclease P1 hydrolysis. The resulting nucleoside 5′-monophosphate mixture was separated by thin-layer chromatography on cellulose plates in solvent A. Position of markers are indicated.
For the actual experiment the tRNA splicing intermediates—the tRNA halves and the linear intron—were incubated with recombinant P. aerophilum RtcB, with M. kandleri S20 cell extract, or—as a control reaction—with T4 polynucleotide kinase/3′-phosphatase and T4 RNA ligase 1. The formed circular introns were cut out of a preparative 12.5% denaturating polyacrylamide gel, recovered by passive elution, and used for a nuclease P1 digest. The P1 hydrolysate was separated by thin-layer chromatography on cellulose plates in solvent A (iso-butyric acid/conc. ammonia/H2O = 57.7/3.8/38.5 [vol/vol/vol]). The labeled [32P]NMPs were visualized by PhosphorImager, and their migration pattern was compared to the unlabeled marker NMPs mixture. For the lane of the M. kandleri extract, as well as for recombinant P. aerophilum RtcB a spot for labeled p*U besides p*A in a ratio 1∶4 is visible—as determined by PhosphorImager integration of the intensity of the spots. This shows that the P. aerophilum RtcB tRNA ligase is the real archaeal 3′-P RNL that directly incorporates the 2′,3′ cyclic phosphate into the newly formed 3′,5′-phosphodiester bond. In agreement with earlier results (11), we confirm that the 3′-P RNA ligation pathway is dominant in archaeal extracts.
Structure of RtcB Guides Mutational Analysis.
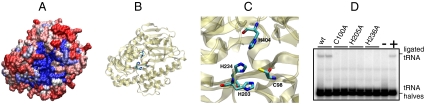
A search of the protein structure database revealed that the structure of the P. horikoshii RtcB ortholog was solved as part of a structural genomics project (14). Previous analysis of the sequence (17) and structure (12, 14) of RtcB suggested a conserved hydrophilic pocket on the surface of the protein (Fig. 4A) might harbor a metal ion binding site, which was predicted to consist of residues Cys98, His203, His234, His404 (12) (Fig. 4 B and C). Because the presence of Cys and His residues in such a combination suggested a Zn2+ binding site (12), we tested if Zn2+ was the metal required for activity. Addition of Zn2+ at physiological concentrations (7 μM) facilitated RtcB-catalyzed RNA ligation in vitro, and 0.5 mM ZnCl2 was added to all subsequent enzyme preparations.
Fig. 4.
Crystal structure of P. horikoshii RtcB. (A) Surface model of the P. horikoshii RtcB crystal structure (taken from ref. 14). Sequence alignment of more than 50 archaeal and 40 eukaryotic RtcB proteins was used for plotting the sequence conservation from high (blue) to low (red) in the surface model. (B) The proposed active side residues forming a Zn2+ binding motif (12) are highlighted in the cartoon representation. (C) Closeup display of the putative Zn2+ coordinating cleft with the putative Zn2+-ligand amino acids (C98, H203, H234, H404). (D) tRNA ligase-activity assay of P. aerophilum wild-type and mutant (C100A, H205A, H236A; these residues correspond to C98, H203, H234 in the P. horikoshii sequence) MBP-RtcB enzymes. The tRNA splicing intermediates were incubated with the indicated P. aerophilum RtcB enzymes, without RtcB (lane −) or with the combined action of T4 polynucleotide kinase/3′-phosphatase and T4 RNA ligase 1 (lane +). The reaction products were analyzed by denaturating PAGE and subsequent PhosphorImager visualization.
The requirement of Zn2+ suggested the residues in the conserved cleft were essential for activity. In order to confirm the location and nature of the putative active site, we constructed three P. aerophilum RtcB mutants (Cys100Ala, His205Ala, and His236Ala). The resulting mutant enzymes were all RNA ligase inactive in vitro (Fig. 4D), and the data are consistent with the importance of Zn2+ coordination for the RNA ligase activity of RtcB.
RtcB Is Conserved in All Three Domains of Life.
RtcB is highly conserved in a wide range of organisms from all three domains of life. It is found in all known archaea as well as in all eukaryotes except fungi and vascular plants. Fungi encode the class I 5′-P RNL, whereas vascular plants have the class II 5′-P RNL (3). The conservation of RtcB in vertebrates implicates their RtcB ortholog as the yet unidentified 3′-P RNL.
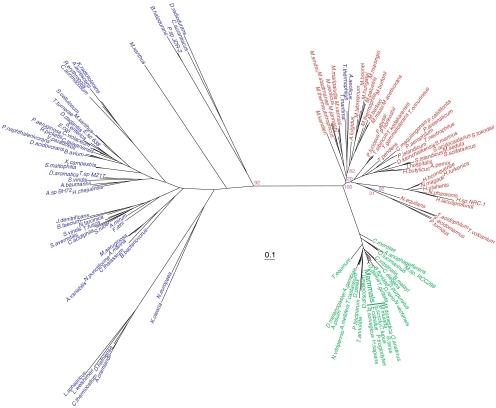
Given the unknown role of RtcB in bacteria and eukaryotes, we applied phylogenetic analysis to determine the extent to which horizontal gene transfer from archaea to bacteria and eukaryotes might have shaped the evolution of RtcB (Fig. 5). Interestingly, the phylogeny revealed three distinct types of RtcBs falling into the taxonomic domain level divisions with the archaea and eukaryotic enzymes showing more similarity to each other than to their phylogenetically well-separated bacterial counterparts. Because RtcB lacks paralogs, we were unable to root the phylogeny, but the tree indicates that RtcB emerged at the time of the last universal common ancestor and that gene has evolved mostly by vertical inheritance with significant gene loss in bacteria and to a lesser extent in eukaryotes. There are some incongruities in the RtcB phylogeny presented and established taxonomy within the domains such as the grouping of the Halobacterium, Thermococci, and Thermoplasmata with the Crenarchaea and not the Euryarchaea. The high degree of sequence conservation of archaeal RtcBs may obscure the phylogenetic signal leading to such discrepancies.
Fig. 5.
Phylogeny of RtcB. Unrooted phylogenetic tree of RtcB sequences from archaeal (red), bacterial (purple), and eukaryotic (green) sources. Scale bar represents 0.1 changes/site. For clarity, only bootstrap values for the branch points discussed in the text are shown.
The major exception to the three-domain phylogeny is the grouping of the Thermus thermophilus, Aquifex aeolicus, and Thermotoga maritima RtcB orthologs with the methanogenic archaea, suggesting these bacteria may have acquired RtcB via horizontal gene transfer from an ancestral archaeon.
Discussion
Almost 30 years ago, a 3′-P RNL activity was detected in HeLa cells (7); however, the enzyme responsible for this activity has remained a mystery. In the present study, we purified the 3′-P RNL activity from M. kandleri cell extract and identified the protein as RtcB (encoded by MK1682) using mass spectroscopy. Recombinant P. aerophilum RtcB showed Zn2+ dependent 3′-P RNL activity, and mutagenesis implicated Cys100, His205, and His236 as critical active site constituents.
RtcB is not only encoded in all known archaea but also in most eukaryotes and a variety of bacteria (Fig. 5). In bacteria, group I introns in pre-tRNAs self-splice and ligate the exon halves together to form the mature tRNA molecules required for translation (18). The presence of RtcB, therefore, in a significant number of bacteria is unexpected. As noted above, three bacterial representatives (T. thermophilus, A. aeolicus, and T. maritima) actually encode the archaeal type RtcB, and we would speculate that the RtcB in these bacteria should function similarly to the archaeal RtcB presented here, whereas genuine bacterial RtcBs might function on a different substrate.
The E. coli RtcB is reported as ligase inactive in vitro (17) but this could be due to the lack of Zn2+ in the reaction mixture, which we have found to be essential for archaeal RtcB 3′-P RNL activity (Fig. 2). In the E. coli genome, rtcB is in an operon with two other genes, rtcA and rtcR (17). RtcR is a σ54-dependent regulator and regulates the expression of rtcA and rtcB (17). RtcA is found in bacteria, archaea, and eukaryotes including humans (17, 19–21). It is an RNA 3′-terminal phosphate cyclase that converts a 3′-phosphate at the ends of RNA molecules to a 2′,3′-cyclic phosphodiester (19), which is a substrate of RtcB. Under laboratory conditions, both rtcA (17) and rtcB (22) are dispensable in E. coli. RtcA and RtcB may serve together in a repair capacity or some alternative nonessential function in E. coli and other bacteria.
Phylogenetic analysis suggests the last common ancestor of archaea and eukaryotes possessed an RtcB that functioned similarly to archaeal RtcB characterized here (Fig. 5), and the conservation of RtcB implicates it as the eukaryotic 3′-P RNL. Intriguingly, RtcB is absent in Saccharomyces cerevisiae. Yeast lack a 3′-P RNL activity, using the class I 5′-P RNL exclusively to ligate tRNA halves (23). RtcB is also absent in vascular plants that use the class II 5′-P RNL (5, 24). Reliance on the 5′-P ligation pathway in fungi and plants may have made RtcB dispensable and subject to gene loss in these lineages. For unknown reasons, animals have retained both ligation pathways (7, 25).
The human 3′-P RNL has been associated with a 160-kDa molecular weight complex (8). The human homolog of RtcB (HSPC117) forms a complex (210 kDa) with DDX1 (a putative DEAD box RNA helicase), FAM98B, and CGI-99 (26, 27); these data are consistent with RtcB serving as the 3′-P RNL in humans. Human RtcB is also posttranslationally modified (phosphorylation of Ser439 and acetylation of Lys496) (28, 29). To be active, human RtcB may need to be both modified and part of a complex. These two factors may be the reason it has been challenging to isolate a 3′-P-RNL from HeLa cells (8). As is the case for many homologous enzymatic pathways found in both archaea and eukaryotes [e.g., RNaseP (30)], the 3′-P ligation pathway in archaea may represent a more simplified version of the one found in eukaryotes.
This archaeal 3′-P RNL may be a more general RNA splicing ligase, that—in addition to tRNA—splices ribosomal RNA introns (10, 31, 32) and even mRNA (for the Cbf5 protein) (33, 34). Our analysis of the archaeal RtcB enzyme will undoubtedly shed light on the role of RtcB orthologs in bacteria and also finally lead to the characterization of RtcB as the elusive 3′P-RNL in humans and other eukaryotes.
Materials and Methods
General.
Oligonucleotide synthesis and DNA sequencing were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. [α32P]ATP (3,000 Ci/mmol) was obtained from PerkinElmer.
Plasmids.
The M. jannaschii RNA splicing endonuclease (Mjan endA; MJ1424) was cloned into pET28a with an N-terminal His6-tag. The coding region of the maltose binding protein together with the multiple cloning site of the vector pMAL c2x was cloned into the NcoI and HinDIII site of pBAD Myc-His A resulting in MBP-pBAD Myc-His A. The archaeal RNA ligase candidate RtcB from P. aerophilum (PAE0998) was amplified from genomic DNA and cloned into the EcoRI and SalI site of MBP-pBAD Myc-His A.
Protein Expression and Purification.
Mjan endA was transformed in E. coli BL21(DE3). Cells were grown at 37 °C to an A600 of 0.8, induced with 0.5 mM IPTG, further incubated at 30 °C for 3 h, and then spun down and resuspended in Ni-NTA buffer (20 mM Tris-HCl, pH 8, 500 mM NaCl, 3 mM MgCl2, 10 mM imidazole, 10 mM 2-mercaptoethanol). After sonification the cell debris and the incusion bodies were spun down and extracted with Ni-NTA buffer containing 4 M urea. The clarified extract of the inclusion bodies was loaded on Ni-NTA resin and washed with Ni-NTA buffer containing 2 M urea, Ni-NTA buffer with 40 mM imidazole before the Mjan end A protein was eluted with Ni-NTA buffer with 0.5 M imidazole.
P. aerophilum RtcB was transformed into E. coli Rosetta (DE3) pLysS (Novagen). Cells were grown to A600 of 0.8 at 37 °C, cooled on ice for 15 min, induced with 0.1% arabinose, incubated at 16 °C for 15 h, then spun down and resuspended in MBP buffer (20 mM Tris-HCl, pH 7.4, 0.2 M NaCl, 3 mM MgCl2, 10 mM 2-mercaptoethanol). After sonication the clarified cell lysate was loaded on amylose resin and washed with MBP buffer. Bound MBP-RtcB fusion protein was eluted with MBP buffer containing 10 mM maltose.
Purification of the Archaeal RNA Ligase from M. kandleri.
All operations were carried out at 4 °C. M. kandleri cells (14 g) were resuspended in 50 mL buffer A (20 mM Tris-HCl, pH 7, 3 mM MgCl2, 10 mM 2-mercaptoethanol, 0.2 mM PMSF) with Roche Complete protease inhibitor cocktail mix without EDTA and broken by sonication. An S20 extract was obtained after centrifugation at 20,000 × g for 20 min and then desalted by Sephadex G25 gelfiltration with buffer A.
The desalted S20 extract was loaded onto a Mono S HR 5/5 column (1 mL). Bound proteins were eluted in a salt gradient from 0–1 M NaCl in buffer A. Active fractions eluting at 450 mM NaCl were concentrated (to 150 μL) using Amicon Ultra Ultracel centrifugal filter device with a molecular weight cutoff of 10 kDa.
A Superdex 75 HR 10/30 column—equilibrated with buffer B (20 mM Tris-HCl, pH 8, 3 mM MgCl2, 500 mM NaCl, 10 mM 2-mercaptoethanol, 0.5 % ε-aminocaproic acid)—was loaded with the concentrated active Mono S elution fraction. Use of ϵ-aminocaproic acid in the chromatographic buffer (35) gives a sharp RtcB activity peak with a native molecular weight of 39 kDa. Active fractions were pooled and desalted against buffer C (20 mM Tris-HCl, pH 8.5, 3 mM MgCl2, 10 mM 2-mercaptoethanol, 0.5% ε-aminocaproic acid) by Sephadex G25 filtration.
The desalted active fractions from the Superdex 75 column were loaded on a Mono Q HR 5/5 (1 mL) column equilibrated in buffer C. Bound proteins were eluted in a linear salt gradient of 0–500 mM NaCl in buffer C. The RNA ligase activity eluted at 450 mM NaCl and correlated with a single protein of 55 kDa in a Coomassie colloidal blue stained SDS-PAGE (Fig. 1).
Preparation of tRNA Splicing Intermediates and RNA Ligase-Activity Assay.
A chimeric intron-containing pre-tRNA with the mature domain of plant tRNATyr and the anticodon and a short version of the intron from M. jannaschii tRNATrp was transcribed by T7 RNA polymerase as described (5). The cleavage of the intron-containing pre-tRNA with the M. jannaschii RNA splicing endonuclease was performed in 20 μL with 10 mM Tris-HCl, pH 7.6, 0.1 M KCl, 10 mM MgCl2, 1 mM DTT, 40 μM spermine and 1 μg recombinant enzyme. After incubation at 65 °C for 15 min, the ribonucleic acids were phenol/chloroform extracted, ethanol precipitated, and directly used for the RNA ligase-activity assay.
In vitro RNA ligation assays were performed in 20-μL reactions containing 10 mM Tris-HCl, pH 7.5, 0.1 M KCl, 6 mM MgCl2, 0.3 mM spermine, 5 mM 2-mercaptoethanol, 40 fmol (40,000 cpm) of tRNA splicing intermediates, 1 mM ATP except otherwise noted, and 2 μL of chromatographic fractions. After incubation at 37 °C for 1 h, the reaction mixtures were phenol/chloroform extracted, ethanol precipitated, and separated in a 12.5% polyacrylamide/8 M urea gel. The radioactive labeled ribonucleic acids were visualized by PhosphorImager analysis.
Analysis of the Ligation Product.
Complete digestion of the circular intron with nuclease P1 was carried out in a 10-μL reaction with 50 mM NH4OAc, pH 5.3, 2 μg carrier tRNA, and 100 ng nuclease P1. After incubation for 2 h at 50 °C, the reaction mix was dried in the Speed-Vac and dissolved in 1 μL NMP mix (10 mg/mL each of AMP, GMP, CMP, and UMP). Separation of the unlabeled and labeled nucleotide mix was performed by thin-layer chromatography on 20 × 20 cm cellulose plates using solvent A (iso-butyric acid/conc. ammonia/H2O = 57.7/3.8/38.5, [vol/vol/vol]). The radioactive label was visualized and quantified by PhosphorImager analysis. The unlabeled NMPs were visualized under UV light at 254 nm.
Phylogenetic Analysis.
RtcB sequences were downloaded from the National Center for Biotechnology Information nonredundant database. Sequence alignments were made and then manually refined in Geneious 4.7.4 prior to phylogenetic calculations as. Phylogenetic analyses were as described (36, 37) using Phyml (38).
Supplementary Material
Acknowledgments.
We thank Karl Stetter (University of Regensburg, Regensburg, Germany) and Imke Schröder (University of California, Los Angeles, CA) for gifts of Methanopyrus kandleri und Pyrobaculum aerophilum cells, respectively. We are indebted to Patrick O’Donoghue and Jiqiang Ling for stimulating discussions. M.E. was a Feodor Lynen Fellow of the Alexander von Humboldt Foundation (Bonn, Germany). This work was supported by National Institutes of Health Grant GM22854 (to D.S.) and Grant RR011823 (to J.R.Y.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018307108/-/DCSupplemental.
References
- 1.Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 2.Peebles CL, Gegenheimer P, Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- 3.Mori T, et al. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol Biol Cell. 2010;21:3722–3734. doi: 10.1091/mbc.E10-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci USA. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LK, Schwer B, Englert M, Beier H, Shuman S. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res. 2006;34:517–527. doi: 10.1093/nar/gkj441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 8.Perkins KK, Furneaux H, Hurwitz J. Isolation and characterization of an RNA ligase from HeLa cells. Proc Natl Acad Sci USA. 1985;82:684–688. doi: 10.1073/pnas.82.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes I, Gupta R. RNA splicing ligase activity in the archaeon Haloferax volcanii. Biochem Biophys Res Commun. 1997;237:588–594. doi: 10.1006/bbrc.1997.7193. [DOI] [PubMed] [Google Scholar]
- 10.Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988;54:693–703. doi: 10.1016/s0092-8674(88)80014-x. [DOI] [PubMed] [Google Scholar]
- 11.Zofallova L, Guo Y, Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal K, Mande SC. Exploiting 3D structural templates for detection of metal-binding sites in protein structures. Proteins. 2008;70:1206–1218. doi: 10.1002/prot.21601. [DOI] [PubMed] [Google Scholar]
- 13.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 14.Okada C, Maegawa Y, Yao M, Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 15.Kanai A, et al. Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus. RNA. 2009;15:420–431. doi: 10.1261/rna.1122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torchia C, Takagi Y, Ho CK. Archaeal RNA ligase is a homodimeric protein that catalyzes intramolecular ligation of single-stranded RNA and DNA. Nucleic Acids Res. 2008;36:6218–6227. doi: 10.1093/nar/gkn602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J Biol Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- 18.Reinhold-Hurek B, Shub DA. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 19.Genschik P, Billy E, Swianiewicz M, Filipowicz W. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka N, Shuman S. Structure-activity relationships in human RNA 3′-phosphate cyclase. RNA. 2009;15:1865–1874. doi: 10.1261/rna.1771509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka N, Smith P, Shuman S. Structure of the RNA 3′-phosphate cyclase-adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure. 2010;18:449–457. doi: 10.1016/j.str.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 23.Phizicky EM, Consaul SA, Nehrke KW, Abelson J. Yeast tRNA ligase mutants are nonviable and accumulate tRNA splicing intermediates. J Biol Chem. 1992;267:4577–4582. [PubMed] [Google Scholar]
- 24.Wang LK, Schwer B, Shuman S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA. 2006;12:2126–2134. doi: 10.1261/rna.271706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zillmann M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Mol Cell Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 27.Trowitzsch S. Göttingen, Germany: Georg-August-Universität; 2008. Functional and structural investigation of spliceosomal snRNPs. PhD thesis. [Google Scholar]
- 28.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 29.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouzuma Y, et al. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem Biophys Res Commun. 2003;306:666–673. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 31.Kjems J, Jensen J, Olesen T, Garrett RA. Comparison of transfer RNA and ribosomal RNA intron splicing in the extreme thermophile and archaebacterium Desulfurococcus mobilis. Can J Microbiol. 1989;35:210–214. doi: 10.1139/m89-033. [DOI] [PubMed] [Google Scholar]
- 32.Kjems J, Garrett RA. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc Natl Acad Sci USA. 1991;88:439–443. doi: 10.1073/pnas.88.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe Y, et al. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- 34.Yokobori S, et al. Gain and loss of an intron in a protein-coding gene in Archaea: The case of an archaeal RNA pseudouridine synthase gene. BMC Evol Biol. 2009;9:198. doi: 10.1186/1471-2148-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laska S, Kletzin A. Improved purification of the membrane-bound hydrogenase-sulfur-reductase complex from thermophilic archaea using epsilon-aminocaproic acid-containing chromatography buffers. J Chromatogr B. 2000;737:151–160. doi: 10.1016/s0378-4347(99)00362-x. [DOI] [PubMed] [Google Scholar]
- 36.Nureki O, et al. Structure of an archaeal non-discriminating glutamyl-tRNA synthetase: A missing link in the evolution of Gln-tRNAGln formation. Nucleic Acids Res. 2010;38:7286–7297. doi: 10.1093/nar/gkq605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinemann IU, et al. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci USA. 2009;106:21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45:667–671. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.