Abstract
Numerous studies have determined that the establishment of Sir protein-dependent transcriptional silencing in yeast requires progression through the cell cycle. In our study we examined the cell cycle requirement for the establishment of silencing at the HML and HMR loci using strains bearing conditional or inducible SIR3 alleles. Consistent with prior reports, we observed that establishing silencing at HMR required progression through the cell cycle. Unexpectedly, we found that the HML locus is far less dependent on cell cycle progression to establish silencing. Seeking cis-acting elements that could account for this difference, we found that deletion of a tRNA gene that serves as a chromatin boundary at HMR abolishes the cell cycle progression requirement at this locus, while insertion of sequences containing this tRNA gene adjacent to HML imposes dependence on cell cycle progression for the full establishment of silencing. Our results indicate that the cell cycle progression requirement is not a property intrinsic to the formation of heterochromatin in yeast, but is instead a cis-limited, locus-specific phenomenon. We show that inactivation of the Scc1 cohesin also abolishes the requirement for cell cycle progression and test models based on a possible link between the tRNA gene and cohesin association.
GENE silencing in yeast is required to control the transcription of key regulatory genes affecting determination of cell type. The silent mating type loci, HML and HMR, contain genes that are kept transcriptionally inactive until transposed to the MAT locus via a regulated, gene conversion event. A weaker but mechanistically similar form of silencing affects genes present near yeast telomeres (Gottschling et al. 1990). Silencing is mediated by the Sir protein complex, which is recruited by sequence-specific DNA binding factors such as Rap1. Sir2 deacetylation of histones H3 and H4 increases the affinity of Sir3 and Sir4 for histone tails (Hecht et al. 1996; Liou et al. 2005); reiterative deacetylation and binding of the complex provides a model for how Sir-dependent spreading can spread from a nucleation site. The efficiency of silencing is aided by an epigenetic mechanism, in which a previously silenced locus has a greater probability of being silenced in the succeeding generation (Pillus and Rine 1989; Mahoney et al. 1991).
Many studies have demonstrated that the establishment of Sir protein-dependent silencing in yeast requires progression through the cell cycle (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001; Lau et al. 2002; Martins-Taylor et al. 2004). Initial reports focusing on the establishment of silencing at HMR using strains expressing a temperature-sensitive Sir3 protein indicated that silencing is principally established in S phase (Miller and Nasmyth 1984), a conclusion in agreement with later studies that used an inducible Sir1 gene to examine the establishment of silencing at HMR (Fox et al. 1997; Kirchmaier and Rine 2001; Li et al. 2001). A subsequent study using the conditional sir3-8 strain concluded that progression through both S and M phases was needed to establish silencing at HMR, but that silencing was largely accomplished in M phase (Lau et al. 2002). Finally, a strain bearing an inducible SIR3 gene was used to assess the establishment of silencing at yeast telomeres; in this case it was found that passage through mitosis was necessary and sufficient to silence a telomere-linked reporter gene (Martins-Taylor et al. 2004).
The direct contribution that cell cycle progression makes to the establishment of silencing has not been determined, but in an insightful study it was found that blocking the transcription of the SCC1 cohesin gene led to silencing of HMR earlier in the cell cycle, and that expression of an uncleavable Scc1 protein decreased the ability to establish silencing (Lau et al. 2002). In the telomere system it was found that deletion of the HTZ1 gene, coding for the histone H2A variant H2A.Z, abolished the requirement for cell cycle progression, and that H2A.Z was displaced from chromatin during mitosis, prior to the establishment of silencing (Martins-Taylor et al. 2011).
While each of these studies consistently observed a requirement for cell cycle progression, they elicited different conclusions about the timing of silencing. These differences could reflect the methods used to observe the establishment of silencing and/or the specific locus studied. We reasoned that understanding the nature of these loci and system-specific differences would provide insights into the general nature of the cell cycle progression requirement. We have examined the cell cycle requirement for the establishment of silencing at the HML and HMR loci using strains bearing conditional or inducible SIR3 alleles. Surprisingly, we find that the HML locus is far less dependent on cell cycle progression to establish silencing than is HMR. Seeking cis-acting elements that could account for this difference, we find that deletion of a tRNA gene that serves as a chromatin boundary at HMR abolishes the cell cycle progression requirement at this locus, while addition of this sequence next to HML imposes dependence on cell cycle progression for the establishment of silencing. Our results indicate that the cell cycle progression requirement is not a property intrinsic to the formation of heterochromatin in yeast, but instead a cis-limited, locus-specific phenomenon.
MATERIALS AND METHODS
Media:
For mating type loci silencing experiments in the inducible system, cultures were grown at 30° in YPraffinose media (1% Bacto yeast extract, 2% Bacto peptone extract, and 2% raffinose). To induce expression of the GAL-SIR3 construct, galactose was added to YPraffinose media to 2%. For mating type loci silencing experiments in the conditional system, cultures were grown at 23° or 37° in YPD media (1% Bacto yeast extract, 2% Bacto peptone extract, and 2% dextrose).
Strains:
Yeast strains used in this study are listed in Table 1. Most gene or locus deletions were constructed by PCR-mediated gene deletion (Wach et al. 1994), using MX-series plasmids as templates (Goldstein and McCusker 1999). Unmarked deletions of PPR1 and SIR3 were made in YSH893 using the recyclable CaURA3MX3 allele from template pAG61 (Goldstein et al. 1999). To construct YSH956, the tRNA gene present downstream of HMR was deleted from YSH893 using the “delitto perfetto” method (Storici et al. 2001). A URA3-KANMX cassette amplified from the pCORE plasmid was integrated adjacent to HMR, deleting the tRNA gene. A PCR fragment from yeast strain ROY1681, which lacks the tRNA gene (Donze et al. 1999), was transformed into these strains, and candidates were screened for displacement of the URA3-KAN cassette. The resulting strain contains an unmarked deletion of sequences 295,481–295,580 [Saccharomyces Genome Database (SGD) coordinates) (Donze and Kamakaka 2001). A similar approach was used to make an unmarked insertion of the same tRNA gene sequences downstream of HML to create strain YSH993. In this strain, a 300-bp fragment containing the tRNA gene (SGD sequences 295,330–295,630) was inserted ∼450 bp downstream of HML-I at position 15,350 (SGD coordinates), placing it in a similar position and orientation as it is found at HMR.
TABLE 1.
Strains
| Strain | Genotype | Source |
|---|---|---|
| YSH505 | ade2Δ∷hisG his3Δ200 met15Δ0 trp1Δ0 ura3Δ0 Δppr1∷HIS3 trp1∷GAL-SIR3-TRP1 URA3-TEL-VR | Martins-Taylor et al. (2004) |
| YSH801 | ade2Δ∷hisG his3Δ200 met15Δ0 trp1Δ0 ura3Δ0 Δppr1∷HIS3 trp1∷GAL-SIR3-TRP1 URA3-TEL-VR Δsir3∷NAT-MX Δhml∷KAN-MX Δmat∷HPH-MX | |
| YSH811 | ade2Δ∷hisG his3Δ200 met15Δ0 trp1Δ0 ura3Δ0 Δppr1∷HIS3 trp1∷GAL-SIR3-TRP1 URA3-TEL-VR Δsir3∷NAT-MX Δmat∷HPH-MX | |
| YSH872 | adeΔ2∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1∷HIS3 URA3-TEL-VR Δsir3∷NAT-MX Δmat∷HPH-MX | |
| YSH494 | ade2 lys1 his5 leu2 can1 Δsir3∷LEU2 ura3∷URA3-sir3-8 | Holmes and Broach (1996) |
| YSH829 | ade2 lys1 his5 leu2 can1 Δsir3∷LEU2 ura3∷URA3-sir3-8 Δmat∷HPH-MX | |
| YSH854 | ade2 lys1 his5 leu2 can1 Δsir3∷LEU2 ura3∷URA3-sir3-8 Δmat∷HPH-MX Δhmr∷NAT-MX | |
| YSH958 | hml∷pURA3-YFP hmr∷pURA3-CFP Δsir3∷LEU2 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Xu et al. (2006) |
| YSH967 | hml∷pURA3-YFP hmr∷pURA3-CFP Δsir3∷LEU2 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sir3-8-TRP1 | |
| YSH968 | hml∷pURA3-YFP hmr∷pURA3-CFP Δsir3∷LEU2 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 GAL-SIR3-TRP1 | |
| YSH461 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 | Brachmann et al. (1998) |
| YSH893 | YSH461; Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX | |
| YSH956 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX HMR-I-ΔtDNA(Thr) | |
| YSH973 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX HMR-I-ΔtDNA(Thr) Δhml∷KAN-MX | |
| YSH993 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX HML-I-tDNA(Thr) | |
| YSH839 | ade2 lys1 his5 leu2 can1 Δsir3∷LEU2 ura3∷URA3-sir3-8 Δmat∷HPH-MX Δhml∷NAT-MX Δhtz1∷KAN-MX | |
| YSH849 | ade2 lys1 his5 leu2 can1 Δsir3∷LEU2 ura3∷URA3-sir3-8 Δmat∷HPH-MX Δsas2∷NAT-MX | |
| YSH942 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 ura3Δ0 Δppr1∷HIS3 URA3-TELVR trp1Δ63∷GAL10p-SIR3-TRP1 Δscc1∷scc1-73-LEU2 Δmat∷HPH-MX Δsir3∷NAT-MX | |
| YSH1016 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 *(trp1Δ63) ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX SCC1-3HA-KAN-MX | |
| YSH1017 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HPH-MX HMRΔtDNA(Thr) SCC1-3HA-KAN-MX | |
| YSH1018 | ade2Δ∷hisG his3Δ200 leu2Δ0 met15Δ0 *(trp1Δ63) ura3Δ0 Δppr1 Δsir3 trp1Δ63∷GAL10p-SIR3-TRP1 Δmat∷HYG HML-I-tDNA(Thr) SCC1-3HA-KAN-MX |
To create strain YSH549 the SCC1 gene was replaced with a DNA fragment containing the scc1-73 allele (amplified from strain KN5832, provided by Kim Nasmyth) and the LEU2 gene from pKMT1, a LEU2 vector based on the MX-series vectors (Wach et al. 1994; Goldstein and McCusker 1999). YSH942 is congenic with YSH549, except that SIR3 and MAT were deleted using NAT1MX and KANMX drug resistant markers, respectively (Goldstein and McCusker 1999). To study Scc1 localization using ChIP, SCC1 was epitope tagged in strains YSH893, YSH956, and YSH993, using a 3HA-KAN cassette (Knop et al. 1999) creating strains YSH1016, YSH1017, and YSH1018, respectively.
A galactose-inducible SIR3 gene was integrated into YSH958 at the TRP1 locus using plasmid pAR83 (Holmes et al. 1997) to create YSH968. A temperature-sensitive SIR3 allele (Miller and Nasmyth 1984) was cloned into pRS404 at the BamHI–SacI site to create pSH146. This plasmid was cut with EcoRV and then transformed into YSH958 to integrate sir3-8 at TRP1, creating YSH967. Levels of the sir3-8 protein in strains grown at permissive temperature (PT) are similar to wild-type Sir3 levels (Stone et al. 2000).
Cell cycle blocks:
Cell cycle blocks were performed as described (Martins-Taylor et al. 2004). α-Factor (10 μg/ml) or nocodazole (15 μg/ml) was used to block cells in G1 or G2/M, respectively. For some G2/M experiments, benomyl (15 μg/ml) was added to the culture after 3 hr of blocking with nocodazole to prevent release from the G2/M block. For the G1 experiments in the sir3-8 temperature-sensitive system, hydroxyurea was added to cells 20 min before they were shifted to 37°. Unless noted, cells exhibited at least a 90% arrest in the cell cycle, as determined by microscopic examination of cell morphology. FACS analysis conducted on selected cultures confirmed that cells did not escape G2/M blocks during the time course of our experiments, but indicated that a small percentage of cells occasionally escaped G1 blocks at later time points (see supporting information, Figure S1). However, the percentage of cells in G1 did not drop below 80% in these experiments. We note that direct comparisons between a1 and α1 expression were always made using the same cell cultures.
RT–PCR:
RNA extraction, cDNA synthesis, and PCR were performed as previously described (Martins-Taylor et al. 2004). Primers specific for α1, a1, CFP, and YFP were used, while ACT1 served as the internal control. Results from 5% acrylamide gels were stained using Sybr gold dye (Invitrogen), and the gels were scanned using a Storm 840 PhosphorImager (GE Healthcare). Each band was quantified using ImageQuant TL (GE Healthcare). Control experiments were performed for each primer set to ensure that detection of message was within the linear range. Identical results were achieved in at least three independent experiments and in repeated determinations from RNA collected from individual experiments.
RESULTS
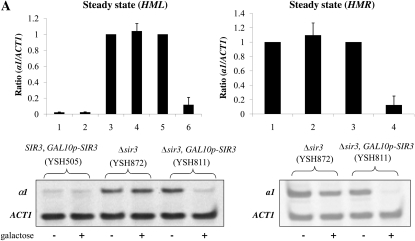
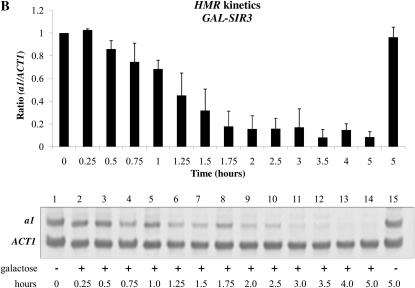
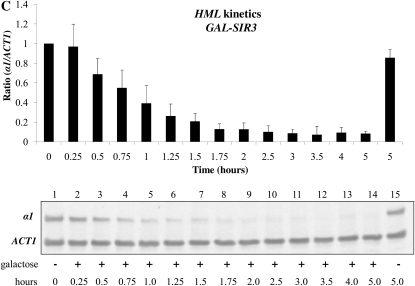
Prior studies designed to examine the cell cycle progression requirements for the establishment of silencing in yeast came to different conclusions about the timing of the establishment event. To investigate whether these differences reflect the alternate means of inducing silencing, or indicate locus-specific requirements, we examined the establishment of silencing at HML and HMR, using both inducible and conditional silencing systems. We first examined the establishment of silencing using a strain in which the sole source of Sir3 is from a single galactose-inducible SIR3 allele. In the control experiment shown in the first panel of Figure 1A, we show the steady state levels of a1 message transcribed from the HML locus in strains bearing the wild-type SIR3 gene, a strain lacking SIR3 genes, and a strain bearing a galactose-inducible SIR3 gene. We find that α1 message is tightly regulated by galactose addition in our experimental strain (lanes 5 and 6). The second panel of Figure 1A shows a similar galactose-dependent repression of a1 message transcribed from HMR (lanes 3 and 4). This experiment demonstrates that silencing is efficiently established in cells grown to steady state (long-term log phase growth) in the indicated conditions. We next examined the kinetics of repression at HMR and HML following induction of Sir3 expression. Repression of the HML locus appears to occur with somewhat faster kinetics (Figure 1C); we observe greater repression compared to HMR at each time point tested. We note that in these and subsequent experiments, mRNA from HMR and HML was measured from the same cell cultures at the same time points.
Figure 1.—
Inducible silencing at HML and HMR. (A) Steady state repression at HML and HMR. Strains YSH505, YSH872, and YSH811 were grown to steady state in media containing raffinose or raffinose with galactose. Levels of α1, a1, and ACT1 mRNA were measured by reverse transcriptase–PCR (RT–PCR). Expression at HML and HMR was normalized to ACT1 and expressed as a ratio relative to the uninduced (no galactose) control. In this and all succeeding figures the cumulative results of at least three independent experiments are shown in the graph, while a representative gel scan from a single experiment is shown. (B) Kinetics of repression at the HMR locus. A culture of strain YSH811 was grown to log phase in YPraffinose media; at time 0, galactose was added to 2%. RNA was collected at the indicated time points and the levels of a1 and ACT1 message were measured by RT–PCR. (C) Kinetics of repression at the HML locus. α1 message was measured from RNA obtained from the same cell cultures described in B.
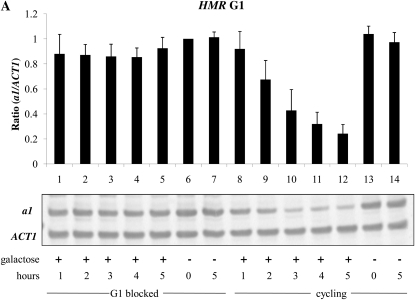
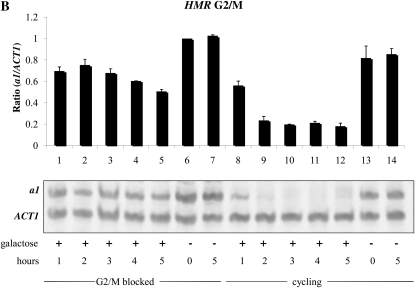
We then asked whether the establishment of silencing in this strain required cell cycle progression. The approach in this and subsequent experiments was similar; we first blocked cells in the cell cycle using α-factor to arrest cultures in G1 phase, or nocodazole to block cells at the G2/M boundary. We then added galactose to induce Sir3 expression. At subsequent time points, we analyzed the mRNA levels from HML and HMR. In each experiment mRNA levels were also measured in parallel cultures that were allowed to continue progression through the cell cycle. The ability to establish silencing at the HMR locus in G1-arrested cells was examined in the experiment shown in Figure 2A. We find that silencing cannot be efficiently established at this block; as seen in lanes 3, 4, and 5, addition of galactose for up to 5 hr had little effect on overall a1 message levels, while repression is clearly established in the cycling control (e.g., lane 12). We observe a somewhat different pattern when conducting a similar experiment in cells blocked in G2/M; in this case we observe moderate silencing of HMR over this time course, although it is less than that seen in the cycling controls (Figure 2B, lanes 1–5).
Figure 2.—
Establishment of silencing at HML and HMR upon induction of Sir3. (A) Establishment of silencing is not observed at HMR at G1. YSH801 was grown to log phase in YPraffinose media. Half of this culture was blocked in G1 phase by addition of α-factor. Galactose was then added to one-half of the α-factor arrested cells. As a control, galactose was also added to the unblocked cycling cells. RNA was isolated at the indicated time points and levels of a1 and ACT1 were measured by RT–PCR. (B) Moderate silencing at HMR is observed in G2/M blocked cells. Strain YSH811 was grown to log phase in raffinose-containing media and subject to the same experimental design as described in A, except that nocodazole was used to block cells at G2/M. (C) Establishment of silencing is observed at HML at G2/M. α1 message was measured in the same cell cultures described in B.
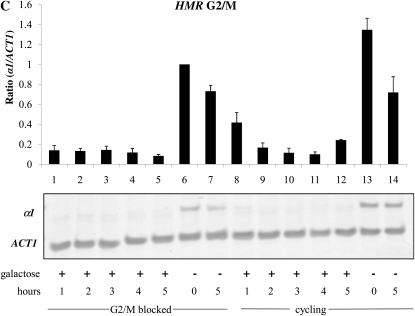
Since repression of the HML locus is required for cells to respond to α-factor, we were unable to examine the establishment of silencing at HML in G1 blocked cells, but could examine this in nocodazole-blocked cultures. In contrast to our HMR result, we observe a rapid and complete repression of α1 message from HML in this experiment (Figure 2C, lanes 1–5). Thus, we find that HMR exhibits a differential ability to permit the establishment of silencing depending on cell cycle position, and that cell cycle progression is not required to establish silencing at HML.
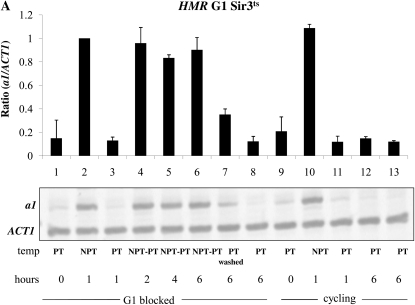
The initial observation that the establishment of silencing required cell cycle progression was made using strains bearing a conditional allele of the SIR3 gene (Miller and Nasmyth 1984). To further explore potential differences in HML and HMR's ability to establish silencing, we compared them in a strain with the temperature-sensitive SIR3 allele. For these experiments we blocked cells in G1 or G2/M, shifted cultures to the nonpermissive temperature (NPT) for 1 hr, sufficient to cause a loss of silencing, and then shifted cultures back to the permissive temperature. Control cultures were maintained at the permissive temperature, and not shifted. mRNA was analyzed at subsequent time points.
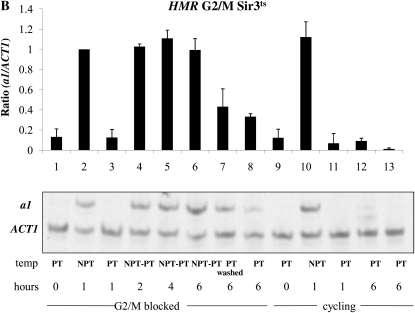
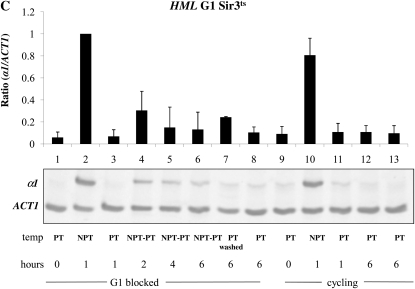
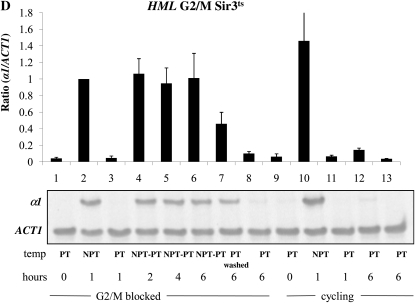
The strains we used for these experiments lack the MAT locus and are therefore sensitive to α-factor when HML is silenced. A loss of silencing at HML caused by a shift to the nonpermissive temperature should eventually lead to insensitivity to α-factor; therefore, we also added hydroxyurea to the media following the G1 block, which will cause any cells escaping the arrest to block in early S phase. Thus, our experiment is similar but not identical to prior experiments that examined the ability to silence the HMR locus in G1 arrested cells (Miller and Nasmyth 1984; Lau et al. 2002). Nonetheless, we observe the same result: we fail to observe a restoration of silencing at HMR in cells held at G1/early S (Figure 3A, lanes 4–6), but do see a decrease in a1 message in control cycling cells (Figure 3A, lanes 11–13) or in cells allowed to escape from the cell cycle block (Figure 3A, lane 7). We went on to conduct a similar experiment in G2/M-blocked cells, achieving the same result: arrested cells cannot regain silencing, but in parallel cycling cells silencing is efficiently restored (Figure 3B). Using the same RNA samples from our HMR experiments, we next examined the transcriptional state of the HML locus. Surprisingly, we again observe rapid and complete silencing of HML message at the G1/S block (Figure 3C). Therefore, in these conditions HML is not subject to the cell cycle progression requirement to establish silencing. However, when we assayed transcription from HML in G2/M-blocked cells subjected to the same temperature shifts we found that HML is unable to reestablish silencing (Figure 3D).
Figure 3.—
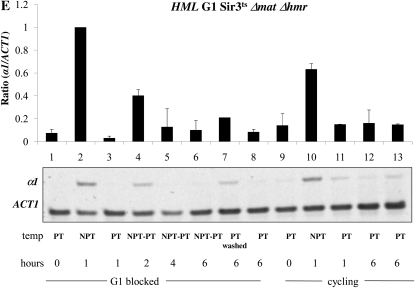
Establishment of silencing at HML and HMR in a conditional sir3-8 strain. (A) The establishment of silencing is not observed at HMR at G1. Strain YSH829 was grown to log phase in low pH YPD. α-Factor was added to one-half of the culture to arrest cells in G1. Following G1 arrest, hydroxyurea was added; after an additional 20-min incubation, the culture was further divided and one-half of the arrested cells were shifted to the nonpermissive temperature (NPT, 37°) for 1 hr (lane 2). Cells were then shifted back to the permissive temperature (PT, 23°) and cells were harvested every 2 hr for 6 hr (lanes 4–6). The unblocked cycling cells that served as the control were treated in a similar manner. In independent control experiments, cells shifted from 37° to 23° exhibited full repression of HML and HMR within 4 hr (Figure S3). The lane 7 control culture was blocked in G1 and then washed to remove α-factor and hydroxyurea to allow resumption of cell cycle progress. The lane 8 control was blocked in G1 but maintained at 23° throughout the experiment. (B) The establishment of silencing is not observed at HMR at G2/M. Strain YSH829 was subject to the same experimental design as that described in panel A, except that nocodazole was used to block cells at G2/M. (C) The establishment of silencing is observed at HML at G1. α1 message was measured from RNA obtained from the same cell culture as described in A. (D) The establishment of silencing is not observed at HML at G2/M. α1 message was measured from RNA obtained from the same cell culture described in B. (E) Silencing at the HML locus does not require HMR expression. An experiment identical to that described in C was performed on strain YSH854 (sir3-8 Δmat∷HYG Δhmr∷NAT).
When the sir3-8 strain is grown at the nonpermissive temperature, both a1 and α2 proteins are expressed, due to loss of silencing at HMR and HML. An a1-α2 heterodimer may decrease expression of the α1 gene at HML (Klar et al. 1981; Nasmyth et al. 1981; Siliciano and Tatchell 1984). To determine whether the repression of HML we observe at G1 is influenced by the a1-α2 heterodimer, we repeated this G1 experiment in strain YSH854, a Δmat Δhmr strain lacking the a1 or a2 genes. As shown in Figure 3E, we observe a similar reduction in α1 message on shifting the cells to the PT from the NPT (lanes 4–6). Thus, a1-α2 regulation is not responsible for the repression that we observe at HML.
Our results indicate a significant difference in the ability of HML and HMR to establish silencing in the absence of cell cycle progression. We attempted to define significant cis- or trans-acting factors that could account for their different sensitivity to cell cycle progression (Figure 4). Each of the HM loci contains divergently transcribed genes; α1 and α2 are transcribed at HML while a1 and a2 are transcribed at HMR. Differences in the promoters or coding sequences could determine sensitivity to cell cycle progression. To test this possibility we used a strain in which the α and a genes at HML and HMR were replaced with the YFP and CFP genes, respectively, each under the control of the URA3 promoter (Xu et al. 2006). We introduced the sir3-8 allele into this strain and repeated our determination of the influence of cell cycle progression. We find that the pattern of silencing at HMR is largely unchanged; in the absence of cell cycle progression, only a mild increase in silencing was observed (Figure 5A, lanes 4–6). At HML we observe a significant degree of silencing, but it is incomplete compared to the cycling control (Figure 5B, lanes 4–6) or compared to the strain containing the wild-type α-genes at HML (Figure 3C). From these experiments we conclude that the genes and promoters do not impose the requirement for cell cycle progression on HMR, but some feature of these sequences may partially protect HML from this requirement. Due to the inefficient induction of GAL promoter sequences in this strain, we were unable to conduct parallel experiments using the inducible SIR3 allele.
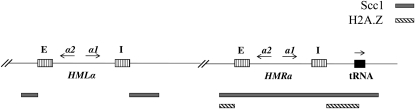
Figure 4.—
Significant cis-elements and trans-acting factors associated with the HML and HMR loci.
Figure 5.—
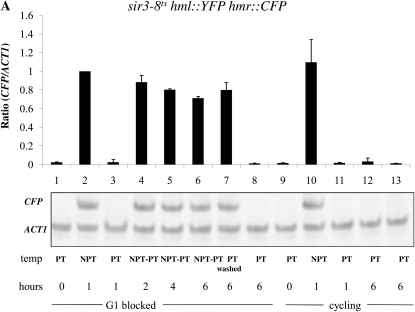
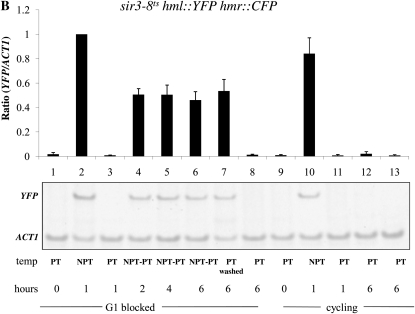
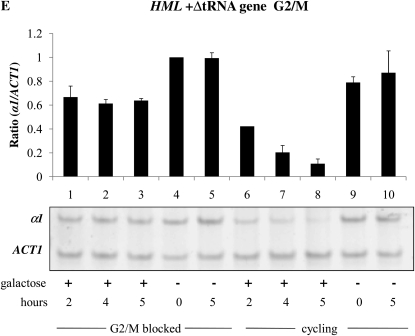
Effect of cis-elements on the establishment of silencing at HML and HMR. The establishment of silencing was assessed in YSH967, a sir3-8 hml∷pURA3-YFP hmr∷pURA3-CFP strain, as described in the Figure 3 legend. (A) Silencing at the HMR locus in G1-blocked cells was assayed by measuring the level of CFP RNA by RT–PCR. (B) Silencing at the HML locus was assayed by measuring, using the same levels of YFP mRNA, which used the same samples for the experiment described in A. (C) Establishment of silencing at HMR at G1 in cells lacking the adjacent tRNA gene. Strain YSH973 was subject to the same experimental design as that described in Figure 2A. (D) Establishment of silencing at HMR at G2/M in cells lacking the adjacent tRNA gene. Strain YSH956 was subject to the same experimental design as that described in C, except that nocodazole was used to block cells at G2/M. (E) Establishment of silencing at HML in a strain bearing a downstream tRNA gene insertion. Silencing of HML in strain YSH993 was assessed using a similar experimental design as that described for Figure 2B, except that nocodazole was used to block cells at G2/M.
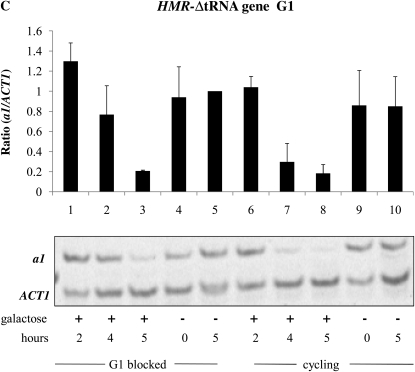
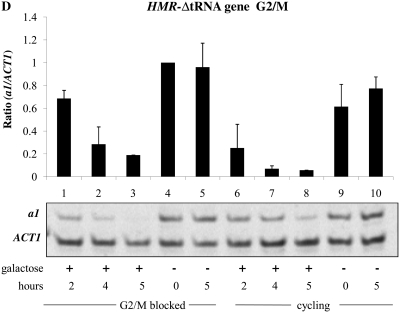
A tRNA gene adjacent to the HMR locus has been shown to act as a barrier to the spread of heterochromatin from HMR (Donze et al. 1999; Lynch and Rusche 2010) and also to be necessary for recruitment of the Scc1 cohesin to the HMR locus (Chang et al. 2005; Dubey and Gartenberg 2007). Scc1 has been identified as an inhibitor of the establishment of silencing at HMR (Lau et al. 2002). No equivalent cis-element is known to exist at the HML locus. We examined the establishment of silencing in strains with precise unmarked deletions of this tRNA gene. Using the inducible Sir3 system, we observed that in strains lacking the tRNA gene the HMR locus is now efficiently silenced in G1- and G2/M-blocked cells (Figure 5, C and D, lanes 1–3) as compared to the wild-type strain (Figure 2, A and B, respectively). Thus, loss of the tRNA gene abolished the requirement for cell cycle progression at HMR.
A DNA sequence encompassing a tRNA gene is necessary to confer cell cycle-dependent establishment of silencing on HMR. To determine whether these sequences are sufficient to confer this regulation, we inserted the tRNA gene downstream of the HML-I sequences in the same relative position and orientation. Insertion of this sequence did not affect the ability to repress HML in cycling cells; however, in this strain the HMLα1 gene could now only be partially repressed in cells blocked at G2/M (Figure 5E, lanes 1–3). Thus, inserting this tRNA gene downstream of the HML locus imposes cell cycle dependence at this locus in the inducible system.
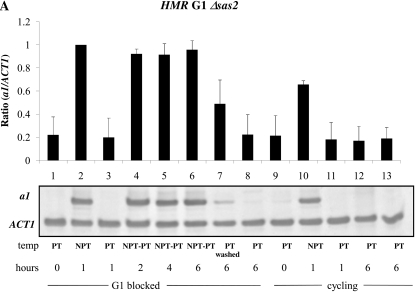
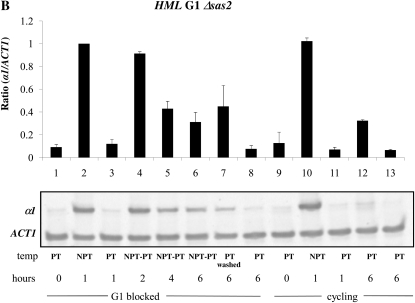
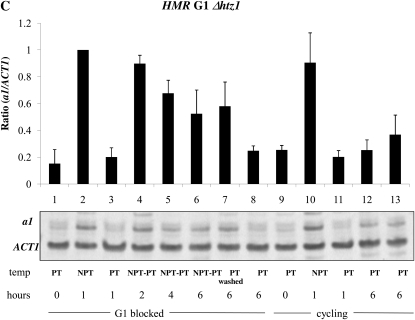
We were also interested in identifying trans-acting factors that differentially regulated cell cycle-dependent silencing at HML and HMR. Prior genome-wide studies found that the histone variant H2A.Z localizes to the boundaries of the HMR locus (Meneghini et al. 2003), but is relatively absent at HML, while Scc1 is localized throughout the HMR locus, but only at the boundaries of the HML locus (Glynn et al. 2004; Lengronne et al. 2004) (Figure 4). We examined the timing of silencing in a strain bearing a conditional allele of SCC1, and in strains lacking the HTZ1 or SAS2 genes, which code for factors shown to affect the timing of the establishment of silencing at yeast telomeres (Martins-Taylor et al. 2011). We observed that deleting SAS2 in the conditional Sir3 strain did not have an effect on the establishment of silencing at HMR or HML (Figure 6, A and B, lanes 4–6). Because strains lacking H2A.Z did not exhibit a robust block at G1 with α-factor, we deleted HML in Δhtz1 strains to efficiently block cells in G1. Loss of H2A.Z in the conditional Sir3 strain relieved to some degree the cell cycle requirement for the establishment of silencing at G1 phase at the HMR locus (Figure 6C, lanes 4–6).
Figure 6.—
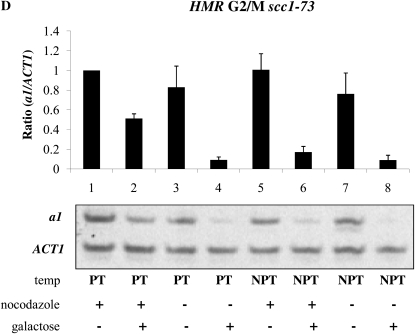
Effect of trans-acting factors on the establishment of silencing at the mating type loci. (A) Establishment of silencing at HMR at G1 in a Δsas2 strain. Strain YSH849 was grown to log phase in low pH YPD and subjected to the same experimental design as described for Figure 3A. (B) Establishment of silencing at HML at G1 in a Δsas2 strain. α1 message was measured from RNA obtained from the same cell culture as described in A. (C) Partial silencing is observed at HMR at G1 in a Δhtz1 strain. Silencing at HMR was measured in strain YSH839 as described in the Figure 3A legend. (D) Inactivating Scc1 allows establishment of silencing at HMR in G2/M phase. Strain YSH942 was grown to log phase in low pH YPD. Nocodazole was added to one-half of the culture to arrest cells in G2/M; the other half of the culture served as the unblocked cycling control. Following arrest the culture was further divided and one-half of the arrested cells were shifted to the nonpermissive temperature for 2 hr to inactivate Scc1-73. Cultures were further divided and galactose was added to one-half to induce SIR3 expression. The unblocked cycling cells that served as the control were treated in the same way.
To assess the influence of Scc1 on the establishment of silencing, we introduced the temperature-sensitive scc1-73 allele into the inducible Sir3 strain. In this strain, silencing was efficiently established in cycling cells upon galactose induction (Figure 6D). Cells blocked at G2/M with nocodazole at the permissive temperature exhibited partial silencing upon galactose induction (Figure 6D, lane 2), while cells blocked and shifted to the nonpermissive temperature were fully silenced upon galactose induction (Figure 6D, lane 6). Therefore, Scc1 acts as a cell cycle-dependent inhibitor of silencing at HMR.
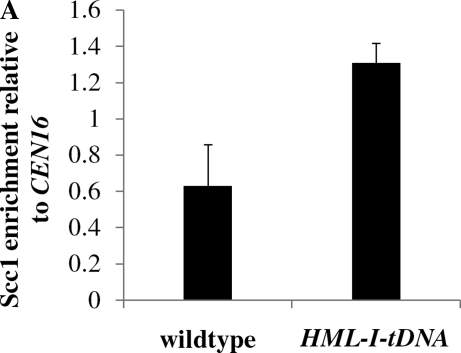
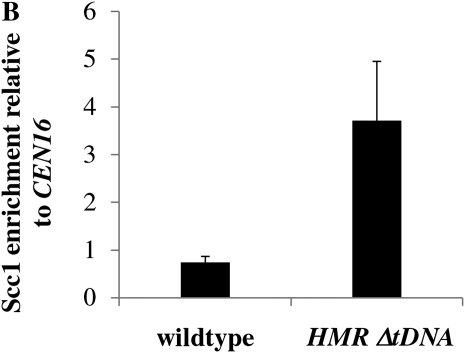
We speculated that the tRNA gene adjacent to HMR might control the establishment of silencing by increasing the recruitment of Scc1 (Dubey and Gartenberg 2007). In this model, inserting the tRNA gene downstream of HML might increase Scc1 localization at the HMLα1 and α2 genes. To test this prediction, we performed chromatin immunoprecipitation (ChIP) assays. We tagged Scc1 with a 3HA epitope to create strain YSH1017, in which the tRNA gene was deleted at HMR, and strain YSH1018, in which the tRNA gene was inserted adjacent to HML. The localization of Scc1 in these strains was compared to that in a strain in which this tRNA gene was not manipulated (YSH1016). Strains were grown to log phase in noninducing raffinose media and cells harvested to perform ChIP. Scc1 enrichment at the mating type loci was determined relative to CEN16, an Scc1 binding site. Consistent with our model, we observe that inserting the tRNA gene downstream of HML increases Scc1 localization at the HMLα1 gene (Figure 7A). However, contrary to our expectations, we found that in the absence of the adjacent tRNA gene there is an increase in Scc1 localization at HMR (Figure 7B).
Figure 7.—
A tRNA gene influences Scc1 association at HML and HMR. (A) Downstream insertion of a tRNA gene at HML increases Scc1 association with the α1 gene. Chromatin immunoprecipitation (ChIP) assays were performed on YSH1016 (wild type) and YSH1018 (HML-I-tDNA) grown to log phase in noninducing raffinose media. Relative enrichment at the α1 coding sequence in both strains was expressed relative to CEN16, a control locus that binds Scc1. (B) Elimination of an adjacent tRNA gene increases Scc1 association with HMR a1. ChIP assays were performed on YSH1016 (wild type) and YSH1017 (HMR ΔtRNA gene) grown to log phase in noninducing raffinose media. Relative enrichment at the a1 coding sequence in both strains was expressed relative to CEN16, a control locus that binds Scc1.
DISCUSSION
Numerous experiments spanning many independent studies established that cell cycle progression influences the establishment of transcriptional silencing in yeast cells (Miller and Nasmyth 1984; Fox et al. 1997; Kirchmaier and Rine 2001, 2006; Li et al. 2001; Martins-Taylor et al. 2004). The specific influence of cell cycle progression has not been determined. Possibilities include regulation of the core silencing machinery; however, thus far there is no evidence for cell cycle-dependent regulation of Sir protein levels or activity. An alternative model is that the chromatin substrates targeted by Sir proteins vary in their potential to be silenced as a function of the cell cycle. Broadly, this could be due to a cell cycle-dependent association of factors that permit or encourage the establishment of silencing or the cell cycle-dependent removal of inhibitors of silencing. Prior studies suggested that the Scc1 cohesin might be such an inhibitor (Lau et al. 2002), although a direct role for Scc1 has not yet been demonstrated. In addition, elimination of the histone variant H2A.Z abolishes the requirement for cell cycle progression in an inducible model of silencing at yeast telomeres (Martins-Taylor et al. 2011).
Results from this study indicate that cell cycle progression is not a general requirement for the establishment of silencing in budding yeast. While our experiments faithfully reproduce past findings indicating such a requirement for establishing silencing at HMR, we find that the establishment of silencing at HML is far less affected by progression though the cell cycle. This lack of a general influence of cell cycle progression suggests that this requirement is not due to intrinsic changes in the activity of the Sir protein complex. Instead, it suggests a locus-specific regulation that likely involves factors independent of the core silencing machinery. We found evidence for two distinct cis-acting elements that modulate or exert the effects of cell cycle progression. First, we observe that exchanging the genes and promoters at HML with an unrelated gene and promoter increases HML's dependence on cell cycle progression for the establishment of silencing. We have not identified the specific sequence feature responsible for this effect; one candidate would be a Rap1 binding site in the promoter of the α1 and α2 genes that does not exist at the HMR locus. This binding site promotes increased transcription of the α1 and α2 genes at MAT, but has also been defined as a “proto-silencer” (Boscheron et al. 1996) and improves the stability of silencing at HML (Cheng and Gartenberg 2000). Thus, the effect we observe could be due to a decrease in the intrinsic promoter strength at HML, or may be due to the loss of an element that promotes more efficient silencing.
We also observed that elimination of a tRNA gene adjacent to HMR abolishes the cell cycle progression requirement for the establishment of silencing, while insertion of this tRNA gene adjacent to HML increases dependence on cell cycle progression for transcriptional repression. Prior studies have determined that several tRNA genes located upstream of RNA polymerase II transcribed genes act to reduce the transcription of these genes (Hull et al. 1994), while others can act as barriers to protect genes from regulation exerted on adjacent transcription units (Simms et al. 2004). In particular, the tRNA gene located adjacent to HMR acts as a barrier to protect nearby genes from the spread of heterochromatin (Donze et al. 1999; Donze and Kamakaka 2001). A recent study found that this tRNA gene also enhances the association of Sir proteins at HMR, particularly under conditions of reduced deacetylase activity of Sir2 (Lynch and Rusche 2010). Finally, the elimination of sequences that included this tRNA gene reduced the association of cohesins at HMR by approximately twofold (Dubey and Gartenberg 2007). Coupled with our observation that eliminating the function of Scc1 also allowed silencing of HMR in the absence of cell cycle progression, we speculated that cohesin molecules were the mediators of cell cycle-dependent regulation of silencing, and that the primary influence of the tRNA gene was in influencing cohesin association. Consistent with this hypothesis, we observed an increase in cohesin association at HML in strains bearing the tRNA gene adjacent to HML, which accompanied an increased dependence on cell cycle progression to establish silencing. However, we also observed an increase in cohesin association at HMR when we deleted the tRNA gene, seemingly contradicting both prior reports and our model. A notable difference between our experiment and the prior report is that while we deleted an 80-bp DNA segment that comprised the tRNA gene, the prior study's deletions included both the tRNA gene and adjacent Ty1 sequences (Dubey and Gartenberg 2007). These sequences adjacent to the tRNA gene bind Scc1 (Laloraya et al. 2000), even in the absence of the tRNA gene (Dubey and Gartenberg 2007); thus, the Ty1 element present in our strain may be sufficient to recruit cohesins to HMR in the absence of the tRNA gene.
Our inactivation experiment using the conditional allele of Scc1, along with prior experiments using a noncleavable version of Scc1 (Lau et al. 2002), indicate that cohesins have a causal role in inhibiting the establishment of silencing at HMR. Does the increase in cohesin association at HMR upon deletion of the tRNA gene suggest that this role is not direct? One possibility is that the tRNA gene does not directly recruit cohesins, but instead acts to constrain their mobility on chromatin. In this model the absence of the tRNA could allow increased migration of cohesin into the HMR locus in Δsir3 strains, and also lead to increased displacement of cohesins upon Sir3 induction. Prior studies have suggested that cohesins can “slide” along chromosomes, possibly pushed along by a transcribing RNA polymerase (Lengronne et al. 2004). This model predicts that in some conditions Sir proteins can successfully compete with an inhibitor of silencing at HMR. Such a competition is suggested by comparing our results in the sir3-8 strain to those observed in the GAL-SIR3 strain. While silencing cannot be established at HML or HMR in G2/M phase in the sir3-8 strain, it can be partially established at HMR and fully established at HML in the GAL-SIR3 strain, in which Sir3 levels are four to five times that in wild-type strains (Figure S2).
It is possible or likely that this tRNA gene could also act to influence the establishment of silencing in addition to or independent of affecting cohesin association. For instance, recent studies have suggested that tRNA genes cluster within the nucleolus (Kendall et al. 2000; Krogan et al. 2004; Wang et al. 2005; Haeusler et al. 2008) and that this clustering is mediated by the association of condensins with the tRNA genes (Haeusler et al. 2008). Conditional mutations in the subunits of the condensin complex not only showed a loss of tRNA gene nucleolar localization but they also reduced tRNA gene-mediated silencing (Haeusler et al. 2008). Interestingly, specific alleles of condensin genes have also been reported to have mild silencing defects (Bhalla et al. 2002).
During the final review of this study, Ren et al. (2010) published data also demonstrating that the HML locus is significantly less dependent on cell cycle progression for the establishment of silencing compared to HMR. Independent experiments in which they exchanged the HML and HMR promoters, or introduced ectopic promoters at HML and HMR, convincingly demonstrated that promoter strength per se influences the dependence on cell cycle progression for the establishment of silencing (Ren et al. 2010). Our result, indicating a decreased dependence on cell cycle progression at HML when we introduced an ectopic gene, is consistent with these results. In addition, our observation that HML and HMR continue to exhibit significantly different requirements for cell cycle progression even when they contain the same promoter and coding sequences indicates sequences independent of the core gene promoters also influence the cell cycle-dependent establishment of silencing; this report indicates that the tRNA gene adjacent to HMR is one such sequence.
Overall our results suggest that a cell cycle-regulated relaxation of a chromatin barrier regulates the establishment of silencing at HMR. In an independent study we have come to similar conclusions studying the establishment of silencing at yeast telomeres, where we observe that a mitosis-specific displacement of the histone variant H2A.Z, a factor that acts as a barrier to the spread of telomeric heterochromatin (Meneghini et al. 2003), correlates with the onset of silencing (Martins-Taylor et al. 2011). These results suggest that there are windows in the cell cycle, possibly related to the dynamic changes in chromosome structure as cells traverse mitosis, that allow resetting of chromatin barriers and transcription states.
Acknowledgments
We thank Rohinton Kamakaka, James Broach, and Kim Nasmyth for yeast strains and members of the Holmes lab for valuable discussions. This work was supported by a grant from the National Science Foundation (MCB-0617986) to S.G.H.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124099/DC1.
References
- Bhalla, N., S. Biggins and A. W. Murray, 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheron, C., L. Maillet, S. Marcand, M. Tsai-Pflugfelder, S. M. Gasser et al., 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15 2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Chang, C. R., C. S. Wu, Y. Hom and M. R. Gartenberg, 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, T., and M. R. Gartenberg, 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14 452–463. [PMC free article] [PubMed] [Google Scholar]
- Donze, D., and R. T. Kamakaka, 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, R. N., and M. R. Gartenberg, 2007. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 21 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C., A. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276 1547–1551. [DOI] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H.-G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2 e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 1541–1553. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., X. Pan and J. H. McCusker, 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15 507–511. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell 63 751–762. [DOI] [PubMed] [Google Scholar]
- Haeusler, R. A., M. Pratt-Hyatt, P. D. Good, T. A. Gipson and D. R. Engelke, 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22 2204–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, A., S. Strahl-Bolsinger and M. Grunstein, 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383 92–96. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., and J. R. Broach, 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10 1021–1032. [DOI] [PubMed] [Google Scholar]
- Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh et al., 1997. Hyperactivation of the silencer proteins Sir2p and Sir3p causes chromosome loss. Genetics 145 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, M. W., J. Erickson, M. Johnston and D. R. Engelke, 1994. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 14 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall, A., M. W. Hull, E. Bertrand, P. D. Good, R. H. Singer et al., 2000. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc. Natl. Acad. Sci. USA 97 13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291 646–650. [DOI] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2006. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar, A. J., J. N. Strathern, J. R. Broach and J. B. Hicks, 1981. Regulation of transcription in expressed and unexpressed mating type cassettes of yeast. Nature 289 239–244. [DOI] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15 963–972. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, C. Sawa et al., 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya, S., V. Guacci and D. Koshland, 2000. Chromosomal addresses of the cohesin component Mcd1p. J..Cell. Biol. 151 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne, A., Y. Katou, S. Mori, S. Yokobayashi, G. P. Kelly et al., 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291 650–653. [DOI] [PubMed] [Google Scholar]
- Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz and D. Moazed, 2005. Assembly of the SIR Complex and Its Regulation by O-Acetyl-ADP-Ribose, a Product of NAD-Dependent Histone Deacetylation. Cell 121 515–527. [DOI] [PubMed] [Google Scholar]
- Lynch, P. J., and L. N. Rusche, 2010. An auxiliary silencer and a boundary element maintain high levels of silencing proteins at HMR in Saccharomyces cerevisiae. Genetics 185 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, D. J., R. Marquardt, G. J. Shei, A. B. Rose and J. R. Broach, 1991. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5 605–615. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor, K., M. L. Dula and S. G. Holmes, 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor, K., U. Sharma, T. Rozario and S. G. Holmes, 2011. H2A.Z (Htz1) controls the cell-cycle-dependent establishment of transcriptional silencing at Saccharomyces cerevisiae telomeres. Genetics 187 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M. D., M. Wu and H. D. Madhani, 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 725–736. [DOI] [PubMed] [Google Scholar]
- Miller, A., and K. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312 247–251. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. A., K. Tatchell, B. D. Hall, C. Astell and M. Smith, 1981. A position effect in the control of transcription at yeast mating type loci. Nature 289 244–250. [DOI] [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcription states in S. cerevisiae. Cell 59 637–647. [DOI] [PubMed] [Google Scholar]
- Ren, J., C. L. Wang and R. Sternglanz, 2010. Promoter strength influences the S phase requirement for establishment of silencing at the Saccharomyces cerevisiae silent mating type loci. Genetics 186 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano, P. G., and K. Tatchell, 1984. Transcription and regulatory signals at the mating type locus in yeast. Cell 37 969–978. [DOI] [PubMed] [Google Scholar]
- Simms, T. A., E. C. Miller, N. P. Buisson, N. Jambunathan and D. Donze, 2004. The Saccharomyces cerevisiae TRT2 tRNAThr gene upstream of STE6 is a barrier to repression in MATalpha cells and exerts a potential tRNA position effect in MATa cells. Nucleic Acids Res. 32 5206–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, E. M., C. Reifsnyder, M. McVey, B. Gazo and L. Pillus, 2000. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici, F., L. K. Lewis and M. A. Resnick, 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19 773–776. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang, B. D., D. Eyre, M. Basrai, M. Lichten and A. Strunnikov, 2005. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 25 7216–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, E. Y., K. A. Zawadzki and J. R. Broach, 2006. Single-cell observations reveal intermediate transcriptional silencing states. Mol. Cell 23 219–229. [DOI] [PubMed] [Google Scholar]