Abstract
APOBEC3G (A3G) is an antiviral protein that binds RNA and single-stranded DNA (ssDNA). The oligomerization state of A3G is likely to be influenced by these nucleic acid interactions. We applied the power of nanoimaging atomic force microscopy technology to characterize the role of ssDNA in A3G oligomerization. We used recombinant human A3G prepared from HEK-293 cells and specially designed DNA substrates that enable free A3G to be distinguished unambiguously from DNA-bound protein complexes. This DNA substrate can be likened to a molecular ruler because it consists of a 235-bp double-stranded DNA visual tag spliced to a 69-nucleotide ssDNA substrate. This hybrid substrate enabled us to use volume measurements to determine A3G stoichiometry in both free and ssDNA-bound states. We observed that free A3G is primarily monomeric, whereas ssDNA-complexed A3G is mostly dimeric. A3G stoichiometry increased slightly with the addition of Mg2+, but dimers still predominated when Mg2+ was depleted. A His-248/His-250 Zn2+-mediated intermolecular bridge was observed in a catalytic domain crystal structure (Protein Data Bank code 3IR2); however, atomic force microscopy analyses showed that the stoichiometry of the A3G-ssDNA complexes changed insignificantly when these residues were mutated to Ala. We conclude that A3G exchanges between oligomeric forms in solution with monomers predominating and that this equilibrium shifts toward dimerization upon binding ssDNA.
Keywords: Atomic Force Microscopy, DNA-binding Protein, DNA-Protein Interaction, HIV, Protein-DNA Interaction, Single Molecule Biophysics
Introduction
APOBEC3G (apolipoprotein B mRNA-editing catalytic-like protein 3G; A3G)2 is a single-stranded DNA (ssDNA) cytosine deaminase that is known for its capacity to restrict the replication of Vif-deficient HIV-1 (reviewed recently in Refs. 1–3). First, for this to occur, A3G must bind RNA to gain access to assembling HIV-1 Gag ribonucleoprotein complexes (4–11). Second, during HIV-1 reverse transcription, A3G must bind viral single-stranded cDNA to catalyze cytosine-to-uracil deamination events that ultimately manifest as genomic strand guanine-to-adenine hypermutations (12–15). In addition to compromising the genetic integrity of the virus through cDNA deamination, A3G is also capable of binding viral RNA and directly impeding reverse transcription (reviewed in Ref. 16).
Although recent biochemical and structural studies have provided some clues as to how A3G may bind ssDNA, the stoichiometric nature of the A3G-ssDNA complex is still unknown (17–26). Atomic force microscopy (AFM) enables one to examine numerous biophysical and structural properties of various protein-DNA complexes at single-molecule levels and nanometer resolution (e.g. Refs. 28–32 and references therein). In the only prior A3G AFM reports, Chelico et al. (25, 27) attempted to characterize the stoichiometry of A3G bound to ssDNA. This work suggested that A3G forms stable complexes with ssDNA. Large oligomers appeared to be involved in complex formation, and the stoichiometry of these complexes appeared to be influenced by salt concentrations and divalent cations. However, the lack of internal size standards and the inherent flexibility of ssDNA prevented the unambiguous determination of A3G-ssDNA complexes from substrate-free A3G complexes.
Here, we use AFM and a hybrid DNA substrate (a molecular ruler) to clearly differentiate between free A3G and ssDNA-bound complexes. The hybrid DNA substrate is composed of a 235-bp DNA duplex spliced to a 69-nucleotide-long 5′-ssDNA tail. Both the duplex and the tail could be visualized with our instrumentation, and A3G was located exclusively within the ssDNA region of the hybrid substrate. Comparisons with the tetrameric Escherichia coli single-strand DNA-binding protein (SSB) and precise protein volume measurements enabled us to determine that A3G binds ssDNA substrates predominantly as a dimer. At the same time, unbound protein was primarily monomeric. The stoichiometry of the A3G-ssDNA complexes was unchanged in the absence of Mg2+ cations. Replacement of Mg2+ cations with Zn2+ cations slightly increased the yield of tetramers, but dimers were still more abundant overall. An H248A/H250A mutant predicted to lack a zinc-mediated intermolecular bridge observed recently in a crystal structure (Protein Data Bank code 3IR2 (19)) was also primarily dimeric in ssDNA complexes with fewer higher order oligomers. These findings suggest a mechanism for A3G assembly on ssDNA and clarify the model HIV restriction by A3G in which dimeric A3G is the form of the protein that deanimates viral cDNA and results in guanine-to-adenine hypermutations.
MATERIALS AND METHODS
A3G-Myc-His Purification
HEK-293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Denville Scientific, Inc.), 50 units/ml penicillin, and 50 μg/ml streptomycin solution (Invitrogen). The pcDNA3.1-A3G-Myc-His expression construct has been described (18). A3G-Myc-His or A3G-H248A/H250A-Myc-His (19) was expressed by transiently transfecting semiconfluent HEK-293T cells as directed by the manufacturer (TransIT-LTI, Mirus Bio). 1 × 108 cells were harvested, washed with PBS, and resuspended in 10 ml of cell lysis buffer (25 mm HEPES, pH 7.4, 150 mm NaCl, 0.5% Triton X-100, 1 mm EDTA, 1 mm MgCl2, 1 mm ZnCl2, 10% glycerol, and Roche EDTA-free Complete protease inhibitor mixture) supplemented with 50 μg/ml RNase A (Qiagen). The cell suspension was transferred to an ice-cold Dounce homogenizer, homogenized for 10 strokes, and then rotated for 1 h at 4 °C, followed by incubation at 25 °C for 20 min. The lysates were then clarified by centrifugation (12,000 × g, 4 °C, 10 min). NaCl was then added to the lysates to bring the final concentration to 0.8 m. The lysates were mixed with 50 μl of nickel-nitrilotriacetic acid-agarose (Qiagen) by rotating overnight at 4 °C. The suspension was then loaded onto a Poly-Prep chromatography column (Bio-Rad). Following extensive washing with wash buffer (50 mm Tris, pH 8.0, 0.3 m NaCl, 10% glycerol, 0.5% Triton X-100, and 50 mm imidazole), His-tagged proteins were eluted in 6 × 200 μl of elution buffer (50 mm Tris, pH 8.0, 0.3 m NaCl, 10% glycerol, 0.5% Triton X-100, and 150 mm imidazole). Protein purity (>80%) was assessed by SDS-PAGE and Coomassie Blue R-250 staining relative to a BSA standard. Protein concentration was determined by Coomassie Blue R-250 staining and densitometry.
DNA Cytosine-to-Uracil Activity Assays
Recombinant human A3G-Myc-His proteins were subjected to a fluorescence-based single-stranded DNA cytosine deaminase activity assay as described (33). Recombinant A3G-Myc-His was incubated for 2 h at 37 °C with a DNA oligonucleotide, 5′-(6-FAM)-AAA-CCC-TAA-TAG-ATA-ATG-TGA-(TAMRA) (Biosearch Technologies, Inc.). Deamination of the underlined cytosine residue resulted in a uracil, which was excised by uracil DNA glycosylase (New England Biolabs) also in the reaction mixture. The resulting abasic site was subjected to hydrolytic cleavage by incubating reactions for 2 min at 95 °C or adding 0.1 m NaOH. When cleaved, the FAM and TAMRA labels were physically separated, FRET diminished, and FAM fluorescence increased. Fluorescence was measured using the 490 nm channel on the LightCycler 480 instrument (Roche Applied Science) or LJL Analyst AD (LJL BioSystems, Inc.).
Gel Shift Experiments
The indicated concentration of purified A3G-Myc-His or A3G-H248A/H250A-Myc-His was incubated with 0.07 μm 43-nucleotide (nt) ssDNA substrate (biotin-ATTATTATTATTCCAATGGATTTATTTATTTATTTATTTATTT-fluorescein; IDT) in binding buffer containing 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm MgCl2, 0.5 mm EDTA, 0.5 mm DTT, and 4% glycerol for 30 min at 37 °C. After incubation, the reaction mixtures were mixed with 10× gel loading buffer (250 mm Tris-HCl, pH 7.5, 0.2% bromphenol blue, and 40% glycerol) and separated using a 6% native Tris borate/EDTA gel in 0.5× Tris borate/EDTA at 4 °C. The resulting gels were imaged with a Storm 840 Phosphor-Chemifluorescence Workstation (GE Healthcare).
DNA Substrate Preparation
The hybrid DNA substrate consisted of a 5′-ending 69-nt ssDNA region attached to a 235-bp double-stranded DNA (dsDNA) fragment. First, a 796-bp dsDNA fragment was obtained by PCR using the pUC19 plasmid DNA vector as a template and primers 5′-GCT TTC CAG TCG GGA AAC CT and 5′-CAG CGG TGG TTT GTT TGC C. The PCR product was phenol/chloroform-purified and digested with NlaIII restriction enzyme. A 210-bp fragment was purified from 2% agarose gel using the QIAquick gel extraction kit (Qiagen). The concentration of DNA was determined by measuring the absorbance at 260 nm. The single-stranded region of the DNA substrate was created by ligating a synthetic 94-mer containing 69-nt sequence for protein binding (5′-AA AGA GAA AGT GAA ACC CAA AGA ATG AAA ACC CAA ATG TTA GAA TTG TTA ATG TGT GTG ATG ATG TTG A-3′) via a 21-mer adapter 5′-AAT ATA ATT CCT ACA CGT ATG-3′. The phosphorylated 21-mer was annealed with a 94-mer at a 1:1 molar ratio and ligated via the sticky end of the DNA restriction fragment with T4 polynucleotide DNA ligase at 16 °C overnight with a 30:1 molar excess of the 94-mer annealed with a 21-mer adapter to a 210-bp restriction fragment. The final product, a 235-bp dsDNA with a 69-nt 5′-extension on one end, was fractionated on a 3% native agarose gel (TopVision agarose gel, Fermentas, Inc.) and gel-purified as described above (see image in supplemental Fig. S1).
Preparation of Protein-DNA Complexes
Recombinant A3G or A3G-H248A/H250A was mixed with DNA substrate at a 5:1 protein/DNA ratio in a total volume of 10 ml. The reaction buffer (1× buffer) contained 50 mm HEPES, pH 7.5, 100 mm NaCl, and 1 mm DTT. To study the effect of different divalent cations on the efficiency of complex formation, each of the following cations was added to the reaction mixture: 5 mm Mg2+, 25 mm Zn2+, or 250 mm Zn2+. The reaction mixture was incubated for 10 min at 37 °C, purified using the Montage PCR purification protocol (Millipore Corp., Bedford, MA), and eluted into 50 μl of imaging buffer. Complexes of the DNA with E. coli SSB at various protein/DNA ratios were prepared in 50 mm Tris, pH 7.5, 100 mm NaCl, 5 mm Mg2+, and 1 mm DTT for 10 min at room temperature.
AFM Sample Preparation
The sample preparation procedure used functionalized 1-(3-aminopropyl)silatrane-mica, which was prepared by the treatment of freshly cleaved mica with 1-(3-aminopropyl)silatrane as described (28, 31, 34). The sample (5 μl) was deposited on 1-(3-aminopropyl)silatrane-mica for 2 min, rinsed with deionized water, and dried with argon gas. Images were acquired in tapping mode in air using the Multimode SPM Nanoscope IV system (Veeco, Santa Barbara, CA). Silicon-etched tapping mode probes with nominal spring constants of 42 newtons/m and a resonant frequency of 320 Hz were used (Veeco).
Data Analysis
For each type of protein-ssDNA complex, data were obtained for the length of the DNA fragments, the volume of the protein, and the yields of the various complexes using FemtoScan Online (Advanced Technologies Center, Moscow, Russia). The detailed measurement of the protein volume in the complex has been described (35). Briefly, the protein was approximated as a segment of the sphere, and the volume was calculated from the cross-section measurements of the images. The data for the volume measurements were summarized as histograms using Origin 6.0 (OriginLab, Northampton, MA). The volume of A3G protein was converted into mass in kilodaltons using the volume data for the complexes of the same DNA substrate with E. coli SSB that binds to ssDNA as a tetramer.
Sucrose Density Gradients
The basic protocol for gradient analysis was as described (36, 37). Protein lysates for gradient experiments were harvested from 5 million HeLa cells 48 h after transient transfection. The cytoplasmic fraction was collected by incubation in low-salt buffer (10 mm KCl), followed by passage through a 28-gauge syringe and low-speed centrifugation to remove the nuclei. The nuclear fraction was collected from the pelleted nuclei treated with high-salt buffer (600 mm KCl). Both fractions were centrifuged at high speed to remove debris and dialyzed into loading buffer (100 mm KCl). 10–40% ultracentrifuge glycerol gradients were prepared on a Gradient Master machine (BioComp), and the samples were loaded on top of the gradients. The samples were centrifuged for 17 h at 32,000 rpm in an SW-41 rotor. The equilibrated gradients were divided into 500-μl fractions and used for immunoblotting.
Localization Experiments
Microscopy experiments were performed as described (38). Briefly, HeLa or HEK-293T cells on Lab-Tek chambered cover glasses (Nunc) were transfected with A3G-enhanced GFP or A3G-H248A/H250A-enhanced GFP (constructed by site-directed mutagenesis) and incubated overnight. Before imaging, the cells were treated with PBS with 0.1% Hoechst dye to stain the nuclei. A DeltaVision deconvolution microscope (Applied Precision) at ×40 magnification was used to collect the images, and deconvolution was performed using DeltaVision softWoRx software (Applied Precision). Images were cropped with Photoshop and assembled in Adobe Illustrator.
RESULTS
A3G Purification and Activity
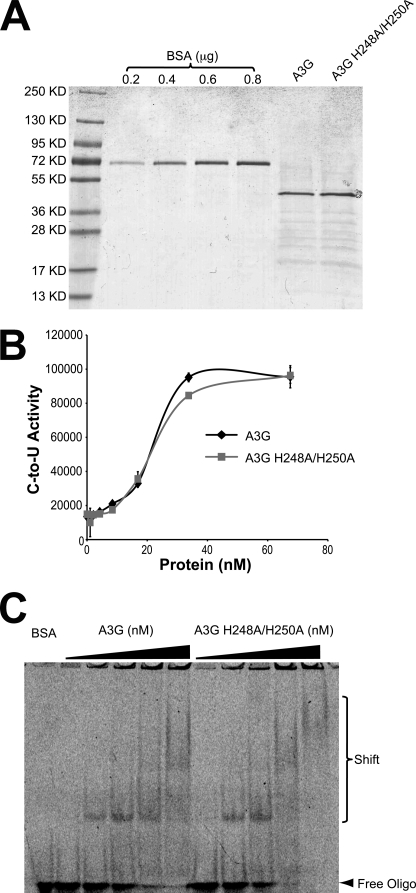
A3G-Myc-His was isolated from HEK-293T cells and found to be >80% pure by denaturing gel fractionation and Coomassie Blue staining (Fig. 1A). An H248A/H250A mutant protein produced in parallel was similarly pure. These two enzymes had indistinguishable activity profiles using a fluorescence-based cytosine-to-uracil DNA deaminase assay (Fig. 1B). Both enzymes were also capable of binding a 43-nucleotide ssDNA, as evidenced by similar mobility shift patterns during native polyacrylamide gel electrophoresis (Fig. 1C). These activity and substrate binding data are consistent with prior functional studies showing that His-248 and His-250 are dispensable for A3G to restrict Vif-deficient HIV-1 (19). However, prior studies still leave open the possibility that these two histidines may contribute to the formation of higher order A3G oligomers that have been observed in living cells (39), crude cell lysates (40–42), and recombinant protein complexes (23, 24, 26, 43).
FIGURE 1.
APOBEC3G purification and activity. A, image of Coomassie Blue-stained A3G and A3G-H248A/H250A-Myc-His proteins purified from HEK-293T cells. A BSA standard is shown for comparison. B, DNA deamination activity of the indicated proteins (arbitrary fluorescent units). C, EMSA for the indicated proteins bound to a 43-nt ssDNA.
Design of the Hybrid DNA Substrate
AFM is capable of providing important information on the characteristics of protein and DNA complexes such as the DNA bend angle, protein position, and protein stoichiometry (31). However, the application of AFM imaging methods to A3G has been complicated by the fact that the preferred substrate for this enzyme is ssDNA rather than dsDNA (12, 18, 23, 26). ssDNA is much more flexible than dsDNA and can form secondary structures (44). For instance, individual ssDNA complexes have been shown to appear on AFM images as globular features of various sizes, and it is problematic to separate free protein from complexes with similar globular shapes (45).
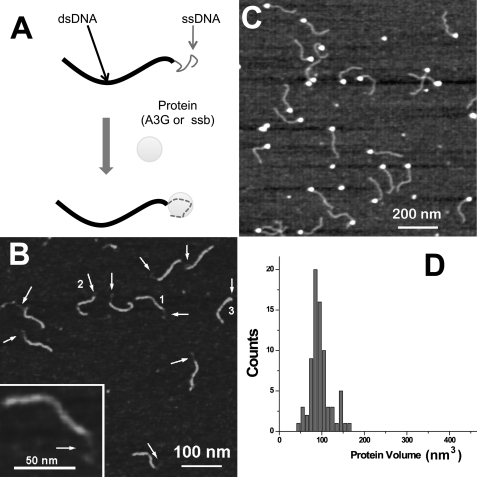
To avoid these potential problems and enable unambiguous identification and characterization of A3G-DNA complexes, we have developed a novel DNA template. This design consists of a 69-nt ssDNA spliced onto the end of a 235-bp dsDNA fragment (Fig. 2A). The ssDNA is a potential deamination substrate, and the dsDNA functions as an imaging tag. The binding of A3G to this substrate is anticipated to occur at the single-stranded end, so complex formation would be detected as the appearance of a globular feature (a blob) at one end of the molecule. A typical AFM image of the hybrid DNA is shown in Fig. 2B, and the dsDNA and ssDNA regions are distinguished readily. The protrusions are the most typical features on these images, although compacted structures appearing as globular features at ends are also seen (indicated with arrows and numbers). One of these molecules with a protrusion with extended morphology is shown in the inset to Fig. 2B (type 1). The contour lengths of DNA tags were very close to the expected length of the double-stranded section of the DNA (supplemental Fig. S2A).
FIGURE 2.
Novel AFM approach for studying protein-DNA interactions. A, a hybrid DNA substrate consisting of a dsDNA region (thick black line) and a ssDNA tail (thin grey line) can be complexed with A3G or SSB and imaged by AFM. B, AFM image of the hybrid DNA substrate. Single-stranded regions are indicated with arrows. Numbers 1, 2, and 3 point to the morphologies of these regions in extended (1), compact (2), and globular (3) forms (1 is enlarged in the inset), respectively. Z-scale is 1.5 nm. C, AFM image of hybrid DNA substrate complexed with E. coli SSB. Complexes appear as clearly identified bright blobs attached to the single-stranded end of the hybrid DNA substrate. The yield of complexes is 100%, reflecting the high affinity of SSB for ssDNA. Z-scale is 1.5 nm. D, volume measurements of SSB-ssDNA complexes. The histogram width is narrow, indicating the formation of homogeneous complexes (n = 77).
System Calibrations with SSB and ssDNA
We first tested our experimental system using the E. coli SSB. This protein binds very specifically to ssDNA as a tetramer (46), and it has been thoroughly characterized by various structural techniques, including single-molecule analysis (47). SSB was complexed with the hybrid DNA substrate and imaged with AFM. Fig. 2C shows one of the typical AFM images of the SSB-DNA complex. A common feature of these images is the appearance of a bright globular structure at one end of the hybrid substrate that we interpret as protein molecules bound to ssDNA. At the selected protein/DNA ratio (25:1), very few free DNA molecules were observed. Similarly to the studies of complexes of proteins with dsDNA (29–31), we used the same formula for calculating the volume for SSB. The distribution of SSB volume measurements is very narrow, suggesting that the stoichiometry of the protein is uniform (Fig. 2D). Prior studies have demonstrated that SSB binds ssDNA as a tetramer (4 × 18.8 kDa), so the 100-nm3 protein volume observed at the distribution maximum could be assigned a net molecular mass of 75.2 kDa.
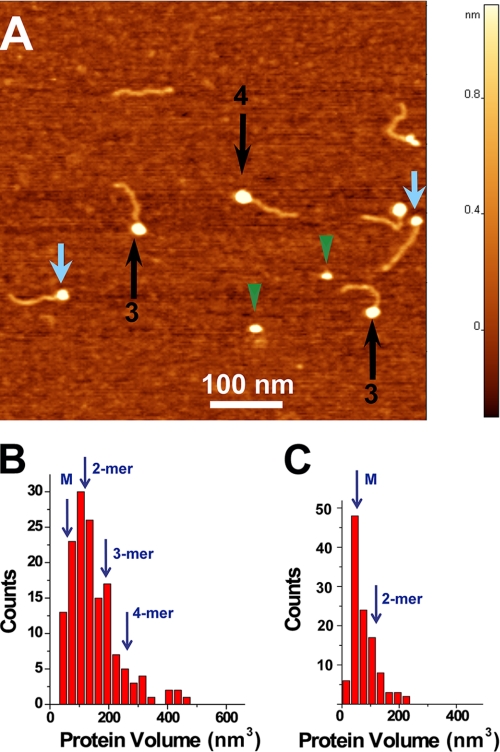
A3G Dimers and Oligomers Complex with ssDNA
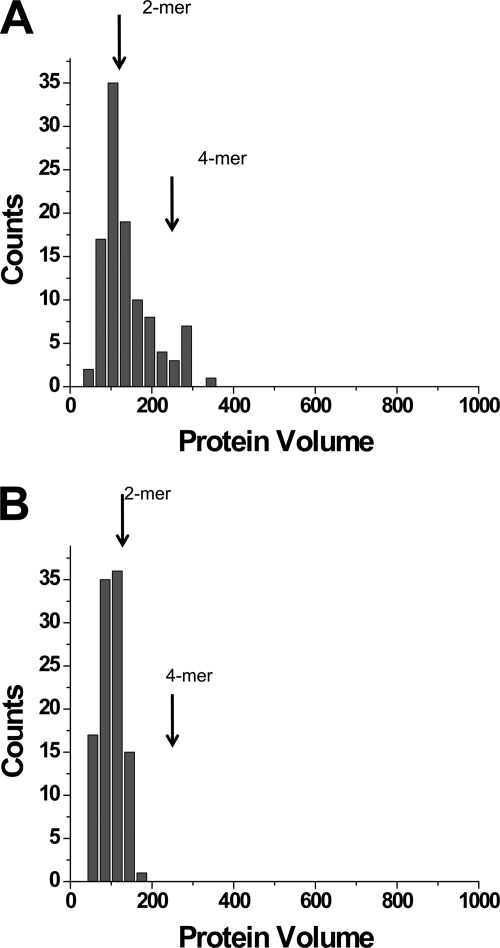
The same hybrid DNA substrate was complexed with A3G and imaged by AFM. One of the typical images of such complexes formed in the presence of Mg2+ is shown in Fig. 3A. Similar to the data for SSB, the DNA fragments with bright blobs attached to one end of the substrate appear in the images with a yield of ∼82%. However, in contrast with the SSB-ssDNA complexes, the sizes of the globular features varied considerably, suggesting that A3G stoichiometry is not constant within the complexes. Small and large globular complexes are indicated with blue and black arrows, respectively (Fig. 3A). The apparent range of protein stoichiometries was supported by volume measurements. The results for the volumes for the end-bound complexes are shown in Fig. 3B. Compared with the histogram for SSB, the histogram for A3G-DNA complexes is wider. Based on the fact that SSB binds DNA as a tetramer, the conversion coefficient from protein volume to molecular mass is equal to 1.3. The estimated volumes for monomeric (49.4 kDa), dimeric, trimeric, and tetrameric forms as indicated with arrows above the histogram in Fig. 3B are 64, 128, 192, and 256 nm3, respectively. Thus, the stoichiometry of A3G ranges primarily between monomers and tetramers, with the maximum corresponding to dimers.
FIGURE 3.
AFM images of wild-type A3G bound to single-stranded DNA. A, AFM image of the complex of the DNA ruler with A3G protein. Black and blue arrows indicate complexes with dimers and monomers of A3G, respectively. Green arrowheads point to free protein. B, histogram summarizing the volume measurements for A3G-ssDNA complexes (n = 143). C, histogram summarizing the volume measurements for unbound (free) A3G (n = 112).
We also imaged free protein; a few examples are indicated with green arrowheads in Fig. 3A. The combined volume data in Fig. 3B indicate that nonbound A3G is predominantly monomeric. Thus, our data suggest that A3G oligomerizes upon binding ssDNA substrate and that oligomerization is skewed toward the formation of dimers.
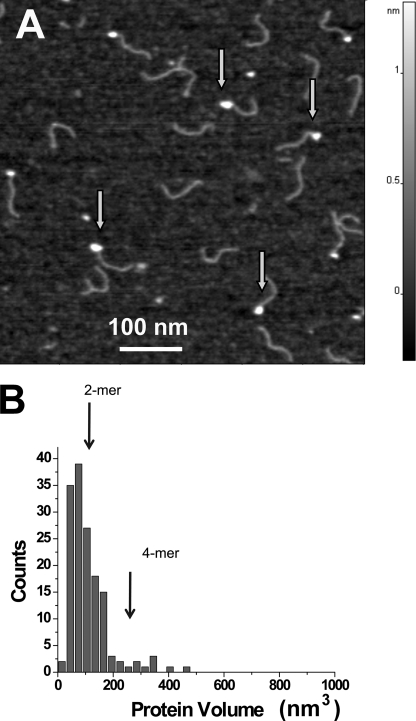
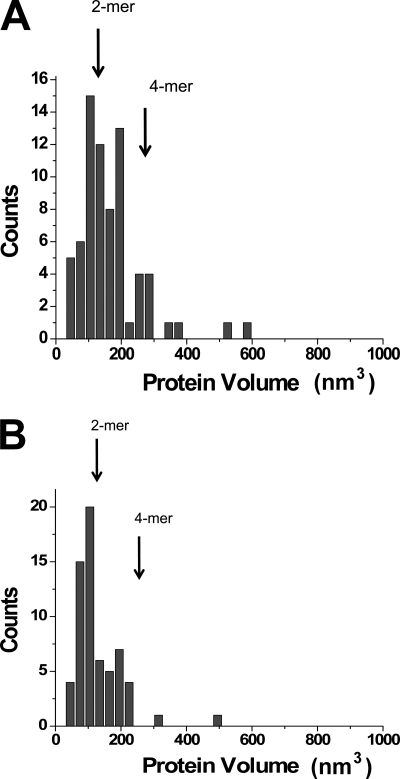
To test the hypothesis that the zinc-mediated bridge observed in a crystal structure between two individual A3G molecules contributes to protein oligomerization upon DNA binding, we performed similar experiments with the A3G-H248A/H250A protein (Fig. 1). The AFM image of the A3G-H248A/H250A mutant in the presence of Mg2+ is shown in Fig. 4A. Similar to wild-type A3G, a strong preference for ssDNA ends was observed, although the yield of complexes dropped almost 3-fold (33 versus 82% for wild-type A3G). The volume measurement data are shown in Fig. 4B. Compared with wild-type A3G, dimers also constitute the predominant morphology of the protein, but there is a clear tendency toward fewer trimers and tetramers (i.e. fewer larger oligomeric forms were observed).
FIGURE 4.
AFM images of A3G-H248A/H250A bound to ssDNA. A, representative AFM image. The end-bound complexes are indicated with arrows (image size, 600 nm). B, histogram of the measured volumes of the A3G-H248A/H250A-ssDNA complexes. Vertical arrows indicate the expected volume for the dimer (2-mer) and tetramer (4-mer) (n = 150).
Effect of Salt on A3G Oligomerization
A3G has a Zn2+-binding pocket coordinated by two α-helices (α2 and α3) within its catalytic domain (19, 21). In addition to its essential role in catalysis, we wondered whether this single Zn2+ cation might also be involved in directly binding ssDNA. To test this hypothesis, we performed AFM analysis of A3G-ssDNA complexes in the presence of excess Zn2+ cations. The experiments were performed with wild-type A3G and A3G-H248A/H250A. Hybrid A3G-DNA complexes were formed in the presence of 25 mm Zn2+ (see images in supplemental Figs. S3 and S4), but yields were 20 times less than those formed in the presence of Mg2+ cations. The results of the volume measurements are summarized for wild-type A3G and the H248A/H250A mutant in Fig. 5 (A and B, respectively). The stoichiometry of the complexes was in the range between the monomer and tetramer, with a maximum yield for dimers. Qualitatively, these distributions are similar to ones obtained for the buffer containing Mg2+ cations, although some decrease in the population of monomers and tetramers, especially for the mutant, was noticed. A 10-fold increase in the concentration of Zn2+ cations to 250 mm completely prevented the formation of the complexes (data not shown). Therefore, the results suggest that Zn2+ cations do not change the protein oligomerization pattern in the complexes with DNA but substantially decrease their stability. We applied independent t test criteria to the data on the volume measurements to evaluate the effect of types of cations on the protein volume. Supplemental Table S1 shows the results of statistical analysis of the data obtained with and without Mg2+ cations. Supplemental Table S2 shows a similar analysis for the complexes obtained with Zn2+ and Mg2+ cations. According to these analyses, there is no statistically significant difference for both pairs of the data sets.
FIGURE 5.
AFM results for volume measurements of the complexes formed in the presence of 25 mm Zn2+ cations by WT A3G (A) and the A3G-H248 mutant (B). The number of complexes analyzed were n = 71 (A) and n = 78 (B).
To test the effect of Mg2+ cations on the formation of the complexes, we performed the AFM analysis in the buffer without Mg2+ cations. The yields of the complexes under these conditions for both proteins were ∼3 times lower than in the presence of Mg2+ cations: 30% for wild-type A3G (82% in 5 mm Mg2+) and 10% for its mutant (33% in 5 mm Mg2+). The results of the volume measurements for the complexes made by both types of A3G are shown in Fig. 6. These analyses revealed a substantial drop of tetramers, suggesting that Mg2+ cations stabilize the formation of higher oligomers of A3G.
FIGURE 6.
AFM data for the complexes formed in the absence of divalent cations by WT A3G (A) and the A3G-H248 mutant (B). 105 complexes were analyzed for both samples.
DISCUSSION
The combination of sensitive AFM techniques, a novel hybrid DNA ruler, and purified A3G from human cells has enabled the first images of this HIV restriction factor bound to ssDNA. A3G is predominantly dimeric when bound to ssDNA, in contrast to native protein, which appears mostly monomeric. Some higher order oligomers are also observed in ssDNA complexes, and their existence is influenced by His-248 and His-250, which coordinate zinc and form a intermolecular bridge in the A3G catalytic domain crystal lattice (19). Oligomerization is also influenced by Zn2+ and Mg2+, suggesting a role for divalent cations. Overall, our data are consistent with a model in which ssDNA-bound A3G is dimeric and enzymatically active.
Our observations are concordant with prior studies demonstrating A3G oligomerization in living cells and in vitro (4, 7, 8, 17, 19–22, 24, 25, 40, 42, 43, 48). However, although mutation of His-248 and His-250 reduces the number of higher order A3G-ssDNA oligomeric complexes observed by AFM (and diminishes the overall yield), these residues are dispensable for cytoplasmic localization, P body formation, and high molecular mass formation in living cells (supplemental Fig. S5 and data not shown). Taken together, these observations suggest that RNA binding, other A3G surfaces (independent of His-248 and His-250), and potentially heterologous protein-protein interactions may regulate A3G oligomerization in vivo.
Our data help distinguish between two models for A3G-ssDNA complex formation. In the first, a dimer may form when two monomers bind adjacent to one another on ssDNA. Alternatively, dimers may first form in solution and then engage ssDNA. In the framework of the first model, monomer binding will be the primary mode at low protein concentrations, and the number of dimeric complexes will grow as the protein concentration increases. The second model predicts that the distribution of complexes depends on the binding constant for each type of the complex, so the yield of dimers will be higher than for the monomers if the dimers form more stable complexes than monomers. Our data are in line with the second model. Indeed, the data in Fig. 3B indicate that the yield of dimers is considerably higher than that of monomers. These observations were made under nonsaturating conditions, and the yield of complexes was 82%. We performed additional experiments in which the protein/DNA ratio was decreased an additional 5-fold (supplemental Fig. S6). According to these data, the major product is still a dimer with a low yield of monomers. A 4-fold drop in the yield of complexes was observed under these conditions. In experiments in which divalent cations were removed (Fig. 6), the yield of complexes dropped 3 times, but the monomeric complexes formed with the same low efficiency. Altogether, the data support the second model for A3G complex formation in which A3G dimers form much more stable complexes than monomers.
Although AFM measures the sizes of the sample in three dimensions, enabling one to measure the volume of the sample, a number of variables such as the tip convolution effect and the conversion of the image volume to the sample volume and eventually to the molecular mass require careful calibration (30, 31). To address this issue, we used the well characterized E. coli SSB as a calibration standard. Similarly to A3G, SSB binds specifically to ssDNA. Various studies including x-ray crystallography (46) have shown that SSB binds ssDNA as a tetramer, and it is therefore an appropriate standard for converting A3G volume data to molecular masses. Importantly, the maxima on histograms for SSB and A3G complexes almost coincide (Figs. 2B and 3B); therefore, as shown in Fig. 3, the A3G dimers are the most representative oligomeric forms. The population of trimers and tetramers is low. Importantly, unbound protein is primarily in the monomeric state, suggesting that the interaction with ssDNA changes the stoichiometry of A3G. The observed dimeric stoichiometry of A3G bound to the ssDNA substrate is concordant with two recent studies (42, 43).
However, our data contrast with the only prior AFM studies of A3G-ssDNA complexes, which indicated that dimeric A3G is a minor species (40% monomer, 14% dimer, and 46% higher order oligomers in the presence of 5 mm MgCl2) (25, 27). These prior studies also suggested that salt (Mg2+ cations) facilitates the formation of the higher order oligomers. A possible explanation for this discrepancy is that Chelico et al. (25, 27) used short ssDNA oligonucleotides, which may be difficult to distinguish from free protein complexes. Our hybrid DNA substrate in which ssDNA is attached to one end of a dsDNA fragment eliminates this ambiguity because A3G-bound complexes are identified as bright blobs located at the end of the DNA duplex. Importantly, the morphology of free ends is entirely different from the morphology of the complexes (Figs. 2B and 3A). Another potential variable is protein source: Chelico et al. used E. coli- or baculovirus-expressed A3G, whereas we used A3G prepared from human cells (Fig. 1).
The role of divalent ions in A3G complex formation and its stoichiometry is another finding. The comparison of effects of Mg2+ and Zn2+ cations indicated that A3G stoichiometry in the ssDNA-bound complexes remains essentially the same, mostly dimeric regardless of the type of divalent cation. However, the yield of complexes depends on the cation type, so the replacement of Mg2+ cations with Zn2+ leads to an almost 20-fold decrease in complex yield. At the same time, complete removal of Mg2+ cations does not have such a dramatic effect, although a 3-fold decrease in complex yield was observed. According to a recent model for A3G dimerization (42), the charge distribution over the dimer surface is not uniform. Rather, a positive charge is clustered in a rather narrow area of the monomer-monomer interface, and this region may interact with negatively charged RNA. If electrostatics are the predominant forces that stabilize the A3G-ssDNA complexes, we should observe a decrease in the yield of the complexes at elevated ionic strengths. Mg2+ cations effectively shield electrostatic interactions decreasing the Debye radius; therefore, in the presence of Mg2+ cations, the yield of complexes should go down. Our data show the opposite effect, suggesting that the interaction of A3G with ssDNA does not have a pure strong electrostatic nature. A dramatic drop in the yield of the complexes upon the addition of Zn2+ cations (25 mm) suggests that the addition of this cation induces conformational changes in the dimer that may not be favorable for binding ssDNA.
The atomic structure of the A3G-ssDNA complex has eluded efforts to date, although indirect methods have yielded considerable information about this interaction. Gel shift studies showed that at least 16 nucleotides are needed for the formation of stable complexes (26). Biochemical studies suggested that A3G does not remain stably bound to ssDNA but that it translocates randomly by a series of jumps and slides and/or intersegmental transfers (23, 27). A3G also has preferred reaction polarity favoring deamination toward the 5′-end of ssDNA substrates. The preferred target site is 5′-CCC with a strong bias toward the third cytosine, and deamination rarely occurs near the 3′-end of substrates. This leads to a so-called deamination “dead zone” (27). Given these properties and the 69-nt length of the ssDNA portion of our hybrid DNA substrate, we anticipated seeing both A3G bound to ssDNA and a visible fraction of unbound ssDNA. However, this scenario was not observed because ssDNA was apparent only in free hybrid substrates. Our observations suggest that dimeric A3G somehow occupies most of the 69-nt ssDNA region. We speculate that ssDNA wraps around the A3G dimer as it does around the SSB tetramer (46, 47) because both complexes yielded morphologically similar AFM images.
Supplementary Material
Acknowledgments
We thank S. Conner for sharing instrumentation and M. Kotler, H. Matsuo, J. Mueller, and C. Schiffer for comments on the manuscript. N. Hoffman contributed to the early stages of this work.
This work was supported, in whole or in part, by National Institutes of Health Grants AI064046 (to R. S. H.) and P01 GM091743 (to R. S. H. and Y. L. L.). This work was also supported by National Science Foundation Grant PHY-0615590 (to Y. L. L.), NATO Grant SFP-983204 (to Y. L. L.), a Nebraska Research Initiative Grant (to Y. L. L.), and a National Science Foundation doctoral scholarship (to L. L.). The AFM experiments were performed in the University of Nebraska Nanoimaging Core Facility with the use of AFM instruments purchased with National Institutes of Health Support Grant SIG 1S10 SS023400.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- A3G
- APOBEC3G
- ssDNA
- single-stranded DNA
- AFM
- atomic force microscopy
- SSB
- single-strand DNA-binding protein
- dsDNA
- double-stranded DNA
- nt
- nucleotide.
REFERENCES
- 1. Hultquist J. F., Harris R. S. (2009) Future Virol. 4, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malim M. H. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Strebel K., Luban J., Jeang K. T. (2009) BMC Med. 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. (2004) J. Biol. Chem. 279, 33177–33184 [DOI] [PubMed] [Google Scholar]
- 5. Svarovskaia E. S., Xu H., Mbisa J. L., Barr R., Gorelick R. J., Ono A., Freed E. O., Hu W. S., Pathak V. K. (2004) J. Biol. Chem. 279, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 6. Popik W., Alce T. M. (2004) J. Biol. Chem. 279, 704–712 [DOI] [PubMed] [Google Scholar]
- 7. Douaisi M., Dussart S., Courcoul M., Bessou G., Vigne R., Decroly E. (2004) Biochem. Biophys. Res. Commun. 321, 566–573 [DOI] [PubMed] [Google Scholar]
- 8. Schäfer A., Bogerd H. P., Cullen B. R. (2004) Virology 328, 163–168 [DOI] [PubMed] [Google Scholar]
- 9. Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X. F. (2004) J. Virol. 78, 11841–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zennou V., Perez-Caballero D., Göttlinger H., Bieniasz P. D. (2004) J. Virol. 78, 12058–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khan M. A., Kao S., Miyagi E., Takeuchi H., Goila-Gaur R., Opi S., Gipson C. L., Parslow T. G., Ly H., Strebel K. (2005) J. Virol. 79, 5870–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 13. Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 14. Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442 [DOI] [PubMed] [Google Scholar]
- 16. Holmes R. K., Malim M. H., Bishop K. N. (2007) Trends Biochem. Sci. 32, 118–128 [DOI] [PubMed] [Google Scholar]
- 17. Harjes E., Gross P. J., Chen K. M., Lu Y., Shindo K., Nowarski R., Gross J. D., Kotler M., Harris R. S., Matsuo H. (2009) J. Mol. Biol. 389, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. (2008) Nat. Struct. Mol. Biol. 15, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 19. Shandilya S. M., Nalam M. N., Nalivaika E. A., Gross P. J., Valesano J. C., Shindo K., Li M., Munson M., Royer W. E., Harjes E., Kono T., Matsuo H., Harris R. S., Somasundaran M., Schiffer C. A. (2010) Structure 18, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 21. Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., Katahira M. (2009) EMBO J. 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 24. Wedekind J. E., Gillilan R., Janda A., Krucinska J., Salter J. D., Bennett R. P., Raina J., Smith H. C. (2006) J. Biol. Chem. 281, 38122–38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chelico L., Prochnow C., Erie D. A., Chen X. S., Goodman M. F. (2010) J. Biol. Chem. 285, 16195–16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iwatani Y., Takeuchi H., Strebel K., Levin J. G. (2006) J. Virol. 80, 5992–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chelico L., Sacho E. J., Erie D. A., Goodman M. F. (2008) J. Biol. Chem. 283, 13780–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lyubchenko Y. L. (2004) Cell Biochem. Biophys. 41, 75–98 [DOI] [PubMed] [Google Scholar]
- 29. Lushnikov A. Y., Brown B. A., 2nd, Oussatcheva E. A., Potaman V. N., Sinden R. R., Lyubchenko Y. L. (2004) Nucleic Acids Res. 32, 4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lushnikov A. Y., Potaman V. N., Oussatcheva E. A., Sinden R. R., Lyubchenko Y. L. (2006) Biochemistry 45, 152–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lyubchenko Y. L., Shlyakhtenko L. S. (2009) Methods 47, 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hansma H. G., Laney D. E., Bezanilla M., Sinsheimer R. L., Hansma P. K. (1995) Biophys. J. 68, 1672–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shlyakhtenko L. S., Gall A. A., Filonov A., Cerovac Z., Lushnikov A., Lyubchenko Y. L. (2003) Ultramicroscopy 97, 279–287 [DOI] [PubMed] [Google Scholar]
- 35. Shlyakhtenko L. S., Gilmore J., Portillo A., Tamulaitis G., Siksnys V., Lyubchenko Y. L. (2007) Biochemistry 46, 11128–11136 [DOI] [PubMed] [Google Scholar]
- 36. Kataoka N., Dreyfuss G. (2008) Methods Mol. Biol. 488, 357–365 [DOI] [PubMed] [Google Scholar]
- 37. Abmayr S. M., Yao T., Parmely T., Workman J. L. (2006) Curr. Protoc. Mol. Biol. Chapter 12, Unit 12.1 [DOI] [PubMed] [Google Scholar]
- 38. Stenglein M. D., Matsuo H., Harris R. S. (2008) J. Virol. 82, 9591–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wichroski M. J., Robb G. B., Rana T. M. (2006) PLoS Pathog. 2, e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chiu Y. L., Soros V. B., Kreisberg J. F., Stopak K., Yonemoto W., Greene W. C. (2005) Nature 435, 108–114 [DOI] [PubMed] [Google Scholar]
- 41. Wang X., Dolan P. T., Dang Y., Zheng Y. H. (2007) J. Biol. Chem. 282, 1585–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. (2009) PLoS Pathog. 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salter J. D., Krucinska J., Raina J., Smith H. C., Wedekind J. E. (2009) Biochemistry 48, 10685–10687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hamon L., Pastre D., Dupaigne P., Le Breton C., Le Cam E., Piètrement O. (2007) Nucleic Acids Res. 35, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brar S. S., Sacho E. J., Tessmer I., Croteau D. L., Erie D. A., Diaz M. (2008) DNA Repair 7, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raghunathan S., Ricard C. S., Lohman T. M., Waksman G. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6652–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roy R., Kozlov A. G., Lohman T. M., Ha T. (2009) Nature 461, 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Conticello S. G. (2008) Genome Biol 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.