Abstract
The building of nuclear bodies after mitosis is a coordinated event crucial for nuclear organization and function. The nucleolus is assembled during early G1 phase. Here, two periods (early G1a and early G1b) have been defined. During these periods, the nucleolar compartments (DFC, GC) corresponding to different steps of ribosome biogenesis are progressively assembled. In telophase, rDNA transcription is first activated and PNBs (reservoirs of nucleolar processing proteins) are formed. The traffic of the processing proteins between incipient nucleoli and PNBs was analyzed using photoactivation. We demonstrate that the DFC protein fibrillarin passes from one incipient nucleolus to other nucleoli but not to PNBs, and that the GC proteins, B23/NPM and Nop52, shuttle between PNBs and incipient nucleoli. This difference in traffic suggests a way of regulating assembly first of DFC and then of GC. The time of residency of GC proteins is high in incipient nucleoli compared to interphase nuclei, it decreases in LMB-treated early G1a cells impairing the assembly of GC. Because the assembly of the nucleolus and that of the Cajal body at the exit from mitosis are both sensitive to CRM1 activity, we discuss the fact that assembly of GC and/or its interaction with DFC in early G1a depends on shuttling between PNBs and NORs in a manner dependent on Cajal body assembly.
Key words: nucleolus, PNBs, Cajal body, traffic, CRM1, living cell, photoactivation, cell cycle, EM structure
Introduction
At each cell cycle, the nucleus is reorganized from inherited material as it passes through mitosis. At exit from mitosis, beside chromosome decondensation and nuclear envelope formation, the nuclear domains (reviewed in ref. 1) that will be active during interphase are reconstituted. This is a crucial period during which cross talk between different domains is established to coordinate the nuclear functions. For example, it was proposed that the assembly of nucleolus and the interchromatin granules (also called speckles) is linked.2,3
The multiple steps of ribosome biogenesis occur in the nucleolus. They include transcription of ribosomal genes (rDNA), processing and modifications of precursor ribosomal RNAs (pre-rRNAs) and assembly with ribosomal proteins.4 The nucleolar organization in three compartments, fibrillar center (FC), dense fibrillar component (DFC) and granular component (GC) reflects the vectorial progression of ribosome biogenesis from DFC (early processing) to GC (late processing), and it is impaired by blockage at any step of the process.5–7 The nucleolus is also a plurifunctional domain involved in assembly of RNPs and a target for viral proteins and viral RNAs.8–11
The assembly of the nucleolus starts during telophase by activation of rDNA transcription of several nucleolar organizer regions (NORs) and by progressive recruitment of early and late processing proteins on rRNA transcripts. In parallel, prenucleolar bodies (PNBs) are assembled.12–16 The PNBs contain nucleolar processing proteins, pre-rRNAs and snoRNAs.17 Even though PNBs are formed in all animal and plant cells, their role in the dynamics of DFC and GC assembly is still unknown.
It is possible to observe and measure the motion of proteins or RNP complexes in living cells. Using these approaches it was demonstrated that macromolecules within the nucleus are highly mobile by a combination of passive diffusion and high-affinity binding sites.18–20 It was also established that the assembly of nuclear bodies after mitosis is strictly ordered.14,21–24 It is now important to characterize the dynamics of the complexes during the building of the functional domains integrated in the nuclear network. We examined the role of the PNB step in the kinetics of DFC and GC assembly. Using photoactivation,25 we demonstrate that traffic between transcription sites and PNBs is unidirectional for fibrillarin (DFC marker) and is at equilibrium between these two sites for B23/NPM and Nop52 (GC markers). It was previously reported that parallels exist in the pathway of reassembly of Cajal bodies (CB) and nucleoli.23 Here we propose that CB assembly and/or function is necessary during early G1 for efficient formation of the nucleolar GC and that the assembly of both nucleolus and CB is CRM1-dependent.
Results
Two periods during nucleolar assembly: early G1a and early G1b.
During nucleolar assembly, the recruitment of GFP-tagged nucleolar processing proteins on sites of rDNA transcription, i.e., in NORs, was analyzed in living HeLa cells. Recruitment in the 6 active NORs starts in telophase concomitantly to the formation of numerous PNBs. Thirty to 90 minutes later, the fluorescence in incipient nucleoli increases while PNB fluorescence decreases (Fig. 1A). Thus proteins are globally transferred from PNBs to incipient nucleoli. The transfer lasts 15–20 minutes for early processing proteins localized in DFC (GFP-fibrillarin) while the recruitment of late processing proteins localized in GC (GFP-B23/NPM, GFP-Nop52 or GFP-Bop1) lasts at least 90 minutes. Remarkably, early and late proteins are in the same PNBs16 but DFC proteins are recruited more rapidly than GC proteins indicating a differential sorting from PNBs and a different steady state in incipient nucleoli.
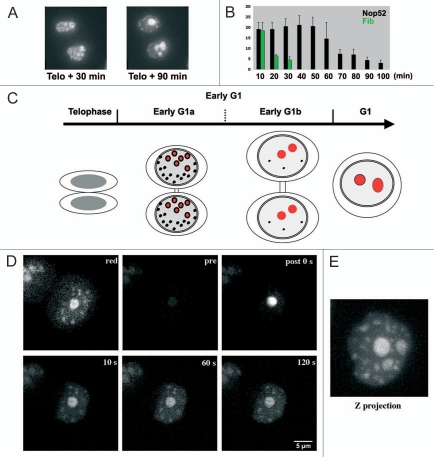
Figure 1.
Nucleolar assembly after telophase. (A) Time-lapse observation of living cells expressing GFP-Nop52. Thirty minutes after telophase Nop52 localizes in incipient nucleoli (large structures) and PNBs (small foci). After 90 minutes the incipient nucleoli are fused and the number of PNBs is low. (B) Number of PNBs evaluated in Z projections on time-lapse images. The presence of fibrillarin is limited in time (green), and the presence of Nop52 (black) is almost stable for nearly 1 hour and then decreases. X axis = time (min); Y axis = number of PNBs. (C) Schematic representation of nucleolar assembly based on time lapse observations. After telophase and before cytokinesis, 2 periods are observed in early G1 cells. Red: incipient nucleoli; black: PNBs. (D) Photoactivation of PAGFPB23/NPM in incipient nucleolus of an early G1 cell. The DsRed-Nop52 (red) shows one incipient nucleolus and several PNBs; PAGFP-B23/NPM (pre) is hardly visible. After activation (post 0 s), the PAGFP-B23/NPM is visible and then PNBs are labeled (10, 60, 120 seconds). (E) Z projection 2 min after activation showing the distribution of PAGFP-B23/NPM of one incipient nucleolus in all incipient nucleoli and PNBs.
After telophase, 20–25 PNBs containing both fibrillarin and B23/NPM were identified in Z projections. Twenty minutes later, the number of PNBs containing fibrillarin is reduced while the number of PNBs containing B23/NPM is stable for 60 minutes and then decreases during ∼50 minutes (Fig. 1B). During this second period, contact and fusion between incipient nucleoli is observed in daughter cells still connected by a cytoplasmic bridge (Fig. 1C). In parallel chromatin decondenses and consequently the nuclear volume progressively increases. The end of this period is characterized by the presence of 2 or 3 large nucleoli and the absence or low number of PNBs. These features reveal two periods in nucleolar assembly that will be named hereafter early G1a and early G1b (Fig. 1C).
During nucleolar assembly, late processing proteins shuttle between sites of rDNA transcription and PNBs.
The great number of PNBs, their persistence for up to 120 minutes after telophase and the difference between early and late processing proteins suggest a role of the PNBs in the traffic of nucleolar proteins. One possibility is that the PNBs are involved in the progressive delivery of processing complexes onto sites of rDNA transcription. To analyze the traffic between PNBs and NORs, the nucleolar proteins were fused to photoactivatable (PA) GFP.25 The position of the region of interest (ROI) for activation is established by the presence of a protein partner tagged in red. The PAGFP-B23/NPM is activated in an incipient nucleolus identified by the presence of DsRed-Nop52 (Fig. 1D, red). Several PNBs containing DsRed-Nop52 are also visible in the optical section. After activation (Fig. 1D and compare pre and post 0 s), the PAGFP-B23/NPM signal is high in the nucleolus, and then decreases as expected of proteins trafficking between nucleolus and nucleoplasm. Surprisingly, activated PAGFP-B23/NPM is detected in PNBs 5–10 seconds after activation (Fig. 1D). The uptake of B23/NPM in PNBs is observed in all PNBs containing mDsRed-Nop52 visible in the focal plane. This indicates that feedback of activated molecules occurs between incipient nucleoli and PNBs. The projection of the Z series of images demonstrates the presence of activated PAGFP-B23/NPM in each incipient nucleolus and all PNBs (Fig. 1E). PAGFP-B23/NPM is detected in all PNBs visible in red (163 in 8 cells) indicating that this redistribution occurs in the entire nuclear volume. Thus in early G1, B23/NPM molecules undergo rapid exchanges between the incipient nucleoli and can still be taken up in PNBs.
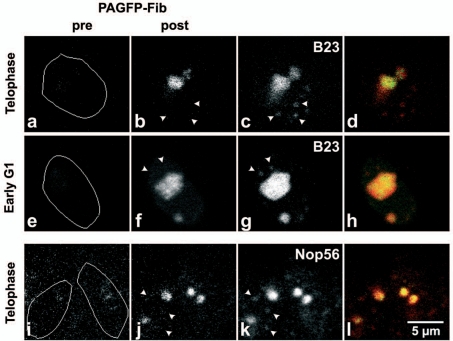
Photoactivation of PAGFP-Nop52 in one incipient nucleolus demonstrates that the uptake of Nop52 in PNBs is similar to that of B23/NPM. The traffic of PAGFP-Nop52 from the nucleolus to PNBs was observed in the entire nuclear volume of 8 early G1a cells, by colocalization with coexpressed DsRed-B23/NPM in Z series of images (data not shown). On the contrary PAGFP-fibrillarin was not detected in PNBs containing mDsRed-B23/NPM (Fig. 2b–d, f–h) or mRFP-Nop56 signal (Fig. 2j–l). However, the redistribution of PAGFP-fibrillarin was observed in each incipient nucleolus of early G1 cells (verified for 50 nucleoli in 10 cells). At this period of the cell cycle, while sharing of fibrillarin between the incipient nucleoli reveals nucleolar-nucleoplasm traffic, the time of residency of fibrillarin in PNBs does not allow its detection (Fig. S1).
Figure 2.
Photoactivation of PAGFP-fibrillarin in incipient nucleolus of telophase and early-G1 cells. The DsRed-B23/NPM (c and d) and RFP-Nop56 (k and l) show incipient nucleoli and several PNBs (arrowheads); PAGFP-fibrillarin (pre) is hardly visible. 2 min after activation (post), the PAGFP-Fib is visible in activated incipient nucleoli and neighbouring incipient nucleoli (b, f and j) but not in PNBs (arrowheads). This is confirmed by the superimposition of green and red signals because only a red signal is visible in PNBs (d, h and l).
In conclusion, the recruitment of two late processing proteins in an incipient nucleolus implies bidirectional traffic between PNBs and the nucleolus. This indicates that the uptake of the late rRNA processing proteins in PNBs is still possible in early G1 and could be a way of regulating GC building during nucleolar assembly. The balance between uptake in incipient nucleoli and PNBs must ultimately be in favor of nucleoli as nucleolar assembly progresses.
LMB disturbs shuttling between incipient nucleoli and PNBs.
In a screen of drugs capable of modifying the assembly of nucleoli at exit from mitosis, we discovered that Leptomycin B (LMB) increases the number of early G1a cells (Fig. 3A and B). Typically these LMB-treated cells, connected by a cytoplasmic bridge with a mid-body, present ∼5–6 incipient nucleoli and PNBs containing GFP-B23/NPM, GFP-Nop52 or GFP-Bop1 (Fig. 3C). On the contrary the presence of fibrillarin-GFP in PNBs was not observed (not shown) confirming that PAGFP-fibrillarin activated in incipient nucleoli does not feed back into PNBs (see Fig. 7 the cap nucleolar distribution of fibrillarin).
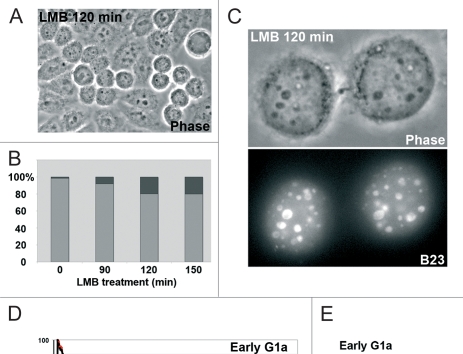
Figure 3.
LMB blocks the cell cycle in early G1. (A) Phase contrast of unsynchronized HeLa cells treated for 2 hours with LMB. The number of early G1 cells (doublet linked by a cytoplasm bridge) is high. (B) Percentage of early G1 (black) depending on the duration of the LMB treatment compared to interphase (grey). The percentage increases with duration of treatment and can be maintained for up to 6 hours. (C) Cells blocked in early G1 after 120 minutes of LMB. In nuclei several large dark structures (phase contrast) containing B23 (GFP-B23 labeling) correspond to incipient nucleoli. In addition B23 is observed in many PNBs. The cytoplasmic bridge with the mid-body is visible in upper image. (D) Traffic of B23/NPM measured by photoactivation in early G1a for 2 minutes. Relative intensity in the activated incipient nucleoli, in control black curve and in LMB-treated cells (2 h) red curve (mean of 8 CTR and 10 LMB-treated cells). (E) Comparison of the t1/2 of B23/NPM traffic in incipient nucleoli of early G1 and interphase nucleoli in control (CTR) and LMB-treated (LMB) cells.
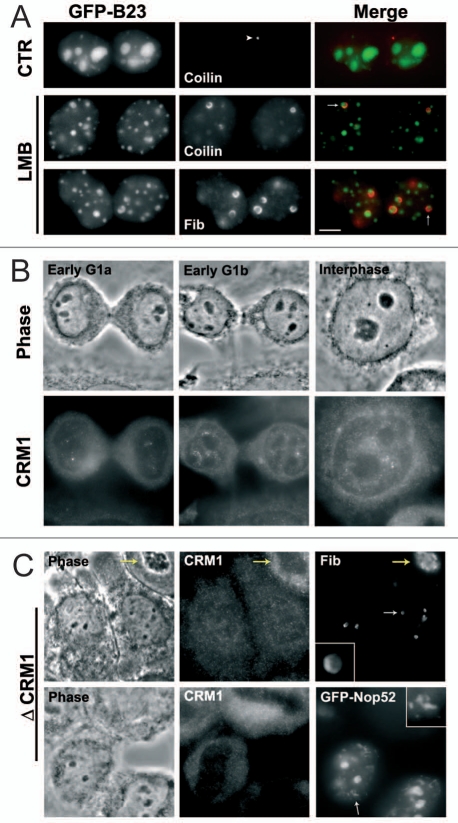
Figure 7.
Knock-down of CRM1 reproduces the LMB effect. (A) In early G1 cells, LMB induces the localization of coilin in the cap (arrow) at the periphery of the incipient nucleoli. In control (CTR) cells, coilin (arrowhead) is localized in small foci. Bar: 5 µm. (B) Immunodetection of CRM1 as small nuclear foci already visible in early G1a nuclei. (C) Early G1 cells in which siRNAs against CRM1 were efficient (Δ CRM1): low CRM1 signal with antibodies compared to CRM1 positive cell in upper right corner (yellow arrow). Fibrillarin is in the cap (enlargment) at the periphery of the incipient nucleoli (white arrow); Nop52 is found both in incipient nucleoli and in PNBs (white arrow and enlargment).
LMB is a potent inhibitor of CRM1-mediated nuclear export26 and of CRM1-dependent intranuclear traffic.27,28 Impaired recruitment of late processing proteins induced by LMB could be the consequence of a modified traffic, i.e., a modified balance between uptake in incipient nucleoli and feedback into PNBs. To verify this possibility, the traffic in incipient nucleoli in control and LMB-treated early G1a cells was measured (Fig. 3D). The exchange dynamics was estimated by the t1/2 of fluorescence decay (time after activation at which 50% of the initial fluorescence is lost). The t1/2 of PAGFP-B23/NPM estimated by photoactivation is 31.79 seconds in incipient nucleoli, compared to 12.30 seconds in the nucleolus of interphase control cells (Fig. 3E). We propose that the long time of residency of B23/NPM in incipient nucleoli is a specific feature of GC formation and of progressive disappearence of PNBs. The residence time of B23 in incipient nucleoli is LMB-sensitive as revealed by the t1/2 of 8.34 seconds measured in LMB-treated cells compared to control early G1a cells (Fig. 3D and E). Conversely, the t1/2 in interphase nucleoli is not or only slightly LMB-sensitive (t1/2 of 9.71 +/− 2.06 seconds versus 12.30 +/− 2.83 seconds). We show that LMB modifies the t1/2 of B23/NPM in incipient nucleoli and maintains the feedback towards PNBs for a long period (Fig. 3C–E). Consequently in control conditions, the residence time of the late processing proteins certainly participates in GC assembly of incipient nucleoli.
The initial target of LMB is not rDNA transcription.
To better understand when LMB interferes with nucleolar assembly, time-lapse imaging was used. After 2 hours of LMB treatment, two couples of cells are analyzed, one in telophase and the other in early G1a (Fig. 4, left and right cells respectively). The telophase cells progress to early G1a in 20 minutes before being arrested similarly to their neighbors, the arrested early G1a visible at the beginning of the time-lapse imaging. Together, these observations indicate that LMB-treated cells pass through mitosis, that nucleolar assembly is impaired after telophase and the cells are blocked in early G1a.
Figure 4.
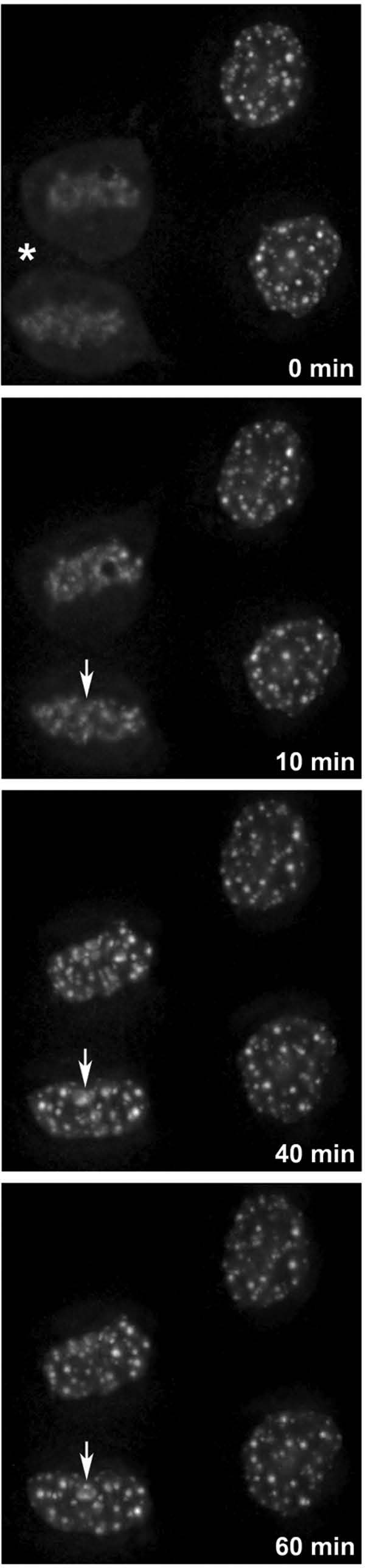
LMB blocks cells in early G1a. Cell cycle progression during LBM treatment observed by time lapse (GFP-B23/NPM cells). At 0 minutes, telophase (asterisk) cells are visible after 2 hours of LMB, and blocked doublet of early G1 cells is visible on the right side. After 10 minutes PNB-containing B23/NPM are visible but not the incipient nucleolus (arrow) that will be visible after 40 minutes. After 60 minutes the telophase cells are blocked in early G1a.
Arrest of transcription could explain impairment of nucleolar assembly and maintenance of PNBs similarly to what happens when rDNA transcription is blocked by actinomycin D.29 In interphase cells, 2 hours of LMB treatment have no effect on BrUTP incorporation in nucleoli (Fig. 5 Nu) and treatments prolonged for up to 6 hours do not induce nucleolar segregation, the typical reorganization process when rDNA transcription is blocked.30 Thus rDNA transcription is not blocked by LMB during interphase but could be specifically blocked in early G1 since rDNA transcription is activated at telophase.15,31,32 In LMB-treated early G1a cells, the BrUTP incorporation demonstrated that the transcription is activated in NORs (Fig. 5 arrows). However the GC nucleolar formation is impaired (Fig. 5B23). This is not because telophase/early G1 cells do not progress beyond early interphase. Other features of G1 nucleus formation occur normally as the appropriate relocalization of mitotic proteins discussed below. Thus the blockage of rDNA transcription by LMB is not the initial cause of the abnormal nucleolar assembly at exit from mitosis.
Figure 5.
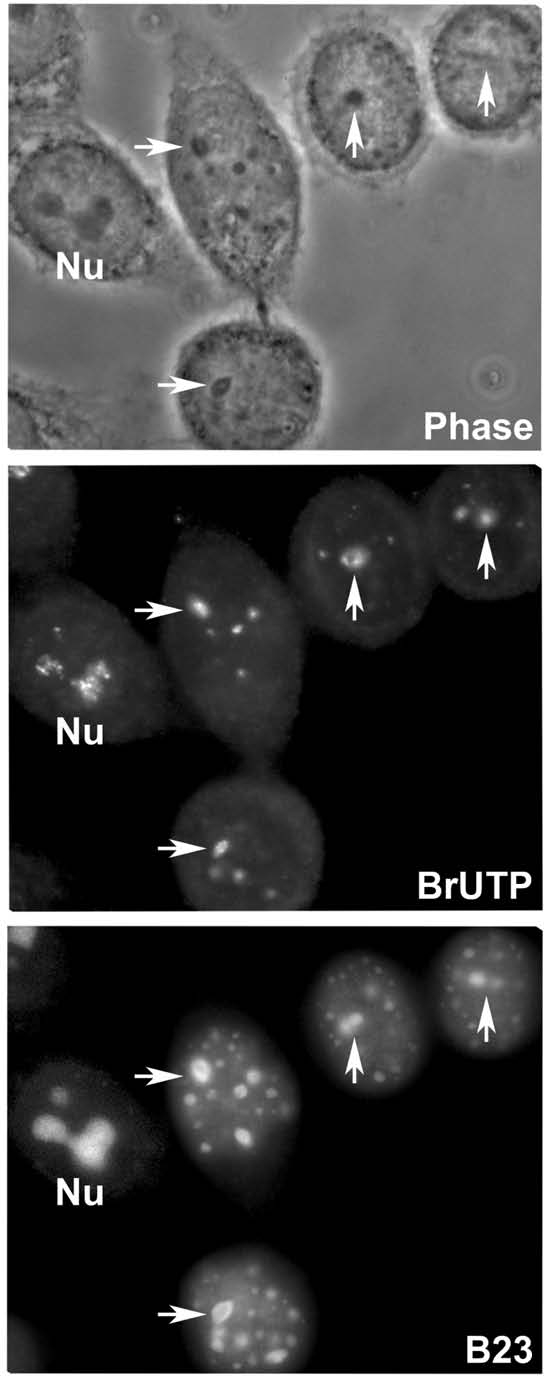
Progressive decrease of rDNA transcription under LMB. Two doublets of early G1a and one interphase (left) observed in phase contrast (phase). rDNA transcription (BrUTP) is detected in nucleoli of an interphase cell (Nu) and in incipient nucleoli indicated by arrows. GFP-B23/NPM is visible in the nucleolus (Nu), incipient nucleoli (arrows) and PNBs (small foci).
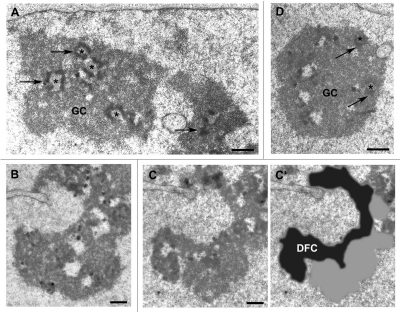
Impaired nucleolar assembly could be due to progressive decrease of rDNA transcription with partial DFC formation. To examine this hypothesis, the nucleolar organization of telophase, early G1 and interphase LMB-treated cells was examined by electron microscopy. Immediately after adding LMB, the phases of the cell cycle were identified and the cells localized on coverslip grids.33 After 2 hours of LMB treatment, the cells were fixed and flat embedded in situ. The grid imprinted on the block surface permits cell identification. In LMB-treated interphase cells, the 3 canonical nucleolar components, i.e., FC, DFC and GC are similar to those of control cells (Fig. 6A). Cells treated during telophase progress to early G1 but the DFC observed on serial sections forms a ribbon at the nucleolar periphery and typical GC-containing granules are not visible (Fig. 6B–C'). The DFC is dark, with a texture different from that of the pseudo GC in close contact. This organization reflects the nucleolar cap observed by light microscopy (see Fig. 7A). In cells treated during early G1 the granules are visible in the GC (Fig. 6D). In addition, the nucleolar organization visible in Figure 6D demonstrates that the nucleolar component can be assembled in LMB-treated cells in the last part of early G1. In conclusion, the ultrastructural organization suggests a defect in the coordination between DFC and GC specifically in LMB-treated telophase cells (compare Fig. 6B–D).
Figure 6.
Ultrastructural organization of nucleoli undergoing 2 hours of LMB treatment. (A) Nucleolar organization is not modified during interphase. Fibrillar centers (asterisks), DFC (arrows) and granular component (GC). Bar: 1 µm. (B–C') Nucleolar organization is impaired in telophase. (B and C) correspond to 2 serial sections of the same nucleolus distant by 0.4 µm. The DFC is formed but with contrasted aggregates. (C') is the super-imposition of the DFC contour in black recognized by its dark and fibrillar texture, and in grey the GC without granule and loosely organized. Bar: 0.5 µm. (D) The GC is assembled in early G1 cells. Fibrillar centers (asterisks), DFC (arrows). Bar: 1 µm.
During early G1a, the assembly of the nucleolus is CRM1-dependent.
To characterize the effect of LMB on nucleolar assembly, several possibilities were considered and tested. We checked the exit of mitosis by the localization of markers differently distributed in mitosis and interphase (data not shown). Exit from mitosis of LMB-treated cells was confirmed by the absence of MPM2 mitosis-specific phosphoproteins in the nuclei34,35 and by the absence of cyclin B1 in early G1 nuclei as expected after destruction in metaphase.36 We verified that mitotic phosphorylation of B23/NPM on Thr199,37 is released in LMB-treated cells. GFP-PP1γ phosphatase was detected in PNBs and incipient nucleoli in LMB-treated as well as in control cells using the permanent cell line expressing GFP-PP1γ (gift of L. Trinckle-Mulcahy; data not shown). These localisations and Thr199 dephosphorylation of B23/NPM indicate that the activity of the phosphatase PP1γ is not inhibited by LMB.38 Thus, it is not a blockage of mitotic exit, and other players could be involved.
In control early G1a, small foci containing both coilin and fibrillarin indicate that CB assembly starts at this period as previously reported.39 This early CB formation was also observed using time-lapse imaging of YFP-coilin.40 In LMB-treated early G1a cells, coilin is observed in nucleoli of arrested early G1a cells whereas coilin-foci are not detected in the nucleoplasm (Fig. 7A). Coilin is localized in the external part of the incipient nucleolus at the border of the fibrillarin cap (Figs. 7A and S2). On the contrary in interphase cells treated with LMB, coilin is not translocated to the nucleoli (Fig. S2 arrowheads). These observations confirm that LMB disturbs CB assembly in early G1.39
To characterize the role of LMB during early G1a, we analysed the distribution of its target, the CRM1/exportin, a protein involved in transport during interphase and localization of proteins during mitosis.41 In early G1a, G1b (long cytoplasm bridge) and interphase nuclei, CRM1 is observed in a few nuclear foci (Fig. 7B). Since, during interphase the presence of CRM1 in CB is well established,27,28,42 CRM1 could already be in the CB in early G1a and b (Fig. 7B). The effect of RNA-interference-mediated CRM1 depletion was then investigated on both nucleolar assembly and the presence of CB in early G1. The sequence chosen for siRNAs was previously found efficient.43 CRM1-depleted cells were identified by the low level of CRM1 signal in the cytoplasm (Fig. 7C). In these early G1 cells, CB foci were not visible and incipient nucleoli presented a disturbed organization. Fibrillarin is localized in the nucleolar cap as it is also after LMB treatment (Fig. 7C upper row) and the PNBs containing late processing proteins are numerous as illustrated for Nop52 (Fig. 7C lower row). These observations therefore demonstrate that the assembly of both nucleolus and CB is CRM1-dependent at this period of the cell cycle.
Discussion
The nucleolus during interphase.
In the nucleolus the different steps of ribosome biogenesis are linked and include transcription by the RNA polymerase I, the cleavage steps and the modification of the transcripts by snoRNA complexes. It is proposed that the production of ribosomes depends on a kinetic balance between the rate of rRNA synthesis and the rates of rRNA folding and maturation.44 DFC is generated by elongation of rDNA transcription and the first cleavage-modification steps of pre-rRNAs. Late cleavage-processing steps and assembly of the large ribosome subunits generate the GC.4,7 The nucleolar organization is modified by inhibition of rDNA transcription producing the typical segregated organization.30,45,46 It was also demonstrated that the caseine kinase 2 (CK2) inhibitor DRB and the cyclin-dependent kinase (CDK) inhibitor roscovitine disrupt the link between DFC and GC.15,47,48 It is possible to disturb DFC and GC organization in the yeast nucleolus by disrupting the nucleolar trimethyl guanosine synthase 1 (Tgs1p) required for pre-rRNA processing.49 These different examples of nucleolar disorganization (not an exhaustive list) are not specific of a particular period of the cell cycle contrary to the inhibition of CRM1 that impairs nucleolar function only during early G1a. Our results also raise the possibility that the fibrillar centers combined with the DFC are really a single domain and are more autonomous from the GC than may have been appreciated previously, i.e., not in the transit of nascent rRNAs and pre-ribosomes but rather as architectural design and assembly sites. Although this concept would require additional investigation, perhaps in a phyletic/evolutionary context.
Nucleolar assembly.
After telophase, two sites in nuclei concentrate the late nucleolar processing proteins, i.e., the PNBs and the incipient nucleoli. Based on our observations, feedback between these two sites appears to control recruitment in GC. Pre-rRNAs are present in both sites and could therefore explain the binding affinity of late processing proteins in both sites. The pre-rRNAs in PNBs are synthesized during prophase and stabilized after one cleavage.29 The link between nucleolar function and the presence of PNBs was established by inhibiting rDNA transcription or pre-rRNA processing while maintaining rDNA transcription.15,29 In both cases, treatment with either actinomycin D or roscovitine, maintains the PNBs and impairs nucleolar assembly.
We have observed here that the time of residency of GC molecules is higher in incipient than in interphase nucleoli (respectively t1/2 of 31.7 seconds and 12.3 seconds). In incipient nucleoli the establishment of the vectorial process of ribosome assembly generated first DFC and then GC as demonstrated in several models.5,50,51 The connection between DFC and GC depends on the pre-rRNA modifications49 and could explain the time of residency of GC proteins during nucleolar assembly. This hypothesis is also supported by the decrease of the time of residency (t1/2 of 8.3 seconds) corresponding to a lower binding affinity and/or a lower number of binding sites when the LMB blocks GC formation. In this case late processing proteins such as B23/NPM and Nop52 transit through incipient nucleoli but in a segregated organization illustrating impaired ribosome assembly.
Nucleolus assembly and CRM1.
Two periods are defined during nucleolar assembly but only one is LMB-sensitive. The increasing number of arrested early G1a cells with increase in duration of LMB treatment indicates that cells progress through the cell cycle before being stopped. Upon CRM1 depletion and LMB treatment, the PNBs are maintained and the binding of late processing proteins in incipient nucleoli is decreased. This low binding inhibits GC assembly blocking pre-rRNA processing as suggested by the absence of granules in GC. We propose that this is due to incomplete or absence of modifications of pre-rRNAs for the following reasons. Normally, CB formation occurs rapidly after mitosis39,40 and CRM1 associates with precursor and mature U3 snoRNAs52 and localizes in CBs.27,28,42 Because we observed that CBs are not assembled in early G1a undergoing LMB treatment, this absence most probably impairs snoRNA formation52,53 and/or the activity of snoRNAs54 preventing pre-rRNA processing. In this case, the presence of unmodified pre-rRNAs in incipient nucleoli could easily explain the persistence of an equilibrium between PNBs and incipient nucleoli in LMB-treated cells and during CRM1 depletion (Fig. S3). Indeed, we previously demonstrated that the 45S pre-rRNAs present in PNBs are synthesized during prophase,29 when rDNA transcription is maintained but the nucleolus is disassembled.55 These 45S pre-rRNAs are not processed and most probably not modified as CB remnants are described in prophase.23 An alternative could be that lack of targeting of snoRNPs to nascent rRNAs is involved because this targeting is CRM1-dependent.27 In this case, the inhibition of CRM1 should not impaired the nucleolar assembly only during the first part of early G1. In conclusion we propose that the assembly of both CB and nucleolus is CRM1-dependent and that the assembly of a functional CB is necessary to coordinate the early and late steps of ribosome biogenesis and to make GC formation possible.
Materials and Methods
Cell culture.
HeLa cells were cultured without antibiotics in Minimum Essential Medium (MEM) with GlutaMAX™ (Invitrogen) supplemented with 10% fetal calf serum and non-essential amino acids at 37°C in 5% CO2. Permanently established HeLa cell lines expressing GFP-Nop52, GFP-B23, GFP-Bop and GFP-fibrillarin were already described.14,16 The GFP-PP1γ cell line was a gift of Laura Trinckle.24 The LMB (Calbiochem) stored at −80°C in small aliquots, was used at a concentration of 30 nM.
The siRNA sequence (5′-TGT GGT GAA TTG CTT ATA C-3′) was used because it is efficient against CRM1 expression.43 Cells were transfected twice at interval of 24 hours with 10 or 50 nM siRNA duplex using the Effectene transfection kit (Qiagen). Control cells were treated in parallel with non-specific siRNA duplex. Cells were fixed 34 hours after the first transfection.
Antibodies and plasmids.
The antibodies were used at the following dilutions: 1/100 CRM1/Exportin (BD Biosciences), 1/400 coilin (BD Biosciences), 1/100 (Thr199) phospho-NPM (Cell signaling technology), 1/100 cyclin B1 (Santa Cruz Biotechnology), 1/200 MPM2-mitotic phosphoproteins (Upstate Biotechnology), 1/100 fibrillarin (Abcam). Secondary antibodies conjugated to FITC or Texas Red were from Jackson Immuno Research Laboratories.
To construct B23-PAGFP, Nop52-PAGFP and PAGFP-fibrillarin, plasmid pEGFP-N1 or plasmid pEGFP-C1 (Clontech Laboratory, USA) was used after replacing the EGFP cassette by the PAGFP sequence, a gift of J. Lippincott-Schwartz.25 Full-length B23 was prepared by PCR amplification from full-length human cDNA (clone IMAGES 5575414) as described.56 The full-length fibrillarin cDNA was a gift of J.P. Aris, and the fibrillarin-GFP and Nop52-GFP constructs have been described.14 For colocalization, co-transfections with the constructs already described DsRed-B23,56 DsRed-Nop52,16 or mRFP-Nop56,56 were used.
Immunofluorescence.
The cells were fixed in 4% paraformaldehyde for 20 minutes at room temperature (RT) and permeabilized with 0.5% Triton X-100 for 5 minutes at RT. They were then washed with PBS and incubated with primary antibodies at RT for 60 minutes. The antibodies were revealed with Texas Red-and/or FITC-conjugated secondary antibodies. Alternatively, the cells were fixed with methanol for 20 minutes at −20°C, air-dried for 5 minutes and rehydrated with PBS for 5 minutes before incubation with antibodies. They were then incubated 5 minutes with DAPI to visualize DNA and mounted with the antifading solution AF1 (Citifluor). Fluorescent microscopy was performed using a CCD camera Leitz DMRB.
Run-on transcription was performed as described by Moore and Ringertz57 and adapted by Roussel et al.32 Briefly, the cells were prefixed 5 minutes at 4°C in ethanol/acetone (v/v), washed and incubated in the run-on buffer Tris-HCl pH 7.9 (100 mM), MgCl2 (12 mM), saccharose (150 mM), β-mercaptoethanol (12 mM) containing ATP, GTP, CTP (0.6 mM, MBI Fermentas) and Br-UTP (0.12 mM, Sigma-Aldrich) during 15 minutes. They were postfixed 20 minutes at RT in paraformaldehyde and permeabilized 5 minutes with 0.5% Triton X-100. Cells were incubated first with mouse Br-UTP antibodies 60 minutes, then with Texas Red-conjugated mouse antibodies 60 minutes.
Time-lapse microscopy.
The cells were grown on glass coverslips, mounted in a Ludin observation chamber (LIS) filled with complete medium supplemented with 10 mM Hepes, pH 7.4. Live-cell 4-D microscopy was carried out using a Leica DM IRBE microscope, equipped with a piezoelectric translator (PIFOC, PI, Germany). Stacks of 20 to 25 images with a Z-step of 0.3 µm were acquired every 60 seconds with a 100x PlanApo N.A. 1.4 oil immersion objective in stream mode (Metamorph) using a 5 MHz Micromax 782Y interline CCD camera (Roper Instruments, France) as described.58 Fluorescence images were then deconvolved using iterative deconvolution with a measured point spread function (PSF).58 Images and movies were mounted using the free downloadable software ImageJ, and the maximum intensity projection was chosen in both cases.
Photoactivation.
Cells were seeded for 24 hours in a Petri dish containing a 32 mm coverslip and transfected with the PAGFP plasmids using the Effectene Transfection Reagent (Qiagen, USA). Cells were observed 36 hours after transfection by inserting the coverslip into a cell chamber maintained at 37°C on a Leica TCS SP2 AOBS confocal microscope. A 63X, 1.32 oil lens was used and pixel sampling was set at 100 nm/pixel. During the observation to avoid background fluorescence, F12 medium without phenol red, vitamin B12, riboflavin, L-glutamine and fetal calf serum was used. LMB (30 nM) was added to the culture medium before and during the observation.
Activation of PAGFP was induced by a single laser pulse at 405 nm using 15–20% of the power of a 50 mW laser diode GaN (Coherent) during 300 mseconds. After activation, the GFP signal was collected on single sections (0.6 µm thick) at 500 mseconds intervals during 2 minutes. Before and after activation, Z series of GFP and either DsRed or mRFP signals were collected. To minimize photobleaching when recording the signals at 488 nm, only one image with 10% of the power of a 20 mW argon laser was used. The focal photoactivation mode was used59,60 and stacks of images in the Z axis before and after photoactivation were taken. Recent improvements of the image registration speed would make it possible in the future, to obtain 3D registration in the millisecond scale that would be necessary to analyze in 3D protein movements in nuclei.
Electron microscopy.
HeLa cells were grown on microgrid coverslips (CELLocate, Eppendorf), the alphanumeric labeling of the grids facilitating the location of individual cells by light microscopy. Immediatly after adding the LMB, the phases of the cell cycle (interphase, telophase and early G1) were identified and then the cells were allowed to progress in the cell cycle, at 37°C. After 2 hours, the cells were treated in situ according to published procedures.33 Briefly, cells were fixed in 2% (w/v) paraformaldehyde, 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 at 4°C for 30 minutes, rinsed, post-fixed in 1% (w/v) osmium tetroxide in the same buffer, dehydrated and embedded in Epon 812. After resin polymerization, the embedded cells were separated from the microgrid coverslips carrying the imprint of the alphanumeric labeling. The cell cycle phases previously located by light microscopy were serially sectioned parallel to the plane of the growing cells. Ultrathin sections were conventionally contrasted with uranyl acetate (10 minutes) followed by lead citrate (5 minutes) and examined in a Philips CM12 electron microscope.
Supplementary Material
Acknowledgements
We thank L. Trinckle-Mulcahy for the GFP-PP1γ cell line and T. Piolot and C. Chamot for their help in the imagery platform. We are grateful to A.-L. Haenni for critical reading of the manuscript. This work was supported in part by the “Centre National de la Recherche Scientifique” and the “Association pour la Recherche sur le Cancer”.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11334
References
- 1.Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 2.Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, et al. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol. 2004;167:51–63. doi: 10.1083/jcb.200404120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Verdun D. The nucleolus: a model for the organization of nuclear functions. Histochem Cell Biol. 2006;126:135–148. doi: 10.1007/s00418-006-0212-3. [DOI] [PubMed] [Google Scholar]
- 5.Gébrane-Younís J, Sirri V, Junéra HR, Roussel P, Hernandez-Verdun D. Diekmann. Visions of the cell nucleus: ASP CA; 2005. Nucleolus: An essential nuclear domain; pp. 120–135. PHaS, ed. [Google Scholar]
- 6.Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- 7.Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 9.Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiscox JA. RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol. 2007;5:119–127. doi: 10.1038/nrmicro1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez-Garcia LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dundr M, Misteli T, Olson MO. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol. 2000;150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savino TM, Gebrane-Younes J, De Mey J, Sibarita JB, Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirri V, Hernandez-Verdun D, Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol. 2002;156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angelier N, Tramier M, Louvet E, Coppey-Moisan M, Savino TM, De Mey JR, et al. Tracking the Interactions of rRNA Processing Proteins during Nucleolar Assembly in Living Cells. Mol Biol Cell. 2005;16:2862–2871. doi: 10.1091/mbc.E05-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimario PJ. Cell and molecular biology of nucleolar assembly and disassembly. Int Rev Cytol. 2004;239:99–178. doi: 10.1016/S0074-7696(04)39003-0. [DOI] [PubMed] [Google Scholar]
- 18.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 19.Carmo-Fonseca M, Platani M, Swedlow JR. Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 2002;12:491–495. doi: 10.1016/s0962-8924(02)02387-5. [DOI] [PubMed] [Google Scholar]
- 20.Janicki SM, Spector DL. Nuclear choreography: interpretations from living cells. Curr Opin Cell Biol. 2003;15:149–157. doi: 10.1016/s0955-0674(03)00012-7. [DOI] [PubMed] [Google Scholar]
- 21.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, et al. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, et al. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci. 2003;116:2039–2050. doi: 10.1242/jcs.00400. [DOI] [PubMed] [Google Scholar]
- 24.Trinkle-Mulcahy L, Sleeman JE, Lamond AI. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- 25.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 26.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 27.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci. 2007;120:1540–1550. doi: 10.1242/jcs.001529. [DOI] [PubMed] [Google Scholar]
- 29.Dousset T, Wang C, Verheggen C, Chen D, Hernandez-Verdun D, Huang S. Initiation of nucleolar assembly is independent of RNA polmerase I transcription. Mol Biol Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125:127–137. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 31.Fomproix N, Gebrane-Younes J, Hernandez-Verdun D. Effects of anti-fibrillarin antibodies on building of functional nucleoli at the end of mitosis. J Cell Sci. 1998;111:359–372. doi: 10.1242/jcs.111.3.359. [DOI] [PubMed] [Google Scholar]
- 32.Roussel P, Andre C, Comai L, Hernandez-Verdun D. The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133:235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gébrane-Younís J, Fomproix N, Hernandez-Verdun D. When rDNA transcription is arrested during mitosis, UBF is still associated with non-condensed rDNA. J Cell Sci. 1997;110:2429–2440. doi: 10.1242/jcs.110.19.2429. [DOI] [PubMed] [Google Scholar]
- 34.Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skoufias DA, Indorato RL, Lacroix F, Panopoulos A, Margolis RL. Mitosis persists in the absence of Cdk1 activity when proteolysis or protein phosphatase activity is suppressed. J Cell Biol. 2007;179:671–685. doi: 10.1083/jcb.200704117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 37.Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negi SS, Olson MO. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J Cell Sci. 2006;119:3676–3685. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- 41.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Ospina JK, Gonsalvez GB, Bednenko J, Darzynkiewicz E, Gerace L, Matera AG. Cross-talk between snurportin1 subdomains. Mol Biol Cell. 2005;16:4660–4671. doi: 10.1091/mbc.E05-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knauer SK, Bier C, Habtemichael N, Stauber RH. The Survivin-Crm1 interaction is essential for chromosome passenger complex localization and function. EMBO rep. 2006;7:1259–1265. doi: 10.1038/sj.embor.7400824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, et al. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadjiolov AA. The nucleolus and ribosome biogenesis. Wien, New-York: Springer-Verlag; 1985. [Google Scholar]
- 46.Puvion-Dutilleul F, Puvion E, Bachellerie J-P. Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with 5′ETS leader probe. Chromosoma. 1997;105:496–505. doi: 10.1007/BF02510486. [DOI] [PubMed] [Google Scholar]
- 47.Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- 48.Louvet E, Junera HR, Le Panse S, Hernandez-Verdun D. Dynamics and compartmentation of the nucleolar processing machinery. Exp Cell Res. 2005;304:457–470. doi: 10.1016/j.yexcr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Colau G, Thiry M, Leduc V, Bordonne R, Lafontaine DL. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol Cell Biol. 2004;24:7976–7986. doi: 10.1128/MCB.24.18.7976-7986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez-Verdun D, Bourgeois CA, Bouteille M. Simultaneous nucleologenesis in daughter cells during late telophase. Bio Cell. 1980;37:1–4. [Google Scholar]
- 51.Verheggen C, Le Panse S, Almouzni G, Hernandez-Verdun D. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis. J Cell Biol. 1998;142:1167–1180. doi: 10.1083/jcb.142.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 53.Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, et al. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gautier T, Robert-Nicoud M, Guilly MN, Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J Cell Sci. 1992;102:729–737. doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- 56.Lechertier T, Sirri V, Hernandez-Verdun D, Roussel P. A B23-interacting sequence as a tool to visualize protein interactions in a cellular context. J Cell Sci. 2007;120:265–275. doi: 10.1242/jcs.03345. [DOI] [PubMed] [Google Scholar]
- 57.Moore GPM, Ringertz NR. Localization of DNA-dependent RNA polymerase activities in fixed human fibroblasts by autoradiography. Exp Cell Res. 1973;76:223–228. doi: 10.1016/0014-4827(73)90439-4. [DOI] [PubMed] [Google Scholar]
- 58.Louvet E, Tramier M, Angelier N, Hernandez-Verdun D. Time-lapse Microscopy and Fluorescence Resonance Energy Transfer to Analyze the Dynamics and Interactions of Nucleolar Proteins in Living Cells. Methods Mol Biol. 2008;463:123–135. doi: 10.1007/978-1-59745-406-3_9. [DOI] [PubMed] [Google Scholar]
- 59.Deryusheva S, Gall JG. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 2004;101:4810–4814. doi: 10.1073/pnas.0401106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Politz JC, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14:4805–4812. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.