Abstract
Proteins of the cytohesin/Arno/Grp1 family of Arf activators are positive regulators of the insulin-signaling pathway and control various remodeling events at the plasma membrane. Arno has a catalytic Sec7 domain, which promotes GDP to GTP exchange on Arf, followed by a pleckstrin homology (PH) domain. Previous studies have revealed two functions of the PH domain: inhibition of the Sec7 domain and membrane targeting. Interestingly, the Arno PH domain interacts not only with a phosphoinositide (phosphatidylinositol 4,5-bisphosphate or phosphatidylinositol 3,4,5-trisphosphate) but also with an activating Arf family member, such as Arf6 or Arl4. Using the full-length membrane-bound forms of Arf1 and Arf6 instead of soluble forms, we show here that the membrane environment dramatically affects the mechanism of Arno activation. First, Arf6-GTP stimulates Arno at nanomolar concentrations on liposomes compared with micromolar concentrations in solution. Second, mutations in the PH domain that abolish interaction with Arf6-GTP render Arno completely inactive when exchange reactions are reconstituted on liposomes but have no effect on Arno activity in solution. Third, Arno is activated by its own product Arf1-GTP in addition to a distinct activating Arf isoform. Consequently, Arno activity is strongly modulated by competition with Arf effectors. These results show that Arno behaves as a bistable switch, having an absolute requirement for activation by an Arf protein but, once triggered, becoming highly active through the positive feedback effect of Arf1-GTP. This property of Arno might provide an explanation for its function in signaling pathways that, once triggered, must move forward decisively.
Keywords: G Proteins, Intracellular Trafficking, Kinetics, Liposomes, Phosphatidylinositol, Guanine Nucleotide Exchange Factor, PH Domain, Sec7 Domain, Positive Feedback Loop
Introduction
Guanine nucleotide exchange factors (GEFs)3 promote the exchange of GDP by GTP on small G proteins (1). In many GEFs, the catalytic domain responsible for the nucleotide exchange activity is flanked by domains that promote binding to cellular membranes. Membrane targeting is favorable for the exchange reaction because most small G proteins are anchored to lipid membranes. In addition, GEFs integrate inputs of different nature through quaternary conformational changes. The combination of membrane translocation and quaternary conformational changes is a recurrent theme in signaling molecules that interact with small G proteins (2–10). However, only a few examples have been dissected so far, given the difficulty in handling multidomain proteins and lipid-modified G proteins on reconstituted membranes.
Arno proteins (Arno, cytohesin, and Grp1) are the simplest GEFs for Arfs, a subfamily of small G proteins involved in membrane traffic (11–13). Arno contains a N-terminal coiled-coil region, a central Sec7 domain and a C-terminal pleckstrin homology (PH) domain. Numerous studies suggest a clear division of labor between the Sec7 domain, which is responsible for the exchange activity, and the PH domain, which promotes membrane recruitment. The Sec7 domain of Arno is among the most efficient GEFs in vitro (kcat/Km ∼106 m−1 s−1), and the mechanism by which it expels GDP from Arf1 is known in great detail (14–16). The interaction of the PH domain of Arno with lipids is also well understood. It contains a basic pocket for phosphoinositides, and splice variants that differ by a unique glycine residue are either specific for phosphatidylinositol 3,4,5-trisphosphate (PIP3) or bind equally well to phosphatidylinositol 4,5-bisphosphate (PIP2) or PIP3 (17–19). In addition, a polybasic region downstream of the PH domain interacts with negatively charged lipids, such as phosphatidylserine (PS) (20–22).
The respective roles of the Sec7 and PH domains of Arno in exchange activity and membrane binding are clearly established. However, their biochemical properties lead to a dilemma when considered in a cellular context. The PH domain binds to a combination of lipids (PS + PIP2 or PIP3) that is found at the plasma membrane (18, 19), whereas the Sec7 domain is much more active on Arf1, localized predominantly to the Golgi, than on Arf6, which is found at the cell periphery (17, 23). To explain this discrepancy, one hypothesis is that the preference of Arno for Arf1 versus Arf6 in vitro is counteracted by additional factors in the cell and notably by subcellular localization effects (24). Alternatively, Arno might activate Arf1 at the plasma membrane under specific circumstances. For instance, it has been reported that Arf1 translocates to the plasma membrane in response to insulin or EGF and also has a role in some endocytic events (25–28). In any case, tight control mechanisms are required to prevent random Arf1 activation.
Recent studies reveal two types of mechanism that, in addition to the recognition of phosphoinositides, could regulate Arno: (i) autoinhibition of the Sec7 domain by the PH domain and (ii) binding of Arf6-GTP of Arl4-GTP to the Arno PH domain (29–33). A crystal structure of the Sec7-PH tandem of Grp1 reveals that this very close Arno homologue adopts an autoinhibited conformation in solution, where a C-terminal helix downstream of the PH domain covers the Arf-binding site of the Sec7 domain (30). This implies that Arno family members open at the surface of lipid membranes through quaternary conformational changes to become active. A second unexpected finding is that the PH domain of Arno interacts not only with lipids but also with some Arf proteins (Arf6 and Arl4) in the GTP-bound conformation (29, 31–33). Thus, Arno has a dual character, being not only an exchange factor for Arf proteins through its Sec7 domain but also an Arf effector through its PH domain.
Collectively, these findings raise novel hypotheses for the spatiotemporal activation of Arf proteins by Arno. The interaction of its PH domain with defined Arf subtypes suggests cascades, akin to Rab cascades (34), in which Arno could serve to change the repertoire of Arf subtypes bound to a membrane. The fact that Arno adopts an autoinhibited conformation suggests that its response to phosphoinositides, PS, and Arf subtypes could be highly synergistic because these membrane determinants could cooperate in opening the structure. These putative mechanisms, which are intimately linked to the membrane environment, have been previously inferred from experiments using minimal soluble components (lipid polar headgroups and truncated forms of Arfs) (30, 31). Here we have studied the mutual regulation between full-length Arno and two myristoylated Arf subtypes, Arf1 and Arf6, at the surface of model liposomes. Our results reveal that Arno is very sensitive to the surface density of not only Arf6-GTP but also its own product, Arf1-GTP, and competes with Arf effectors. These results suggest a model in which Arno behaves as a bistable switch, mobilizing a large amount of Arf molecules only under specific circumstances. This model should help in understanding the physiological function of Arno.
EXPERIMENTAL PROCEDURES
Proteins
Full-length human Arno (“three-glycine” form) and its PH domain (amino acids 261–400) were expressed in a pET-8c vector and purified by nickel-NTA chromatography (Qiagen). After elution with imidazole, Arno was dialyzed against 20 mm Tris, pH 7.2, 100 mm NaCl (500 mm in the case of the PH domain), 1 mm MgCl2, 1 mm DTT, and 10% glycerol, followed by ultracentrifugation to remove aggregates. Mutants were generated with the QuikChange kit (Stratagene). Myristoylated Arf1-GDP was purified from Escherichia coli co-expressing bovine Arf1 and N-myristoyltransferase through ammonium sulfate precipitation, DEAE chromatography, and MonoS chromatography (35). Myristoylated Arf6 with a C-terminal hexahistidine tag was purified by nickel-NTA chromatography. In pilot experiments, we compared this form with a non-tagged version of myristoylated Arf6-GTP (36) and observed similar activation of Arno on liposomes. Chromatography on a MonoQ column after protein denaturation showed that purified Arf1 was in the GDP-bound state, whereas Arf6 was in the GTP-bound state. The effect of GDP or GTP addition on the fluorescence of the two proteins confirmed this difference.
Liposomes
Egg phosphatidylcholine, liver phosphatidylethanolamine, brain PS, and brain phosphoinositides were from Avanti Polar Lipids. All liposomes contained 20 mol % phosphatidylethanolamine, 0 or 30 mol % PS, and the indicated percentage of PIP2 or phosphatidylinositol 4-phosphate (up to 5 mol %). The remaining lipid was phosphatidylcholine. Lipids in chloroform were first mixed in pear-shaped glassware. The glassware was attached to a rotary evaporator and immersed in a water bath at 34 °C for 5 min before evaporation. This step improves PIP2 homogenization with other lipids (37). A lipid film was produced by rapid evaporation of chloroform under vacuum. The lipid film was resuspended in 50 mm Hepes, 120 mm potassium acetate, pH 7.2. The suspension (4 mm lipids) was submitted to five cycles of freezing/thawing and extruded through polycarbonate filters (pore size = 0.1 μm). Liposomes were stored at room temperature and used within 2 days.
Nucleotide Exchange Assays
Nucleotide exchange on Arf1 was followed by tryptophan fluorescence (excitation, 297.5 nm; emission, 340 nm) or by FRET between Arf1 and mant-nucleotides (excitation, 297.5 nm; emission, 455 nm). The fluorimeter was equipped with injection and stirring instruments. Experiments were performed at 37 °C in 50 mm Hepes, 120 mm potassium acetate, 1 mm MgCl2, 5 μm CaCl2, 100 μm EGTA, 1 mm DTT, pH 7.2. Calculations suggest that, at the protein/lipid ratio used in the experiments, the majority of the liposome surface should remain accessible. Therefore, the kinetics should not be affected by crowding effects. An Arf molecule (4.8 × 3.8 × 3.7 nm3) occupies a surface of ∼15 nm2 corresponding to a monolayer of ∼20 lipids. At the highest concentration of Arf1 and Arf6 used here (0.4 and 0.9 μm, respectively) the two proteins should cover ∼30 μm lipids (i.e. 30% of the liposome surface, assuming that half of the lipids are in the external leaflet (total lipid concentration = 200 μm)).
Data Analysis
The activation of Arf1 by Arno on liposomes follows complex kinetics because of the involvement of a positive feedback loop. The rate constant shown in the plots corresponds to the initial rate, which was determined from the slope of the initial fluorescence increase or from a semilog plot. When Arf6-GTP was present, the reaction followed first order kinetics, and the rate constant was determined from a monoexponential fit. For very slow reactions, we took into account the slow decay in fluorescence that arises from photobleaching. Most experiments were performed on liposomes containing limiting amounts of PIP2 (0.5 or 1 mol %). We noticed large variations (up to 5-fold) in the absolute activity of wild-type Arno on different preparations of liposomes with the same limiting amount of PIP2 despite the use of protocols to improve its distribution in liposomes (37). The origin of these variations is not well understood but is probably linked to the liposomes because the kinetics of [Δ17]Arf1 activation in solution in the presence of Arno was very reproducible (±10%). Note that on a relative scale, the variations observed between different batches of liposomes were minor compared with the overall range of Arno activity on liposomes of different composition (3 orders of magnitude; see supplemental Fig. S1A). Each figure panel presents kinetics recordings performed the same day with the same liposome batch. When independent experiments were compared, the exchange activity of wild-type Arno was arbitrarily set at 1.
Flotation Experiments
Protein binding to liposome was assessed by a flotation assay (38). The protein of interest (1 μm) was incubated at 37 °C, for 15 min with liposomes (1 mm, containing 0.2% fluorescent lipid 4-nitrobenzo-2-oxa-1,3-diazole-phosphatidylethanolamine) in the same buffer as that used in fluorescence experiments. Thereafter, the suspension was adjusted to 33% (w/v) sucrose and overlaid with two cushions of decreasing density (25 and 0% sucrose, respectively). The sample was centrifuged at 55,000 rpm in a swing rotor (Beckman TLS 55) for 1 or 2 h. The bottom, middle, and top fractions were collected using a Hamilton syringe and analyzed by SDS-PAGE using Sypro Orange staining. 4-Nitrobenzo-2-oxa-1,3-diazole fluorescence of the various fractions showed that about 95% of the lipids was recovered in the top fraction.
Cell Transfection
Arno with mCherry at the N terminus was constructed in a pmCherryC1 vector. Arf1 with a GFP at the C terminus was constructed in a pEGFP-N1 vector. Arf6 with an HA tag at the C terminus was in a pcDNA3 vector (gift of Michel Franco). Telomerase-immortalized human retinal pigment epithelial (RPE) cells were grown in DMEM plus glutaMAX medium (Invitrogen) supplemented with 10% serum and antibiotics. Cells were transfected with Lipofectamine 2000 reagent and processed for immunofluorescence 18 h after transfection. Cells were fixed in 3% paraformaldehyde during 25 min in PBS, treated with 0.5% saponin in PBS, and then treated with 10% horse serum, 0.05% saponin in PBS. Arf6-HA was revealed by an anti-HA 3F10 antibody (Roche Applied Science) and an anti-rat Fluoprobe642 antibody (Interchim). The cells were examined under a Leica SP5 confocal microscope. For morphological analysis, we focused on cells expressing comparable levels of wild-type and mutated forms of Arno. Three independent experiments were used to compare the distribution of Arf1-GFP in cells cotransfected with wild-type Arno or the triple AAE mutant, and about 300–400 cells were examined in each experiment.
RESULTS
The Exchange Activity of Arno on Membranes Cannot Be Explained by Current Models
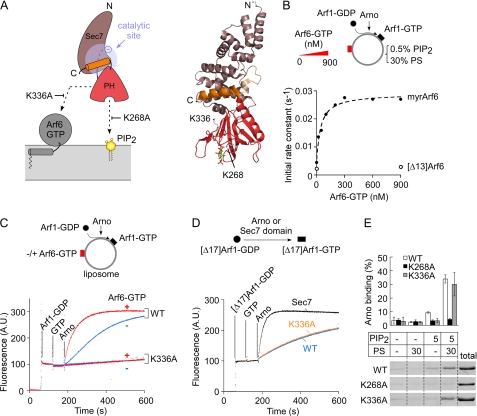
Fig. 1A shows a schematic representation of the Sec7-PH tandem of Arno based on the crystal structure of the close homologue Grp1, which is 85.5% identical in primary sequence with Arno (30). In this conformation, Arno cannot promote nucleotide exchange on Arf because the catalytic site of the Sec7 domain is covered by the C-terminal helix. However, as schematized in Fig. 1A, this conformation is compatible with membrane recruitment; the phosphoinositide-binding site of the PH domain is accessible, and residues of the PH domain (e.g. Lys336) that are believed to contact Arf6-GTP or Arl4-GTP (29, 31, 32) are solvent-exposed. Moreover, the relative position of these two binding sites seems compatible with a dual interaction; the PH domain could simultaneously engage a “bottom” contact with PIP2 and a “side” contact with membrane-bound Arf6-GTP. How these contacts cooperate in opening Arno structure is not known, but these considerations suggest that a membrane surface should provide a favorable environment for the stimulatory effect of Arf6-GTP on Arno.
FIGURE 1.
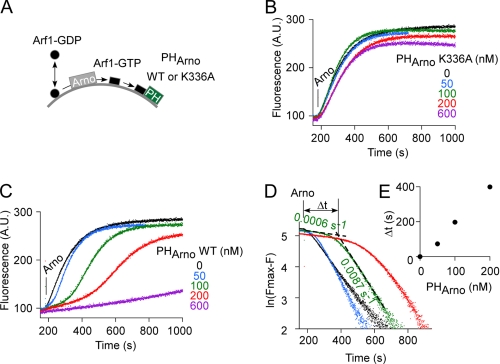
The K336A mutation preserves the basic biochemical properties of Arno but abolishes its activity on liposomes regardless of the presence of Arf6-GTP. A, schematic representation of the Sec7-PH tandem of Arno according to the crystal structure of the close homologue Grp1 (30) (Protein Data Bank entry 2R0D). A possible orientation of the Sec7-PH tandem toward a lipid membrane is shown. B, dose-response curve for the effect of Arf6-GTP or [Δ13]Arf6-GTP on the activity of Arno in the presence of liposomes containing 30 mol % PS, 0.5 mol % PIP2. The hyperbolic fit gives a half-effect of 43 ± 10 nm for Arf6-GTP. C, time course of GDP to GTP exchange on myristoylated Arf1 (400 nm) catalyzed by wild type Arno or the K336A mutant (7.5 nm each) on liposomes. The liposomes (200 μm lipids) contained 1% PIP2 and 30% PS and were supplemented (red) or not (blue) with 300 nm myristoylated Arf6-GTP. D, time course of GDP to GTP exchange on [Δ17]Arf1 (1 μm) in solution catalyzed by wild-type Arno, [K336A]Arno, or the Sec7 domain of Arno (each at 100 nm). E, binding of wild type Arno or mutants (1 μm) to liposomes (1 mm lipids) with the indicated percentage of anionic lipids. Error bars, S.E. A.U., arbitrary units.
To test the effect of the lipid membrane environment on the activity of Arno, we performed nucleotide exchange reactions on liposomes of defined composition using myristoylated Arf1-GDP as a substrate of Arno and myristoylated Arf6-GTP as a potential activator. The three-glycine variant of Arno, which recognizes PIP2, was used in all experiments (17–19). We followed GDP to GTP exchange on Arf1 by tryptophan fluorescence. Because Arf6 was already in the GTP-bound conformation, it did not contribute to the fluorescence change, and its fluorescence level was subtracted for clarity. Fig. 1B and supplemental Fig. S1 show three sets of experiments aimed at testing the effect of Arf6-GTP on the activity of full-length Arno in a minimal membrane environment. Overall, these experiments show that Arf6-GTP acts in synergy with anionic lipids (supplemental Fig. S1, A and B) and that Arf6-GTP is a much more potent activator of Arno on lipid membranes than in solution. On liposomes containing 0.5 mol % PIP2 and 30 mol % PS, the half-maximal stimulatory effect of Arf6-GTP was 43 ± 10 nm (Fig. 1B). This value corresponds to a 300-fold increase in efficiency compared with what has been reported for [Δ12]Arf6-GTP in solution (half-stimulatory effect of 14 μm) (30). In line with this, [Δ13]Arf6-GTP at submicromolar levels had no effect on Arno in our liposome system (Fig. 1B). We conclude that the membrane environment strongly facilitates the stimulatory effect of Arf6-GTP on Arno.
The strong synergy between membrane-bound Arf6-GTP and phosphoinositides were compatible with the model of Fig. 1A. However, other observations were more disconcerting. Fig. 1C compares exchange reactions performed with wild-type Arno and with a mutant (K336A) lacking a lysine of the PH domain critical for Arf6-GTP binding (31). The liposomes contained 30 mol % PS and 1 mol % PIP2. Under these conditions, wild-type Arno displayed robust exchange activity on Arf1, which was further increased 6-fold by the addition of Arf6-GTP. If, as schematized in Fig. 1A, Lys336 of Arno was solely involved in Arf6-GTP binding, the K336A mutation should abolish the stimulatory effect of Arf6-GTP (red traces) without changing the activity level of Arno in the absence of Arf6-GTP (blue traces). Surprisingly, however, the K336A mutation rendered Arno inactive regardless of the presence of Arf6-GTP (Fig. 1C). This observation was puzzling because the K336A mutant was indistinguishable from wild type Arno in other biochemical assays (Fig. 1, D and E). Although mostly autoinhibited, Arno displays some activity in solution on a soluble form of Arf1 ([Δ17]Arf1). This activity is detectable at higher concentration than on liposomes (100 and 7.5 nm, respectively) and gives, when compared with the activity of the isolated Sec7 domain, an estimate of the inhibitory constraint imposed by the PH domain (30). In solution, both wild-type Arno and the K336A mutant displayed 7% of the exchange activity of the isolated Sec7 domain (Fig. 1D), suggesting that the mutation did not induce gross structural defects in Arno. In line with this, we observed no difference between wild-type Arno and the K336A mutant by size exclusion chromatography and limited proteolysis (supplemental Fig. S2). Liposome-binding experiments also showed that the K336A mutant bound to PIP2 plus PS liposomes as well as wild-type Arno (Fig. 1E). In conclusion, the K336A mutation is puzzling because it totally prevents Arno from activating Arf1 on a membrane surface, whereas it affects neither the interaction of Arno with lipids nor the level of autoinhibition as tested in solution.
The surprising behavior of Arno K336A prompted us to examine other mutants. We focused on three surfaces of Arno PH domain (Fig. 2A). Region 1 corresponds to the phosphoinositide-binding site (18). Region 2 includes K336 and other neighboring residues, such as Ile303, that are important for the interaction of the isolated PH domain with Arf6 and Arl4 (29, 31, 32). Region 3 is opposite to region 2 but forms a binding site for Arf1-GTP in the PH domain of ARHGAP21, the sole PH domain whose structure in complex with an Arf protein has been solved (39). As a control, we used the E156K mutant, which lacks the glutamic finger of the Sec7 domain that serves to expel GDP from Arf1 (14–16).
FIGURE 2.
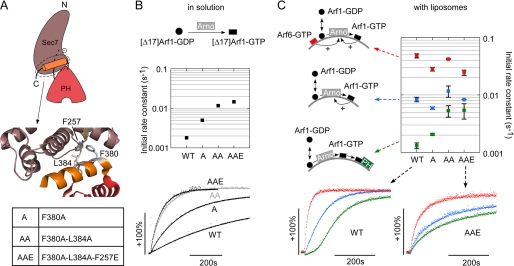
The region around Lys336 in the PH domain plays an essential role in the exchange activity of Arno at the surface of liposomes. A, three regions of the PH domain of Arno were selected for mutagenesis and are shown in the autoinhibited structure of the close Arno homologue Grp1 (Protein Data Bank entry 2R0D) (30). For simplicity, the amino acid numbering of Arno is used. B and C, activity measurements. Kinetics experiments in solution (B) were performed with 1 μm [Δ17]Arf1-GDP, 40 μm GTP, and 100 nm Arno. Kinetics experiments on membranes (C) were performed with 400 nm Arf1-GDP, 40 μm GTP, and 7.5 nm Arno and with (red bars) or without (blue bars) Arf6-GTP (300 nm). The liposomes (200 μm lipids) contained 30% PS and 1% PIP2. The initial rates in B and C were normalized to the values observed for wild-type Arno in solution and on liposomes, respectively. Data are represented as mean ± S.E. (error bars) (n = 3). All values below 5% of the activity of wild-type Arno are roughly equal to the rate of spontaneous exchange (e.g. see Fig. 1C). The large error bar in this region indicates the difficulty in measuring very slow activation kinetics by tryptophan fluorescence.
In agreement with DiNitto et al. (30), all mutants harboring mutations in region 1, 2, or 3 behaved as wild-type Arno when tested on [Δ17]Arf1-GDP in solution, suggesting similar levels of autoinhibition (Fig. 2B). When these mutants were tested on Arf1 in the presence of liposomes displaying a favorable composition (30 mol % PS + 1 mol % PIP2 ± Arf6-GTP), marked differences emerged (Fig. 2C). Mutants in region 3 (E313A and D315A) were indistinguishable from wild-type Arno, whereas mutants in region 2 (I303E, K336A, K336R, and Y290A) except for the conservative mutant Y290F showed a strong defect in activity. Importantly, no mutation in region 2 specifically affected the response of Arno to Arf6-GTP. Instead, we observed a general decrease in activity, and two mutants, I303E and K336A, appeared as inactive as E156K lacking the catalytic glutamic finger or K268A defective in PIP2 binding (Fig. 2C). These results emphasized the key but enigmatic role of region 2 in the activity of Arno at the surface of lipid membranes.
Arno Is Activated by Arf1-GTP through a Positive Feedback Loop
We noticed that, in the absence of Arf6-GTP and at a moderate percentage of PIP2, the kinetics of Arf1 activation by Arno displayed a slight sigmoidal shape (blue trace in Fig. 1C). To explain this observation, we envisaged that Arno could be activated by a positive feedback loop in which new Arf1-GTP molecules produced by the exchange reaction activate in turn the exchange factor. This model, which resembles that of Sos, a Ras GEF with no homology to Arno (7, 40), was appealing because it offered an explanation for the key role of region 2 of Arno PH domain; if the robust activity of Arno on liposomes involves not only the recognition of anionic lipids but also a feedback effect of Arf1-GTP, mutations in Arno that were expected to solely reduce its sensitivity to exogenously added Arf6-GTP should also prevent the feedback effect of Arf1-GTP.
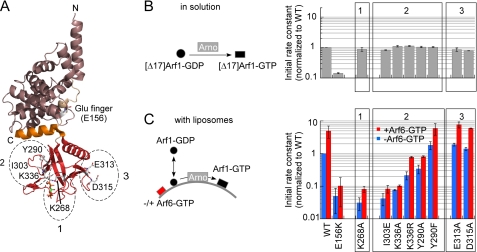
To test the positive feedback loop hypothesis, we used a two-stage protocol (Fig. 3). During the first stage, we activated increasing amounts of Arf1-GDP (0–400 nm) in the presence of Arno, liposomes, and GTP. Once this first pool of Arf1 was activated, we added a second pool of Arf1-GDP (400 nm). Strikingly, the rate of activation of this new pool increased up to 13-fold with the amount of Arf1-GTP that had formed during the first stage. In addition, the characteristic sigmoidal curve observed in the absence of preformed Arf1-GTP disappeared when Arf1-GTP was present (Fig. 3, inset). Therefore, Arf1-GTP, the product of the exchange reaction, apparently favors the activity of Arno toward its substrate, Arf1-GDP. Interestingly, we also noticed that the shape of the activation kinetics became more sigmoidal when we decreased the liposome concentration while keeping the protein concentration constant (supplemental Fig. S3). This observation, which suggests that Arno is sensitive to the surface density rather than the volume concentration of Arf1-GTP, highlights the interfacial character of the exchange reaction.
FIGURE 3.
Arno is activated by a positive feedback loop at the surface of liposomes. To test the influence of membrane-bound Arf1-GTP on the activity of Arno, a two-stage protocol was used. In the first stage, various amounts of Arf1 were activated by Arno (7.5 nm) and GTP (40 μm) at the surface of liposomes (200 μm) containing 1% PIP2 and 30% PS. In the second stage, 400 nm Arf1-GDP was further added. The rate of activation of this second pool of Arf1 increased with the amount of Arf1-GTP that had formed during the first stage. In addition, the activation kinetics loses its sigmoidal shape and becomes more exponential (inset). A.U., arbitrary units.
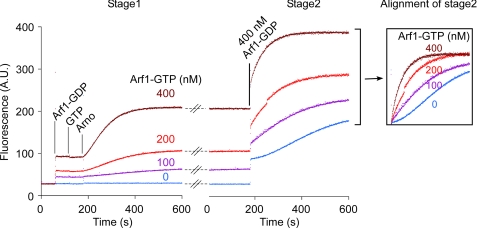
A prediction of the positive feedback loop model is that futile GDP to GDP exchange on Arf1 by Arno should be less favorable than GDP to GTP exchange (7). To test this prediction, we used mant-derivatives of GDP and GTP and followed their binding to Arf1 (initially in the GDP-bound state) by FRET between Arf1 tryptophans and the mant moiety. Mant-GTP bound about 4 times faster than mant-GDP when exchange reactions were conducted with full-length Arno, Arf1-GDP, and liposomes (Fig. 4A, black curves). In contrast, the two nucleotides showed similar binding kinetics when reactions were conducted on liposomes using EDTA to stimulate GDP release (Fig. 4A, red curves) or in solution using Arno Sec7 domain and [Δ17]Arf1-GDP (supplemental Fig. S4). It should be noted that the comparison between mant-GDP and mant-GTP binding kinetics is complex because Arf1-GTP is tightly bound to liposomes in contrast to Arf1-GDP. Thus, the presence of liposomes favors the formation of the GTP-bound form at the expense of the GDP-bound form, an effect that does not occur when a soluble form of Arf1 is used. In addition, reactions on interfaces are extremely sensitive to the rate by which enzymes associate with and dissociate from the membrane (“scooting” versus “hopping” modes (41)). How the membrane context interferes with the dissociation of Arno when nucleotide exchange in Arf1 occurs in the forward or backward direction is difficult to predict (42). Therefore, the above results are compatible with the positive feedback loop model but do not prove it in an unequivocal manner.
FIGURE 4.
Modulation of the exchange activity of Arno on liposomes by the nature of the added nucleotide and by the addition of an Arf1 effector. A, binding of mant-GDP or mant-GTP (7.5 μm) to Arf1-GDP (1 μm) upon the addition of Arno (7.5 or 15 nm) or EDTA (2 mm) in the presence of liposomes (200 μm) containing 1% PIP2 and 30% PS. Mant-GTP binds more rapidly than mant-GDP when the reaction is catalyzed by Arno. B, time course of Arf1-GDP (400 nm) activation by Arno (7.5 nm) on liposomes containing 1% PIP2 and 30% PS in the presence of the C-terminal region of the golgin GMAP-210 (GMAPC-long) or a slightly shorter form that does not interact with Arf1-GTP (GMAPC-short). A.U., arbitrary units.
To circumvent these difficulties, we tested the feedback loop model by a third strategy. We performed exchange reactions in the presence of Arf effectors. By binding to Arf1-GTP, they should prevent Arf1-GTP from stimulating Arno. This strategy does not have the previously mentioned caveats, because it permits the control of surface density of free Arf1-GTP without changing the total protein and lipid concentrations and without changing the direction of the reaction (in contrast to the first and the second strategies, respectively). As an effector, we first chose the C-terminal region of the coiled-coil protein GMAP-210, which forms a stoichiometric complex with Arf1-GTP at the surface of liposomes (43). This construct (GMAPC-long) decreased 10-fold the rate of Arf1 activation by Arno on liposomes, whereas a slightly shorter construct that does not bind to Arf1-GTP on liposomes (GMAPC-short), had almost no effect (Fig. 4B). GMAPC-long affected neither the activation of [Δ17]Arf1 by Arno in solution (supplemental Fig. S5A) nor the activation of Arf1 by EDTA on liposomes (supplemental Fig. S5B).
In another set of experiments, we used the isolated PH domain of Arno as a competitor (Fig. 5A). Under these conditions, strong inhibition of the exchange reaction was observed, an effect that was abolished by the K336A mutation (Fig. 5, compare B and C). These observations suggest that the PH domain inhibited the exchange reaction by specifically titrating Arf1-GTP but not PIP2, which was present in excess over the PH domain (2 μm and 0–0.6 μm, respectively). Interestingly, Arno PH domain also caused a marked distortion of the activation kinetics; the exchange reaction started very slowly and, after a lag phase, switched abruptly to a 15-fold faster regime. This second phase had a rate close to that observed in the absence of the PH domain (Fig. 5D). Because the duration of the lag phase increased according to the amount of PH domain added (Fig. 5E), we concluded that the PH domain titrated newly formed Arf1-GTP molecules at the expense of Arno. Once Arf1-GTP surpassed the buffering capacity of the PH domain, it could activate Arno.
FIGURE 5.
The activation of Arf1 by Arno is delayed in the presence of Arno PH domain. A, schematic diagram of the experiment. B and C, fluorescence recordings. The liposomes (200 μm) contained 1 mol % PIP2 and 30 mol % PS and were supplemented with increasing amounts of Arno PH domain (B, K336 mutant; C, wild type). The fluorescence level of the PH domain was subtracted. Arf1 (400 nm) activation was induced by the addition of 40 μm GTP and 7.5 nm Arno. D, semi-log representation of the experiments shown in C. E, plot of slow phase duration as a function of PH domain concentration. A.U., arbitrary units.
The PH domain of Arno and GMAPC-long seem to act in a similar way: they bind to newly produced Arf1-GTP molecules, thereby reducing the stimulatory effect of Arf1-GTP on Arno. However, these two effectors might have different affinities for Arf1-GTP. Moreover, the PH domain has also the advantage of interacting with PIP2. As a consequence, the PH domain probably titrates Arf1-GTP at the surface of PIP2-containing liposomes better than GMAPC-long. This tighter titration explains the abrupt burst in the activity of Arno that was observed when Arf1-GTP exceeded the amount of PH domain present, which contrasted with the smoother change that was observed when GMAPC-long was present (compare Figs. 4B and 5C).
Communication between the PH Domain and the Sec7 Domain
The above experiments show that the robust activity of Arno on liposomes containing PS and PIP2 is “contaminated” by an Arf6-GTP-like effect of the Arf1-GTP that is being formed during the exchange reaction. Together with the lack of activity on liposomes of Arno mutants in region 2 of the PH domain (Fig. 2C), these experiments suggest that in all circumstances, binding of a GTP-bound Arf molecule to region 2 of Arno PH domain is essential for Arno activation.
Region 2 is remote from the interface with the Sec7 domain. Therefore, a binding event at this region could activate Arno only if coupled to disruption of inhibitory contacts between the Sec7 domain and the C-terminal helix (Fig. 6A). We reasoned that weakening such inhibitory contacts could make Arno less dependent on the presence of Arf6-GTP and on the feedback effect of Arf1-GTP. To test this hypothesis, we selected three hydrophobic residues at the heart of the Sec7-PH domain interface (Phe380, Leu384, and Phe257) and mutated them sequentially such as to gradually weaken the interdomain contact (Fig. 6A).
FIGURE 6.
Alleviating the inhibitory contact between the Sec7 domain and the PH domain decreases the requirement for Arf1-GTP or Arf6-GTP. A, close up view of the interdomain region of Arno. Three bulky hydrophobic residues were sequentially mutated to make this region more flexible. B, kinetics experiments in solution with 1 μm [Δ17]Arf1-GDP, 40 μm GTP, and 100 nm Arno or the indicated single, double, or triple mutant. C, kinetics experiments on liposomes with 400 nm Arf1-GDP, 40 μm GTP, 7.5 nm Arno, and 200 μm lipids containing 30% PS and 0.5% PIP2. Blue traces, liposomes only; red traces, liposomes with Arf6-GTP (300 nm); green traces, liposomes with Arno PH domain (150 nm). The characteristic lag phase observed for wild-type Arno essentially disappeared with the mutants, suggesting that the requirement for Arf1-GTP is, at least partially, bypassed. Data are represented as mean ± S.E. (n = 3).
The exchange activity of the various mutants in solution varied according to the number of mutations; the simple (F380A), double (F380A-L384A), and triple (F380A-L384A-F257E) mutants were 2.7-, 6.5-, and 8-fold more active on [Δ17]Arf1 than wild-type Arno, respectively (Fig. 6B). These results were in agreement with the autoinhibitory mechanism described previously (30). Next, we performed GDP to GTP exchange reactions on Arf1 in the presence of liposomes. Each mutant was tested under three conditions: with PS + PIP2 liposomes, with the same liposomes supplemented with Arf6-GTP, or with the same liposomes supplemented with the PH domain of Arno. This last condition should reveal “bypass” mutants (i.e. mutants displaying robust initial activity because their activation is not conditioned by the accumulation of free Arf1-GTP at the liposome surface). The results are summarized in Fig. 6C, which also compares typical kinetics for wild-type Arno and for the triple F380A/L384A/F257E mutant. In the presence of the PH domain, all interdomain mutants showed higher initial activity than wild-type Arno. Strikingly, the time course of Arf1 activation showed no lag phase, in contrast with what was observed with wild-type Arno (compare the green traces in Fig. 6C). When the exchange reaction was performed on naked liposomes or on liposomes supplemented with Arf6-GTP, the various mutants did not differ markedly from wild-type Arno; they kept a significant sensitivity to Arf6-GTP, suggesting that the binding site for Arf-GTP (region 2 of the PH domain) is not affected by the interdomain mutations.
Perturbation of Arf1 Localization in Cells Expressing a “Loose” Arno Mutant
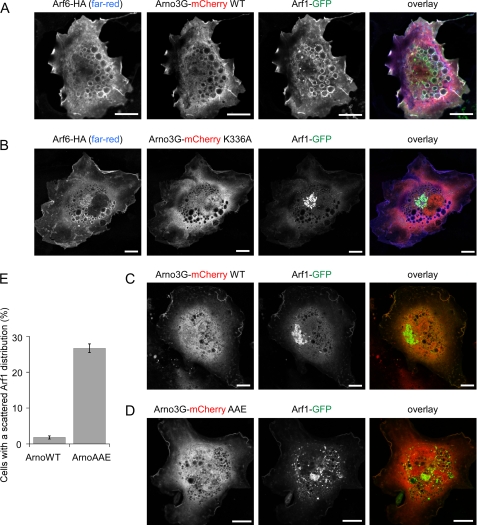
The results shown in Fig. 6 suggest that the critical priming step whereby Arno senses through its PH domain the presence of effector-free Arf-GTP (Arf6 or even Arf1) can be bypassed, at least partially, by loosening the autoinhibitory conformation of the Sec7-PH tandem. We tested this model by performing co-expression experiments in RPE1 cells. Arno was N-terminally fused with the red fluorescent protein mCherry, whereas Arf1 and Arf6 were C-terminally fused with GFP or an HA tag, respectively. In HeLa cells, coexpression of Arf6 with Arno diverts Arf1 from the Golgi to the plasma membrane and large vacuolar structures (31). This key observation, which led to the Arf cascade model, could be reproduced in RPE1 cells (Fig. 7A). When Arno displayed the K336A mutation (Fig. 7B) or when wild-type Arno was expressed in the absence of Arf6 (Fig. 7C), Arf1-GFP maintained a characteristic perinuclear Golgi localization. A different picture was observed in many cells expressing a “loose” interdomain Arno mutant (F380A/L384A/F257E); here, patches of intense Arf1-GFP staining were found scattered within the cell (Fig. 7, D and E). Some of these elements contained GM130, whose localization was severely perturbed (supplemental Fig. S6), indicating that expression of mutant Arno leads to Golgi dispersal. A similar phenotype is observed when the function of the Golgi Arf1 GEF, GBF1, is inhibited (44), suggesting that the expression of unregulated mutant Arno activates Arf1 at the expense of endogenous Golgi Arf1 GEFs. These results are also consistent with those of a previous study (45), in which effects on the structure of the Golgi were observed in cells expressing a much higher level of wild-type Arno. We conclude that the tight autoinhibitory conformation of Arno is key for preventing Arno from indiscriminately activating Arf1 without the guidance effect of Arf6. Further investigation will be needed, however, to assess the relative contribution of Arf6 and Arf1 as activators and/or substrates of Arno in a cellular context.
FIGURE 7.
Effect of a “loose” interdomain mutant of Arno on the subcellular distribution of Arf1-GFP. A and B, in RPE1 cells, coexpression of Arno (N-terminally tagged with mCherry) with Arf6-HA diverted Arf1-GFP from the Golgi to the plasma membrane and to vacuolar structures, whereas Arno-mCherry K336A had no effect. C and D, without overexpressed Arf6 wild type, Arno does not affect the Golgi localization of Arf1-GFP, whereas the triple interdomain mutant (F380A/L384A/F257E) promotes the formation of dispersed Arf1-GFP-labeled structures. E, percentage of cells transfected as in C and D with an aberrant Arf1-GFP distribution. Scale bar, 10 μm. Error bars, S.E.
DISCUSSION
Arno and its homologues have been instrumental for structural studies (15, 16, 18, 30). In contrast, their functions are not clear cut because they do not fit well with simple models whereby a bona fide GEF is active on one Arf subtype at one subcellular location and for one given function. The recent discovery that Arno proteins are not only GEFs for Arf proteins but also Arf effectors suggests complex circuits involving several Arf subtype (31–33). Our study on model liposomes reveals that such circuits display remarkable kinetics features. These features might help in understanding the function of Arno proteins in a cellular context.
Our analysis focuses on a circuit where Arno is allowed to sense, through its PH domain, Arf6-GTP on an artificial membrane and to promote GDP to GTP exchange on Arf1 through its Sec7 domain. This cascade has been described recently in transfected cells (31). Because other cascades involving different Arf subtypes are possible (32, 33), a complete analysis would require the testing of all combinations of Arf proteins both as an input and as an output. The difficulty in purifying some myristoylated Arf-like species (e.g. Arl4) makes this aim challenging. Given the better affinity of the PH domain of Arno for Arf6 versus Arf1 (31) and the higher exchange activity of its Sec7 domain on Arf1 versus Arf6 (23), the pathway studied here should be considered as a reasonable but not unique model. However, the underlying mechanisms probably apply for any Arf/Arno cascade because they are based on the general properties of Arno. Note also that the “2G” variant of Arno, which was not studied here and which is subject to an additional layer of control by the rare lipid PIP3, is also sensitive to Arf6-GTP (31, 32). As such, it should be also regulated by the various mechanisms reported here.
Arf6-GTP is a very potent activator of Arno on synthetic membranes. The half-maximum effect (43 ± 10 nm; Fig. 1B) is 300-fold lower than the value reported for a truncated form of Arf6 in solution (30) and corresponds to a surface concentration of ∼0.05 mol % (43 nm protein versus 100 μm accessible lipid). Assuming that an Arf molecule occupies a patch of 20–25 lipids, this implies that Arno is sensitive to the coverage of 1% of the membrane surface by Arf6-GTP. Through its PH domain, Arno is thus a very good surface sensor of both anionic lipids and Arf6-GTP and integrates these inputs in a combinatorial manner (supplemental Fig. S1).
In the absence of Arf6-GTP, Arno is very active on liposomes, provided that the levels of PS and PIP2 are high enough (supplemental Fig. S1) (11). This longstanding observation has remained puzzling because these lipids are present on the plasma membrane, which is generally poor in Arf1 under resting conditions. Our experiments suggest that this situation corresponds to a more complex mechanism than initially thought. The high exchange activity of Arno on anionic liposomes lacking Arf6-GTP seems to be due to a feedback effect of Arf1-GTP (Fig. 3). The importance of the positive feedback loop in the functioning of Arno is highlighted by the very specific defect induced by some mutations. Arno mutants in region 2 of the PH domain (e.g. K336A) appear normal when tested for basic properties (Figs. 1, D and E, and 2B and supplemental Fig. S2), yet they are essentially inert when tested for their exchange activity on liposomes (Figs. 1C and 2C) and for their effect on the subcellular localization of Arf1 (Fig. 7B). Thus, in any circumstance, it seems that the activation of Arno is contingent on the presence of a stimulatory Arf-GTP molecule. This strict requirement is also underscored in Fig. 6C, where, despite the presence of PS and PIP2, the activity of wild-type Arno varied 50-fold depending on whether Arf6-GTP was present or newly formed Arf1-GTP molecules were diverted from Arno. Note that a full and quantitative explanation of these variations awaits further investigation because they depend on many equilibrium and kinetics constants, which are presently unknown (e.g. Kd values for the interaction of Arf1-GTP or Arf6-GTP with various effectors and Arno).
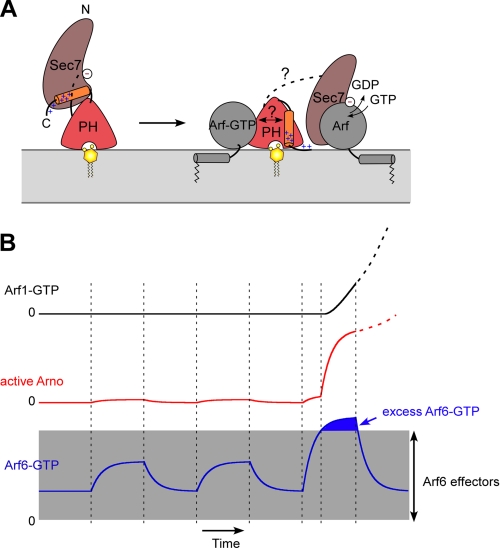
The mechanism by which Arno opens at the membrane surface remains to be determined. The structural analysis of Arno and our study suggest a pathway starting from the binding of Arf1/6-GTP to region 2 of the PH domain and resulting in disruption of inhibitory contacts between the Sec7 and PH domains (Fig. 8A). The synergy between Arf6-GTP and anionic lipids (supplemental Fig. S1) suggests that this pathway combines with a “lipid” pathway that starts with binding of region 1 of the PH domain to phosphoinositides and then involves displacement of the autoinhibitory region through the movement of positive charges of the C terminus toward the negatively charged membrane surface. These interdependent conformational changes allow Arno to vary its activity 500-fold, depending on the proteins and lipids present on the membrane (supplemental Fig. S1). Note that this model does not integrate the role of the coiled-coil region of Arno, which mediates homodimerization in vitro but which may interact with other proteins in vivo (28, 46–51). However, experiments with a truncated form of Arno lacking the coiled-coil region show that this form keeps all features of full-length Arno. This includes (i) stimulation by nanomolar concentration of Arf6-GTP, (ii) sigmoidal kinetics, and (iii) inhibition by Arno PH domain (data not shown).
FIGURE 8.
Structural and functional aspects of Arno activation on lipid membranes. A, proposed conformational changes in Arno at the surface of lipid membranes. The switch to an active membrane-bound conformation depends not only on the presence of anionic lipids (e.g. PS + PIP2) but also on the presence of a free Arf-GTP molecule. B, model of Arno response. In resting cells, Arno remains inert because no membrane contains both anionic lipids and free Arf-GTP. Notably, the plasma membrane, which contains PS and PIP2, might not be suitable for Arno activation when active Arf species (e.g. Arf6) are engaged in interaction with classical effectors and for constitutive functions. Ignition of Arno requires a burst of active Arf6 or Arl4 at the plasma membrane that exceeds the buffering capacity of effectors. After this initiation step, Arno is engaged in a sustained self-activating pathway through the feedback effect of newly formed Arf-GTP molecules (e.g. Arf1) and remains active even if some initial inputs disappear. The proposed circuit is not restricted to Arf6 and Arf1 and could also apply when Arno is activated by a different Arf subtype (e.g. Arl4).
Positive feedback loops are frequently observed when cells change their behavior because they assure that a given reaction moves forward decisively (52). This property might help to better delineate the functions of Arno. This exchange factor, which exists only in metazoa, does not seem to fulfill constitutive membrane traffic functions. Instead, Arno participates in cell migration (53, 54), phagocytosis (55), macropinocytosis (31), and organism growth controlled by insulin (56) (i.e. events that require the sudden remodeling of an unusually large amount of cellular membrane). We propose that the functioning of Arno in a switch-like manner through the combinatory effects of the positive feedback loop and competition by effectors is adapted to these functions. In resting cells, the levels of Arf-GTP subtypes on membranes probably vary within well defined limits to sustain elementary functions, such as PIP2 synthesis at the plasma membrane through Arf6 or COPI vesicle formation at the Golgi apparatus through Arf1. Under these conditions, Arno would remain dormant due to its autoinhibitory conformation and the competition effect of Arf effectors. In line with this, Arno is generally found in the cytosol despite bearing a PH domain adapted to the plasma membrane. Ignition of Arno probably requires a large burst of Arf6-GTP or Arl4-GTP (i.e. exceeding the amounts of effectors) at the plasma membrane, which is rich in PS and PIP2 (Fig. 8B). Then, by activating new Arf molecules and notably Arf1, which is much more abundant than Arf6 (57), Arno could sustain its own activity thanks to the feedback effect of Arf1-GTP and became more permissive with regard to Arf6-GTP.
In the proposed circuit, neither the PH domain nor the Sec7 domain needs to display an absolute specificity. The preference of the PH domain for Arf6 over Arf1 and the preference of the Sec7 domain for Arf1 over Arf6 should simply ensure gradual enrichment in Arf1-GTP versus Arf6-GTP. The cascade might be even more robust if the specificity of the two domains of Arno is not too pronounced because different pathways could sequentially occur (e.g. Arf6 to Arf6, Arf6 to Arf1, and last Arf1 to Arf1). However, in any circumstance, stimulation of Arno requires the presence of PS and phosphoinositides (supplemental Fig. S1). This should prevent Arno from being activated by Arf1-GTP at the Golgi. Note that the model fits also quite well with the general idea that Arf subtypes work in pairs in cells, with specific combinations displaying a unique profile of activities (58).
Our model also highlights the difficulty in studying Arno in vivo. First, the fact that Arno not only activates Arf proteins but is also activated by them complicates the interpretation of studies using overexpressed proteins (including the present work). Any Arno pathway should be exquisitely sensitive to the relative amounts of expressed proteins. Second, bioprobes engineered to detect a given Arf-GTP species might paradoxically inhibit the production of this species if Arno is the GEF involved because they should prevent the feedback effect of Arf-GTP.
In conclusion, the behavior of Arno on artificial liposomes with two Arf species and effectors suggests that this exchange factor is tuned to sense the presence of an unusual excess of activated Arf6 (or Arl4) at the plasma membrane and to convert this input into a sustained autoamplified reaction through Arf1. Because other exchange factors combine two binding sites for small G proteins, one regulatory and one catalytic (34, 40), the mechanisms described here might be quite general.
Supplementary Material
Acknowledgments
We thank V. Morello for purified GMAP-210 proteins, B. Goud for RPE1 cells, N. Leroudier and F. Brau for technical support, and M. Coppey-Moisan for the gift of the mCherry plasmid. We thank J. Bigay, G. Drin, and M. Franco for comments on the manuscript and all members of the laboratory for help.
This work was supported by Agence Nationale de la Recherche Grant ANR-08-BLAN-0060-01.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- GEF
- guanine nucleotide exchange factor
- PS
- phosphatidylserine
- PH
- pleckstrin homology
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP3
- phosphatidylinositol 3,4,5-trisphosphate
- mant
- methylanthraniloyl.
REFERENCES
- 1. Bos J. L., Rehmann H., Wittinghofer A. (2007) Cell 129, 865–877 [DOI] [PubMed] [Google Scholar]
- 2. Canagarajah B., Leskow F. C., Ho J. Y., Mischak H., Saidi L. F., Kazanietz M. G., Hurley J. H. (2004) Cell 119, 407–418 [DOI] [PubMed] [Google Scholar]
- 3. Li P., Martins I. R., Amarasinghe G. K., Rosen M. K. (2008) Nat. Struct. Mol. Biol. 15, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prehoda K. E., Scott J. A., Mullins R. D., Lim W. A. (2000) Science 290, 801–806 [DOI] [PubMed] [Google Scholar]
- 5. Rehmann H., Das J., Knipscheer P., Wittinghofer A., Bos J. L. (2006) Nature 439, 625–628 [DOI] [PubMed] [Google Scholar]
- 6. Kam J. L., Miura K., Jackson T. R., Gruschus J., Roller P., Stauffer S., Clark J., Aneja R., Randazzo P. A. (2000) J. Biol. Chem. 275, 9653–9663 [DOI] [PubMed] [Google Scholar]
- 7. Gureasko J., Galush W. J., Boykevisch S., Sondermann H., Bar-Sagi D., Groves J. T., Kuriyan J. (2008) Nat. Struct. Mol. Biol. 15, 452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lebensohn A. M., Kirschner M. W. (2009) Mol. Cell 36, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groves J. T., Kuriyan J. (2010) Nat. Struct. Mol. Biol. 17, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chardin P., Paris S., Antonny B., Robineau S., Béraud-Dufour S., Jackson C. L., Chabre M. (1996) Nature 384, 481–484 [DOI] [PubMed] [Google Scholar]
- 12. Moss J., Vaughan M. (2002) Arch. Biochem. Biophys. 397, 156–161 [DOI] [PubMed] [Google Scholar]
- 13. Jackson C. L., Casanova J. E. (2000) Trends Cell Biol. 10, 60–67 [DOI] [PubMed] [Google Scholar]
- 14. Béraud-Dufour S., Robineau S., Chardin P., Paris S., Chabre M., Cherfils J., Antonny B. (1998) EMBO J. 17, 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldberg J. (1998) Cell 95, 237–248 [DOI] [PubMed] [Google Scholar]
- 16. Renault L., Guibert B., Cherfils J. (2003) Nature 426, 525–530 [DOI] [PubMed] [Google Scholar]
- 17. Ogasawara M., Kim S. C., Adamik R., Togawa A., Ferrans V. J., Takeda K., Kirby M., Moss J., Vaughan M. (2000) J. Biol. Chem. 275, 3221–3230 [DOI] [PubMed] [Google Scholar]
- 18. Cronin T. C., DiNitto J. P., Czech M. P., Lambright D. G. (2004) EMBO J. 23, 3711–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klarlund J. K., Tsiaras W., Holik J. J., Chawla A., Czech M. P. (2000) J. Biol. Chem. 275, 32816–32821 [DOI] [PubMed] [Google Scholar]
- 20. Nagel W., Schilcher P., Zeitlmann L., Kolanus W. (1998) Mol. Biol. Cell 9, 1981–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Macia E., Paris S., Chabre M. (2000) Biochemistry 39, 5893–5901 [DOI] [PubMed] [Google Scholar]
- 22. Santy L. C., Frank S. R., Hatfield J. C., Casanova J. E. (1999) Curr. Biol. 9, 1173–1176 [DOI] [PubMed] [Google Scholar]
- 23. Macia E., Chabre M., Franco M. (2001) J. Biol. Chem. 276, 24925–24930 [DOI] [PubMed] [Google Scholar]
- 24. Frank S., Upender S., Hansen S. H., Casanova J. E. (1998) J. Biol. Chem. 273, 23–27 [DOI] [PubMed] [Google Scholar]
- 25. Li H. S., Shome K., Rojas R., Rizzo M. A., Vasudevan C., Fluharty E., Santy L. C., Casanova J. E., Romero G. (2003) BMC Cell Biol. 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boulay P. L., Cotton M., Melançon P., Claing A. (2008) J. Biol. Chem. 283, 36425–36434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumari S., Mayor S. (2008) Nat. Cell Biol. 10, 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim J., Zhou M., Veenstra T. D., Morrison D. K. (2010) Genes Dev. 24, 1496–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Várnai P., Bondeva T., Tamás P., Tóth B., Buday L., Hunyady L., Balla T. (2005) J. Cell Sci. 118, 4879–4888 [DOI] [PubMed] [Google Scholar]
- 30. DiNitto J. P., Delprato A., Gabe, Lee M. T., Cronin T. C., Huang S., Guilherme A., Czech M. P., Lambright D. G. (2007) Mol. Cell 28, 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. (2007) Mol. Biol. Cell 18, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hofmann I., Thompson A., Sanderson C. M., Munro S. (2007) Curr. Biol. 17, 711–716 [DOI] [PubMed] [Google Scholar]
- 33. Li C. C., Chiang T. C., Wu T. S., Pacheco-Rodriguez G., Moss J., Lee F. J. (2007) Mol. Biol. Cell 18, 4420–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grosshans B. L., Ortiz D., Novick P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Franco M., Chardin P., Chabre M., Paris S. (1995) J. Biol. Chem. 270, 1337–1341 [DOI] [PubMed] [Google Scholar]
- 36. Chavrier P., Franco M. (2001) Methods Enzymol. 329, 272–279 [DOI] [PubMed] [Google Scholar]
- 37. Wang J., Gambhir A., Hangyás-Mihályné G., Murray D., Golebiewska U., McLaughlin S. (2002) J. Biol. Chem. 277, 34401–34412 [DOI] [PubMed] [Google Scholar]
- 38. Bigay J., Casella J. F., Drin G., Mesmin B., Antonny B. (2005) EMBO J. 24, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ménétrey J., Perderiset M., Cicolari J., Dubois T., Elkhatib N., El Khadali F., Franco M., Chavrier P., Houdusse A. (2007) EMBO J. 26, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boykevisch S., Zhao C., Sondermann H., Philippidou P., Halegoua S., Kuriyan J., Bar-Sagi D. (2006) Curr. Biol. 16, 2173–2179 [DOI] [PubMed] [Google Scholar]
- 41. Berg O. G., Yu B. Z., Rogers J., Jain M. K. (1991) Biochemistry 30, 7283–7297 [DOI] [PubMed] [Google Scholar]
- 42. Alberty R. A. (1953) J. Am Chem. Soc. 75, 1928–1932 [Google Scholar]
- 43. Drin G., Morello V., Casella J. F., Gounon P., Antonny B. (2008) Science 320, 670–673 [DOI] [PubMed] [Google Scholar]
- 44. García-Mata R., Szul T., Alvarez C., Sztul E. (2003) Mol. Biol. Cell 14, 2250–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Franco M., Boretto J., Robineau S., Monier S., Goud B., Chardin P., Chavrier P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9926–9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. White D. T., McShea K. M., Attar M. A., Santy L. C. (2010) Mol. Biol. Cell 21, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venkateswarlu K. (2003) J. Biol. Chem. 278, 43460–43469 [DOI] [PubMed] [Google Scholar]
- 48. Shmuel M., Santy L. C., Frank S., Avrahami D., Casanova J. E., Altschuler Y. (2006) J. Biol. Chem. 281, 13300–13308 [DOI] [PubMed] [Google Scholar]
- 49. Mansour M., Lee S. Y., Pohajdak B. (2002) J. Biol. Chem. 277, 32302–32309 [DOI] [PubMed] [Google Scholar]
- 50. Nevrivy D. J., Peterson V. J., Avram D., Ishmael J. E., Hansen S. G., Dowell P., Hruby D. E., Dawson M. I., Leid M. (2000) J. Biol. Chem. 275, 16827–16836 [DOI] [PubMed] [Google Scholar]
- 51. Boehm T., Hofer S., Winklehner P., Kellersch B., Geiger C., Trockenbacher A., Neyer S., Fiegl H., Ebner S., Ivarsson L., Schneider R., Kremmer E., Heufler C., Kolanus W. (2003) EMBO J. 22, 1014–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brandman O., Meyer T. (2008) Science 322, 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Santy L. C., Casanova J. E. (2001) J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Viaud J., Zeghouf M., Barelli H., Zeeh J. C., Padilla A., Guibert B., Chardin P., Royer C. A., Cherfils J., Chavanieu A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10370–10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Beemiller P., Hoppe A. D., Swanson J. A. (2006) PLoS Biol. 4, e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fuss B., Becker T., Zinke I., Hoch M. (2006) Nature 444, 945–948 [DOI] [PubMed] [Google Scholar]
- 57. El-Annan J., Brown D., Breton S., Bourgoin S., Ausiello D. A., Marshansky V. (2004) Am. J. Physiol. Cell Physiol. 286, C768–C778 [DOI] [PubMed] [Google Scholar]
- 58. Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. (2005) Mol. Biol. Cell 16, 4495–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.