Abstract
Telomere maintenance is essential for protecting chromosome ends. Aberrations in telomere length have been implicated in cancer and aging. Telomere elongation by human telomerase is inhibited in cis by the telomeric protein TRF1 and its associated proteins. However, the link between TRF1 and inhibition of telomerase elongation of telomeres remains elusive because TRF1 has no direct effect on telomerase activity. We have previously identified one Pin2/TRF1-interacting protein, PinX1, that has the unique property of directly binding and inhibiting telomerase catalytic activity (Zhou, X. Z., and Lu, K. P. (2001) Cell 107, 347–359). However, nothing is known about the role of the PinX1-TRF1 interaction in the regulation of telomere maintenance. By identifying functional domains and key amino acid residues in PinX1 and TRF1 responsible for the PinX1-TRF1 interaction, we show that the TRF homology domain of TRF1 interacts with a minimal 20-amino acid sequence of PinX1 via hydrophilic and hydrophobic interactions. Significantly, either disrupting this interaction by mutating the critical Leu-291 residue in PinX1 or knocking down endogenous TRF1 by RNAi abolishes the ability of PinX1 to localize to telomeres and to inhibit telomere elongation in cells even though neither has any effect on telomerase activity per se. Thus, the telomerase inhibitor PinX1 is recruited to telomeres by TRF1 and provides a critical link between TRF1 and telomerase inhibition to prevent telomere elongation and help maintain telomere homeostasis.
Keywords: Chromatin Regulation, DNA-binding Protein, Enzyme Inhibitors, Protein-Protein Interactions, Telomere
Introduction
Telomeres cap the ends of linear chromosomes, which are composed of repetitive sequences and associated proteins that are necessary for telomeric maintenance and regulation (1–5). Telomeric sequences can be extended by telomerase, a unique reverse transcriptase composed of the catalytic subunit TERT and the RNA template TER (6–16). Although TER is constitutively expressed in most cells, TERT is expressed at very low levels or not at all in most normal human cells with the exception of highly proliferative tissues (17–19). Therefore, telomeres shorten during each cell division due to the end replication problem of DNA polymerase as well as postreplication processing by unidentified nucleases or helicases (20–23). Telomere shortening to a critical length and/or the uncapping of the telomere leads to replicative senescence or crisis (24–31).
Telomerase activation has been well documented as one of the necessary factors for cell immortalization and transformation. It is reactivated in most human cancer cells (17, 18, 32, 33) and is not only critical for transforming primary human cells (34) but is also sufficient to allow transformed cells to escape from crisis (35–38). In fact, many oncogenes and tumor suppressors regulate transcription of TERT (39–42). Furthermore, ablation of telomerase shortens telomeres and limits cell proliferation (43–45). It confers resistance to tumorigenesis in mice (46–51), although a lack of telomerase in mice over five consecutive generations leads to oncogenesis especially in the p53 mutant genetic background (43, 45, 49, 52, 53). Although telomerase activation allows telomere extension, it does not lead to unlimited telomere elongation; telomeres are stably maintained within a relatively narrow size distribution in telomerase-positive cells (32, 54, 55), although massive overexpression of TERT and TER can elongate telomeres to beyond physiological lengths (56). These results indicate that the ability of telomerase to elongate telomeres is further regulated under physiological conditions.
Preserving optimal telomere length is crucial for cells as excessive telomere attrition can result in genomic instability and chromosome fusions. In yeast, telomere maintenance is regulated by a number of telomere proteins, including RAP1, Pif1, Cdc13, Taz1, and Pot1 (57–67). In humans, telomere surveillance is carried out in cis by telomere-associated proteins, including six major players that form a protective complex termed shelterin (5). At the base of the complex are two telomeric DNA binding factors, TRF14 and TRF2, that act as scaffolds to recruit other proteins to telomeres. Although TRF2 protects telomere integrity possibly by forming T-loop structures, TRF1 maintains telomeres at reasonable lengths (5, 55, 68, 69).
It has been proposed that TRF1 acts in cis to progressively block telomerase access and telomere elongation as increased numbers of TRF1 molecules bind to telomeres. Consistent with this idea is that TRF1 and its interacting proteins, including Tankyrase, Tin2, Pot1, TPP1, and Fbx4, have been shown to regulate telomere length (5, 66, 69–85). However, because neither TRF1 nor these associated proteins affect telomerase activity in vitro or in vivo, it is not fully understood how TRF1 performs this crucial function. It has been shown that the TRF1 complex brings Pot1 to the single-stranded DNA 3′ overhang of telomeres to physically block telomerase from accessing telomeres (73, 79). On the other hand, more recent studies indicate that during telomere extension, the Pot1-TPP1 complex does not prevent telomerase from elongating telomeres but, instead, serves rather as a processivity factor for telomerase to promote telomere elongation (84, 85). These results suggest that Pot1 or Pot1 complexes that are recruited by TRF1 to telomere ends have at least two potentially opposing functions; that is, blocking telomerase from accessing telomeres to inhibit telomere elongation and increasing telomerase processivity to elongate telomeres during telomere extension. However, it remains unclear how these two opposite functions are coordinated. Therefore, the link between TRF1 and telomerase inhibition remains elusive.
We isolated TRF1 as Pin2 in the same screen for Pin1 (86, 87). Both Pin1 and TRF1/Pin2 are important for mitotic regulation (87–91), and Pin1 regulates TRF1 function in telomere maintenance and aging (92). We have identified new TRF1-interacting proteins, including PinX1–4, to elucidate important TRF1 function in telomere maintenance and mitosis (80, 92–94).
Unlike all other TRF1-interacting proteins, PinX1 has the unique property of directly interacting with the telomerase catalytic component, TERT, and potently inhibiting telomerase catalytic activity (93). Furthermore, PinX1 overexpression in human cancer cells shortens telomeres, induces crisis, and inhibits tumorigenicity, whereas PinX1 depletion increases telomerase activity, elongates telomeres, and enhances tumorigenicity. The ability of PinX1 to inhibit telomere elongation has been shown to be highly conserved in other organisms such as yeast and rats (95–98). These results indicate that PinX1 is a potent telomerase inhibitor that inhibits telomere elongation. However, it is not known how PinX1 is recruited to telomeres, where telomere elongation by telomerase action takes place. Similarly, even though PinX1 was identified as a TRF1-interacting protein (93), nothing is known about whether this interaction has any role in telomere regulation.
To address these questions, we first mapped the minimal domains responsible for the PinX1-TRF1 interaction and then identified a single PinX1 point mutation, L291E, that is able to completely abolish the ability of PinX1 to interact with TRF1 but does not affect its ability to bind and inhibit telomerase. Comparing the subcellular localization and telomere function of PinX1 and its TRF1 binding mutant uncovered that TRF1 is required for recruiting PinX1 to telomeres as well as for PinX1 to inhibit telomere elongation in vivo. The biological significance of these structure-function analyses was further confirmed by our demonstration that PinX1 fails to localize to telomeres and to inhibit telomere elongation when endogenous TRF1 is knocked down. These results demonstrate for the first time that the telomerase inhibitor PinX1 is recruited to telomeres by TRF1 and provides a crucial link between TRF1 and inhibition of telomere elongation by telomerase to help maintain telomere homeostasis.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Recombinant Protein Purification
Truncation and point mutants of PinX1 and TRF1 were generated by PCR mutagenesis procedures and confirmed by sequencing. N-terminal GST-tagged proteins were generated by subcloning cDNAs encoding PinX1, TRF1, or their mutants into a pGEX vector. The resulting fusion proteins were expressed in BL21 Escherichia coli cells and purified by glutathione beads, as described (87, 93). Briefly, cells were pelleted and lysed in buffer containing 50 mm Tris-Cl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1% Triton, 5% glycerol, lysozyme, and protease inhibitors. After sonication, lysates were centrifuged, and glutathione-agarose beads were added to the supernatant followed by incubation at 4 °C for 2 h. The beads were then washed, and protein was eluted with 30 mm glutathione. Proteins were then dialyzed in 10 mm HEPES, pH 7.5, 10 mm NaCl, 1 mm DTT, 0.5 mm EDTA, pH 8.0, 0.5 mm EGTA, pH 8.0, 5% glycerol, and protease inhibitors before use.
GST Pulldown, Immunoprecipitation, and Immunoblot Analyses
GST pulldown and immunoprecipitation assays were performed as described (93). Relevant proteins were expressed in vitro using a TNT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]Met or in human cell lines by transient or stable transfection. Proteins were then incubated at 4 °C for 1 h with 1 μm GST fusion proteins in lysis buffer. Protein A/G beads or glutathione-agarose beads were then added followed by additional incubation for 1 h at 4 °C. Beads were then collected by centrifugation, washed multiple times, and boiled in SDS sample buffer. Samples were then separated by electrophoresis on acrylamide gels and transferred by semidry transfer to PVDF membranes for immunoblot analysis.
Protein Localization and Fluorescence Microscopy
The subcellular localization of proteins was determined as described previously (87, 93). HT1080 cells seeded on glass coverslips were transiently transfected with GFP-PinX1 and RFP-TRF1. At 16 h post-transfection, cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Nonidet P-40/PBS, and DAPI-stained. Coverslips were mounted using Fluoromount-G (SouthernBiotech) and visualized by fluorescence microscopy using a Zeiss microscope.
TRF1 Knockdown Cell Lines or Pools
Two different TRF1 RNAi sequences and vectors were used. The TRF1 RNAi retroviral construct (CGCTTGCCAGTTGAGAACGATA) and control vector pSM2C were purchased from Open Biosystems followed by generating TRF1 RNAi retroviral stable cell lines as described (80). Plasmids were transfected into Pheonix packaging cells, and viral supernatants were harvested 48 h after transfection followed by infection of HT1080 cells. After selection with antibiotics and limiting dilution, multiple independent cell lines were isolated and checked for protein expression by immunoblotting analysis with anti-TRF1 antibodies. For generating stable TRF1 RNAi lentiviral cell pools, PinX1 stable or vector cell lines were infected with lentiviruses produced from the TRF1 shRNA lentiviral vector containing sense sequence CCCTTGATGCACAGTTTGAAA or the pLKO.1 control vector (Open Biosystems). Lentiviral production and infection was performed as suggested by the RNAi Consortium. Briefly, 293T cells were transfected with the pLKO.1 shRNA or control construct along with CMV-D8.9 viral packaging vector and CMV-VSVG viral envelope vector. Media was replaced 18 h post-transfection, and viral supernatants were harvested after an additional 24 h. Target cells were infected with viral supernatant and subsequently selected with 2 μg/ml puromycin, as described (99). Knockdown of TRF1 protein levels was confirmed by immunoblotting analysis.
Stable Cells Overexpressing PinX1 or Its Mutants
HA epitope-tagged PinX1 or mutants were stably transfected into HT1080 fibrosarcoma cells in multiple independent experiments using the vector as a control followed by protein expression confirmation using immunoblotting analysis with anti-HA antibodies (Covance) as described (93). Cells were seeded at 5 × 105 cells per 10-cm culture dish, then passaged and collected every fourth day for telomerase activity and telomere length studies.
Telomerase Activity and Telomere Length Assays
Telomerase activity was assayed using the TRAPeze Telomerase detection kit (Millipore) according to manufacturer's instructions and as described (93). Briefly, cells were lysed with CHAPS lysis buffer and centrifuged, and the telomerase-containing supernatant was collected. To assay the effects of purified PinX1 proteins on telomerase activity, recombinant proteins were incubated with cell lysates containing telomerase for 10 min at 4 °C before setting up reactions for telomere extension by PCR. Telomerase products were separated on 10% non-denaturing polyacrylamide gels, stained with SYBR Green I Nucleic Acid Stain (Molecular Probes), and visualized.
Telomere restriction fragment (TRF) length at various population doublings was measured, as described previously (93). Genomic DNA was isolated by phenol extraction and digested with HinfI and RsaI restriction enzymes at 37 °C overnight. DNA concentrations were determined by fluorometry before samples were separated on 0.8% agarose gels. Gels were dried at room temperature, and in-gel hybridization was performed with a (TTAGGG)3 telomeric DNA probe. Average telomere restriction fragment length was calculated by quantifying the hybridization signals using ImageQuant.
RESULTS
Multiple Charged Residues and Leu-291 in PinX1 Are Essential for the PinX1-TRF1 Interaction
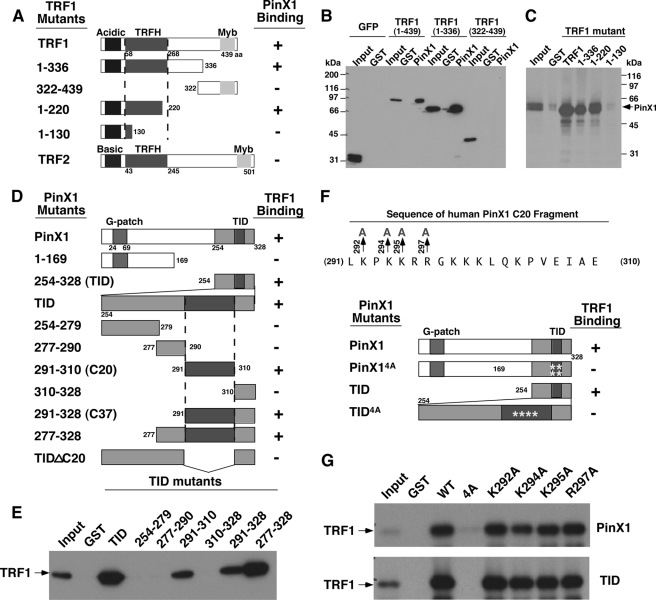
Although PinX1 was originally identified as a TRF1-interacting protein (93), nothing is known about whether this interaction is important for telomere regulation. To address this question, we started by searching for the domains responsible for the TRF1-PinX1 interaction by generating systematic truncations of TRF1 (Fig. 1A) and PinX1 (Fig. 1D) followed by various binding assays, performed as previously described (93). GST-PinX1 bound to GFP-TRF1 fragments encompassing residues 1–439 and 1–336 but not 322–439, which were expressed in mammalian cells (Fig. 1B). Conversely, GST-TRF1 fragments encompassing residues 1–439, 1–336, and 1–220, but not 1–130, bound to PinX1 that was synthesized by in vitro transcription and translation (Fig. 1C). These results indicate that the PinX1 binding domain is located in the TRF homology (TRFH) domain of TRF1, as shown for several other TRF1-binding proteins (71, 80, 100). Importantly, this PinX1 and TRF1 interaction is highly specific because PinX1 completely failed to bind to TRF2 (Fig. 1A), even though the TRFH domains of TRF1 and TRF2 adopt almost identical three-dimensional structures (100, 101).
FIGURE 1.
The identification of the minimal domains responsible for the PinX1 and TRF1 interaction. A–C, PinX1 binds to the TRFH domain of TRF1. A series of TRF1 truncation mutants (A) were expressed in cells as GFP fusion proteins followed by GST pulldown with GST-PinX1 or control GST (B) or expressed in bacteria and purified as GST fusion proteins followed by GST pulldown with in vitro synthesized 35S-PinX1 (C). PinX1 binding of TRF1 mutants or TRF2 was summarized in A. D and E, a 20-amino acid domain (C20) in PinX1 is necessary and sufficient for binding to TRF1. A series of PinX1 truncation mutants (D) was expressed in bacteria and purified as GST fusion proteins followed by GST pulldown with in vitro synthesized 35S-TRF1 (E). TRF1 binding of PinX1 mutants was summarized in D. F and G, the mutation of multiple positively charged residues in C20 is required to abolish the PinX1-TRF1 interaction. The C20 fragment of PinX1 contains several positively charged residues, including Lys-292, Lys-294, Lys-295, and Arg-297 (F). Full-length PinX1 or its TID containing Ala substitutions of these residues individually or altogether were expressed in bacteria and purified as GST fusion proteins followed by GST pulldown with in vitro synthesized 35S-TRF1 (G). Note that the mutation of these charged residues individually in full-length PinX1 or its TID was not sufficient to disrupt the PinX1-TRF1 interaction.
Upon further examination of the TRF1 binding domain in PinX1, we confirmed our previous findings that the TRF1 binding domain is located at the C-terminal 75 amino acids of PinX1 (Fig. 1, D and E), which we previously named TID for the TRF1-interacting domain and telomerase-inhibitory domain (93). To further define the TRF1 binding sequence in PinX1, we generated numerous smaller fragments of the TID and tested their binding with TRF1 as described (93). All GST-PinX1 mutant proteins containing residues 291–310 with or without any N-terminal or C-terminal flanking sequences were able to bind to TRF1, whereas mutant proteins lacking this domain could not (Fig. 1, D and E). These results indicate that the 20-amino acid fragment (C20) from residue 291 to 310 of PinX1 is both necessary and sufficient to mediate the interaction with TRF1.
The C20 fragment of PinX1 contains several positively charged residues (Fig. 1F) that are often involved in protein-protein interactions, suggesting they might be important for interacting with TRF1. To test this possibility, we generated PinX1 mutants replacing four positively charged residues with Ala either individually (K292A, K294A, K295A, or R297A) or altogether (4A) (Fig. 1F) and assayed their ability to interact with TRF1. Although all PinX1 mutants containing a single Ala substitution at these four residues still bound to TRF1 (Figs. 1G and 2A), all 4A mutants in various PinX1 truncation fragments completely failed to interact with TRF1 (Fig. 1, F and G, and 2, A and B). These results indicate that the mutation of these multiple charged residues in the C20 fragment disrupts the ability of PinX1 to bind TRF1.
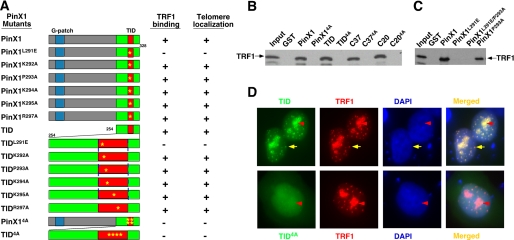
FIGURE 2.
TRF1 binding and telomere localization of a large panel of PinX1 mutants. A, TRF1 binding and telomere localization of single point mutants or quadruple Ala substitution mutants (K292A, K294A, K295A, and R297A) in different PinX1 fragments were summarized, respectively. B and C, examples of TRF1 binding of selected PinX1 mutants are shown. Selected single point mutations or quadruple Ala mutation (K292A, K294A, K295A, and R297A) were introduced into full-length PinX1 or various PinX1 fragments, and mutant proteins were expressed in bacteria and purified as GST fusion proteins followed by GST pulldown with in vitro synthesized 35S-TRF1. The amino acid residues of PinX1 truncation mutants are from 254 to 328 (TID), from 291 to 328 (C37), and from 291 to 310 (C20). D, examples of subcellular localization of TID or TID4A with TRF1 are shown. The localization of these mutant proteins were determined by co-transfecting HT1080 cells overnight with vectors expressing RFP-tagged TRF1 and GFP-tagged wild-type full-length PinX1 or its TID with or without mutations followed by subjecting them to fluorescence microscopy (D). Yellow arrows point to colocalization of TRF1 and TID at telomeres.
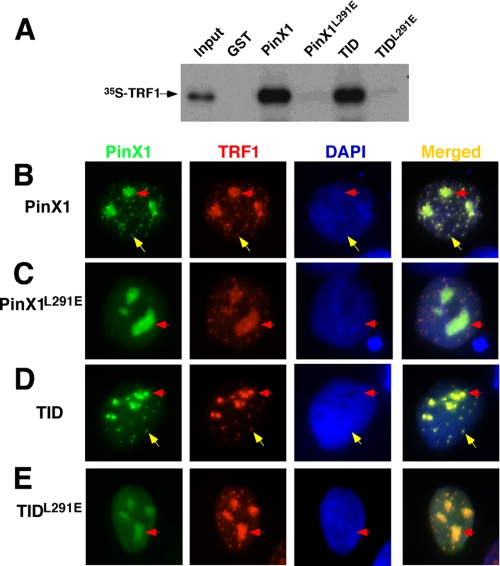
Interestingly, recent co-crystal structural analyses independently reveal that the TRFH domain of TRF1 interacts with proteins such as TIN2 via a hydrophobic interaction with a common LXP motif and that a Leu to Glu mutation is sufficient to disrupt their interaction in peptide binding assays in vitro (100). To examine the importance of Leu-291 and Pro-293 in PinX1 for mediating the TRF1-PinX1 interaction, we mutated Leu-291 to glutamic acid (L291E) and Pro-293 to alanine (P293A) individually or together (L291E/P293A) in PinX1. Although the P293A mutant bound well to TRF1, neither the L291E nor L291E/P293A mutant could interact with TRF1 regardless of whether they were in full-length PinX1 or its TID (Figs. 2, A and C, and 3A). These results indicate that a single L291E point mutation is sufficient to completely disrupt the ability of PinX1 to bind TRF1, whereas quadruple mutations (K292A, K294A, K295A, and R297A) in the hydrophilic interaction are required, although we could not completely rule out the possibility that PinX1 quadruple mutant could simply be misfolded. Taken together, these analyses indicate that the TRFH domain of TRF1 specifically recognizes a 20-amino acid sequence (residues 291–297) of PinX1 likely by both hydrophobic as well as hydrophilic interactions.
FIGURE 3.
Disrupting PinX1 binding to TRF1 by the L291E mutation abolishes the ability of PinX1 to localize to telomeres in human cells. A, the L291E mutation completely abolishes the ability of either full-lengthPinX1 or its TID to interact with TRF1. The L291E mutation was introduced into full-length PinX1 or its TID, and mutant proteins were expressed in bacteria and purified as GST fusion proteins followed by GST pulldown with in vitro synthesized 35S-TRF1. B–E, the L291E mutation completely abolishes the ability of either full-length PinX1 or its TID to localize to telomeres. HT1080 cells were co-transfected overnight with vectors expressing RFP-tagged TRF1 and GFP-tagged wild-type full-length PinX1 (B) or its TID (D) or their L291E point mutants (C and E) followed by subjecting them to fluorescence microscopy. Yellow arrows point to colocalization at telomeres, whereas red arrows point to colocalization at nucleoli.
The PinX1-TRF1 Interaction Is Required for Recruiting PinX1 to Telomeres
After having defined the TRF1-PinX1 interaction, the key question is whether this interaction has any biological importance. We have previously shown that PinX1 and its 75-residue C-terminal TRF1 binding TID are localized to telomeres when co-expressed with TRF1 in cells and that overexpression of PinX1 or TID induces progressive telomere shortening in telomerase-positive cells, with the TID being much more potent in inducing rapid telomere shortening and cell senescence (93). However, it is unknown how PinX1 is recruited to telomeres where inhibition of telomere elongation occurs and whether the TRF1-PinX1 interaction plays any role in regulating PinX1 function. To address these questions, we examined the role of the TRF1-PinX1 interaction for PinX1 or TID to localize to telomeres and to induce telomere shortening independent of their ability to inhibit telomerase activity in cells.
To examine whether PinX1 or TID colocalizes with TRF1 at telomeres in vivo, we overexpressed GFP-PinX1 or -TID in human cells with RFP-TRF1 followed by immunofluorescence microscopy. As well documented previously (55, 87, 93, 102), GFP-PinX1 and RFP-TRF1 colocalized in a punctated pattern in the nucleus that is characteristic of telomeres (Fig. 3B, yellow arrows) as well as in nucleoli (Fig. 3B, red arrows). Furthermore, GFP-TID and RFP-TRF1 also colocalized to telomeres in addition to nucleoli (Figs. 2D and 3D). These results indicate that both PinX1 and TID colocalize with TRF1 at telomeres.
To understand whether this telomere colocalization is mediated by the TRF1-PinX1 interaction, we determined the subcellular localization of a large number of PinX1 mutants containing mutations that disrupt its hydrophilic or hydrophobic interaction with TRF1. All the PinX1 or TID mutants that bound to TRF1 localized to telomeres when co-expressed with TRF1 in cells (Fig. 2A). In contrast, all TRF1 or TID mutants that failed to bind TRF1, including PinX1L291E and PinX14A or TIDL291E and TID4A, were unable to localize to telomeres, although they remained localized to nucleoli (Figs. 2, A and D, 3, C and E). These results indicate that telomere localization of PinX1 is mediated by its direct interaction with TRF1.
Leu-291 in PinX1 Is Not Essential for PinX1 to Interact with TERT and Inhibit Telomerase Activity
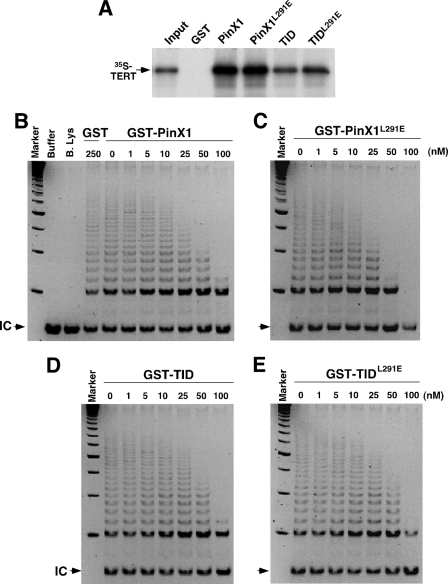
To further determine the importance of the TRF1-PinX1 interaction and, specifically, the recruitment of PinX1 to telomeres for PinX1 and TID to inhibit telomere elongation, we required TRF1 binding mutants of PinX1 and TID that are fully active in inhibiting telomerase activity. Therefore, we examined whether the TRF1 binding mutations in PinX1 or TID affected their ability to bind TERT or inhibit telomerase activity. Although the quadruple Ala substitution of the charged residues (4A mutation) reduced the ability of PinX1 and TID to bind to TERT and to inhibit telomerase activity (data not shown), the L291E point mutation had no effect on the ability of PinX1 or TID to bind or inhibit telomerase (Fig. 4). Compared with PinX1, the C-terminal TID had reduced binding to TERT (Fig. 4A), likely due to the fact that the N-terminal domain of PinX1 also contributes to TERT binding, as shown before (93). More importantly, PinX1L291E and TIDL291E bound to TERT at similar levels as their wild-type counterparts (Fig. 4A). Furthermore, PinX1L291E and TIDL291E inhibited telomerase activity as potently as, if not more than, their wild-type counterparts when detected using the TRAP assay in vitro (Fig. 4, B–E). These results indicate that the failure of PinX1L291E and TIDL291E to localize to telomeres is due to their inability to interact with TRF1. Hence, the TRF1-PinX1 interaction is required for PinX1 to localize to telomeres.
FIGURE 4.
The L291E point mutation in PinX1 does not affect the ability of PinX1 or its TID to interact TERT and inhibit telomerase activity. A, the L291E mutation does not affect the ability of PinX1 or TID to interact with human TERT. Glutathione beads bound to control GST, GST-PinX1, -PinX1L291E, -TID (D) or -TIDL291E were incubated with 35S-hTERT synthesized by in vitro transcription and translation. After washing, bound 35S-hTERT was separated on SDS-containing gels followed by autoradiography. B–E, the L291E mutation does not affect the ability of PinX1 to inhibit telomerase activity. Different concentrations of GST or GST-PinX1 (B), -PinX1L291E, (C), -TID (D), or -TIDL291E (E) were incubated with telomerase extracts for 10 min before the TRAP assay. Buffer alone or boiled lysates (B. Lys) were used in some assays as controls. Arrows point to the internal control (IC) for PCR amplification.
The TRF1-PinX1 Interaction Is Required for PinX1 or TID to Efficiently Inhibit Telomere Elongation in Human Cells
The above results indicate that the L291E mutation completely disrupts the ability of PinX1 or TID to interact with TRF1 but does not affect the ability of PinX1 or TID to interact with TERT and to inhibit telomerase activity. This allows us to specifically study the role of PinX1 recruitment to telomeres and understand the spatial significance of the protein while retaining its telomerase inhibitory function.
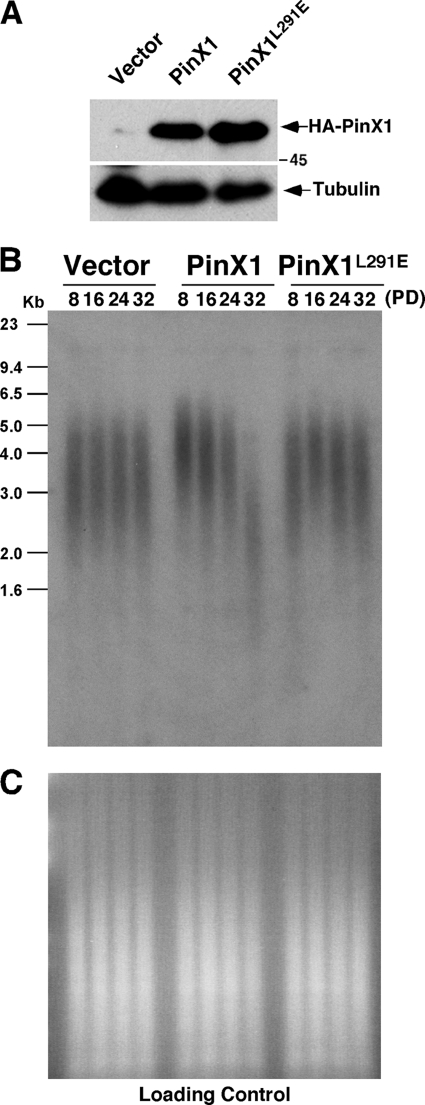
Therefore, we stably transfected telomerase-positive human HT1080 cells with the construct expressing HA-PinX1 or the L291E mutants and confirmed protein expression using immunoblotting with anti-HA antibody (Fig. 5A). These stable cells were continuously passaged in culture and periodically collected to assay telomere restriction fragments lengths at various population doublings (PDs) (55, 71, 93). As shown previously (93), overexpression of PinX1 led to gradual and progressive telomere shortening, which was detected by increased mobility of telomere restriction fragments and a decrease in the intensity of the hybridization signal in multiple independent experiments (Fig. 5B, data not shown). In contrast, there was no obvious change in the mobility of telomere restriction fragments or in the intensity of the hybridization signal in cells stably overexpressing the L291E mutant at various PDs (Fig. 5B, data not shown). This difference was not due to DNA loading as shown by staining gels with ethidium bromide before hybridization (Fig. 5C). These results indicate that the L291E mutation abolishes the ability of PinX1 to induce telomere shortening.
FIGURE 5.
PinX1-TRF1 interaction is required for PinX1 to inhibit telomere elongation in human cells. A, shown is an establishment of HT1080 cells stably expressing HA-PinX1, PinX1L291E, or control vector. HT1080 cells were stably transfected with the control vector or a vector expressing HA-PinX1 or its L291E mutant followed by detecting protein expression using immunoblot with anti-HA antibody. B and C, the L291E mutation abolishes the ability of PinX1 to induce telomere shortening in human cells. Stable cells were maintained continuously in culture and harvested at various PDs as indicated, and genomic DNA was isolated and digested with HinfI and RsaI followed by in-gel hybridization with a TTAGGG repeat probe (B). Before hybridization, gels were stained with ethidium bromide to check loading of genomic DNA (C).
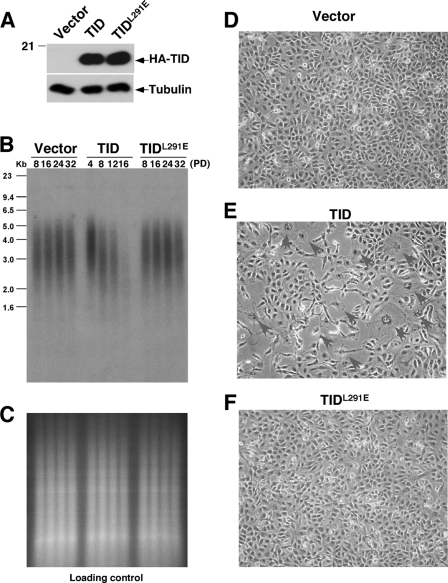
To further confirm these findings, we also generated stable human cells expressing HA-TID or the HA-TIDL291E mutants (Fig. 6A) because the TID induces telomere shortening more potently than the full-length protein and abruptly causes cellular senescence by PD 16 (93). Indeed, stable overexpression of TID was more potent than the full-length protein in inducing telomere shortening (Figs. 6B and 5B). However TIDL291E completely failed to induce obvious telomere shortening in multiple independent experiments (Fig. 6B, data not shown), which was not due to DNA loading (Fig. 6C). Furthermore, as shown previously (93), TID-overexpressing cells experienced growth arrest and exhibited increased cell size, flattened cell morphology, and positive staining for the senescence-associated β-galactosidase and could not grow beyond PD 16 (Fig. 6E). In sharp contrast, this phenotype was not observed in vector or TIDL291E cell lines, which could be continually passaged to PD 32 and which did not exhibit any growth impairment or senescence (Fig. 6, D and F). Taken together, these results indicate that although the L291E point mutation does not affect the ability of PinX1 or TID to inhibit telomerase activity, it completely abolishes the ability of PinX1 or TID to interact with TRF1, to localize to telomeres, and to induce telomere shortening and cell senescence.
FIGURE 6.
PinX1-TRF1 interaction is required for TID to inhibit telomere elongation and induce the senescence phenotype in human cells. A, establishment of HT1080 cells stably expressing HA-TID, TIDL291E, or control vector is shown. HT1080 cells were stably transfected with the control vector or a vector expressing HA-TID or its L291E mutant in two independent experiments followed by detecting protein expression using immunoblot with anti-HA antibody. B and C, the L291E mutation abolishes the ability of TID to induce telomere shortening in human cells. Stable cells were maintained continuously in culture and harvested at various PDs as indicated, and genomic DNA was isolated and digested with HinfI and RsaI followed by in-gel hybridization with a TTAGGG repeat probe (B). Before hybridization, gels were stained with ethidium bromide to check loading of genomic DNA (C). D–F, the L291E mutation abolishes the ability of TID to induce the senescence phenotype in human cells. HT1080 cell lines stably transfected with the control vector (D) or expressing similar levels of TID (E) and TIDL291E (F) were fixed at 16 PDs and then subjected to senescence-associated β-galactosidase staining followed by microscopy.
TRF1 Knockdown Completely Abolishes the Ability of PinX1 to Localize to Telomeres and Inhibit Telomere Elongation in Human Cells
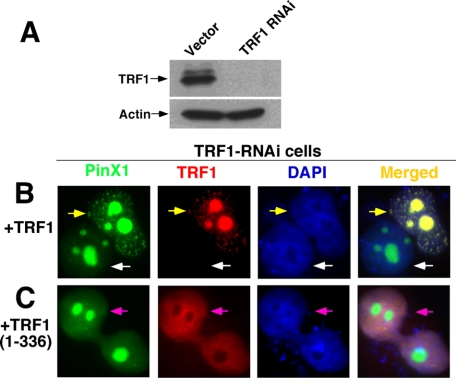
The above structure-function analyses suggest that TRF1 might be essential for recruiting PinX1 to telomeres to negatively regulate telomere elongation. To examine this possibility further, we knocked down endogenous TRF1 in cells using RNAi and then determined its effects on the ability of PinX1 to localize to telomeres and to induce telomere shortening. Localization of PinX1 in a TRF1 knockdown setting was accomplished by stably infecting HT1080 cells with TRF1-specific RNAi or control retroviruses followed by examining the subcellular localization of PinX1 in the presence or absence of re-expressed TRF1. In contrast to control cells, TRF1 protein was not detectable in TRF1 RNAi-stable cells (Fig. 7A). These TRF1-depleted cells were then co-transfected with GFP-PinX1 and either RFP-TRF1 or the RFP-TRF1-(1–336) mutant that is able to bind PinX1 (Fig. 1, A and B) but unable to localize to telomeres due to the lack of a DNA binding Myb domain (55, 87, 89, 90, 103). If telomeric localization of PinX1 is dependent on TRF1 recruitment, we would expect the mutant RFP-TRF1-(1–336) to be unable to guide GFP-PinX1 to telomeres. As expected, GFP-PinX1 failed to localize to telomeres in TRF1-depleted cells that did not express RFP-TRF1 and instead remained in nucleoli (Fig. 7B, white arrows). More importantly, however, telomere localization of GFP-PinX1 was re-established in TRF1-depleted cells upon re-expression of RFP-TRF1 (Fig. 7B, yellow arrows) but not its mutant TRF1-(1–336) (Fig. 7C). Together with the findings that disrupting the TRF1-PinX1 interaction abolishes PinX1 localization to telomeres (Fig. 2A), these results further emphasize that the presence of TRF1 is necessary for targeting PinX1 to telomeres.
FIGURE 7.
TRF1 knockdown abolishes the ability of PinX1 to localize to telomeres, which can be rescued by re-expression of TRF1 but not its mutant lacking the telomeric DNA binding domain. HT1080 cells were transfected with TRF1 RNAi, and stable clones were selected followed by immunoblotting analysis with anti-TRF1 antibodies to confirm depletion of TRF1 (A). TRF1-depleted cells were then transiently transfected with GFP-PinX1 and RFP-TRF1 (B) or RPF-TRF1-(1–336) mutant (C) overnight and fixed followed by subjection to fluorescence microscopy. Please note that PinX1 failed to localize to telomeres in TRF1-depleted cells (white arrows) or in TRF1-depleted cells that re-expressed the RPF-TRF1-(1–336) mutant lacking the telomeric DNA binding domain (pink arrows) but was able to localize to telomeres in TRF1-depleted cells that re-expressed wild-type RPF-TRF1 (yellow arrows).
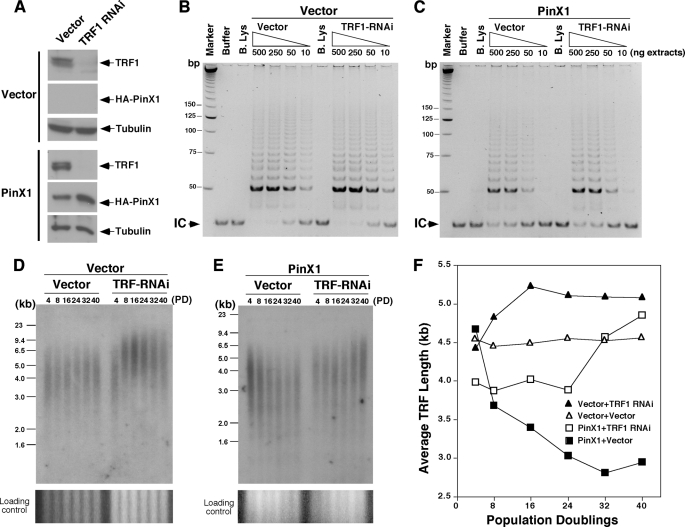
To determine whether TRF1 knockdown affects the ability of PinX1 to inhibit telomere elongation, HT1080 cells stably expressing PinX1 or the vector control, which were previously established (93), were infected with lentiviruses expressing a different TRF1 RNAi sequence or control vector followed by selection for stable cell pools. As compared with control viruses, the TRF1 RNAi viruses significantly reduced TRF1 expression both in PinX1-overexpressing cells and vector control cells (Fig. 8A). In vector control cells, TRF1 knockdown did not affect telomerase activity (Fig. 8B) but led to gradual and progressive telomere elongation (Fig. 8, D and F). These results confirm that the TRF1 RNAi effectively knocks down TRF1 expression in cells and also produces the expected phenotype of a steady increase in telomere length without affecting telomerase activity, as shown previously (5, 55, 68, 69).
FIGURE 8.
Knockdown of TRF1 completely abolishes the ability of PinX1 to inhibit telomere elongation in human cells. A, establishment of stable HT1080 cell pools expressing PinX1 with or without TRF1 knockdown is shown. HT1080 stable cells expressing HA-PinX1 or control vector at PD 1 were infected with lentiviruses expressing TRF1 RNAi or vector control virus. After selection, levels of TRF1 in stable cell pools were determined by immunoblot with anti-TRF1 antibodies. B and C, TRF1 knockdown does not affect the ability of PinX1 to inhibit telomerase activity in human cells. Stable HT1080 cell pools that expressed HA-PinX1 (C) or control vector (B) and TRF1 RNAi or vector control virus were harvested at eight PDs, and telomerase-containing fractions were prepared followed by subjecting different amounts of proteins as indicated to the TRAP assay. Buffer alone or boiled lysates (B. Lys) were used in some assays as controls. Arrows point to the internal control (IC) for PCR amplification. D–F, TRF1 knockdown completely abolishes the ability of PinX1 to inhibit telomere elongation in human cells. Stable HT1080 cell pools that expressed HA-PinX1 (E) or control vector (D) and TRF1 RNAi or vector control virus were maintained continuously in culture and harvested at various PD as indicated followed by telomere genomic Southern blotting analysis (D and E). Average telomere restriction fragment length versus PD number was quantified using ImageQuant (F).
Significantly, TRF1 knockdown in PinX1-overexpressing cells had no effect on telomerase activity but completely abolished the ability of PinX1 to inhibit telomere elongation. As shown previously (93), telomerase activity in PinX1-overexpressing cells was significantly lower than that in vector control cells due to the telomerase inhibitory activity of PinX1. This reduction in telomerase activity was not affected by TRF1 knockdown (Fig. 8C), confirming that TRF1 does not affect telomerase activity in cells (5, 55, 68, 69). Importantly, infecting PinX1-overexpressing cells with control viruses resulted in gradual and progressive telomere shortening (Fig. 8, E and F), validating the findings that overexpression of PinX1 in HT1080 cells inhibits telomerase activity and leads to telomere shortening (93). However, when TRF1 was knocked down in PinX1-overexpressing cells, telomere lengths were not changed in the first 24 PDs and were then followed by gradual telomere elongation (Fig. 8, E and F). The telomere elongation at late PDs is likely due to the long term depletion of TRF1, further substantiating that PinX1 acts downstream of TRF1 and is unable to inhibit telomere elongation when endogenous TRF1 is depleted. These results indicate that TRF1 knockdown has no effect on the ability of PinX1 to inhibit telomerase activity but abolishes the ability of PinX1 to localize to telomeres and to inhibit telomere elongation (Fig. 8), phenotypes that are almost the same as those induced by disrupting the TRF1-PinX1 interaction by the L291E PinX1 mutation (Figs. 5 and 6). Taken together, this study has not only identified residues important for PinX1 and TRF1 interactions but also uncovered the essential role of this interaction for recruiting PinX1 to telomeres and to inhibit telomere elongation in vivo.
DISCUSSION
Although PinX1 was originally identified as a TRF1-interacting protein, nothing is known about the biological function of this interaction. In this report we first performed extensive structure-function analysis to define the PinX1-TRF1 interaction and then used this information to dissect the biological function of the PinX1-TRF1 interaction in regulating telomere maintenance. We found that the TRFH domain of TRF1 specifically recognizes a 20-amino acid sequence (residues 291–310) of PinX1 by both hydrophobic as well as hydrophilic interactions (Fig. 1). Moreover, a single PinX1 point mutation in the hydrophobic interaction or the quadruple mutations in the hydrophilic interaction completely disrupts the ability of PinX1 or its TID to bind to TRF1 and to localize to telomeres in cells (Figs. 2 and 3). This indicates that the TRF1-PinX1 interaction is essential for recruiting PinX1 to telomeres. More importantly, the L291E point mutation in PinX1 completely abolishes its ability to interact with TRF1 (Figs. 2C and 3, A and B) but does not affect its telomerase inhibitory activity (Fig. 4). This allowed us to demonstrate that disrupting the TRF1-PinX1 interaction alone is sufficient to severely impair the ability of PinX1 or its TID to inhibit telomere elongation in human cells (Figs. 5 and 6). These structure-function analyses demonstrate that the TRF1-PinX1 interaction is required not only for targeting PinX1 to telomeres but also for PinX1 to prevent telomere elongation in cells.
To confirm these findings, we knocked down endogenous TRF1 in cells using RNAi to determine its impact on the ability of PinX1 to localize to telomeres and to inhibit telomere elongation. Indeed, TRF1 knockdown does not affect the ability of PinX1 to inhibit telomerase activity but completely abolishes the ability of PinX1 to localize to telomeres and to inhibit telomere elongation (Figs. 7 and 8), phenotypes that are similar to those induced by disrupting the TRF1-PinX1 interaction by the L291E mutation (Figs. 5 and 6). These functional studies demonstrate for the first time the essential role of the TRF1-PinX1 interaction for recruiting PinX1 to telomeres to inhibit telomere elongation in cells.
Given the unique and potent ability of PinX1 to inhibit telomerase activity and to induce telomere elongation in cells (93, 95), our results suggest that the TRF1-PinX1 interaction affects the loading of PinX1 onto telomeres to prevent telomere elongation. This may provide an additional level of telomerase regulation in conjunction to the physical occlusion from telomeres by Pot1 (73, 79) by supplying a link between TRF1 and telomerase inhibition that contributes toward maintaining telomere homeostasis. TRF1 binds along the duplex part of the telomere and functions as a measuring device to assess telomere length (5, 55, 69, 102). For telomere length homeostasis to be effective, the information about the length of the telomere needs to be relayed from TRF1 to telomerase via other proteins as TRF1 does not directly affect telomerase activity (5, 55). Our data indicate that the telomerase inhibitor PinX1 might be recruited by TRF1 to a telomere to stop telomerase action as the telomere is being elongated and reaches a certain threshold (Fig. 9). The two unique features of PinX1 that allow it to relay information about telomere length to telomerase are that it is a potent telomerase catalytic inhibitor and that its accumulation on telomeres depends on its specific interaction with TRF1.
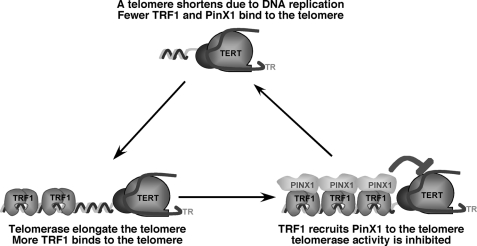
FIGURE 9.
A model for PinX1 as a link between TRF1 and telomerase inhibition. As telomeres are elongated by telomerase, increased TRF1 molecules bind, recruiting more PinX1 for telomerase inhibition. Once telomerase activity is inhibited, telomeres begin to shorten with each round of DNA replication, resulting in fewer TRF1 binding sites, less recruitment of PinX1 to inhibit telomerase, and therefore, re-entry of telomerase. This leads to telomere elongation and the start of another round of this cycle.
In this model (Fig. 9), when telomerase extends a telomere to a certain length, the elongated telomere binds more TRF1, which might in turn recruit more PinX1 to the telomere. Once concentrated locally on telomeres at a high concentration, which might be important because endogenous levels of TERT and PinX1 are rather low,5 simple mass-action might allow PinX1 to more effectively stop telomerase from adding more repeats. Conversely, when a telomere is shortened after each cell division due to the end replication problem (104–106), the shortened telomere contains less TRF1, which might recruit fewer PinX1 or none at all to the telomere. Therefore, the telomere might have a greater chance of being elongated due to telomerase being inhibited less or even not at all. This negative feedback mechanism might help maintain telomeres at a similar median length. Consistent with this model are the current findings that either disrupting the TRF1-PinX1 interaction by mutating the critical Leu-291 residue in PinX1 or knocking down endogenous TRF1 abolishes the ability of PinX1 to localize to telomeres and to inhibit telomere elongation in cells even though neither effects its ability to bind and inhibit telomerase activity. This model can also explain the previous findings that depleting either endogenous PinX1 (93) or endogenous TRF1 (80) or preventing TRF1 from binding to telomeres (55, 69, 70, 72) can all lead to telomere elongation. In contrast, overexpression of PinX1 (93) or TRF1 (55, 80) results in telomere erosion. Moreover, all these effects of TRF1 or PinX1 depend on the presence of telomerase activity in cells (55, 93). Because many other TRF1-interacting proteins have been shown to regulate telomere length (5, 66, 69–85, 93), further experiments are needed to determine how these different TRF1-interacting proteins are coordinated to regulate telomere homeostasis. Nevertheless, our results demonstrate that TRF1 recruits the telomerase inhibitor PinX1 to telomeres to prevent telomere elongation, which may help maintain telomere homeostasis by providing an important link between TRF1 and telomerase inhibition.
In summary, we have shown that TRF1 recognizes PinX1 through both hydrophobic as well as hydrophilic interactions and that this interaction is required for recruiting PinX1 to telomeres and also for PinX1 to prevent telomere elongation, as demonstrated either by mutating the critical Leu-291 residue in PinX1 to disrupt the TRF1-PinX1 interaction or by using RNA interference to knock down endogenous TRF1. These results indicate that the TRF1-mediated recruitment of PinX1 to telomeres helps maintain telomere homeostasis by providing a critical link between TRF1 and telomerase inhibition. Future studies aimed at understanding the structural details of how PinX1 inhibits telomerase action at telomeres, the relationship between PinX1 and other TRF1-interacting proteins, and the importance of PinX1-mediated telomerase regulation in pathological conditions such as cancer should help elucidate the molecular mechanisms of telomere maintenance and the pathological significance of telomerase deregulation.
Acknowledgments
We thank L. Cantley, W. Hahn, and D. Frank for constructive discussions and W. Hahn for constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA122434 (to X. Z. Z.).
X. Z. Zhou, unpublished data.
- TRF1
- telomeric DNA-binding protein 1
- TRF
- telomere restriction fragment
- TRFH
- TRF homology
- Pin2
- protein interacting with NIMA-2
- PinX1
- Pin2/TRF1-interacting protein X1
- PD
- population doubling
- TID
- TRF1-interacting domain or telomerase-inhibitory domain of PinX1
- RFP
- red fluorescent protein
- TRAP
- telomere repeat amplification protocol.
REFERENCES
- 1. Blackburn E. H. (2001) Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 2. Shay J. W., Wright W. E. (2002) Cancer Cell 2, 257–265 [DOI] [PubMed] [Google Scholar]
- 3. Maser R. S., DePinho R. A. (2002) Science 297, 565–569 [DOI] [PubMed] [Google Scholar]
- 4. Cech T. R. (2004) Cell 116, 273–279 [DOI] [PubMed] [Google Scholar]
- 5. de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 6. Greider C. W., Blackburn E. H. (1985) Cell 43, 405–413 [DOI] [PubMed] [Google Scholar]
- 7. Greider C. W., Blackburn E. H. (1989) Nature 337, 331–337 [DOI] [PubMed] [Google Scholar]
- 8. Shippen-Lentz D., Blackburn E. H. (1990) Science 247, 546–552 [DOI] [PubMed] [Google Scholar]
- 9. Yu G. L., Bradley J. D., Attardi L. D., Blackburn E. H. (1990) Nature 344, 126–132 [DOI] [PubMed] [Google Scholar]
- 10. Singer M. S., Gottschling D. E. (1994) Science 266, 404–409 [DOI] [PubMed] [Google Scholar]
- 11. Feng J., Funk W. D., Wang S. S., Weinrich S. L., Avilion A. A., Chiu C. P., Adams R. R., Chang E., Allsopp R. C., Yu J. (1995) Science 269, 1236–1241 [DOI] [PubMed] [Google Scholar]
- 12. Cohn M., Blackburn E. H. (1995) Science 269, 396–400 [DOI] [PubMed] [Google Scholar]
- 13. Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997) Science 276, 561–567 [DOI] [PubMed] [Google Scholar]
- 14. Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., Harley C. B., Cech T. R. (1997) Science 277, 955–959 [DOI] [PubMed] [Google Scholar]
- 15. Counter C. M., Meyerson M., Eaton E. N., Weinberg R. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9202–9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyerson M., Counter C. M., Eaton E. N., Ellisen L. W., Steiner P., Caddle S. D., Ziaugra L., Beijersbergen R. L., Davidoff M. J., Liu Q., Bacchetti S., Haber D. A., Weinberg R. A. (1997) Cell 90, 785–795 [DOI] [PubMed] [Google Scholar]
- 17. Broccoli D., Young J. W., de Lange T. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9082–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor R. S., Ramirez R. D., Ogoshi M., Chaffins M., Piatyszek M. A., Shay J. W. (1996) J. Invest. Dermatol. 106, 759–765 [DOI] [PubMed] [Google Scholar]
- 19. Masutomi K., Yu E. Y., Khurts S., Ben-Porath I., Currier J. L., Metz G. B., Brooks M. W., Kaneko S., Murakami S., DeCaprio J. A., Weinberg R. A., Stewart S. A., Hahn W. C. (2003) Cell 114, 241–253 [DOI] [PubMed] [Google Scholar]
- 20. Watson J. D. (1972) Nat. New Biol. 239, 197–201 [DOI] [PubMed] [Google Scholar]
- 21. Olovnikov A. M. (1973) J. Theor. Biol. 41, 181–190 [DOI] [PubMed] [Google Scholar]
- 22. Hockemeyer D., Sfeir A. J., Shay J. W., Wright W. E., de Lange T. (2005) EMBO J. 24, 2667–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sfeir A. J., Chai W., Shay J. W., Wright W. E. (2005) Mol. Cell 18, 131–138 [DOI] [PubMed] [Google Scholar]
- 24. Hayflick L., Moorhead P. (1961) Exp. Cell Res. 25, 585–621 [DOI] [PubMed] [Google Scholar]
- 25. Goldstein S. (1990) Science 249, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 26. Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., Greider C. W., Harley C. B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10114–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vaziri H., Schächter F., Uchida I., Wei L., Zhu X., Effros R., Cohen D., Harley C. B. (1993) Am. J. Hum. Genet. 52, 661–667 [PMC free article] [PubMed] [Google Scholar]
- 28. Harley C. B., Vaziri H., Counter C. M., Allsopp R. C. (1992) Exp. Gerontol. 27, 375–382 [DOI] [PubMed] [Google Scholar]
- 29. Wright W. E., Shay J. W. (1992) Exp. Gerontol. 27, 383–389 [DOI] [PubMed] [Google Scholar]
- 30. Chin L., Artandi S. E., Shen Q., Tam A., Lee S. L., Gottlieb G. J., Greider C. W., DePinho R. A. (1999) Cell 97, 527–538 [DOI] [PubMed] [Google Scholar]
- 31. Reaper P. M., di Fagagna F., Jackson S. P. (2004) Cell Cycle 3, 543–546 [PubMed] [Google Scholar]
- 32. Counter C. M., Avilion A. A., LeFeuvre C. E., Stewart N. G., Greider C. W., Harley C. B., Bacchetti S. (1992) EMBO J. 11, 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 34. Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. (1999) Nature 400, 464–468 [DOI] [PubMed] [Google Scholar]
- 35. Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., Wright W. E. (1998) Science 279, 349–352 [DOI] [PubMed] [Google Scholar]
- 36. Vaziri H., Benchimol S. (1998) Curr. Biol. 8, 279–282 [DOI] [PubMed] [Google Scholar]
- 37. Halvorsen T. L., Leibowitz G., Levine F. (1999) Mol. Cell. Biol. 19, 1864–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo C., Geverd D., Liao R., Hamad N., Counter C. M., Price D. T. (2001) J. Urol. 166, 694–698 [PubMed] [Google Scholar]
- 39. Wang J., Xie L. Y., Allan S., Beach D., Hannon G. J. (1998) Genes Dev. 12, 1769–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greenberg R. A., O'Hagan R. C., Deng H., Xiao Q., Hann S. R., Adams R. R., Lichtsteiner S., Chin L., Morin G. B., DePinho R. A. (1999) Oncogene 18, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 41. Wu K. J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., Dalla-Favera R. (1999) Nat. Genet. 21, 220–224 [DOI] [PubMed] [Google Scholar]
- 42. Lin S. Y., Elledge S. J. (2003) Cell 113, 881–889 [DOI] [PubMed] [Google Scholar]
- 43. Blasco M. A., Lee H. W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W. (1997) Cell 91, 25–34 [DOI] [PubMed] [Google Scholar]
- 44. Lee H. W., Blasco M. A., Gottlieb G. J., Horner J. W., 2nd, Greider C. W., DePinho R. A. (1998) Nature 392, 569–574 [DOI] [PubMed] [Google Scholar]
- 45. Rudolph K. L., Chang S., Lee H. W., Blasco M., Gottlieb G. J., Greider C., DePinho R. A. (1999) Cell 96, 701–712 [DOI] [PubMed] [Google Scholar]
- 46. Greenberg R. A., Chin L., Femino A., Lee K. H., Gottlieb G. J., Singer R. H., Greider C. W., DePinho R. A. (1999) Cell 97, 515–525 [DOI] [PubMed] [Google Scholar]
- 47. Farazi P. A., Glickman J., Jiang S., Yu A., Rudolph K. L., DePinho R. A. (2003) Cancer Res. 63, 5021–5027 [PubMed] [Google Scholar]
- 48. González-Suárez E., Samper E., Flores J. M., Blasco M. A. (2000) Nat. Genet. 26, 114–117 [DOI] [PubMed] [Google Scholar]
- 49. Rudolph K. L., Millard M., Bosenberg M. W., DePinho R. A. (2001) Nat. Genet. 28, 155–159 [DOI] [PubMed] [Google Scholar]
- 50. Qi L., Strong M. A., Karim B. O., Armanios M., Huso D. L., Greider C. W. (2003) Cancer Res. 63, 8188–8196 [PubMed] [Google Scholar]
- 51. Wong K. K., Maser R. S., Bachoo R. M., Menon J., Carrasco D. R., Gu Y., Alt F. W., DePinho R. A. (2003) Nature 421, 643–648 [DOI] [PubMed] [Google Scholar]
- 52. Artandi S. E., Chang S., Lee S. L., Alson S., Gottlieb G. J., Chin L., DePinho R. A. (2000) Nature 406, 641–645 [DOI] [PubMed] [Google Scholar]
- 53. O'Hagan R. C., Chang S., Maser R. S., Mohan R., Artandi S. E., Chin L., DePinho R. A. (2002) Cancer Cell 2, 149–155 [DOI] [PubMed] [Google Scholar]
- 54. Shampay J., Szostak J. W., Blackburn E. H. (1984) Nature 310, 154–157 [DOI] [PubMed] [Google Scholar]
- 55. van Steensel B., de Lange T. (1997) Nature 385, 740–743 [DOI] [PubMed] [Google Scholar]
- 56. Cristofari G., Lingner J. (2006) EMBO J. 25, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Conrad M. N., Wright J. H., Wolf A. J., Zakian V. A. (1990) Cell 63, 739–750 [DOI] [PubMed] [Google Scholar]
- 58. Lustig A. J., Kurtz S., Shore D. (1990) Science 250, 549–553 [DOI] [PubMed] [Google Scholar]
- 59. Schulz V. P., Zakian V. A. (1994) Cell 76, 145–155 [DOI] [PubMed] [Google Scholar]
- 60. Krauskopf A., Blackburn E. H. (1996) Nature 383, 354–357 [DOI] [PubMed] [Google Scholar]
- 61. Nugent C. I., Hughes T. R., Lue N. F., Lundblad V. (1996) Science 274, 249–252 [DOI] [PubMed] [Google Scholar]
- 62. Marcand S., Gilson E., Shore D. (1997) Science 275, 986–990 [DOI] [PubMed] [Google Scholar]
- 63. Cooper J. P., Nimmo E. R., Allshire R. C., Cech T. R. (1997) Nature 385, 744–747 [DOI] [PubMed] [Google Scholar]
- 64. Evans S. K., Lundblad V. (1999) Science 286, 117–120 [DOI] [PubMed] [Google Scholar]
- 65. Zhou J., Monson E. K., Teng S. C., Schulz V. P., Zakian V. A. (2000) Science 289, 771–774 [DOI] [PubMed] [Google Scholar]
- 66. Baumann P., Cech T. R. (2001) Science 292, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 67. Boulé J. B., Vega L. R., Zakian V. A. (2005) Nature 438, 57–61 [DOI] [PubMed] [Google Scholar]
- 68. van Steensel B., Smogorzewska A., de Lange T. (1998) Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- 69. Ancelin K., Brunori M., Bauwens S., Koering C. E., Brun C., Ricoul M., Pommier J. P., Sabatier L., Gilson E. (2002) Mol. Cell. Biol. 22, 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smith S., Giriat I., Schmitt A., de Lange T. (1998) Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- 71. Kim S. H., Kaminker P., Campisi J. (1999) Nat. Genet. 23, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Smith S., de Lange T. (2000) Curr. Biol. 10, 1299–1302 [DOI] [PubMed] [Google Scholar]
- 73. Loayza D., De Lange T. (2003) Nature 423, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 74. Colgin L. M., Baran K., Baumann P., Cech T. R., Reddel R. R. (2003) Curr. Biol. 13, 942–946 [DOI] [PubMed] [Google Scholar]
- 75. Smogorzewska A., de Lange T. (2004) Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 76. Ye J. Z., de Lange T. (2004) Nat. Genet. 36, 618–623 [DOI] [PubMed] [Google Scholar]
- 77. Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004) Genes Dev. 18, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004) Nat. Cell Biol. 6, 673–680 [DOI] [PubMed] [Google Scholar]
- 79. Kelleher C., Kurth I., Lingner J. (2005) Mol. Cell. Biol. 25, 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee T. H., Perrem K., Harper J. W., Lu K. P., Zhou X. Z. (2006) J. Biol. Chem. 281, 759–768 [DOI] [PubMed] [Google Scholar]
- 81. Wu L., Multani A. S., He H., Cosme-Blanco W., Deng Y., Deng J. M., Bachilo O., Pathak S., Tahara H., Bailey S. M., Deng Y., Behringer R. R., Chang S. (2006) Cell 126, 49–62 [DOI] [PubMed] [Google Scholar]
- 82. Hockemeyer D., Daniels J. P., Takai H., de Lange T. (2006) Cell 126, 63–77 [DOI] [PubMed] [Google Scholar]
- 83. Hockemeyer D., Palm W., Else T., Daniels J. P., Takai K. K., Ye J. Z., Keegan C. E., de Lange T., Hammer G. D. (2007) Nat. Struct. Mol. Biol. 14, 754–761 [DOI] [PubMed] [Google Scholar]
- 84. Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 85. Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007) Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 86. Lu K. P., Hanes S. D., Hunter T. (1996) Nature 380, 544–547 [DOI] [PubMed] [Google Scholar]
- 87. Shen M., Haggblom C., Vogt M., Hunter T., Lu K. P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lu K. P., Zhou X. Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 904–916 [DOI] [PubMed] [Google Scholar]
- 89. Kishi S., Lu K. P. (2002) J. Biol. Chem. 277, 7420–7429 [DOI] [PubMed] [Google Scholar]
- 90. Nakamura M., Zhou X. Z., Kishi S., Kosugi I., Tsutsui Y., Lu K. P. (2001) Curr. Biol. 11, 1512–1516 [DOI] [PubMed] [Google Scholar]
- 91. Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., Tsai R. Y. (2009) J. Cell Biol. 185, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee T. H., Tun-Kyi A., Shi R., Lim J., Soohoo C., Finn G., Balastik M., Pastorino L., Wulf G., Zhou X. Z., Lu K. P. (2009) Nat. Cell. Biol. 11, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhou X. Z., Lu K. P. (2001) Cell 107, 347–359 [DOI] [PubMed] [Google Scholar]
- 94. Zhou X. Z., Perrem K., Lu K. P. (2003) J. Cell. Biochem. 89, 19–37 [DOI] [PubMed] [Google Scholar]
- 95. Liao C., Zhao M. J., Zhao J., Jia D., Song H., Li Z. P. (2002) World J. Gastroenterol 8, 1050–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lin J., Blackburn E. H. (2004) Genes Dev. 18, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Banik S. S., Counter C. M. (2004) J. Biol. Chem. 279, 51745–51748 [DOI] [PubMed] [Google Scholar]
- 98. Oh B. K., Yoon S. M., Lee C. H., Park Y. N. (2007) Gene 400, 35–43 [DOI] [PubMed] [Google Scholar]
- 99. Lim J., Balastik M., Lee T. H., Nakamura K., Liou Y. C., Sun A., Finn G., Pastorino L., Lee V. M., Lu K. P. (2008) J. Clin. Invest. 118, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen Y., Yang Y., van Overbeek M., Donigian J. R., Baciu P., de Lange T., Lei M. (2008) Science 319, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 101. Fairall L., Chapman L., Moss H., de Lange T., Rhodes D. (2001) Mol. Cell 8, 351–361 [DOI] [PubMed] [Google Scholar]
- 102. Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. (1995) Science 270, 1663–1667 [DOI] [PubMed] [Google Scholar]
- 103. Kishi S., Wulf G., Nakamura M., Lu K. P. (2001) Oncogene 20, 1497–1508 [DOI] [PubMed] [Google Scholar]
- 104. Cooke H. J., Smith B. A. (1986) Cold Spring Harbor Symp. Quant. Biol. 51, 213–219 [DOI] [PubMed] [Google Scholar]
- 105. Harley C. B., Futcher A. B., Greider C. W. (1990) Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 106. Hastie N. D., Dempster M., Dunlop M. G., Thompson A. M., Green D. K., Allshire R. C. (1990) Nature 346, 866–868 [DOI] [PubMed] [Google Scholar]