Abstract
Adenosine monophosphate-activated protein kinase (AMPK) is a conserved sensor of intracellular energy activated in response to low nutrient availability and environmental stress. In a screen for conserved substrates of AMPK, we identified ULK1 and ULK2, mammalian orthologs of the yeast protein kinase Atg1, which is required for autophagy. Genetic analysis of AMPK or ULK1 in mammalian liver and C. elegans revealed a requirement for these kinases in autophagy. In mammals, loss of AMPK or ULK1 resulted in aberrant accumulation of the autophagy adaptor p62 and defective mitophagy. Reconstitution of ULK1-deficient cells with a mutant ULK1 that cannot be phosphorylated by AMPK revealed that such phosphorylation is required for mitochondrial homeostasis and cell survival following starvation. These findings uncover a conserved biochemical mechanism coupling nutrient status with autophagy and cell survival.
A highly conserved sensor of cellular nutrient status found in all eukaryotes is the AMP-activated protein kinase (AMPK). In response to decreases in intracellular ATP, AMPK is activated and serves as a metabolic checkpoint, restoring ATP levels through acute regulation of metabolic enzymes and inhibition of pro-growth anabolic pathways (1). Inactivation of LKB1, the upstream kinase necessary for activation of AMPK under low energy conditions, is a frequent event in several forms of human cancer (2). In addition, LKB1 signaling is required in the liver for the therapeutic effect of metformin, the most prevalent type 2 diabetes drug worldwide, and LKB1 inactivation in murine liver results in a type 2 diabetes like metabolic disease (3). Thus the LKB1-AMPK pathway provides a direct link between tumor suppression and control of cellular and organismal metabolism.
Similar to AMPK activation, the cellular process of autophagy is initiated under nutrient-poor and low-energy conditions as a survival mechanism to ensure availability of critical metabolic intermediates and to eliminate damaged organelles including mitochondria (4). Autophagy is thought to be initiated under nutrient-limited conditions by a conserved kinase complex containing the serine-threonine kinase Atg1 and its associated subunits, Atg13 and Atg17 (5). In mammals this complex is encoded by two Atg1 homologs, ULK1 and ULK2, and the subunits Atg13 and FIP200, which signals to downstream autophagy regulators through still poorly understood mechanisms. In yeast and mammalian cells, Atg1 or ULK1 activity is suppressed under nutrient rich conditions by the TOR (target of rapamycin) complex 1 (TORC1) (6). However, biochemical events that activate Atg1 or ULK1 have not yet been identified.
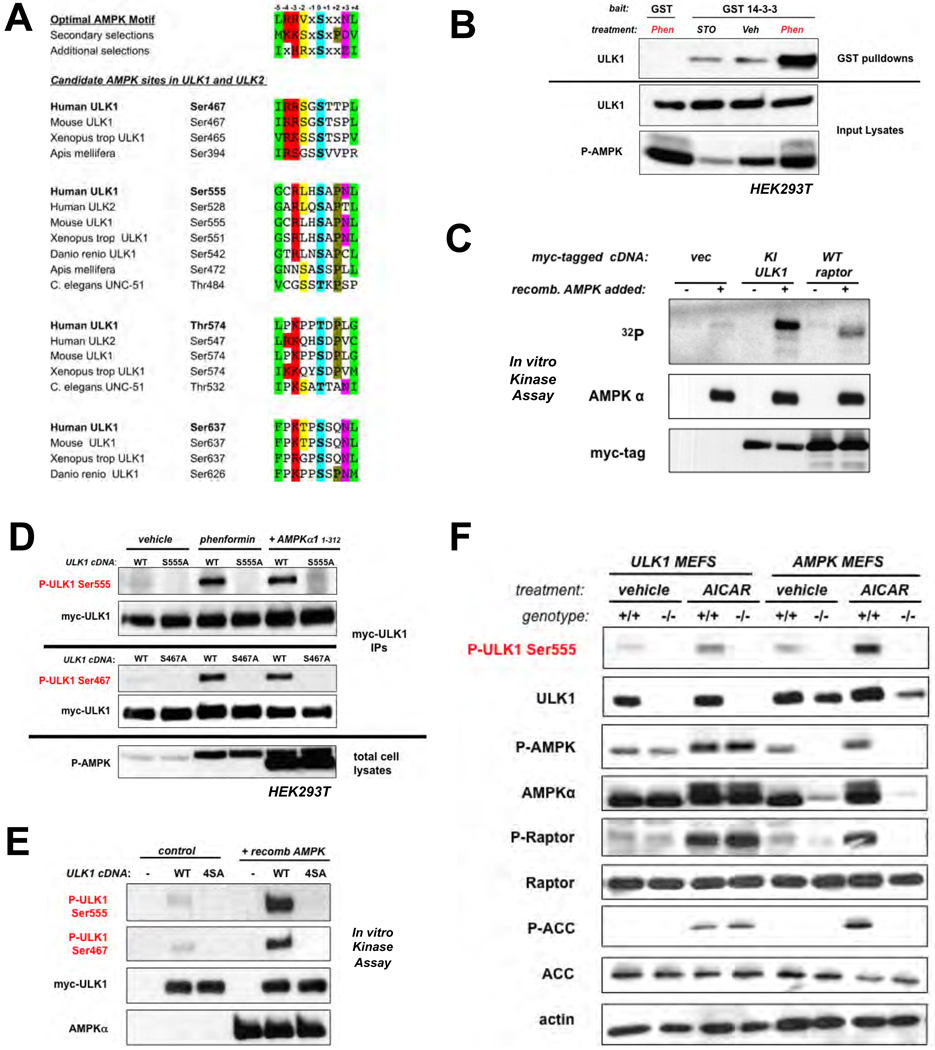
We used a two-part screen to identify substrates of AMPK that mediate its effects on cell growth and metabolism. First, we utilized an optimal AMPK substrate motif (7) to search eukaryotic databases for proteins containing conserved candidate target sites. Many in vivo substrates of AMPK not only conform to this motif, but also bind to the phospho-binding protein 14-3-3 inducibly upon phosphorylation by AMPK. We therefore screened for proteins that bound to recombinant 14-3-3 in wild-type but not AMPK-deficient cells, and only under conditions of energy stress when AMPK would be active. One protein we identified that contained multiple conserved candidate AMPK phosphorylation sites and associated with 14-3-3 in an AMPK-dependent manner was the mammalian Atg1 homolog ULK1 (Fig. 1A,B). ULK1 contains four sites (Ser467, Ser555, Thr574, Ser637) matching the optimal AMPK substrate motif, all of which are conserved in higher eukaryotes. Two of the sites are conserved back to C. elegans (Ser555 and Ser574) and in the mammalian family member ULK2, though not the more distant family members ULK3 and ULK4, which unlike ULK1 and ULK2, are not thought to function in autophagy. Indeed, endogenous AMPK subunits co-immunoprecipitated with ULK1 and ULK2 but not ULK3 (Fig. S1) and AMPK subunits were found in unbiased identifications of proteins co-immunoprecipitating with overexpressed ULK2 (Fig. S2), consistent with recent proteomic analyses (8). To examine ULK1 in vivo phosphorylation sites, we used tandem mass spectrometry on epitope-tagged ULK1 isolated from cells treated with or without the mitochondrial complex I inhibitor phenformin (9). We detected peptides spanning three of the four candidate AMPK sites in ULK1 (Ser555, Thr574, Ser637), and all three were phosphorylated only after phenformin treatment (Fig. S3, S4). To examine whether ULK1 could serve as a direct substrate for AMPK in vitro, we created a kinase-inactive allele (K46I; ref. 10), to remove its autophosphorylation. AMPK phosphorylated ULK1 to a greater extent than an established substrate, Raptor (Fig. 1C, S5), which may reflect the presence of at least four potential AMPK sites in ULK1, as compared to Raptor, which has two reported AMPK sites (7). We generated phospho-specific antibodies against Ser467 and Ser555 of ULK1. Phosphorylation of both sites was induced by phenformin treatment or expression of ULK1 with a constitutively active AMPKα1 allele (11) in the absence of energy stress (Fig. 1D). Purified AMPK also induced phosphorylation at these sites in an in vitro kinase assay, consistent with their direct phosphorylation (Fig. 1E). Using AMPK- and ULK1-deficient primary mouse embryonic fibroblasts (MEFs) or matched control wild-type MEFs, we observed phosphorylation of endogenous ULK1 on Ser555 in an AMPK-dependent manner after treatment of cells with the AMP-mimetic AICAR (Fig. 1F). Notably, the phosphorylation of ULK1 in these cells paralleled that of two bona-fide AMPK substrates, ACC and Raptor (Fig. 1F, S6).
Fig. 1.
ULK1 is a conserved substrate of AMPK. (A) Clustal alignment of four conserved sites in ULK1 and two sites in ULK2 matching the optimal AMPK substrate motif. (B) ULK1 and GST or GST-14-3-3 expression vectors were transfected into human embryonic kidney (HEK)293T cells, and placed in media containing 20µM STO-609 (STO), vehicle (veh), or 5mM phenformin (Phen) for 1h. Cell lysates and GST pulldowns were immunoblotted as indicated. (C) In vitro kinase assays with myc-tagged catalytically inactive (KI: K46I) ULK1 or myc-tagged wild-type raptor which were immunoprecipitated from HEK293T cells and used as substrates for purified active AMPK in the presence of 32P-γ-ATP. (D) HEK293T cells transfected with myc-tagged wild-type ULK1 or indicated Serine-to-alanine ULK1 mutants were treated with either vehicle or 1mM phenformin for 1h, or were co-transfected with a constitutively active AMPKα1 (aa1-312) mammalian expression vector (11). Proteins from lysates were immunoblotted with phospho-specific antibodies as indicated. (E) In vitro kinase assays using myc-ULK1 and purified AMPK as above. Phosphorylation of myc-ULK1 detected by immunoblotting with indicated phospho-specific antibodies. (F) Primary murine embryonic fibroblasts (MEFs) were treated with 2mM AICAR or vehicle for 1h. Lysates immunoblotted as indicated including detection of endogenous ULK1 P-Ser555
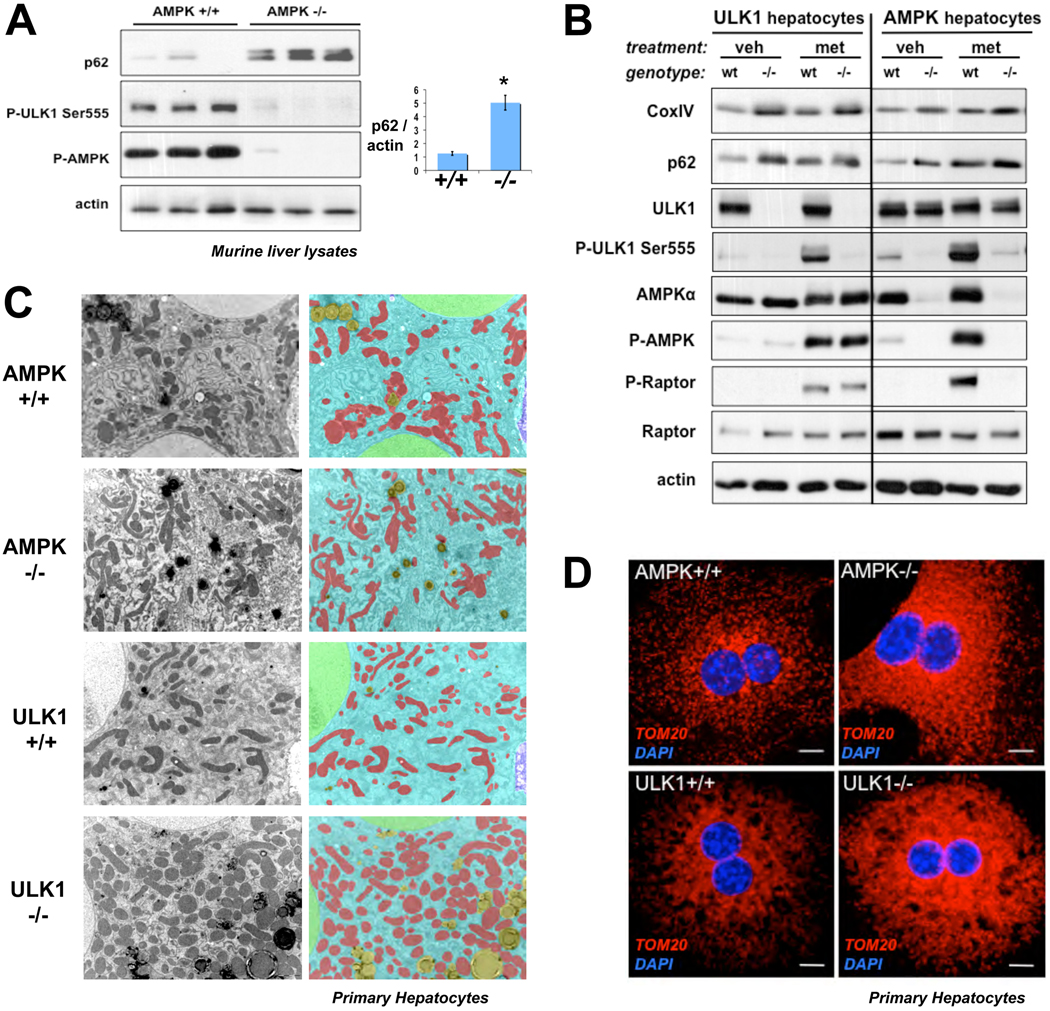
We examined the phenotypic consequences of AMPK- or ULK1-deficiency on markers of autophagy in murine liver and primary hepatocytes. Immunoblot and immunohistochemical analysis of AMPK-deficient livers (12) showed accumulation of the p62 protein (Fig. 2A, S7), whose selective degradation by autophagy has established it as a widely used marker of this process (13). p62 contains a UBA ubiquitin binding domain which mediates binding to ubiquitinated cargo targeted for autophagy mediated degradation (13). Consistent with this function, p62 aggregates colocalized with ubiquitin aggregates in AMPK-deficient livers (Fig. S7). Notably, p62 is recruited to mitochondria targeted for mitophagy, and is involved in mitochondrial aggregation and clearance (14, 15). ULK1-deficient mice exhibit accumulation of defective mitochondria in mature red blood cells, which are normally devoid of mitochondria (16). Given the aberrant accumulation of p62 in the absence of AMPK in mouse liver and the fact that rodent hepatocytes undergo significant mitophagy upon culturing (17), we examined whether AMPK- or ULK1- deficiency in primary hepatocytes might exhibit mitochondrial defects. Protein levels of p62 and the mitochondrial marker protein CoxIV were similarly elevated in lysates from AMPK- or ULK1-deficient hepatocytes cells but not wild-type controls (Fig. 2B, S8). Increased phosphorylation of endogenous ULK1 Ser555 was observed in wild-type but not AMPK-deficient hepatocytes after AMPK activation by metformin treatment (Fig. 2B). Further analysis of the ULK1 and AMPK hepatocytes using transmitting electron microscopy (TEM) revealed elevated levels of abnormal mitochondria, which was analyzed quantitatively using morphometric software (Fig. 2C, right panels). Similar to findings in other autophagy-mutant hepatocytes (18), the number of mitochondria per cell was significantly increased in AMPK- and ULK1-deficient hepatocytes compared to wild-type controls (Fig. S9), also seen by immunocytochemical staining for the mitochondrial membrane protein TOM20 (Fig. 2D).
Fig. 2.
Genetic deficiency for AMPK or ULK1 in murine liver or primary murine hepatocytes results in autophagy defects. (A) Liver lysates from littermate-matched mice were immunoblotted for the indicated antibodies. p62 to actin ratio calculated from densitometry performed on immunoblots. Data shown as mean +/− SEM. * p<.01 (B) Primary hepatocytes derived from ULK1+/+ or ULK1−/− mice or AMPKa1+/−a2L/+ or AMPKa1−/−a2L/L as described in methods were placed in media containing 2mM metformin (met) or vehicle (veh) for 2h. Lysates were immunoblotted with the indicated antibodies. (C) Transmitting electron microscopy (TEM) was performed on primary murine hepatocytes of the indicated genotypes revealing accumulation of mitochondria in both AMPK- and ULK1-deficient cells. Mitochondria pseudocolored RED, cytoplasm BLUE, nuclei GREEN, and lipid droplets YELLOW. (D) Primary murine hepatocytes of the indicated genotypes stained by immunocytochemistry for the mitochondrial marker TOM20 (red) and nuclei (blue). Scale bar, 10 microns.
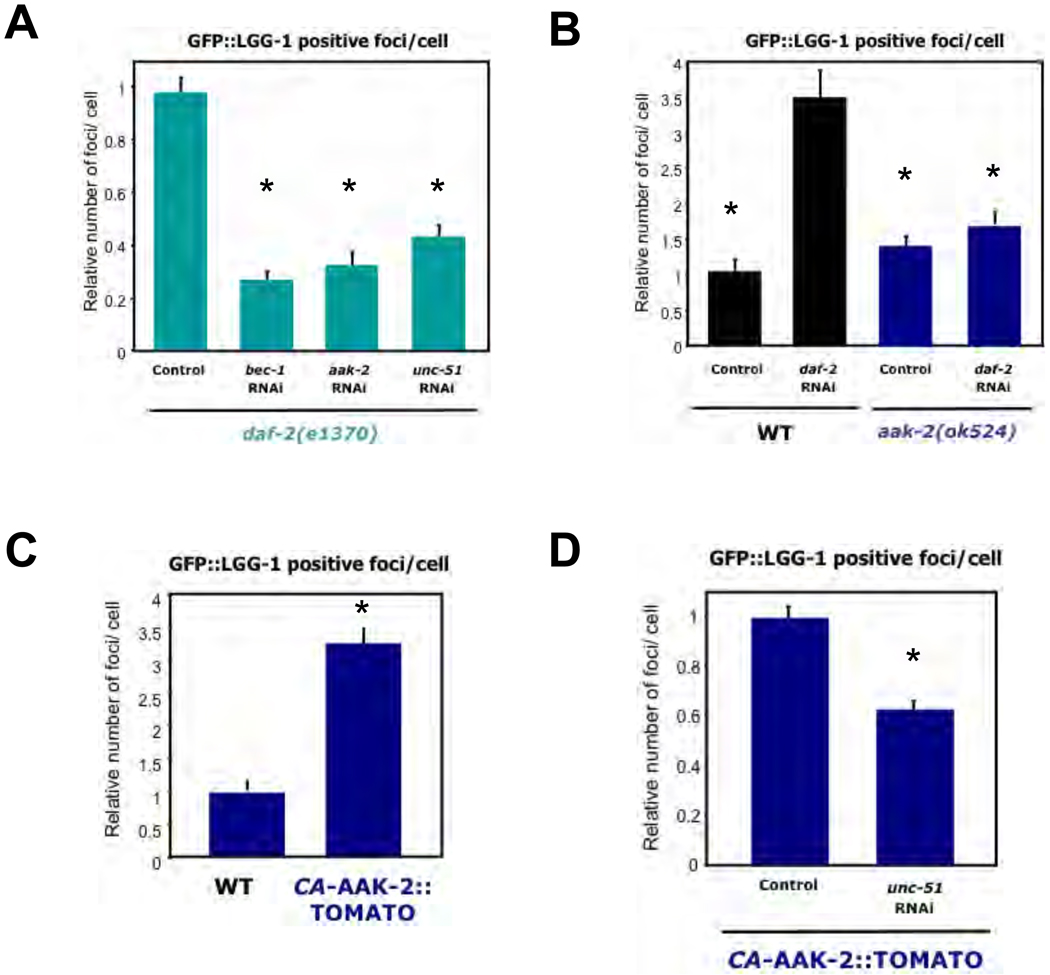
Given the conservation of AMPK sites in ULK1, we examined whether these two proteins play conserved roles in autophagy in the nematode C. elegans. In a reporter assay based on the C. elegans LC3 homolog LGG-1 (19,20), we observed that loss of insulin signaling through genetic mutation (daf-2 (e1370)) or RNAi against the insulin receptor daf-2, resulted in increased numbers of GFP::LGG-1 positive foci in hypodermal seam cells, indicative of increased autophagy and consistent with the established role for insulin signaling in the suppression of autophagy in C. elegans (19–23). daf-2 mutant worms treated with RNAi to aak-2 or unc-51, the AMPK and ULK1 orthologs, respectively, resulted in a decrease in abundance of LGG-1 containing puncta (Fig. 3A). daf-2 RNAi failed to increase the number of LGG-1 positive foci in AMPK-deficient worms (Fig. 3B). These data indicate that both AMPK and ULK1 have critical roles in autophagy induced by reduced insulin signaling in C. elegans. Transgenic worms expressing constitutively active AMPK exhibited a ~3-fold increase in the number of LGG-1 positive foci in seam cells compared to the number of foci in controls (Fig. 3C). The number of LGG-1-positive foci was significantly reduced when these animals were fed unc-51 RNAi (Fig. 3D) [all raw data in Fig. S10]. These observations indicate that AMPK activation is sufficient to induce autophagy in worms, and ULK1 is required for this induction.
Fig. 3.
AMPK is necessary and sufficient for autophagy induction in C. elegans. (A) Insulin receptor daf-2(e1370) mutant worms expressing GFP::LGG-1 (equivalent to GFP-LC3) were treated with control RNAi or RNAi against bec-1 (Beclin), aak-2 (AMPKα2) or unc-51 (ULK1) and the number of LGG-1/LC3-positive puncta per hypodermal seam cell were quantified. (B) aak-2(ok524) mutants or wild-type N2 (WT) animals expressing GFP::LGG-1 were treated with control or daf-2 RNAi and were scored for LGG-1 positive puncta per seam cell. (C) Transgenic worms expressing constitutively active AAK-2 (ref. 11) (amino acids 1-321)::TOMATO (CA-AAK-2::TOMATO(1-321) fusion or controls were analyzed for LGG-1 positive puncta per seam cell. (D) Animals expressing both CA-AAK-2(1-321)::TOMATO and GFP::LGG-1 were treated with control or unc-51 RNAi and scored for LGG-1/LC3-positive puncta per seam cell. All panels show relative counts, see Figure S10 for details. Data shown as mean +/− SEM. * P<.0001
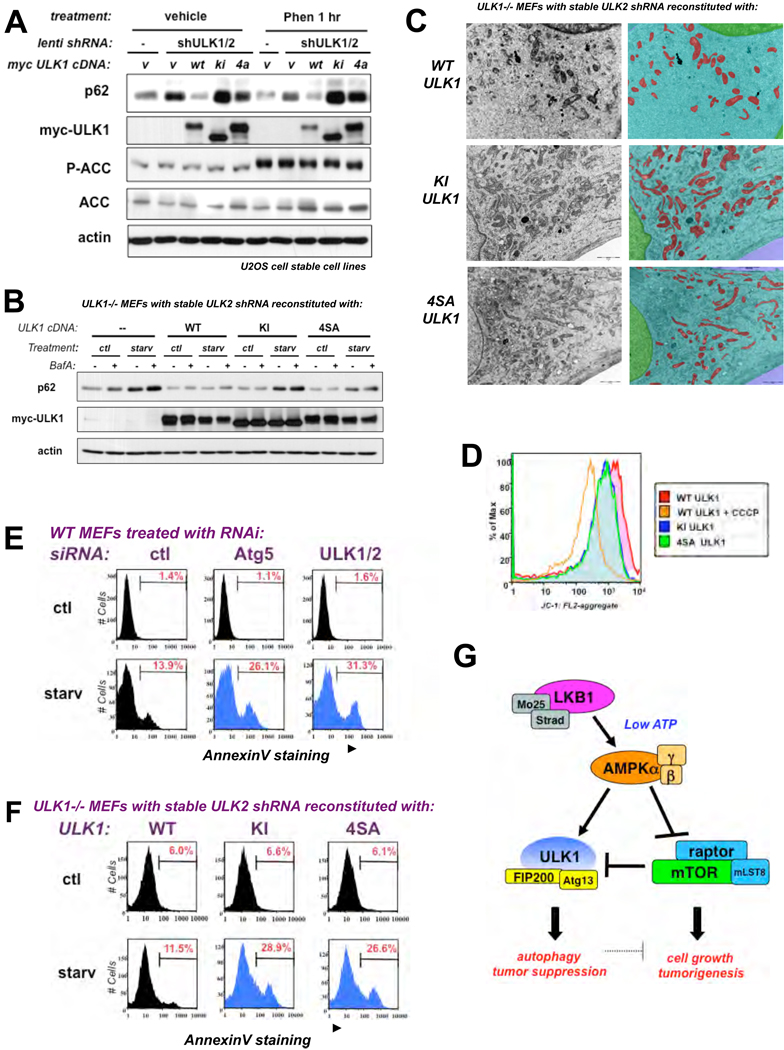
To test whether AMPK phosphorylation of ULK1 is required for ULK1 function, we stably introduced wild-type (WT), catalytically inactive (KI), or the AMPK non-phosphorylatable (4SA) ULK1 cDNA into human osteosarcoma U2OS cells in which we subsequently reduced endogenous ULK1 and ULK2 with lentiviral hairpin shRNAs against each (Fig. S11). U2OS cells stably expressing ULK1 and ULK2 shRNA exhibited increased amounts of p62 indicative of defective autophagy compared to that of parental U2OS cells infected with an empty lentiviral vector (Fig. 4A, compare lane 1 and 2). Stable retroviral reconstitution of a myc-tagged WT ULK1 cDNA, but not the 4SA or KI mutant, restored p62 degradation (Fig. 4A, lanes 3-5; Fig. S12). Furthermore, we reconstituted ULK1−/− MEFs that were also knocked down for endogenous ULK2 (Fig. S13) with WT, KI, or 4SA ULK1 cDNAs and examined the extent of autophagy following placement of these cell lines into starvation media. MEFs deficient for ULK1 and ULK2 contained elevated levels of p62 upon starvation. Cells reconstituted with WT ULK1 had reduced p62 levels, unlike the KI or 4SA-expressing cells which behaved like the ULK-deficient state (Fig. 4B, S14). To test whether the 4SA mutant exhibited effects on mitochondrial homeostasis, we utilized TEM and mitochondrial-selective dyes on the WT, KI, and 4SA ULK1 stably reconstituted ULK-deficient MEFs. TEM and Mitotracker Red staining revealed that the KI- and 4SA-ULK1 expressing cells had altered mitochondrial homeostasis compared WT ULK1 cells, denoted by increases in the overall number and aberrant morphology of mitochondria (Fig. 4C, S18, S19). The altered cristae and aberrant morphology of the mitochondria in the KI- and 4SA-ULK1 reconstituted cells was enhanced upon starvation (Fig. S19). To test whether these mitochondria were functionally impaired, we analyzed the mitochondrial membrane potential with the activity dependent JC-1 dye, which revealed defects in KI- and 4SA- reconstituted MEFs (Fig. 4D).
Fig. 4.
AMPK phosphorylation of ULK1 is required for mitophagy and cell survival upon nutrient deprivation (A) U2OS cells stably expressing mouse wild-type (WT) or catalytically inactive (KI) or AMPK non-phosphorylatable (4SA) ULK1 cDNA or the empty retroviral vector (v) along with a shRNA against endogenous human ULK1 and ULK2 were placed in media containing 5mM phenformin (Phen) or vehicle for 1h. Lysates were immunoblotted as indicated. (B) ULK1−/− MEFs stably expressing WT, KI, or 4SA ULK1 cDNA or the empty retroviral vector (v) along with a shRNA against endogenous ULK2 were placed in EBSS starvation media (starv) or control media (ctl) for 6h in the presence or absence of BafilomycinA (BafA) and immunoblotted as indicated. (C) Cells from (B) analyzed by TEM and Inform morphometric software. Mitochondria pseudocolored RED, cytoplasm BLUE, and nuclei GREEN. (D) Flourescence Activated Cell Sorting (FACS) analysis on cells from (B) which were stained with JC-1 under basal conditions, or with the mitochondrial uncoupler CCCP as a control, to measure mitochondrial membrane potential. Compromised mitochondrial membrane potential is shifted to the left, as observed in cells treated with CCCP. (E) Wild-type (WT) MEFs transfected with 20nM siRNA pools to a universal control (ctl), murine Atg5, or murine ULK1 and ULK2 for 72 hours were then placed in starvation medium (starv) or standard media (ctl) for 12h and cell death was scored by AnnexinV- FACS. (F) Cells from (B) were placed in starvation medium (starv) or standard media (ctl) for 12h and cell death was scored by AnnexinV- FACS. (G) Model for AMPK activation of ULK1 in a two-pronged mechanism via direct phosphorylation of ULK1 and inhibition of mTORC1 suppression of ULK1.
A hallmark of cells defective for autophagy is a predisposition to undergo apoptosis after stress stimuli that normally would activate autophagy to promote cell survival (24). We examined how ULK1/2 deficiency would compare to loss of central downstream autophagic regulator such as Atg5 in terms of requirement for cell survival following starvation. Wild-type MEFs were treated with control, Atg5, or combined ULK1 and ULK2 siRNA and analyzed for effects on cell viability after being placed into starvation conditions. Simultaneous depletion of ULK1 and ULK2 mirrored the magnitude and kinetics of cell death observed with Atg5 loss upon starvation (Fig. 4E, S20). We next investigated whether mutation of the AMPK sites in ULK1 might also mimic ULK1/2 loss of function in this cell survival assay. ULK-deficient MEFs reconstituted with WT, but not KI or 4SA ULK1, restored cell survival after starvation (Fig. 4F). ULK1-deficient cells expressing the KI or 4SA mutant ULK1 showed rates of cell death like WT MEFs treated with Ulk1 and Ulk2 siRNA. Thus, loss of the AMPK sites in ULK1 mimics complete loss of ULK1 and ULK2 in control of cell survival after nutrient deprivation.
Our findings reveal a direct connection between energy sensing and core conserved autophagy proteins. In mammals, phosphorylation of ULK1 by AMPK is required for ULK1 function in the response to nutrient deprivation. As AMPK suppresses mTOR activity and mTOR inhibits ULK1 (25–29), AMPK controls ULK1 via a two-pronged mechanism, ensuring activation only under the appropriate cellular conditions (Fig. 4G, S21). There are a number of physiological and pathological contexts where this pathway is likely to play a critical role (30). Beyond the conserved nature of these signaling events and the role of some autophagy genes as tumor suppressors (24, 31), AMPK is defective in a variety of human cancers bearing inactivating mutations in its upstream kinase LKB1. Thus ULK1 may have a central role in the beneficial effects of the LKB1/AMPK pathway on tumor suppression or in treatment of metabolic disease, as observed here with metformin stimulation of ULK1 phosphorylation in liver and the profound defect in autophagy in AMPK-deficient livers. ULK1-dependent effects on mitochondrial homeostasis and cell survival may represent additional beneficial effects of metformin and other AMPK activators in overall organismal health and lifespan (32).
Supplementary Material
References and Notes
- 1.Hardie DG. Nat Rev Mol Cell Biol. 2007;8:774. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 2.Shackelford DB, Shaw RJ. Nat Rev Cancer. 2009;9:563. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw RJ, et al. Science. 2005;310:1642. [Google Scholar]
- 4.He C, Klionsky DJ. Annu Rev Genet. 2009;43:67. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. Curr Opin Cell Biol. 2010 [Google Scholar]
- 6.Chan EY. Sci Signal. 2009;2:pe51. doi: 10.1126/scisignal.284pe51. [DOI] [PubMed] [Google Scholar]
- 7.Gwinn DM, et al. Mol Cell. 2008;30:214. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrends C, Sowa ME, Gygi SP, Harper JW. Nature. 2010;466:68. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardie DG. Gastroenterology. 2006;131:973. doi: 10.1053/j.gastro.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Chan EY, Longatti A, McKnight NC, Tooze SA. Mol Cell Biol. 2009;29:157. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. J Biol Chem. 1998;273:35347. doi: 10.1074/jbc.273.52.35347. [DOI] [PubMed] [Google Scholar]
- 12.Andreelli F, et al. Endocrinology. 2006;147:2432. doi: 10.1210/en.2005-0898. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, McEwan DG, Novak I, Dikic I. Mol Cell. 2009;34:259. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Geisler S, et al. Nat Cell Biol. 2010;12:119. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 15.Narenda DP, Kane LA, Hauser DN, Fearnley IM, Youle RJ. Autophagy. 2010;6:1. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundu M, et al. Blood. 2008;112:1493. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Enriquez S, Kai Y, Maldonado E, Currin RT, Lemasters JJ. Autophagy. 2009;5:1099. doi: 10.4161/auto.5.8.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Vicente M, et al. Nat Neurosci. 2010;13:567. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melendez A, et al. Science. 2003;301:1387. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 20.Kang C, You YJ, Avery L. Genes Dev. 2007;21:2161. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen M, et al. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia K, et al. Proc Natl Acad Sci U S A. 2009;106:14564. doi: 10.1073/pnas.0813319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hars ES, et al. Autophagy. 2007 Mar-Apr;3:93. [Google Scholar]
- 24.Levine B, Kroemer G. Cell. 2008;132:27. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung CH, et al. Mol Biol Cell. 2009;20:1992. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang YY, Neufeld TP. Mol Biol Cell. 2009;20:2004. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa N, et al. Mol Biol Cell. 2009;20:1981. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamada Y, et al. Mol Cell Biol. 2010;30:1049. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganley IG, et al. J Biol Chem. 2009;284:12297. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lippai M, et al. Autophagy. 2008;4:476. doi: 10.4161/auto.5719. [DOI] [PubMed] [Google Scholar]
- 31.Mathew R, Karantza-Wadsworth V, White E. Nat Rev Cancer. 2007;7:961. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogarty S, Hardie DG. Biochim Biophys Acta. 2010;1804:581. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 33.We thank L. Gerken for mouse colony assistance, F. Esterman for confocal assistance, C. Chu for assistance in scoring LGG-1 assays, M. Wood at The Scripps Research Institute Electron Microscopy facility, X. Yang in the BIDMC Mass spectometry facility, R. Chitta from St. Jude’s proteomic core, K. Lamia for comments on the manuscript. DFE, DMG, and MMM were supported by T32 CMG training grant to UCSD/Salk. DBS was funded by T32 CA009370 to the Salk Institute Center for Cancer Research. Work in the lab of RJS is supported by NIH R01 DK080425, 1P01CA120964, NCI P30CA014195, an American Cancer Society Research Scholar Award, the American Diabetes Association Junior Faculty Award 1-08-JF-47, and HHMI Early Career Scientist Award. JMA is supported by 5P30CA006516-43 and 1P01CA120964-01A. MK is funded by NHBLI K08, Burroughs Welcome Fund, and the American Lebanese Syrian Association. BV is supported by Agence Nationale de la Recherche (ANR). MH is an Ellison Medical Foundation New Scholar in Aging. We also thank the Leona M. and Harry B. Helmsley Charitable Trust for their generous support.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.