Abstract
Despite exciting progress in understanding the Piwi-interacting RNA (piRNA) pathway in the germ line, less is known about this pathway in somatic cells. We showed previously that Piwi, a key component of the piRNA pathway in Drosophila, is regulated in somatic cells by Yb, a novel protein containing an RNA helicase-like motif and a Tudor-like domain. Yb is specifically expressed in gonadal somatic cells and regulates piwi in somatic niche cells to control germ line and somatic stem cell self-renewal. However, the molecular basis of the regulation remains elusive. Here, we report that Yb recruits Armitage (Armi), a putative RNA helicase involved in the piRNA pathway, to the Yb body, a cytoplasmic sphere to which Yb is exclusively localized. Moreover, co-immunoprecipitation experiments show that Yb forms a complex with Armi. In Yb mutants, Armi is dispersed throughout the cytoplasm, and Piwi fails to enter the nucleus and is rarely detectable in the cytoplasm. Furthermore, somatic piRNAs are drastically diminished, and soma-expressing transposons are desilenced. These observations indicate a crucial role of Yb and the Yb body in piRNA biogenesis, possibly by regulating the activity of Armi that controls the entry of Piwi into the nucleus for its function. Finally, we discovered putative endo-siRNAs in the flamenco locus and the Yb dependence of their expression. These observations further implicate a role for Yb in transposon silencing via both the piRNA and endo-siRNA pathways.
Keywords: Cellular Regulation, Protein-Protein Interactions, RNA-binding Protein, RNA Interference (RNAi), Somatic Cell Genetics, Armitage, Drosophila, Piwi, Yb, piRNA
Introduction
The discovery of a complex class of small noncoding RNAs that interact with Piwi proteins, called piRNAs,3 revealed an unexpectedly complex aspect of biology (reviewed in Ref. 1). Both Piwi proteins and piRNAs have been widely regarded as germ line-specific molecules, where they are implicated in germ line specification, germ line stem cell maintenance, meiosis, spermiogenesis, and transposon silencing (2–9). However, there is long-standing evidence indicating that Piwi proteins are also present in somatic cells, such as human CD34+ hematopoietic stem/progenitor cells (10), human gastric epithelial cells (11), and planarian neoblasts (12–14). Among the three Drosophila Piwi proteins (Piwi, Aubergine (Aub), and Argonaute 3 (Ago3)), only Piwi is expressed in somatic cells (5, 15) and has been implicated with somatic functions, especially in epigenetic regulation and early embryogenesis (3, 16–18). It was recently shown that piRNA exists in somatic cells and that the biogenesis of piRNAs in somatic cells uses a primary pathway that does not involve the ping-pong amplification mechanism (17, 19–22).
To investigate the unique aspect of piRNA biogenesis and function in somatic cells, we studied Yb, a gene that is specifically expressed in somatic cells in the Drosophila gonad and regulates piwi in somatic niche cells in controlling both germ line and somatic stem cell self-renewal (23–26). Yb is so far the only gene known to regulate the coordinated self-renewing divisions of two different types of stem cells within an organ (23–25). It achieves this by regulating Piwi and Hedgehog expression in somatic niche cells. Piwi and Hedgehog then mediate two separate signaling pathways downstream of Yb to control the division of germ line and somatic stem cells (the two main constituent types of stem cells in the Drosophila ovary). Loss of Yb function eliminates germ line stem cells and reduces somatic stem cell division, whereas overexpression of Yb increases germ line stem cell number and causes somatic stem cell overproliferation. In addition to its stem cell function, Yb is required for follicle cell differentiation by interacting with the Notch group of cell-cell signaling genes (27), consistent with its expression in all somatic cells in the ovary (26).
The biochemical mechanism by which Yb achieves its regulation of germ line and somatic stem cell division has also begun to be revealed. Yb encodes a protein with localized regions bearing similarity to DEAD/DEAH box RNA helicases and a Tudor domain (23). Recently, we discovered that Yb is localized to a novel cytoplasmic sphere of ∼1.5 μm in diameter, the Yb body, that is exclusively present in the somatic cells in the ovary and testis (26). Because Yb is a Tudor domain-containing protein that genetically interacts with Piwi and frequently associates with RNA-rich spots in follicle cells (26), and because other Tudor domain-containing proteins are known to be involved in the piRNA pathway (28), we examined the possible involvement of Yb in the piRNA pathway. Because Piwi is a nuclear protein but Yb is a cytoplasmic protein, we examined whether Yb regulates Piwi via other piRNA pathway components in the cytoplasm, such as Armitage (Armi), which is expressed in somatic cells.
Among piRNA pathway components, Armi is a putative RNA helicase in the cytoplasm of both germ line and follicle cells in the Drosophila ovary (29). We noticed from the micrographs of Cook et al. (30) that, in follicle cells, Armi is localized to discrete spots that resemble the Yb body. Hence, we examined the potential colocalization and the relationship between Armi and Yb to explore the potential link between Yb and the piRNA pathway. Here, we report that Armi indeed localizes to the Yb body, where it is associated with Yb. In Yb mutants, Armi is mislocalized, and surprisingly, Piwi is drastically reduced in abundance and fails to enter the nucleus in somatic cells. Moreover, the biogenesis of piRNAs and endo-siRNAs in the somatic cells is severely disrupted, and transposons are desilenced. These observations indicate a crucial role of Yb and the Yb body in piRNA biogenesis.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Culture
All strains were grown at 25 °C on standard Drosophila medium. The following fly strains were used. w1118 served as the WT strain. Yb72 was previously described as a Yb-null allele (23); 5×FLAG-Yb transgenic w1118 flies, carrying a fully functional FLAG-Yb gene (26), were used in immunofluorescence staining and co-immunoprecipitation.
Immunohistochemistry
Ovaries were dissected, fixed, and stained as described previously (31). For immunofluorescence staining, the following primary antibodies were used: monoclonal mouse anti-FLAG M2 (1:300; Sigma-Aldrich), polyclonal rabbit anti-FLAG F7425 (1:500; Sigma-Aldrich), polyclonal rabbit anti-Armi-NT1 (1:500; a gift from W. E. Theurkauf (30)), monoclonal mouse anti-AGO3 (1:500; a gift from M. C. Siomi (32)), monoclonal mouse anti-PIWI (1:200; a gift from M. C. Siomi (32)), and polyclonal rabbit anti-Aub (1:1000; a gift from P. Lasko). Alexa Fluor 488- or 568-conjugated goat anti-rabbit and anti-mouse IgG secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and were used at 1:500 dilution. Immunofluorescently labeled samples were also counterstained with DAPI as described previously (31). Images were taken using a Leica TCS SP5 spectral confocal microscope in the sequential scanning mode and then processed using Photoshop (Adobe).
Immunoprecipitation
The ovaries were dissected in ice-cold PBS and homogenized in MCB buffer (100 mm KOAc, 0.1% Triton X-100, 50 mm HEPES, pH 7.4, 2 mm Mg(OAc)2, 10% glycerol, 1 mm DTT, and 1× Complete mini EDTA-free protease inhibitor mixture (Roche Applied Science)). The ovarian lysates were collected by centrifugation at 10,000 × g for 30 min at 4 °C. Immunoprecipitation was performed using mouse anti-FLAG M2 affinity gel (Sigma) or rabbit anti-Armi-NT1 antibody with protein A/G-agarose beads (Santa Cruz Biotechnology). Immunoprecipitates were washed three times with MCB buffer at 4 °C, eluted from the beads in 2× SDS sample buffer, and assayed by Western blot analysis. For Western blotting, the rabbit anti-Armi-NT1 and the monoclonal mouse anti-FLAG M2 antibodies were used at 1:5000 and 1:1000 dilutions, respectively.
Quantitative RT-PCR
RNA was isolated from Drosophila ovaries, and quantification of the transposon transcripts by quantitative RT-PCR was performed as described previously (20). The following primer pairs were used: 412, 5′-CACCGGTTTGGTCGAAAG-3′ (forward) and 5′-GGACATGCCTGGTATTTTGG-3′ (reverse); Accord, 5′-ACAATCCACCAACAGCAACA-3′ (forward) and 5′-AAAAGCCAAAATGTCGGTTG-3′ (reverse); Accord2, 5′-TTGCTTTCGGACTTCGTCTT-3′ (forward) and 5′-TTCCACAACGAAAACAACCA-3′ (reverse); Blood, 5′-TGCCACAGTACCTGATTTCG-3′ (forward) and 5′-GATTCGCCTTTTACGTTTGC-3′ (reverse); Diver, 5′-GGCACCACATAGACACATCG-3′ (forward) and 5′-GTGGTTTGCATAGCCAGGAT-3′ (reverse); Diver2, 5′-CTTCAGCCAGCAAGGAAAAC-3′ (forward) and 5′-CTGGCAGTCGGGTGTAATTT-3′ (reverse); gtwin, 5′-TTCGCACAAGCGATGATAAG-3′ (forward) and 5′-GATTGTTGTACGGCGACCTT-3′ (reverse); gypsy, 5′-GTTCATACCCTTGGTAGTAGC-3′ (forward) and 5′-CAACTTACGCATATGTGAGT-3′ (reverse); gypsy6, 5′-GACAAGGGCATAACCGATACTGTGGA-3′ (forward) and 5′-AATGATTCTGTTCCGGACTTCCGTCT-3′ (reverse); HeT-A, 5′-CGCGCGGAACCCATCTTCAGA-3′ (forward) and 5′-CGCCGCAGTCGTTTGGTGAGT-3′ (reverse); Hopper, 5′-GGCTGGCTTCAACAAAAGAA-3′ (forward) and 5′-GGACTCCCGAAAACGTCATA-3′ (reverse); I-element, 5′-GACCAAATAAAAATAATACGACTTC-3′ (forward) and 5′-AACTAATTGCTGGCTTGTTATG-3′ (reverse); Invader1, 5′-GTACCGTTTTTGAGCCCGTA-3′ (forward) and 5′-AACTACGTTGCCCATTCTGG-3′ (reverse); Max, 5′-TCTAGCCAGTCGAGGCGTAT-3′ (forward) and 5′-TGGAAGAGTGTCGCTTTGTG-3′ (reverse); mdg1, 5′-AACAGAAACGCCAGCAACAGC-3′ (forward) and 5′-CGTTCCCATGTCCGTTGTGAT-3′ (reverse); R1A1, 5′-AATTCCCGAGCTGTGCTAGA-3′ (forward) and 5′-GTCTCAAGGCACCTTTCAGC-3′ (reverse); rp49, 5′-CCGCTTCAAGGGACAGTATCTG-3′ (forward) and 5′-ATCTCGCCGCAGTAAACGC-3′ (reverse); Rt1a, 5′-CCACACAGACTGAGGCAGAA-3′ (forward) and 5′-ACGCATAACTTTCCGGTTTG-3′ (reverse); ZAM, 5′-ACTTGACCTGGATACACTCACAAC-3′ (forward) and 5′-GAGTATTACGGCGACTAGGGATAC-3′.
Small RNA Cloning and Analysis
To prepare small RNA libraries, low-molecular-weight RNA was isolated from the adult ovaries of w1118 and the Yb72/Yb72 mutant using a microRNA isolation kit (Ambion). Small RNAs ranging in size between 16 and 29 nucleotides (nt) (below 2 S rRNA) were gel-purified, and small RNA libraries were prepared using a small RNA sample prep kit (Illumina) according to the alternative v1.5 protocol. The clones were sequenced using Genome Analyzer II. Only sequences perfectly matching the Drosophila melanogaster release 5 genome (excluding Uextra) were analyzed. Libraries were normalized using the total number of miRNAs to allow for cross-analysis between the mutant and control. After removal of abundant cellular noncoding RNA and miRNAs, we mapped 18–32-nt small RNAs to a subset of known piRNA clusters and to the complete collection of D. melanogaster transposable elements (Repbase) (33).
Small RNA Annotation
Over 22 million sequences were obtained for each library (WT and Yb mutant). The 3′-linker was clipped from the sequences, and the clipping rates for the WT and Yb mutant were ∼80 and ∼70%, respectively. The sequences ranging 18–32 nt in size were extracted. The sequences were mapped to the D. melanogaster release 5 genome (excluding Uextra) using perfect match criteria. Only genome-matching sequences were analyzed. Prior to any analysis, sequences that could be mapped to rRNA, tRNA, small nucleolar RNA, snRNA, and noncoding RNA (FlyBase annotation and UCSC RepeatMasker annotation) were removed. Considering that the noncoding RNA annotation is not completed so far, up to two mismatches were allowed with insertion/deletion included. After removal of cellular RNA contamination, we mapped small RNAs to annotated miRNA precursors in miRBase using perfect match criteria. After removal of miRNA sequences, the small RNAs were used for piRNA cluster and transposable element annotation analysis. The number of small RNA sequences obtained in each step is found in supplemental Table S1.
Small RNA Normalization
Although both WT and Yb mutant samples underwent the same preparation procedures, sequencing depths varied between these two samples, which prevented direct comparison of small RNA levels between the two libraries. Because the global trend of the miRNA expression profile between those two libraries was similar (supplemental Table S2), we assumed that the miRNA pathway is unaffected in the Yb mutant and normalized both libraries to the level of total miRNAs. All calculations involved in piRNA cluster analysis and transposon annotation were based on the normalized libraries.
piRNA Cluster Analysis
To assess the effect of Yb mutation on piRNAs from individual clusters, we analyzed the expression of piRNAs from the following 17 known clusters: 15 piRNA clusters from Hannon and co-workers (21), which produce the greatest number of piRNAs according to their data; one piRNA cluster (Traffic Jam) from Siomi and co-workers (19); and one piRNA cluster (OSS2) located in ChrX (11088517–11095451) from Lai and co-workers (22). These 17 piRNA clusters comprise 12 germ line cell-enriched, two intermediate, and three somatic cell-enriched clusters. To avoid the ambiguous assignment of small RNAs, we only consider the uniquely mapped small RNAs, i.e. those that can be perfectly mapped only to one position on the D. melanogaster release 5 genome.
Transposon Analysis
For transposable element analysis, only 23–28-nt small RNAs were selected and mapped to the complete collection of D. melanogaster transposable elements (Repbase) (33). The mapping allows for up to three mismatches without including internal insertions or deletions to account for the strain difference and variation occurrence within elements.
RESULTS
Armi, an RNA Helicase Involved in the piRNA Pathway, is Localized in the Yb Body in Ovarian Follicle Cells
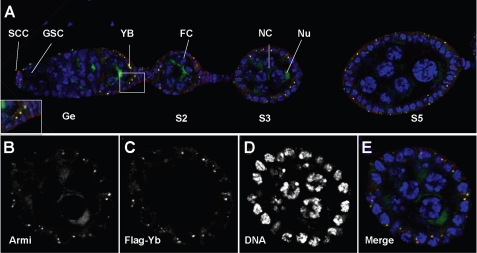
To determine whether Armi and Yb are colocalized to the Yb body, we conducted immuno-colocalization experiments using WT flies carrying an eye color mutation (w1118) and a fully functional 5×FLAG-tagged Yb gene (26). Double labeling of ovaries from these transgenic flies with anti-FLAG and anti-Armi antibodies revealed that Armi not only was enriched in germ line cells as shown previously (30) but also colocalized with the Yb body in somatic cap cells and follicle cells (Fig. 1), as we suspected. The Armi-Yb double staining of the Yb body highlights the somatic cells in both germaria and egg chambers. Every Armi spot colocalizes with Yb, and no Armi spot exists outside the Yb body.
FIGURE 1.
Armitage is localized in the Yb body in ovarian somatic cells. Shown is the immunofluorescence costaining of FLAG-Yb (red) and Armi (green) in a 5×FLAG-Yb transgene ovariole (A) and a stage 4 egg chamber (B–E). DNA was labeled with DAPI (blue). Armi was enriched in the nuage (Nu) in nurse cells (NC) and colocalized with Yb in somatic cells in the Yb body (YB). The inset in A is a magnified view of the boxed area in A. GSC, germ line stem cells; SCC, somatic cap cells; FC, follicle cell; Ge, germarium. S2, S3, and S5 designate stage 2, 3, and 5 egg chambers, respectively.
In contrast to Armi, Piwi was localized to the nucleus in both germ line and somatic cells (supplemental Fig. 1), whereas Ago3 and Aub were enriched in the nuage of germ line cells but were not detectable in somatic cells (supplemental Fig. S1), as reported previously. None of them colocalized with Yb in somatic cells. The complete colocalization of Armi and Yb in ovarian somatic cells indicates that Yb might be involved in the somatic piRNA pathway via Armi as a close interactor.
Yb and Armi Are in the Same Molecular Complex
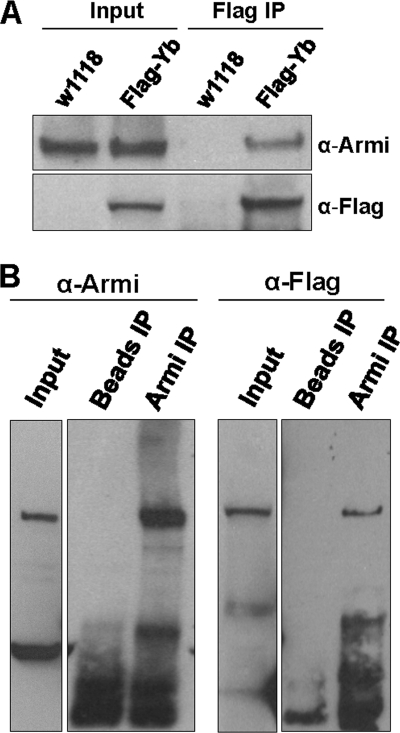
Although the same staining pattern of Armi in follicle cells was observed in an earlier study, it was dismissed as nonspecific labeling because the same staining pattern was detected in Armi1/Armi72.1 mutants (30). This could be due to the fact that Armi1 and Armi72.1 are not protein-null alleles (30). To verify the colocalization of Armi and Yb and to determine whether Armi is in a molecular complex with Yb, we used an anti-FLAG antibody to immunoprecipitate FLAG-Yb complexes from the ovarian lysate of FLAG-Yb transgenic w1118 flies, using nontransgenic w1118 flies as a negative control. Western blot analysis indicated that Armi was co-immunoprecipitated from the lysates of the transgenic flies but not from those of the control flies (Fig. 2A). Hence, Armi was associated with Yb. This result was confirmed by a reciprocal immunoprecipitation of the ovarian lysate from FLAG-Yb transgenic flies with anti-Armi antibody, which co-immunoprecipitated FLAG-tagged Yb (Fig. 2B). These data verified the colocalization of Yb and Armi in the Yb body because Yb is not detectable outside the Yb body (26). Moreover, these results demonstrate that Yb and Armi physically interact in the same molecular complex. This close interaction between Yb and Armi suggests that Yb is likely a component of the piRNA pathway and may be involved in piRNA biogenesis through interaction with Armi.
FIGURE 2.
Armi and Yb are in the same molecular complex. A, co-immunoprecipitation of Armi with FLAG-tagged Yb. The input was the ovarian lysates from FLAG-Yb transgenic and nontransgenic w1118 flies, and the Yb complexes were co-immunoprecipitated with the anti-FLAG antibody. The upper panel was stained for Armi, which was present in both the transgenic and nontransgenic input as well as in the FLAG-immunoprecipitated complex of transgenic flies, but not in nontransgenic flies. The lower panel shows the same blot stained for FLAG-Yb. B, reciprocal co-immunoprecipitation of FLAG-Yb with Armi. The input was the ovarian lysate from FLAG-Yb transgenic females run on separate gels. The Armi-Yb complexes were co-immunoprecipitated with the anti-Armi antibody. The ovarian lysate of FLAG-Yb transgenic flies immunoprecipitated (IP) with beads but without antibody as a negative control. The left panels were stained for endogenous Armi, and the right panels were stained for FLAG-Yb.
piRNA Pathway Components Armi and Piwi, but Not Ago3 or Aub, Are Mislocalized in Yb Mutants
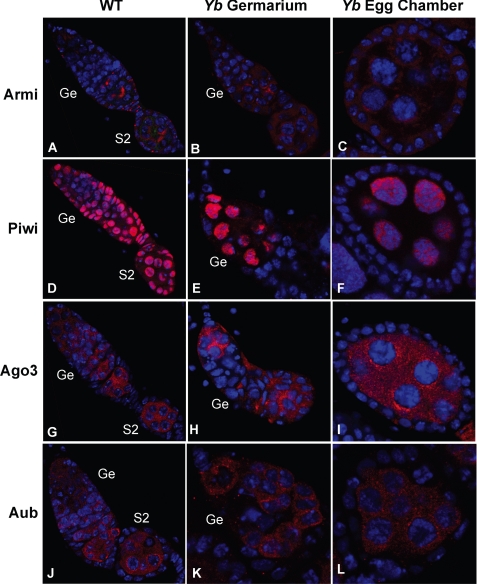
To further examine the potential role of Yb in the piRNA pathway, we examined whether the expression and localization of the piRNA pathway proteins Armi, Piwi, Ago3, and Aub are affected in the Yb mutant background. In contrast to the distinct localization of Armi in the Yb body in the WT ovary (Fig. 3A), Armi was no longer localized to the Yb body in Yb72/Yb72 somatic cells but became evenly distributed in their cytoplasm, even though its germ line localization was unaffected (Fig. 3, B and C). Remarkably, Piwi localization was also drastically affected, specifically in Yb72/Yb72 somatic cells; it was lost from the nucleus and was not significantly above the background in the cytoplasm (Fig. 3, D–F). Expectedly, Ago3 and Aub were still not expressed in the soma, and no detectable change was observed for their staining pattern in the germ line (Fig. 3, G–L). These data suggest that the Yb protein is required for localization of Armi as well as the localization and expression and/or stability of Piwi, specifically in ovarian somatic cells.
FIGURE 3.
piRNA pathway components Armi and Piwi, but not Ago3 or Aub, are mislocalized in the Yb mutant. Shown is the immunofluorescence staining of Armi (A–C), Piwi (D–F), Ago3 (G–I), and Aub (J–L) in WT germaria and early-stage egg chambers (A, D, G, and J) and in Yb mutant germaria (B, E, H, and K) and early-stage egg chambers (C, F, I, and L). Armi and Piwi were mislocalized in somatic cells in the Yb mutant, whereas Ago3 and Aub were not affected. Ge, germarium; S2, stage 2 egg chambers.
Yb Is Required for piRNA Biogenesis in Somatic Cells
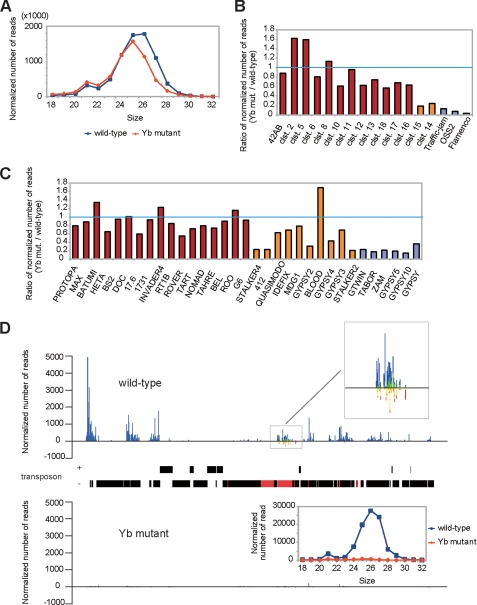
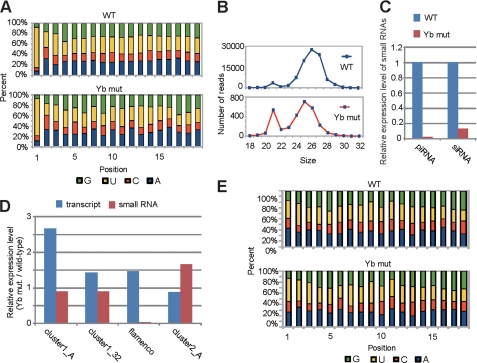
To directly assess the impact of Yb mutation on the piRNA pathway, we sequenced small RNA libraries prepared from an equal quantity of WT (w1118) and Yb72/Yb72 mutant ovaries (supplemental Table S1). The Yb mutant ovaries showed a decrease in the abundance of 26–27-nt piRNAs that were enriched in Piwi-associated piRNAs, which are expressed in both germ line and somatic cells (Fig. 4A) (34). However, the levels of 23–24-nt piRNAs that were enriched in Aub/Ago3-bound piRNAs, which are expressed only in the germ line, were unchanged. Mapping of the small RNAs to known piRNA clusters or retrotransposons (19–22, 34) showed that piRNAs that are expressed in somatic cells were significantly decreased, whereas piRNAs that are expressed in the germ line were much less affected (Fig. 4, B and C). These results suggest that only Piwi-associated somatic piRNAs are affected in the mutant. For example, the flamenco locus is known to generate predominately Piwi-associated piRNAs that are somatically expressed to regulate retrotransposons (34). In Yb mutants, small RNAs mapped to this locus were decreased by >38.4-fold (Figs. 4, B and D, and 5C). However, the flamenco small RNAs that remained in the mutant still showed a bias toward U at the first position (Fig. 5A), which is characteristic of piRNAs. Moreover, the peak of these piRNAs shifted from 26 to 25 nt (Fig. 5B), suggesting the depletion of longer Piwi-associated somatic piRNAs in the mutant and the relative “enrichment” of shorter germ line Aub/Ago3-associated piRNAs in the remaining pool. Together, these results suggest that Yb is essential for the expression of Piwi-associated somatic piRNAs.
FIGURE 4.
Yb is involved in the biogenesis of somatically expressed piRNAs. A, length distribution of small RNAs (excluding miRNAs) in WT and Yb mutant ovaries. The blue lines and red lines show the distribution of small RNAs in the WT and Yb mutant, respectively. Note that the level of 26–27-nt small RNAs that are enriched in Piwi-associated piRNAs was decreased in the Yb mutant. B, expression of piRNA cluster-derived small RNAs in the WT and Yb mutant. The relative expression levels of small RNAs derived from 17 previously reported piRNA clusters (clst.) are shown. Small RNAs uniquely mapped to these clusters were used for analysis. These clusters are grouped into three groups according to the expression preference of their piRNAs (19, 21, 22), with the germ line cell-enriched class colored in red, the intermediate class colored in orange, and the soma-enriched class colored in violet. The soma-enriched class clusters were significantly decreased in Yb mutant ovaries, the intermediate clusters were moderately decreased, and the germ line cell-enriched clusters were essentially unchanged. C, expression of transposon-derived small RNAs in the WT and Yb mutant. Shown are the relative expression levels of piRNAs generated by each transposon. Small RNAs were mapped to the consensus sequence of each transposon, allowing up to three mismatches. These transposons are grouped into three classes according to the expression preference of their piRNAs (21), with the germ line cell-enriched class colored in red, the intermediate class colored in orange, and the soma-enriched class colored in violet. The expression levels of piRNAs from the soma-enriched class were significantly decreased in Yb mutant ovaries, whereas those from the germ line cell-enriched class were essentially unchanged. D, piRNA densities distributed along the flamenco locus in Yb mutant and WT ovaries, with a map of the locus drawn to the scale corresponding precisely to the position of the x axis of the density charts. In addition, the map distinguishes plus-strand from minus-strand and marks the gypsy6 transposons in the flamenco locus in red and the non-gypsy6 transposons in black. Uniquely mapped small RNAs are plotted against the flamenco locus with a window size of 1 nt. The blue lines represent small RNAs produced from the plus-strand, whereas the red lines represent those produced from the minus-strand. The putative endo-siRNAs in the gypsy6 sublocus is enlarged in the upper inset, in which green bars and yellow bars represent 21-nt putative endo-siRNAs from the plus- and minus-strands, respectively. The lower inset shows the length profiles of flamenco-derived small RNAs in the WT (blue) and Yb mutant (red).
FIGURE 5.
Yb is involved in the biogenesis of flamenco-derived piRNAs and endo-siRNAs. A, nucleotide composition of flamenco-derived small RNAs from the WT and Yb mutant. Only small RNAs uniquely mapped to the flamenco locus were analyzed. B, size profiles of flamenco-derived small RNAs in the WT and Yb mutant. The blue curves and red curves represent small RNAs in WT and Yb mutant ovaries, respectively. C, relative expression levels of piRNAs and siRNAs from the flamenco locus in WT and Yb mutant flies. The ratio of the piRNA/siRNA expression level to miRNA expression level (with the WT level set as 1) is shown. Note the drastic decrease in the expression of both flamenco piRNAs and endo-siRNAs in the mutant. D, expression of putative piRNA precursors and small RNAs (including piRNAs and endo-siRNAs) in the WT and Yb mutant. The ratio of the expression level in the Yb mutant to that in the WT is shown. The expression level of putative piRNA precursors was normalized using the level of RP49. The positions and primer sequences analyzed here are found in Ref. 43. E, nucleotide composition of flamenco-derived 21 nt small RNAs from WT and Yb mutant ovaries. Only small RNAs uniquely mapped to the flamenco locus were analyzed.
To narrow down the step(s) of piRNA biogenesis at which Yb is involved, we examined the expression of the putative piRNA precursors of flamenco, with precursors of germ line cluster 1 (clusters 1-A and 1–32) and cluster 2 (cluster 2-A) as negative controls. Although flamenco-derived piRNAs were decreased by 38.4-fold in the Yb mutant (Fig. 5C), its precursor level remained unchanged (Fig. 5D). In contrast, piRNAs derived from the germ line clusters were not decreased in the Yb mutant, even though their precursor levels showed minor fluctuations (<2.7-fold) in the Yb mutant (Fig. 5D). This indicates that Yb is not required for the transcription of the somatic piRNA precursors but is involved in the processing of piRNAs from their precursors and/or the stability of the mature piRNAs. Given that Yb is required for proper expression and localization of Piwi in ovarian somatic cells, it is possible that Yb regulates piRNA biogenesis and/or stability via Piwi.
Yb Appears to Be Required for Endogenous siRNA Biogenesis in Somatic Cells
Endogenous siRNAs are also known to regulate retrotransposons. Among small RNAs from the flamenco locus, we observed a 21-nt peak (Fig. 5B) and wondered whether they were endogenous siRNAs (endo-siRNAs). We therefore selected these small RNAs and analyzed their sequence feature. These small RNAs did not show the first U bias at their 5′-end (Fig. 5E), which is characteristic of retrotransposon-derived endo-siRNAs (22). Moreover, these small RNAs corresponded to the sense and antisense strands at similar frequencies (Fig. 4D), suggesting that they may be the products of a bidirectionally transcribed double-stranded RNA precursor(s). To our knowledge, this is the first report of endo-siRNAs in the flamenco locus. This endo-siRNA cluster corresponded exactly to the gypsy6 transposon (Fig. 4D). Interestingly, in the Yb mutant, endo-siRNAs were decreased by 8.0-fold when normalized against miRNAs (Fig. 5C; see “Experimental Procedures”). This suggests that Yb is required for the generation and/or stability of the endo-siRNAs from the flamenco locus.
Somatic Retrotransposons Are Desilenced in Yb Mutant
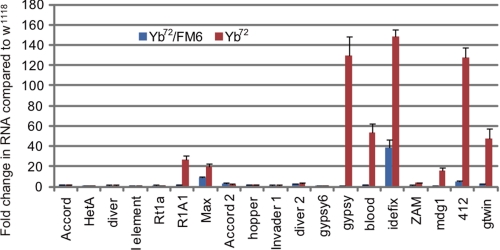
It has been shown that Piwi interacts with the flamenco-derived piRNAs in somatic cells and suppresses somatically active transposons, including gypsy, Idefix, and ZAM (34–37). To further explore the role of Yb in the somatic Piwi-piRNA pathway, we investigated whether Yb is involved in transposon silencing. We examined the expression of 19 transposons in Yb72 homozygous and heterozygous (w1118,Yb72/FM6) control ovaries by quantitative RT-PCR (Fig. 6). Consistent with our immunofluorescence data showing that the Piwi protein was mislocalized from the nucleus of the somatic cell, most Piwi-dependent transposons were up-regulated in the Yb mutant compared with the heterozygous control. Among them, the level of Idefix, gypsy, and 412 RNAs increased most dramatically in the mutant, to ∼140-fold of the WT level. Particularly, the level of the Idefix RNA was increased close to 40-fold, even in the heterozygous control, suggesting that the Yb suppression toward this transposon is dose-sensitive. In contrast, germ line-active transposons were not noticeably affected. However, transposons R1A1 and Max, two members of the Group I germ line-active transposon family that are silenced by Ago3 (20), showed ∼20-fold increases in their expression in the Yb mutant. This mild degree of desilencing of these two germ line transposons could reflect the fact that they are also expressed in somatic cells.
FIGURE 6.
Somatically active transposons are desilenced in Yb mutants. Quantitative RT-PCR was performed to determine the expression of multiple transposons, relative to rp49, in WT (w1118), Yb72/FM6, and Yb mutant ovaries. Transcript levels from WT ovaries were set as 1, and -fold changes are indicated. The error bars indicate S.D.
DISCUSSION
Although exciting progress has been made in understanding the piRNA pathway in the germ line, less is known about this pathway in somatic cells. In this study, we have reported that Yb is a novel component of the somatic piRNA pathway. Because Yb is localized only in the Yb body in somatic cells (26), our results further implicate the Yb body as a key site in the cytoplasm for piRNA biogenesis in ovarian somatic cells.
How is Yb involved in the piRNA pathway? We showed previously that Yb genetically acts upstream of Piwi to regulate its expression in somatic cells (24, 25), yet physically, Armi is the only known piRNA pathway component that colocalizes and physically interacts with Yb (Figs. 1 and 2). While this manuscript was in preparation and under consideration, Olivieri et al. (38) and Saito et al. (39) also reported the Armi-Yb interaction and their role in the piRNA pathway. In addition, Haase et al. (40) reported the role of Armi in Piwi and piRNA expression. These studies verified our observations. It is likely that Yb regulates Piwi via Armi, a RNA helicase (29, 30). It is possible that Yb first recruits Armi to the Yb body, where the Armi-Yb complex, possibly involving other factors, might then serve as a site for the biogenesis and/or loading piRNAs and/or other factors to Piwi. The resulting Piwi-piRNA complex then enters the nucleus to achieve epigenetic regulatory functions (17, 18). Such epigenetic regulation then leads to transposon silencing in somatic cells and niche cell function, as reported previously (19, 23). Without Yb, Armi might fail to facilitate the biogenesis and/or loading of piRNA to Piwi. The unloaded Piwi might then fail to enter the nucleus and is subject to degradation. Of course, other possibilities exist, such as Armi-Yb interaction leading to the regulation of transcription or translation of Piwi. In any case, such regulation is specific to Piwi because Aub and Ago3 are not expressed in somatic cells in the ovary, and Yb mutations do not affect the expression or localization of Aub and Ago3.
Although Yb has long been regarded as a protein required in somatic niche cells to regulate germ line and follicle stem cell division (23, 24), its underlying molecular mechanism remains elusive. Our study reveals a molecular mechanism through which Yb functions in the niche cells: it regulates Piwi activity, possibly via Armi. The activated Piwi then enters the nucleus to epigenetically regulate the niche cell genome (41), which ensures the genome integrity and defines the niche signaling function toward stem cells.
In addition to its involvement in the piRNA pathway, Yb appears to be also involved in the endo-siRNA pathway. The gypsy6 endo-siRNA, which is decreased by 8-fold in the Yb mutant, is from the flamenco cluster. The flamenco cluster generates both endo-siRNAs and piRNAs. It is possible that the precursor transcripts of the piRNA pathway also serve as precursors of the endo-siRNA pathway, either in their original forms or as processed intermediates. Alternatively, piRNAs can target the single-stranded precursor of endo-siRNAs to form mature double-stranded precursors to produce endo-siRNAs. In either case, the dual role of Yb is reminiscent of the possible involvement of Piwi in the miRNA pathway (5). These observations together point to an “inconvenient” fact, i.e. the specificity of Ago and Piwi proteins with respect to the siRNA/miRNA pathway versus piRNA pathway is only in a relative sense, just like the specificity of Drosophila Ago1 and Ago2 for the siRNA versus miRNA pathway is also a relative sense, as shown recently (42). In fact, our data suggest that Yb is required for transposon silencing, likely via both the piRNA and endo-siRNA pathways. Further investigation of the Yb-mediated mechanism should reveal a new dimension of the biogenesis and regulatory function of the piRNA pathway.
Supplementary Material
Acknowledgments
We thank Drs. Haruhiko Siomi, William Theurkauf, and Paul Lasko for providing antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HD33760 and DP1OD006825 (to H. L.). This work was also supported by the Mathers Award (to H. L.) and a fellowship from the Japan Society for the Promotion of Science (to T. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
The small RNA sequences reported in this paper were deposited in Gene Expression Omnibus under accession no. GSE 26407.
- piRNA
- Piwi-associated RNA
- Aub
- Aubergine
- Ago3
- Argonaute 3
- Armi
- Armitage
- miRNA
- microRNA
- nt
- nucleotide(s).
REFERENCES
- 1. Thomson T., Lin H. (2009) Annu. Rev. Cell Dev. Biol. 25, 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin H., Spradling A. C. (1997) Development 124, 2463–2476 [DOI] [PubMed] [Google Scholar]
- 3. Cox D. N., Chao A., Baker J., Chang L., Qiao D., Lin H. (1998) Genes Dev. 12, 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris A. N., Macdonald P. M. (2001) Development 128, 2823–2832 [DOI] [PubMed] [Google Scholar]
- 5. Megosh H. B., Cox D. N., Campbell C., Lin H. (2006) Curr. Biol. 16, 1884–1894 [DOI] [PubMed] [Google Scholar]
- 6. Aravin A. A., Hannon G. J., Brennecke J. (2007) Science 318, 761–764 [DOI] [PubMed] [Google Scholar]
- 7. Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Lin H., Matsuda Y., Nakano T. (2004) Development 131, 839–849 [DOI] [PubMed] [Google Scholar]
- 8. Deng W., Lin H. (2002) Dev. Cell 2, 819–830 [DOI] [PubMed] [Google Scholar]
- 9. Unhavaithaya Y., Hao Y., Beyret E., Yin H., Kuramochi-Miyagawa S., Nakano T., Lin H. (2009) J. Biol. Chem. 284, 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma A. K., Nelson M. C., Brandt J. E., Wessman M., Mahmud N., Weller K. P., Hoffman R. (2001) Blood 97, 426–434 [DOI] [PubMed] [Google Scholar]
- 11. Liu X., Sun Y., Guo J., Ma H., Li J., Dong B., Jin G., Zhang J., Wu J., Meng L., Shou C. (2006) Int. J. Cancer 118, 1922–1929 [DOI] [PubMed] [Google Scholar]
- 12. Palakodeti D., Smielewska M., Lu Y. C., Yeo G. W., Graveley B. R. (2008) RNA 14, 1174–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rossi L., Salvetti A., Lena A., Batistoni R., Deri P., Pugliesi C., Loreti E., Gremigni V. (2006) Dev. Genes Evol. 216, 335–346 [DOI] [PubMed] [Google Scholar]
- 14. Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005) Science 310, 1327–1330 [DOI] [PubMed] [Google Scholar]
- 15. Cox D. N., Chao A., Lin H. (2000) Development 127, 503–514 [DOI] [PubMed] [Google Scholar]
- 16. Pal-Bhadra M., Leibovitch B. A., Gandhi S. G., Rao M., Bhadra U., Birchler J. A., Elgin S. C. (2004) Science 303, 669–672 [DOI] [PubMed] [Google Scholar]
- 17. Yin H., Lin H. (2007) Nature 450, 304–308 [DOI] [PubMed] [Google Scholar]
- 18. Brower-Toland B., Findley S. D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S. C., Lin H. (2007) Genes Dev. 21, 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M. C. (2009) Nature 461, 1296–1299 [DOI] [PubMed] [Google Scholar]
- 20. Li C., Vagin V. V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M. D., Syrzycka M., Honda B. M., Kittler E. L., Zapp M. L., Klattenhoff C., Schulz N., Theurkauf W. E., Weng Z., Zamore P. D. (2009) Cell 137, 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malone C. D., Brennecke J., Dus M., Stark A., McCombie W. R., Sachidanandam R., Hannon G. J. (2009) Cell 137, 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lau N. C., Robine N., Martin R., Chung W. J., Niki Y., Berezikov E., Lai E. C. (2009) Genome Res. 19, 1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. King F. J., Lin H. (1999) Development 126, 1833–1844 [DOI] [PubMed] [Google Scholar]
- 24. King F. J., Szakmary A., Cox D. N., Lin H. (2001) Mol. Cell 7, 497–508 [DOI] [PubMed] [Google Scholar]
- 25. Szakmary A., Cox D. N., Wang Z., Lin H. (2005) Curr. Biol. 15, 171–178 [DOI] [PubMed] [Google Scholar]
- 26. Szakmary A., Reedy M., Qi H., Lin H. (2009) J. Cell Biol. 185, 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson E., Wayne S., Nagoshi R. (1995) Genetics 140, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shoji M., Tanaka T., Hosokawa M., Reuter M., Stark A., Kato Y., Kondoh G., Okawa K., Chujo T., Suzuki T., Hata K., Martin S. L., Noce T., Kuramochi-Miyagawa S., Nakano T., Sasaki H., Pillai R. S., Nakatsuji N., Chuma S. (2009) Dev. Cell 17, 775–787 [DOI] [PubMed] [Google Scholar]
- 29. Tomari Y., Du T., Haley B., Schwarz D. S., Bennett R., Cook H. A., Koppetsch B. S., Theurkauf W. E., Zamore P. D. (2004) Cell 116, 831–841 [DOI] [PubMed] [Google Scholar]
- 30. Cook H. A., Koppetsch B. S., Wu J., Theurkauf W. E. (2004) Cell 116, 817–829 [DOI] [PubMed] [Google Scholar]
- 31. Lin H., Yue L., Spradling A. C. (1994) Development 120, 947–956 [DOI] [PubMed] [Google Scholar]
- 32. Gunawardane L. S., Saito K., Nishida K. M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M. C. (2007) Science 15, 1587–1590 [DOI] [PubMed] [Google Scholar]
- 33. Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005) Cytogenet. Genome Res. 110, 462–467 [DOI] [PubMed] [Google Scholar]
- 34. Brennecke J., Alexei A., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G. J. (2007) Cell 128, 1089–1103 [DOI] [PubMed] [Google Scholar]
- 35. Pélisson A., Song S. U., Prud'homme N., Smith P. A., Bucheton A., Corces V. G. (1994) EMBO J. 13, 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Desset S., Meignin C., Dastugue B., Vaury C. (2003) Genetics 164, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sarot E., Payen-Groschêne G., Bucheton A., Pélisson A. (2004) Genetics 166, 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olivieri D., Sykora M. M., Sachidanandam R., Mechtler K., Brennecke J. (2010) EMBO J. 29, 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K. M., Siomi H., Siomi M. C. (2010) Genes Dev. 24, 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haase A. D., Fenoglio S., Muerdter F., Guzzardo P. M., Czech B., Pappin D. J., Chen C., Gordon A., Hannon G. J. (2010) Genes Dev. 24, 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin H., Yin H. (2008) Cold Spring Harb. Symp. Quant. Biol. 73, 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ghildiyal M., Xu J., Seitz H., Weng Z., Zamore P. D. (2010) RNA 16, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klattenhoff C., Xi H., Li C., Lee S., Xu J., Khurana J. S., Zhang F., Schultz N., Koppetsch B. S., Nowosielska A., Seitz H., Zamore P. D., Weng Z., Theurkauf W. E. (2009) Cell 138, 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.