Abstract
Macroautophagy, a homeostatic process that shuttles cytoplasmic constituents into endosomal and lysosomal compartments, has recently been shown to deliver antigens for presentation on major histocompatibility complex (MHC) class II molecules. Skeletal muscle fibers show a high level of constitutive macroautophagy and express MHC class II molecules upon immune activation. We found that tumor necrosis factor-α (TNF-α), a monokine overexpressed in inflammatory myopathies, led to a marked up-regulation of macroautophagy in skeletal myocytes. Furthermore, TNF-α augmented surface expression of MHC class II molecules in interferon-γ (IFN-γ)-treated myoblasts. The synergistic effect of TNF-α and IFN-γ on the induction of MHC class II surface expression was not reflected by higher intracellular human leukocyte antigen (HLA)-DR levels and was reversed by macroautophagy inhibition, suggesting that TNF-α facilitates antigen processing via macroautophagy for more efficient MHC class II loading. Muscle biopsies from patients with sporadic inclusion body myositis, a well defined myopathy with chronic inflammation, showed that over 20% of fibers that contained autophagosomes costained for MHC class II molecules and that more than 40% of double-positive muscle fibers had contact with CD4+ and CD8+ immune cells. These findings establish a mechanism through which TNF-α regulates both macroautophagy and MHC class II expression and suggest that macroautophagy-mediated antigen presentation contributes to the immunological environment of the inflamed human skeletal muscle.
Keywords: Antigen Processing, Autophagy, Cytokine, Cytokine Action, Immunology
Introduction
Autophagy is a homeostatic process that enables eukaryotic cells to deliver cytoplasmic constituents for lysosomal degradation, for example to recycle nutrients for survival during starvation. In addition to this original function, autophagy has emerged as a key mechanism in orchestrating innate and adaptive immune responses. During macroautophagy, the major route of degradation of cytoplasmic constituents, proteins, and organelles are sequestered inside double-membrane vesicles (autophagosomes) that fuse with lysosomes/late endosomes. In the fusion vesicles, often multivesicular and multilamellar amphisomes, the captured material is degraded, and antigenic fragments are loaded onto major histocompatibility complex (MHC) class II molecules for presentation to CD4+ T cells. In turn, innate and adaptive immune signals are capable of regulating macroautophagy via cytokine secretion and cell contact-dependent mechanisms (1–3).
In vivo analysis of macroautophagy in transgenic mice expressing the GFP-coupled specific autophagosome marker light chain 3 (LC3), one mammalian homologue of yeast autophagy-related gene 8 (Atg8), demonstrated that the regulation of macroautophagy is organ-dependent and that some tissues produce LC3-positive autophagosomes even in the absence of nutrient starvation. Such constitutive autophagosomal activity has been observed in metabolically active tissues such as liver, thymus, and skeletal muscle (4).
Skeletal myocytes are facultative antigen-presenting cells, and they actively participate in local immune reactions (5, 6), which develop spontaneously during the course of autoimmune and infectious muscle diseases (7) or can be induced by immunotherapeutic gene transfer into muscle (8). Up-regulation of macroautophagy and lysosomal genes has been documented at the transcript and protein level in different settings in myopathies associated with immune cell infiltration (9–13). We previously showed that muscle fibers of patients with sporadic inclusion body myositis (sIBM),7 the most common acquired skeletal muscle disease in patients above the age of 50 years, show increased frequencies of autophagosomes in degenerating fibers compared with nonmyopathic muscle or nonvacuolated myopathic controls and that intracellular amyloid precursor protein/β-amyloid colocalized with autophagosomal compartments (10, 11). Notably, colocalization of amyloid precursor protein/β-amyloid and LC3 was associated with up-regulation of MHC class II molecules as well as CD4+ and CD8+ immune cell infiltration. The mechanisms that regulate macroautophagy and MHC expression in the immunological environment of the skeletal muscle are not well understood. We investigated immune-mediated macroautophagy regulation and MHC expression in primary human muscle cells and in skeletal muscle biopsies from patients with sIBM.
MATERIALS AND METHODS
Patients
Under Institutional Review Board-approved protocols, muscle biopsies were used from ten patients with sIBM, who fulfilled the typical clinical and histopathological criteria.
Cell Culture
All cell lines used in this study were routinely cultured in DMEM with 10% FCS, 2 mm glutamine, 110 μg/ml sodium pyruvate, and 2 μg/ml gentamycin. CCL136 cells were purchased from ATCC (Manassas, VA); HaCat cells were a kind gift from Dr. Rajiv Khanna (Brisbane, Australia), and MDAMC cells were a kind gift from Dr. Irene Joab (Paris, France). LC3 fusion constructs and stable transfectants were generated as described previously (14). Primary human skeletal muscle cells from healthy donors were purchased from PromoCell (Heidelberg, Germany) and cultured as well as differentiated into myotubes according to the provider's recommendations. T cells, monocytes, and dendritic cells (DCs) were prepared from peripheral blood mononuclear cells isolated from leukocyte concentrates (buffy coats) from the New York Blood Center. Positive selection for T cells and monocytes/macrophages was performed with anti-CD3 and anti-CD14 MicroBeads from Miltenyi Biotec, respectively. 1 × 106 T cells were cultured in the presence of phytohemagglutinin (1 μg/ml) for 48 h. DCs were generated from CD14-positive cells in RPMI 1640 + 1% single donor plasma + glutamine + gentamycin. Recombinant human IL-4 (L-4, 500 units/ml) and recombinant human GM-CSF (1000 units/ml) were added on days 0, 2, and 4. For maturation, floating immature DCs were transferred to new plates on day 5, and half of the medium was replaced with fresh medium containing the TLR3 agonist poly(I-C) (25 μg/ml). IL-4 was obtained from PeproTech (Rocky Hill, NJ), and GM-CSF was from Immunex (Seattle, WA).
atg12-specific RNA Interference
The following 21-nucleotide siRNA oligonucleotides were used: atg12 sense, 5′-UCAACUUGCUACUACAUGAUdT; atg12 antisense, 5′-UCAUGUAGUAGCAAGUUGAUdT (nucleotides 687–705 of NM_004707). siRNA duplexes were delivered by transfection with Lipofectamine 2000 (Invitrogen) at 30 pmol of siRNA + 1.5 μl of Lipofectamine/well in a 24-well format, and the effect of RNA silencing was analyzed after 2–3 days.
Antibodies
The LC3 antiserum was generated by immunizing two rabbits with the N-terminal peptide LC3(1–15) (MPSEKTFKQRRTFEQR) conjugated to keyhole limpet hemocyanin carrier protein (Cocalico Biologicals, Reamstown, PA). Animals were boosted 5 times (2, 3, 7, 11, and 15 weeks after initial inoculation) and then sacrificed to obtain terminal bleeds. Antiserum collected from one rabbit showed good LC3 reactivity by ELISA and in Western blots and was used at a dilution of 1:2000 for Western blots. A polyclonal rabbit anti-human amyloid precursor protein antibody recognizing an epitope within the C terminus of the protein (AHP663, Serotec, Oxford, UK) was used at a 1:100 dilution. Anti-β-actin antibody was purchased from Sigma. Immunoblots were performed as described previously (14). For immunocytochemistry, we used a monoclonal mouse anti-human LC3-specific antibody (Nanotools, Teningen, Germany). Secondary antibodies were rhodamine red-X-conjugated donkey anti-mouse IgGs (Jackson ImmunoResearch, West Grove, PA). FITC-conjugated TNF-R1 and phycoerythrin-conjugated TNF-R2 antibodies and their respective isotype controls were purchased from R & D Systems (Minneapolis, MN). Phycoerythrin-conjugated HLA-DR and FITC-conjugated HLA-A, -B, and -C and their respective isotype controls were purchased from BD Biosciences. For immunohistochemistry, LC3 was detected by the rabbit polyclonal antiserum (see above) at 1:1000 dilution; for surface MHC class II (DP, DQ, and DR), a mouse monoclonal antibody (clone TDR31.1 from Ancell, Bayport, MN) was used at 10 μg/ml; CD3 was detected by a rat monoclonal antibody (clone CD-12 from Serotec, Düsseldorf, Germany) at 5 μg/ml; CD4 was labeled by a mouse monoclonal antibody (clone 34930 from R & D Systems) at 2 μg/ml; CD8 was stained using a rat monoclonal antibody (clone YTC 182.20 from AbD Serotec, Düsseldorf, Germany) at 5 μg/ml.
Immunocytochemistry and Confocal Microscopy
Cells were grown on microscopy cover glasses in 24-well plates overnight, fixed in 3% paraformaldehyde in PBS for 15 min, and permeabilized in 0.1% Triton X-100 in PBS for 5 min. Cells were blocked for 30 min in blocking buffer (TSA kit from PerkinElmer Life Sciences) + 0.1% saponin. Primary and secondary antibodies were applied in blocking buffer + 0.1% saponin + 5% normal donkey serum for 30–60 min, followed by three 5-min washes in PBS + 0.1% saponin. Finally, cells were stained with DAPI nucleic acid stain (0.5 μg/ml, Invitrogen) for 1 min, and cover glasses were mounted onto microscope slides with Prolong Gold antifade reagent (Invitrogen). All steps were carried out at room temperature. Cells were analyzed with an inverted LSM 510 laser scanning confocal microscope (Zeiss Axiovert 200) with a ×63 or ×100/1.4 N.A. oil immersion lens with a pinhole diameter of 1 airy unit. Pictures were taken with the LSM 510 confocal software (Zeiss).
Immunohistochemistry and Fluorescence Microscopy
For immunohistochemistry, 5-μm frozen sections of muscle biopsy specimens were fixed in 4% paraformaldehyde (LC3) or ice-cold acetone (all other antibodies) for 10 min. Unspecific binding was reduced by 30 min of incubation with 5% bovine serum albumin (BSA) and 3% goat serum (all from Jackson ImmunoResearch, West Grove, PA) in PBS. All primary and secondary reagents were diluted in 1% BSA. Double labeling for LC3 and MHC class II as well as for CD3 and CD4 was followed by secondary goat anti-rabbit IgG-Alexa 594 or anti-mouse IgG-Alexa 488, respectively (Invitrogen). Nuclear counterstaining was performed by DAPI, followed by mounting in Fluoromount G (Electron Microscopy Sciences, Hatfield, PA). Immunofluorescent microscopy and digital photography was performed on a Zeiss Axiophot microscope (Zeiss, Göttingen, Germany), using appropriate filters for green (488 nm), red (594 nm), and blue (350 nm) fluorescence and a cooled CCD digital camera (Retiga 1300, Qimaging, Burnaby, British Columbia, Canada) using the QCapture software. For quantitative analysis of double labeling of CD3 and CD4, five inflammatory fields with a mean of 249 cells were manually counted in 10 muscle biopsies of patients with sIBM.
Flow Cytometry
Primary human myoblasts or CCL136 cells were harvested from tissue culture plates and washed once in PBS, either permeabilized in 0.5% saponin for 10 min at room temperature or left nonpermeabilized, and stained with the directly fluorochrome-labeled antibodies phycoerythrin-conjugated HLA-DR and FITC-conjugated HLA-A, -B, and -C and their respective isotype controls (BD Biosciences) for 30 min at 4 °C. The cells were washed once with PBS and resuspended in 200 μl of FACS buffer prior to FACS analysis. The samples were measured on a BD LSR II flow cytometer (BD Biosciences). Gating and calculations for precursor frequencies were performed with FlowJo software (Tree Star, Ashland, OR).
Luminex Assay
Cell supernatants from DC and T cell cultures were analyzed for 20 cytokines and chemokines using the protein multiplex immunoassay kit (BIOSOURCE) as per the manufacturer's protocol. In brief, multiplex beads were vortexed and sonicated for 30 s, and 25 μl was added to each well and washed two times with wash buffer. The samples were diluted 1:2 with assay diluent and loaded onto a multiscreen BV 96-well filter plate with 50 μl of incubation buffer already added to each well. Serial dilutions of cytokine standards were prepared in parallel and added to the plate. Samples were then incubated on a plate shaker at 600 rpm in the dark at room temperature for 2 h. The plate was applied to a multiscreen vacuum manifold (Millipore) and washed twice with 200 μl of wash buffer. 100 μl of biotinylated anti-human multicytokine reporter was added to each well. The plate was incubated on a plate shaker at 600 rpm in the dark at room temperature for 1 h. The plate was applied to a multiscreen vacuum manifold (Millipore) and washed twice with 200 μl of wash buffer. Streptavidin/phycoerythrin was diluted 1:10 in wash buffer, and 100 μl was added directly to each well. The plate was incubated on a plate shaker at 600 rpm in the dark at room temperature for 30 min. The plate was then applied to the vacuum manifold and washed twice, and each well was resuspended in 100 μl of wash buffer and shaken for 1 min. The assay plate was then transferred to the Bio-Plex Luminex 100 XYP instrument for analysis. Cytokine concentrations were calculated using Bio-Plex Manager 3.0 software with a five-parameter curve-fitting algorithm applied for standard curve calculations.
DNA Fragmentation
Apoptotic DNA fragmentation was assessed as described previously (15, 16). Briefly, cells were fixed by adding 70% (v/v) cold ethanol on ice, washed, and stained with TO-PRO-3 as DNA intercalating agent for 1 h. Hypodiploid and diploid DNA peaks were quantified by flow cytometry.
Statistics
Statistical analyses were performed using commercial software (PRISM 4, GraphPad Software, San Diego). The paired t test was used to compare MHC expression levels and specific DNA fragmentation in cytokine-stimulated muscle cell.
RESULTS
Macroautophagy Is a Constitutively Active Process in Human Skeletal Muscle Cells
To quantify macroautophagy, we made use of the specific autophagosome marker LC3. LC3 is a ubiquitin-like protein that is covalently coupled via its C terminus to a phospholipid in the newly forming inner and outer autophagosomal membranes and thus is specifically incorporated into autophagosomes (17). After fusion of autophagosomes with endosomes or lysosomes, intraluminal LC3 is rapidly degraded by lysosomal proteases. The more autophagosomes that are formed, the more LC3 is degraded in autolysosomes, and therefore, lysosomal turnover of LC3 is a good measure for macroautophagic activity (18). To visualize the lysosomal turnover of LC3 in human skeletal muscle cells by fluorescence microscopy, we transduced a human skeletal muscle cell line (CCL136) with a lentiviral GFP-LC3 fusion construct (14). GFP-LC3-transfected cells were treated with the lysosomal acidification inhibitor chloroquine (CQ) to block lysosomal proteolysis and thereby to visualize the accumulation of GFP-LC3 in autolysosomes.
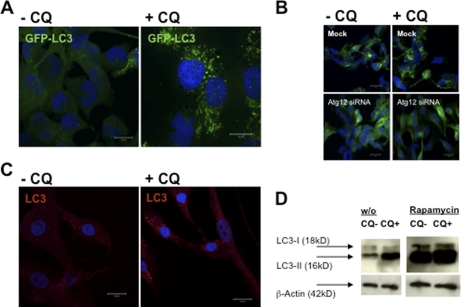
In the steady state (no CQ), only a small fraction of CCL136 cells had GFP-LC3-positive autophagosomes (Fig. 1A). However, GFP-LC3 strongly accumulated in cytosolic vesicles after 10 h of CQ treatment (Fig. 1A, right panel), suggesting that large numbers of GFP-LC3-labeled autophagosomes had formed and fused with lysosomes/late endosomes for GFP-LC3 degradation during the 10-h observation period. The accumulation of GFP-LC3+ vesicles upon CQ treatment was dependent on macroautophagy, because siRNA-mediated silencing of atg12, a gene essential for autophagosome formation, abrogated accumulation of these vesicles (Fig. 1B). Similar to CCL136 cells, primary human myoblasts showed a substantial level of constitutive macroautophagy and a marked accumulation of LC3+ vesicular compartments upon CQ treatment (Fig. 1C).
FIGURE 1.
Macroautophagy is a constitutively active process in human skeletal muscle cells. A, human rhabdomyosarcoma cells (CLL136) were stably transfected with a GFP-LC3 reporter construct and analyzed for GFP-LC3 turnover with and without lysosomal proteolysis blockade due to CQ treatment, 50 μm for 10 h. GFP-LC3 strongly accumulated in ring-shaped and cup-shaped cytosolic vesicles after 10 h of CQ treatment (A, right panel), suggesting that large numbers of GFP-LC3-labeled autophagosomes had formed and fused with lysosomes/late endosomes during the 10-h observation period. Cells were fixed, stained with DAPI nucleic acid stain, and analyzed by confocal microscopy. Scale bars, 20 μm. B, accumulation of GFP-LC3+ vesicles upon CQ treatment was dependent on macroautophagy, because siRNA-mediated silencing of atg12 abrogated accumulation of these vesicles. C, similar to the skeletal muscle cell line, human primary myoblasts stained with a monoclonal antibody specific for LC3 showed a detectable level of constitutive macroautophagy and a marked accumulation of LC3+ vesicular compartments upon CQ treatment. D, autophagosome-associated LC3 (called LC3-II) and free cytosolic LC3 (called LC3-I) can be distinguished by their apparent molecular weights in SDS-PAGE (16 and 18 kDa, respectively), and thus can be quantified separately in anti-LC3 immunoblots. Autophagosome-associated LC3-II strongly accumulated upon CQ treatment, further demonstrating that LC3-II-labeled autophagosomes were constitutively degraded in endosomes and/or lysosomes over the course of 10 h. In addition, inhibition of the mammalian target of rapamycin by rapamycin resulted in a substantial increase in LC3-II expression in both CQ-treated and untreated cells. w/o, without.
Autophagosome-associated LC3 (called LC3-II) and free cytosolic LC3 (called LC3-I) can be distinguished by their apparent molecular weights in SDS-PAGE (16 and 18 kDa, respectively) and thus can be quantified separately in anti-LC3 immunoblots (14, 17). CCL136 muscle cells were cultured in the presence or absence of the lysosomal protease inhibitor CQ for 10 h, and accumulation of LC3-II was quantified by immunoblotting. Autophagosome-associated LC3-II strongly accumulated upon CQ treatment (Fig. 1D), demonstrating that LC3-II-labeled autophagosomes were constitutively degraded in endosomes and/or lysosomes over the course of 10 h. In addition, inhibition of the mammalian target of rapamycin by rapamycin resulted in a substantial increase in LC3-II expression in both CQ-treated and untreated cells (Fig. 1D). Altogether, these experiments confirm that macroautophagy is a constitutively active process in human skeletal muscle cells under nutrient-rich conditions.
TNF-α Selectively Up-regulates Macroautophagy in Skeletal Myocytes
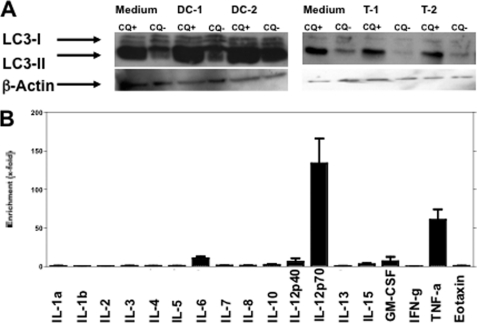
T cells and myeloid DCs are essential components of inflammatory infiltrates in most immune-mediated skeletal muscle diseases (19). To identify immune factors that stimulate macroautophagy in skeletal myocytes, we exposed CCL136 muscle cells to supernatants derived from mitogen-activated polyclonal T cells and mature monocyte-derived DCs. As shown in Fig. 2A, accumulation of LC3-II was observed after treatment with DC-derived supernatants only. We next quantified a set of 20 soluble inflammatory molecules in both DC- and T cell-derived supernatants via Luminex (Fig. 2B). Cytokines that were detected in higher concentrations in DC-derived compared with T cell supernatants included interleukin (IL)-12, IL-15, IL-6, GM-CSF. and TNF-α (Fig. 2B). These candidates together with IFN-γ as a classical proinflammatory cytokine produced by cytotoxic T cells were tested for their ability to induce LC3-II accumulation in CCL136 muscle cells after 24 h by Western blotting. Each cytokine was tested at least three times at a concentration quantified in stimulatory DC-derived supernatants by Luminex as well as at a 1 log higher concentration (Table 1). Among all 10 candidates tested, only TNF-α showed a consistent effect on LC3-II accumulation. Notably, neither IFN-γ nor IP-10 as a downstream molecule induced by IFN-γ augmented LC3-II expression in CCL136 skeletal muscle cells (Fig. 2A, left panel). In contrast, the TNF-α induced increase in LC3-II expression was present in both CQ-treated and untreated cells and was detectable at TNF-α concentrations as low as 0.05 ng/ml (Fig. 3A, middle panel).
FIGURE 2.
Cytokines enriched in supernatants with stimulatory activity on macroautophagy in skeletal myocytes. A, CCL136 muscle cells were incubated for 24 h with supernatants derived from monocyte-derived DCs matured with poly(I-C) as TLR3 agonists (left panel) and polyclonal T cells (T) stimulated with phytohemagglutinin (right panel). Accumulation of LC3-II was observed after treatment with DC- but not with T cell-derived supernatants. DC-1/2 and T-1/2 indicate replicates of identical conditions. B, quantification of inflammatory cytokines enriched in stimulatory DC supernatants compared with nonstimulatory T cell supernatants. IP-10 and MCP-1 levels in DC-derived supernatants exceeded the detection range of the Luminex assay (data not shown).
TABLE 1.
Cytokines tested
| Cytokine | Concentrationa |
|---|---|
| ng/ml | |
| IL-12 | 5 |
| IL-15 | 0.5 |
| IL-6 | 1 |
| IL-8 | 5 |
| IP-10 | 5 |
| IL-1b | 0.1 |
| IL-4 | 1 |
| GM-CSF | 1 |
| TNF-α | 0.5 |
a Concentration in supernatants from monocyte-derived matured dendritic cell cultures was quantified by Luminex and rounded to 0.1, 0.5, 1, and 5 ng/ml. Individual cytokines were tested for their ability to regulate macroautophagy in skeletal myocytes with and without blockade of lysosomal proteolysis by chloroquine treatment. Each cytokine was tested at least three times at a concentration quantified in stimulatory DC-derived supernatant as well as in a 1 log higher concentration. MCP-1 was also found to be enriched in DC-derived cultures but was not tested for autophagy induction. As a classical proinflammatory cytokine, IFN-γ was additionally tested at concentrations of 0.1 and 1 ng/ml.
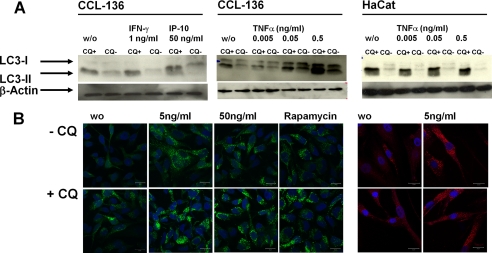
FIGURE 3.
TNF-α selectively up-regulates macroautophagy in skeletal myocytes. A, TNF-α but not IFN-γ or IP-10 induces LC3-II accumulation in CCL136 muscle cells but not in the human keratinocyte cell line HaCat. B, TNF-α induces accumulation of autophagosomes in GFP-LC3+-transfected CCL136 muscle cells (left panel) and in untransfected primary myoblasts stained with a monoclonal antibody specific for LC3 (right panel). w/o, without.
We noticed that TNF-α-induced up-regulation of macroautophagy was not ubiquitously observed in other cell lines. Although the MDAMC breast carcinoma cell line was susceptible to TNF-α mediated macroautophagy regulation (not shown), we could not detect any increase in LC3-II accumulation in the human keratinocyte cell line HaCat following TNF-α treatment (Fig. 3A, right panel). In contrast, TNF-α-induced augmentation of autophagosome formation was confirmed by immunocytochemistry in CCL136 muscle cells as well as in primary human myoblasts (Fig. 3B).
These data indicate that human primary skeletal muscle cells are susceptible to cytokine-mediated macroautophagy regulation. Among all monokines, lymphokines, and chemokines tested, we found that the induction of macroautophagy in skeletal myocytes is primarily mediated by TNF-α.
TNF-α Regulates MHC Expression in IFN-γ-treated Primary Myocytes
Cultured human myoblasts constitutively express classical human MHC class I antigens (i.e. HLA-A, -B, and -C), but lack MHC class II expression. However, MHC class II expression can be induced and MHC class I expression can be augmented in vitro by proinflammatory cytokines such as IFN-γ (5, 6) or in vivo as observed in inflammatory myopathies (20). Thus, activated human muscle cells are equipped with all major constituents necessary for MHC II antigen processing, and they can present exogenous as well as endogenous antigens leading to the stimulation of antigen-specific CD4+ T cells (21–24).
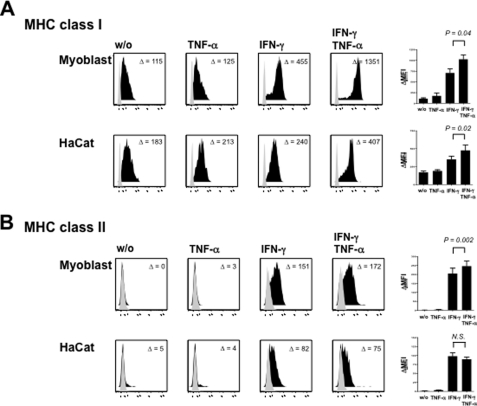
To investigate the effect of TNF-α on MHC regulation in cultured human myoblasts, we exposed primary muscle cell cultures to IFN-γ, TNF-α or both cytokines simultaneously and quantified MHC surface expression by flow cytometry. As expected, human myoblasts lacked constitutive MHC class II expression (HLA-DR), whereas MHC class I molecules (HLA-A, -B, and -C) were detectable at low levels in untreated cultures (Fig. 4). Exposure to TNF-α alone did not show any statistically significant effect on both MHC class I and MHC class II surface levels, although we noticed that expression levels of HLA-A, -B, and -C molecules were slightly higher upon TNF-α. In line with previous data, TNF-α also showed a synergistic effect with IFN-γ on MHC class I expression (8). IFN-γ strongly induced both MHC class I and MHC class II expression. Notably, addition of TNF-α to myoblasts exposed to IFN-γ significantly augmented MHC class II surface expression even though TNF-α alone had no effect on HLA-DR expression levels. The additive effect of TNF-α on MHC class II levels was not detectable in HaCat cells (Fig. 4B), which up-regulated MHC class II surface expression only upon IFN-γ.
FIGURE 4.
TNF-α increases MHC class I and MHC class II surface expression in IFN-γ-treated primary muscle cells. A, myoblasts and HaCat cells constitutively express HLA class I molecules on their cell surface. MHC class I levels are up-regulated by both TNF-α (50 ng/ml) and IFN-γ (100 ng/ml) and both cytokines act synergistically on MHC class I expression in both cell types. B, IFN-γ induces HLA-DR expression in human myoblasts and HaCat cells, which both lack constitutive MHC class II expression. TNF-α alone does not induce HLA-DR expression on both cell types. In contrast, addition of TNF-α to IFN-γ enhances MHC class II surface expression on myocytes but not HaCat cells. Diagrams display means ± S.E. of MHC expression levels from at least four independent experiments on myoblasts and HaCat cells, respectively. Isotype controls are highlighted in gray. Numbers in each individual histogram represent mean fluorescence intensity compared with isotype control (ΔMFI). MHC expression levels were analyzed using the paired t test.
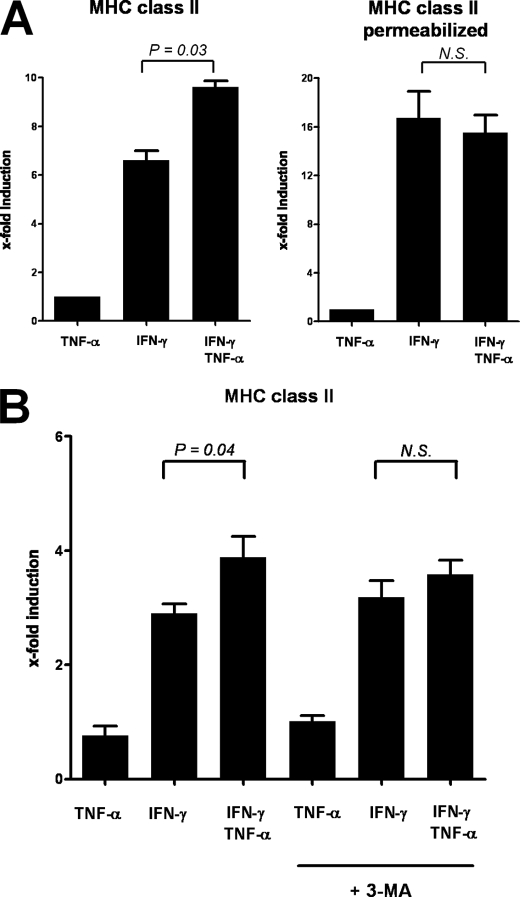
To investigate the hypothesis that TNF-α-mediated up-regulation of macroautophagy contributes to increased MHC class II surface expression levels in IFN-γ-treated myoblasts, we next performed titration experiments that included concentrations of TNF-α that were sufficient to induce autophagosome formation in skeletal myocytes. As shown in supplemental Fig. 1, an additive effect of TNF-α was detectable at 50 ng/ml for MHC class I expression and at 5 ng/ml for MHC class II expression. Furthermore, we addressed whether TNF-α augments HLA-DR synthesis or affected intracellular versus surface HLA-DR compartmentalization in myocytes exposed to IFN-γ. To this end, we compared MHC class II expression levels in permeabilized and nonpermeabilized myoblasts following 48 h of cytokine treatment. As shown in Fig. 5, the synergistic effect of TNF-α on MHC class II expression was lost following permeabilization (Fig. 5A), indicating that TNF-α, unlike IFN-γ, does not induce de novo HLA-DR synthesis but regulates MHC class II expression via translocation of HLA-DR from intracellular compartments to the cell surface. Because we found primary human myoblasts resistant to inhibition of autophagy by siRNA (data not shown), we inhibited macroautophagy by addition of 3-methyladenine (3-MA), which abolished the additive effect of TNF-α on IFN-γ-induced up-regulation of MHC class II surface expression in myoblasts (Fig. 5B).
FIGURE 5.
Synergistic effect of TNF-α on MHC class II regulation is restricted to cell surface expression levels and reversed by inhibition of macroautophagy. Displayed are changes in MHC expression levels in permeabilized and nonpermeabilized myoblasts following 48 h of cytokine exposure compared with controls. A, TNF-α increases HLA-DR expression induced by IFN-γ in myoblasts (left panel). The synergistic effect of TNF-α was not detectable in permeabilized myocytes (right panel). B, pharmacological inhibition of macroautophagy with 3-MA blocked the additive effect of TNF-α on MHC class II surface expression. Displayed are means ± S.E. from four (A) and three (B) independent experiments in which MHC expression levels were simultaneously quantified in both permeabilized versus nonpermeabilized and in 3-MA-treated versus non 3-MA-treated myocytes. MHC expression levels were compared using the paired t test. N.S., not significant.
Altogether, these data show that TNF-α regulates MHC class I and class II expression in IFN-γ-activated myoblasts. Moreover, they suggest that TNF-α increases HLA-DR surface expression via up-regulation of autophagosome formation, possibly via delivery of autophagosome content to MHC class II compartments for efficient MHC class II loading and release of HLA-DR molecules to the cell surface.
Resistance of Skeletal Muscle Cells to TNF-α-induced Cell Death
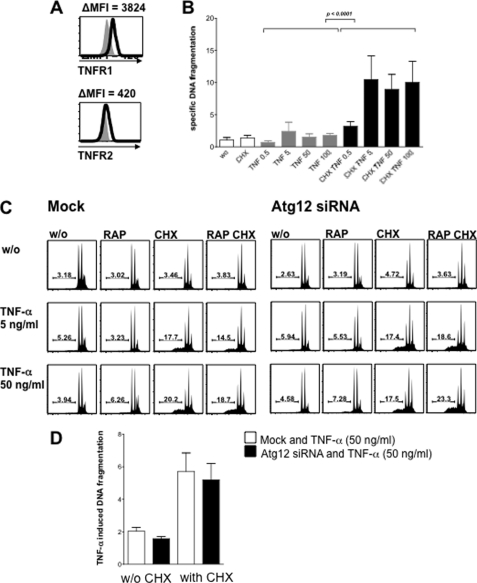
Because macroautophagy has been implicated in protection from cell death (25) (26), we additionally determined whether human skeletal muscle cells are susceptible to receptor-mediated apoptosis following TNF-α stimulation. TNF-R1 and TNF-R2 are expressed on the surface of CCL136 muscle cells (Fig. 6A) and primary human myoblasts (data not shown) (27, 28). In most cell types, TNF-R1 only signals for cell death when the survival pathway is blocked, e.g. by inhibition of protein synthesis (29). Therefore, we treated skeletal muscle cells with increasing concentrations of TNF-α in the presence and absence of the protein synthesis inhibitor cycloheximide (CHX; 1 μg/ml). Apoptotic cell death was analyzed after 24 h by quantifying TO-PRO-3 permeability and specific DNA fragmentation in hypodiploid cells (15, 16). As depicted in Fig. 6B, CCL136 skeletal muscle cells were completely resistant to TNF-α treatment alone but showed a dose-dependent induction of cell death in the presence of TNF-α and CHX. CHX alone did not induce cell death at a concentration of 3 μg/ml or lower. CHX-dependent cell death induction was observed in both CCL136 muscle cells and primary human myoblasts (not shown), indicating that TNF-α requires protein synthesis inhibition to induce apoptotic cell death in human myocytes. We additionally determined whether loss of macroautophagy function due to atg12-specific RNA interference or gain of macroautophagy function by mammalian target of rapamycin inhibition due to rapamycin treatment interferes with the susceptibility for TNF-α-mediated cell death. Neither atg12-specific RNA silencing in CCL136 muscle cells nor rapamycin showed an effect on the susceptibility toward TNF-α-induced apoptosis (Fig. 6, C and D). Thus, although macroautophagy can be induced by TNF-α in skeletal muscle cells, these cells are resistant to TNF receptor-mediated cell death and its macroautophagic regulation.
FIGURE 6.
Resistance of skeletal muscle cells to TNF-α-induced cell death. A, TNF-R1 and TNF-R2 are expressed on the surface of CCL136 muscle cells. B, CCL136 skeletal muscle cells are resistant to TNF-α treatment alone but showed a dose-dependent induction of cell death in the presence of TNF-α and the protein synthesis inhibitor CHX (1 μg/ml). Apoptotic cell death was analyzed after 24 h by quantifying TO-PRO-3 permeability and specific DNA fragmentation in hypodiploid cells. C, macroautophagy function does not interfere with susceptibility to TNF-receptor-mediated cell death in human CCL136 muscle cells. Neither loss of macroautophagic function due to atg12-specific RNA interference nor gain of macroautophagy function by mammalian target of rapamycin inhibition due to rapamycin treatment (RAP, 1 μg/ml) showed any effect on specific DNA fragmentation levels following TNF-α treatment. D, quantification of TNF-α-induced DNA fragmentation in CLL136 cells treated with or without atg12-specific RNA in the presence or absence of CHX averaged from seven independent experiments. Specific DNA fragmentation was compared using the paired t test.
Muscle Fibers from Patients with sIBM Show Colocalization of Autophagosomes and MHC Class II Molecules
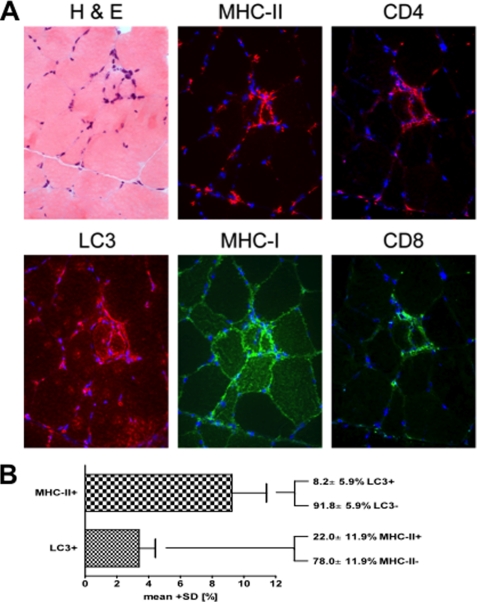
To explore possible implications of macroautophagic processing for antigen presentation via MHC class II molecules in skeletal muscle in vivo, we performed immunohistochemical staining in human biopsy tissue. We chose specimens from five sIBM patients, because LC3-positive macroautophagic processing has previously been demonstrated (11) and a specific immune response with antigen presentation in the muscle has been proposed for this disorder (30). Serial sections were stained by double-immunofluorescence for LC3, MHC class I and II, CD4, CD8, and by hematoxylin/eosin (Fig. 7A), followed by subsequent manual analysis of microphotographs with a total of 1864 muscle fibers. Comparable with previous results (11), LC3 was noted in 3.4 ± 2.1% of the fibers and MHC class II in 9.3 ± 4.4% (Fig. 7B). Subtype analysis revealed that only a minor fraction of 8.2 ± 5.9% of the MHC class II-positive fibers were double positive for LC3. On the other hand, more than one-fifth of the fibers positive for LC3 displayed double labeling for MHC class II (22.0 ± 11.9%). Furthermore, serial staining of all MHC class II/LC3 double-positive fibers revealed that almost half of them had cell-to-cell contact to immune cells positive for CD4 (47.2%), CD8 (43.1%), or both (43.1%; data not shown). A subsequent colocalization analysis showed that 56.4% of CD4+ cells within inflammatory infiltrates costained for CD3. On the other hand, 36.3% of all CD3+ cells were double positive for CD4 (supplemental Fig. 2). These data indicate that in subset of immune cell-surrounded muscle fibers autophagosomes colocalize with MHC class II loading compartments in sIBM patients.
FIGURE 7.
Muscle fibers from patients with sIBM show colocalization of autophagosomes and MHC class II molecules. A, serial and double immunofluorescent staining of a representative skeletal muscle from a patient with sIBM with H&E histochemistry and immunolabeling for LC3, MHC class I and II, CD4, and CD8. Photomicroscopy using a ×40 objective. B, quantitative double-labeling analysis of four patients with sIBM stained for MHC class II and LC3. Values indicate the mean ± S.D. of a total of 1864 skeletal muscle fibers.
DISCUSSION
The aim of this study was to identify inflammatory regulators of macroautophagy in a tissue type that had previously been described to show high levels of constitutive and starvation-induced macroautophagy and to efficiently regulate this pathway during immune-mediated tissue damage (4, 11). We found that TNF-α selectively induced macroautophagy and coacted with IFN-γ in regulating MHC class I and MHC class II expression in skeletal muscle cells.
TNF-α is readily detectable and predominantly expressed in macrophages as well as in the endomysium and perimysium of affected muscle fibers from patients with polymyositis, dermatomyositis, and sIBM (20, 27, 31–33). The monokine mediates inflammation by increasing vascular permeability and endothelial cell adhesiveness as well as via activation of antigen-presenting cells such as dendritic cells and macrophages, which initiate and orchestrate adaptive immune responses. In addition, we identified TNF-α from complex mixtures of inflammatory cytokines as the predominant regulatory factor for macroautophagy in human primary myocytes. In line with our findings, Djavaheri-Mergny et al. (34) previously reported that TNF-α treatment induces macroautophagy in Ewing sarcoma cells lacking NF-κB activation. Our results suggest that TNF-α is one of the major immune regulators of macroautophagy in the skeletal muscle and during inflammatory myopathies.
Cytokines such as interferons and members of the tumor necrosis factor receptor-ligand family such as CD40-CD40L stimulation (35–37) have previously been reported to modulate macroautophagy, primarily in the context of host defense responses to intracellular pathogens. Restriction of HSV-1 infection by macroautophagy in vitro and in vivo was found to be dependent on the type I IFN signaling machinery (38, 39). Type II IFN has been reported to enhance Mycobacterium tuberculosis and Rickettsia conorii degradation by macroautophagy (40, 41) and to induce macroautophagy and mycobacterial clearance through immunity-related GTPases in mice (42). We did not detect any IFN-γ-mediated effect on macroautophagy regulation in human skeletal muscle cells. Thus, proinflammatory cytokines might affect macroautophagy differently in a tissue-dependent manner. Moreover, mouse tissues are probably more susceptible to the IFN-γ-mediated macroautophagy regulation mechanism because their immunity-related GTPases are IFN-γ-inducible, whereas the human immunity-related GTPase is not, suggesting that immune signals that stimulate macroautophagy differ between rodents and man.
Functional implications of our findings lie in the augmenting effect of TNF-α on macroautophagy, autophagic protein degradation, and MHC class II surface expression. Skeletal muscle cells lack constitutive MHC class II expression but strongly up-regulate MHC class II surface expression to present exogenous as well as endogenous antigens to CD4+ T cells following exposure to proinflammatory cytokines such as IFN-γ and TNF-α (21–24). The MHC class II transactivator, a transcriptional coactivator, is the key intermediate that directs constitutive and IFN-γ-inducible expression of MHC class II genes in professional and nonprofessional antigen-presenting cells, respectively (43). It has previously been suggested that TNF-α augments MHC class II expression through a mechanism downstream or independent of class II transactivator induction in nonprofessional antigen-presenting cells (44) and that class II transactivator-independent MHC class II induction mediated by TNF-α promotes expression of endogenous rather than exogenous peptides in immune-privileged sites (45). Our data on TNF-α-mediated macroautophagy induction and MHC class II regulation suggest that TNF-α regulates MHC class II expression levels via delivery of autophagosome content to the MHC class II compartments for efficient MHC class II loading and successive release to the cell surface.
Previous studies demonstrated that inhibition of macroautophagy can promote or prevent apoptosis in the same cell depending on the nature of the death stimulus and subsequent compensatory changes, reflecting the complex interplay between macroautophagy and cell death pathways (25). We found that TNF-α-induced up-regulation of macroautophagy does not alter the susceptibility of skeletal muscle cells to TNF receptor-mediated apoptosis. In line with this observation, several studies failed to detect relevant apoptosis in skeletal muscle fibers of patients with nonmyopathic muscle, myopathies or myocytis, including sIBM (46), suggesting that skeletal muscle fibers appear to be rather resistant to apoptotic cell death.
To address the in vivo relevance of our findings, we studied muscle biopsies from patients with sIBM, a clinically and histopathologically well described entity of chronic muscle inflammation. In this disease, we previously showed that macroautophagic processing is present in skeletal muscle fibers (11). Moreover, it has been suggested that antigen presentation may occur in sIBM (30) as well as under other inflammatory conditions in skeletal muscle (6).
Muscle biopsies from patients with sIBM showed that over 20% of fibers that contained autophagosomes costained for MHC class II molecules and that more than 40% of double-positive fibers had contact with muscle-infiltrating CD4+ and CD8+ immune cells. The majority of CD4+ cells within inflammatory infiltrates costained for CD3. Although CD8+ T cells are the main component of inflammatory infiltrates in sIBM, a number of studies provided clear evidence that CD4+ T cells are not only present but also clonally expanded in sIBM lesional tissue (30, 47–49). Because autophagy promotes MHC class II presentation from intracellular source proteins (50), it is tempting to speculate that TNF-α-mediated macroautophagy induction and MHC class II up-regulation could potentially maintain CD4+ T cell-mediated (auto)-immunity in skeletal muscle via increased local autoantigen presentation. Thus, the immunohistochemical ex vivo analysis in skeletal muscle biopsies supports a functional relevance of our observations that can be related to human disease. The inflammatory component of sIBM is very similar to that of polymyositis and includes an autoimmune attack of muscle fibers by T-cells, which may be activated by muscle fibers themselves (51). Therefore, the mechanisms observed here may not be restricted to sIBM but could also occur in other inflammatory diseases of the skeletal muscle.
In conclusion, our findings establish a mechanism through which TNF-α regulates both macroautophagy and MHC expression in skeletal myocytes and suggest that TNF-α is an important mediator and a potential therapeutic target in T cell-mediated inflammatory myopathies in which macroautophagy is involved.
Supplementary Material
Acknowledgments
We gratefully acknowledge Monica Lee and Monique Gannage (Laboratory of Viral Immunobiology, Rockefeller University, New York) and Can G. Pham (Laboratory of Apoptosis and Cancer Biology, Rockefeller University, New York) for expert technical advice concerning the immunoblot and cell death experiments. We thank Konstanze Barthel (University Medical Center, Göttingen, Germany) for analysis of part of the data and Marinos C. Dalakas (Imperial College, London, UK) for providing patient biopsies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA108609 and R01CA101741 from NCI and Foundation for National Institutes of Health Grand Challenges in Global Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- sIBM
- sporadic inclusion body myositis
- DC
- dendritic cell
- CQ
- chloroquine
- CHX
- cycloheximide
- 3-MA
- 3-methyladenine.
REFERENCES
- 1. Mizushima N., Klionsky D. J. (2007) Annu. Rev. Nutr. 27, 19–40 [DOI] [PubMed] [Google Scholar]
- 2. Klein L., Münz C., Lünemann J. D. (2010) FEBS Lett. 584, 1405–1410 [DOI] [PubMed] [Google Scholar]
- 3. Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hohlfeld R., Engel A. G. (1994) Immunol. Today 15, 269–274 [DOI] [PubMed] [Google Scholar]
- 6. Wiendl H., Hohlfeld R., Kieseier B. C. (2005) Trends Immunol. 26, 373–380 [DOI] [PubMed] [Google Scholar]
- 7. Dalakas M. C., Hohlfeld R. (2003) Lancet 362, 971–982 [DOI] [PubMed] [Google Scholar]
- 8. Wiendl H., Mitsdoerffer M., Hofmeister V., Wischhusen J., Weiss E. H., Dichgans J., Lochmuller H., Hohlfeld R., Melms A., Weller M. (2003) Brain 126, 176–185 [DOI] [PubMed] [Google Scholar]
- 9. Kumamoto T., Ueyama H., Tsumura H., Toyoshima I., Tsuda T. (2004) Acta Neuropathol. 107, 59–65 [DOI] [PubMed] [Google Scholar]
- 10. Lünemann J. D., Schmidt J., Dalakas M. C., Münz C. (2007) Autophagy 3, 384–386 [DOI] [PubMed] [Google Scholar]
- 11. Lünemann J. D., Schmidt J., Schmid D., Barthel K., Wrede A., Dalakas M. C., Münz C. (2007) Ann. Neurol. 61, 476–483 [DOI] [PubMed] [Google Scholar]
- 12. Kimura N., Kumamoto T., Kawamura Y., Himeno T., Nakamura K. I., Ueyama H., Arakawa R. (2007) Pathobiology 74, 169–176 [DOI] [PubMed] [Google Scholar]
- 13. Fujita E., Kouroku Y., Isoai A., Kumagai H., Misutani A., Matsuda C., Hayashi Y. K., Momoi T. (2007) Hum. Mol. Genet. 16, 618–629 [DOI] [PubMed] [Google Scholar]
- 14. Schmid D., Pypaert M., Münz C. (2007) Immunity 26, 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riccardi C., Nicoletti I. (2006) Nat. Protoc. 1, 1458–1461 [DOI] [PubMed] [Google Scholar]
- 16. Lünemann J. D., Waiczies S., Ehrlich S., Wendling U., Seeger B., Kamradt T., Zipp F. (2002) J. Immunol. 168, 4881–4888 [DOI] [PubMed] [Google Scholar]
- 17. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2005) Autophagy 1, 84–91 [DOI] [PubMed] [Google Scholar]
- 19. Greenberg S. A., Pinkus G. S., Amato A. A., Pinkus J. L. (2007) Muscle Nerve 35, 17–23 [DOI] [PubMed] [Google Scholar]
- 20. Schmidt J., Barthel K., Wrede A., Salajegheh M., Bähr M., Dalakas M. C. (2008) Brain 131, 1228–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goebels N., Michaelis D., Wekerle H., Hohlfeld R. (1992) J. Immunol. 149, 661–667 [PubMed] [Google Scholar]
- 22. Michaelis D., Goebels N., Hohlfeld R. (1993) Am. J. Pathol. 143, 1142–1149 [PMC free article] [PubMed] [Google Scholar]
- 23. Warrens A. N., Zhang J. Y., Sidhu S., Watt D. J., Lombardi G., Sewry C. A., Lechler R. I. (1994) Int. Immunol. 6, 847–853 [DOI] [PubMed] [Google Scholar]
- 24. Curnow J., Corlett L., Willcox N., Vincent A. (2001) J. Neuroimmunol. 115, 127–134 [DOI] [PubMed] [Google Scholar]
- 25. Wang Y., Singh R., Massey A. C., Kane S. S., Kaushik S., Grant T., Xiang Y., Cuervo A. M., Czaja M. J. (2008) J. Biol. Chem. 283, 4766–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 27. De Bleecker J. L., Meire V. I., Declercq W., Van Aken E. H. (1999) Neuromuscul. Disord. 9, 239–246 [DOI] [PubMed] [Google Scholar]
- 28. Porter K. E., Turner N. A., O'Regan D. J., Ball S. G. (2004) Cardiovasc. Res. 64, 507–515 [DOI] [PubMed] [Google Scholar]
- 29. Gupta S. (2002) J. Clin. Immunol. 22, 185–194 [DOI] [PubMed] [Google Scholar]
- 30. Schmidt J., Rakocevic G., Raju R., Dalakas M. C. (2004) Brain 127, 1182–1190 [DOI] [PubMed] [Google Scholar]
- 31. Lundberg I., Brengman J. M., Engel A. G. (1995) J. Neuroimmunol. 63, 9–16 [DOI] [PubMed] [Google Scholar]
- 32. Lepidi H., Frances V., Figarella-Branger D., Bartoli C., Machado-Baeta A., Pellissier J. F. (1998) Neuropathol. Appl. Neurobiol. 24, 73–79 [DOI] [PubMed] [Google Scholar]
- 33. Kuru S., Inukai A., Liang Y., Doyu M., Takano A., Sobue G. (2000) Acta Neuropathol. 99, 585–588 [DOI] [PubMed] [Google Scholar]
- 34. Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Souquère S., Pierron G., Codogno P. (2006) J. Biol. Chem. 281, 30373–30382 [DOI] [PubMed] [Google Scholar]
- 35. Mills K. R., Reginato M., Debnath J., Queenan B., Brugge J. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3438–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thorburn J., Moore F., Rao A., Barclay W. W., Thomas L. R., Grant K. W., Cramer S. D., Thorburn A. (2005) Mol. Biol. Cell 16, 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andrade R. M., Wessendarp M., Gubbels M. J., Striepen B., Subauste C. S. (2006) J. Clin. Invest. 116, 2366–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D. A., Levine B. (2007) Cell Host Microbe 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 39. Tallóczy Z., Jiang W., Virgin H. W., 4th., Leib D. A., Scheuner D., Kaufman R. J., Eskelinen E. L., Levine B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 41. Singh S. B., Davis A. S., Taylor G. A., Deretic V. (2006) Science 313, 1438–1441 [DOI] [PubMed] [Google Scholar]
- 42. Taylor G. A., Feng C. G., Sher A. (2004) Nat. Rev. Immunol. 4, 100–109 [DOI] [PubMed] [Google Scholar]
- 43. Steimle V., Siegrist C. A., Mottet A., Lisowska-Grospierre B., Mach B. (1994) Science 265, 106–109 [DOI] [PubMed] [Google Scholar]
- 44. Nikcevich K. M., Piskurich J. F., Hellendall R. P., Wang Y., Ting J. P. (1999) J. Neuroimmunol. 99, 195–204 [DOI] [PubMed] [Google Scholar]
- 45. Arancibia-Cárcamo C. V., Osawa H., Arnett H. A., Háskova Z., George A. J., Ono S. J., Ting J. P., Streilein J. W. (2004) Eur. J. Immunol. 34, 471–480 [DOI] [PubMed] [Google Scholar]
- 46. Fyhr I. M., Lindberg C., Oldfors A. (2002) Acta Neurol. Scand. 105, 403–407 [DOI] [PubMed] [Google Scholar]
- 47. Engel A. G., Arahata K. (1984) Ann. Neurol. 16, 209–215 [DOI] [PubMed] [Google Scholar]
- 48. Pandya J. M., Fasth A. E., Zong M., Arnardottir S., Dani L., Lindroos E., Malmstrom V., Lundberg I. E. (2010) Arthritis Rheum. 62, 3457–3466 [DOI] [PubMed] [Google Scholar]
- 49. Englund P., Wahlström J., Fathi M., Rasmussen E., Grunewald J., Tornling G., Lundberg I. E. (2007) Arthritis Rheum. 56, 372–383 [DOI] [PubMed] [Google Scholar]
- 50. Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., Brock R., Driessen C., Rammensee H. G., Stevanovic S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dalakas M. C. (2006) Nat. Clin. Pract. Rheumatol. 2, 219–227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.