Abstract
Engineered tissues can provide models for imaging and disease progression and the use of such models is becoming increasingly prevalent. While structural characterization of these systems is documented, a combination of biochemical and structural knowledge is often helpful. Here, we apply Fourier transform infrared (FT-IR) spectroscopic imaging to examine an engineered tissue model of melanoma. We first characterize the biochemical properties and spectral changes in different layers of growing skin. Second, we introduce malignant melanocytes to simulate tumor formation and growth. Both cellular changes associated with tumor formation and growth can be observed. In particular, chemical changes associated with tumor-stromal interactions are observed during the course of tumor growth and appear to influence a 50–100 μm region. The development of this analytical approach combining engineered tissue with spectroscopy, imaging and computation will allow for quality control and standardization in tissue engineering and novel scientific insight in cancer progression.
Introduction
A majority of human cancers arise within the skin. Though melanoma comprises less than 4% of diagnosed skin cancers, it causes over 75% of skin cancer-related deaths and its incidence is rapidly increasing.1 Despite decades of scientific research, the sole effective cure is surgical excision of the primary tumor2 when the lesions are smaller than 1 mm.3 The prognosis for a patient with stage I or II melanoma is mainly related to tumor thickness. Hence, early detection is critical in melanoma treatment, and the evolution of the tumor and its penetration into the dermis are key factors to be studied in understanding the disease.4 Melanoma is a tumor of melanocytes, which are a class of cells located at the epidermal-dermal junction. Functionally, they interact with multiple keratinocytes to form an epidermal melanin unit. Hence, melanocytes are morphologically and functionally predisposed to multiple interactions with diverse cell types. These properties lead to two specific difficulties in the scientific study of melanoma progression: the first is that the location of the tumor and interactions with multiple cells5,6 make behavior in traditional, two-dimensional cell culture less realistic. Observations in situ are limited, however, and tumor progression cannot be followed in humans due to ethical concerns. Hence, appropriate model systems are needed. Second, the spatially-distributed nature of the tumor requires the use of imaging techniques but the influence of multiple cells requires biochemical contrast–especially to study subtle molecular changes in early stage disease and its progression.
Two converging trends can now address these issues. The first is the development of model systems to study disease and the second is the development of new imaging technologies. Engineered tissue structures7 are attracting increased interest as a surrogate for human experiments. In particular, compared to traditional, two-dimensional (2D) cell cultures, engineered tissues provide a more realistic, three-dimensional (3D) and controllable model to study a variety of basic cellular processes.8 Their use in studying disease progression has also been proposed but is somewhat limited.9 Due to the precise structuring of the sample, engineered tissue models are also a convenient sample set for developing imaging technology. As opposed to geometrical phantoms, engineered tissues represent a more realistic biological subject10 while their simplified structure is often an acceptable surrogate for the less-accessible, variable and more complex human tissue. Among all human tissues, skin has a relatively simple, laminar structure that is naturally suited to tissue engineering. Consequently, much success has been achieved in engineering skin tissue,11 models are commercially available and have been used for a variety of research and therapeutic applications.12
Imaging methods provide useful insight into tumor structure and its evolution and are actively being developed for clinical use in various modalities. Similarly, there have been efforts to characterize melanoma progression in biochemical terms,13 including understanding the role of cells other than melanocytes in tumor evolution.14 While characterization of tissue and tissue models is routinely accomplished by structural imaging, a biochemical characterization typically requires destruction of the structure, thereby losing spatially specific information. A combination of biochemical and structural knowledge is often helpful and is enabled by the emerging fields of chemical imaging15 and microscopy. While molecular imaging is dye- or probe-based, chemical imaging holistically measures chemical species in the sample using spectroscopy. The desired information is then extracted using computational methods. Among the prominent approaches are vibrational spectroscopic, both Raman and infrared (IR), imaging.16 Fourier transform IR (FT-IR) spectroscopic imaging17 is useful for the analysis of tissue biopsies and has a well-developed instrumentation and data analysis knowledge base.18 Recent compilations19 describe biomedical applications, in general, and several reports specifically describe IR spectroscopic studies on human skin. The skin-focused reports20 examine the structure of skin,21–24 diseases,25–29 dynamics of diffusion30–37 or use it as a model system for studies.38 There are no reports, to our knowledge, on analyzing engineered skin with IR spectroscopic imaging. Similarly, while tumor grades have been related to spectral signatures in the prostate for example,39 we are not aware of reports specifically examining tumor progression in a controlled environment.
Here, we apply FT-IR imaging to the study of melanoma progression in engineered skin. We systematically examine spectral and spatial properties of engineered skin with reference to measurements of human tissue, the spectral recognition of tumor and the changes in each cell type as a function of tumor progression. In particular, we examine the question of stromal involvement in melanoma progression. Though the concept was proposed almost 120 years ago,40 facile methods to measure such a transformation are lacking. By examining both engineered skin and tumor progression, we present new tools that will allow for quality control and standardization of both the structural and biochemical properties of engineered tissues, models to study disease progression and phantoms for imaging.
Materials and experiments
Engineered skin culture
Skin scaffolds (MatTek Inc.) consist of 8–10 layers of normal human epidermal keratinocytes grown on a 1 mm think layer of normal human dermal fibroblasts seeded in collagen gel, which is in turn supported by an inert polymer insert with 0.4 μm pore size. A malignant melanocyte cell line, A375, is mixed with and incorporated into the epidermal layer during cell seeding. The tissue is cultured in MCDB 153 serum free medium, which is renewed every two days.
Engineered skin sample preparation
Each tissue sample is removed from the culture at a specified time and fixed with 10% neutral buffered formalin, followed by gradient dehydration in 70%, 95% and 100% ethanol. The sample is cleared with a 1: 1 xylene/ethanol mixture and then with neat xylene. Subsequently, the sample is embedded in paraffin following standard histological procedure. Embedded tissue is sectioned to 5 μm slice with a SHUR/Sharp™ 4060E Electronic Rotary Microtome (Triangle Biomedical Sciences) and placed on BaF2 substrates.
Human skin
Human skin samples are obtained from Carle Foundation Hospital after institutional board review and appropriate anonymization of sources. Tissues are formalin-fixed and paraffin-embedded before being sectioned, following the same procedure for engineered skin samples. These samples are placed on low-e glass slides.
Haematoxylin and eosin staining
Tissues placed on slides are de-paraffinized using xylene and are subsequently rehydrated using solutions of 100%, 95%, 70% ethanol and deionized water. Slides are then stained with haematoxylin for 5 min and eosin for 30 s with subsequent dehydration in a series of gradient ethanol and xylene. Images of stained tissues are acquired using a VWR VistaVision inverted microscope.
FT-IR imaging: data acquisition and processing
Samples for IR imaging are placed on 1 mm thick BaF2 substrates or low-e glass slides and dehydrated using hexane as previously described.41 IR spectroscopic images of tissue sections are collected using the Perkin-Elmer Spotlight 400 imaging spectrometer. A spatial pixel size of 6.25 μm and a spectral resolution of 4 cm−1 are employed, with 4 scans averaged for each pixel over the entire mid-infrared range. An undersampling ratio of 2 with respect to the He–Ne laser is used in recording the interferogram. Norton-Beer medium apodization is employed with a zero fill factor of 2 during the Fourier transform. An IR background image is acquired with 120 scans co-added at a location on the substrate where no tissue is present. A ratio of the background to tissue spectra is then computed to remove substrate and air contributions to the spectral data. All further computation is done using programs written in-house in ENVI/IDL.
Results
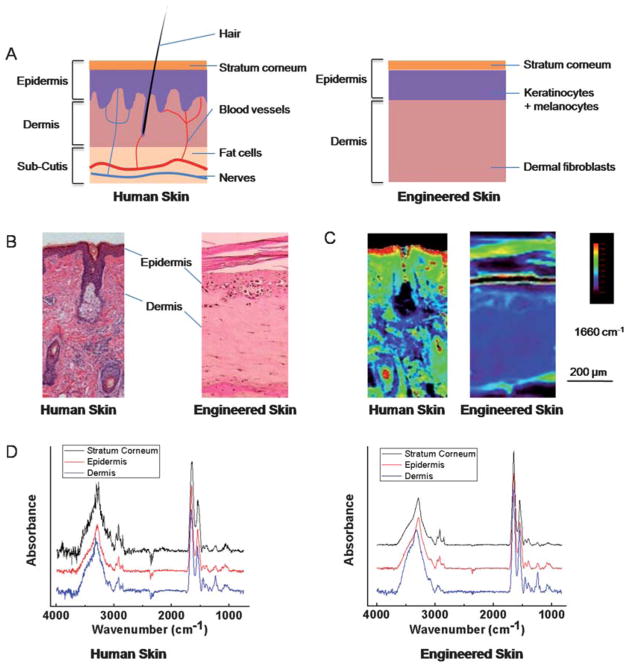
A comparison of the optical microscopy and IR data of human and engineered skin is shown in Fig. 1. As apparent in the structural images, the engineered skin model has simplified structures with only two major layers–epidermis (including its associated stratum corneum structure) and dermis, without complicated multiple sub-layers and various other histological sub-types seen in human skin (Fig. 1A, 1B). Structurally, engineered and human skin have been shown to be similar in use42 and these simplified tissue models have retained many important skin functions. We compared both the structure and the spectral features of the common tissue sub-types between human and engineered skin. The FT-IR imaging data here indicate that the engineered tissues have preserved skin differentiation seen in human skin (Fig. 1C), although there are small chemical composition differences likely due to the simplified structures and differences in cellular density and cell-cell interactions (Fig. 1D).
Fig. 1.
Skin structure and characterizations. (A) Schematic of human (left) and engineered skin (right). The epidermis is located at the skin-air interface while the dermis is located to the bottom of the image and contains various histologic structures. Engineered skin retains the most important components but does not contain accessory structures. (B) Histology of skin is usually deduced manually in tissue stained by Haematoxylin and Eosin (H&E). Epidermis typically stains darker than dermis. (C) Infrared absorbance image from a corresponding serial section of the tissue at 1660 cm−1. Human skin was imaged on a low-e slide in reflectance mode, and engineered skin was imaged on a BaF2 substrate in transmission mode. (D) IR spectra of single pixels from epidermis, dermis and stratum corneum regions of human and engineered skin after piecewise linear baseline subtraction.
Characteristic IR absorption peaks in human skin have been reported in various studies.23,43–45 They have been summarized, as shown in Table 1, for the two major groups under consideration here. Each of the different histological structures in skin will have different distributions of various chemical constituents, namely the abundance and types of lipids, proteins, carbohydrates and nucleic acids. These differences will correspond to differences in specific spectral features, largely in the spectral fingerprint region. Here, we compared spectral features observed in engineered skin with those observed in human tissue in the context of their assignments as reported in the literature (Table 1). Notably, engineered skin samples differ in both intensity and position of spectral features (Fig. 1C), indicating the abundance and environment of some biomolecules are likely different. There are also important histologic differences that make direct comparisons difficult, changes in immortalization of cell lines are probably influential and individual differences probably do exist. For example, the multiple-layer structure of epidermis is not present in engineered skin samples, although stratum corneum, the outmost layer, is indeed developed over the time (Fig. 1B). Notably, many important spectral features are still observed in engineered skin as listed in the table. In summary, spectroscopically, engineered skin is different from tissue derived from humans but is reasonably representative of the spectral properties of skin.
Table 1.
Assignment of IR spectral features in skin. Results are reported from compilations in the literature [ref. 43–45], from other studies referenced here and from analysis of this study’s skin samples, including both clinical and engineered tissue a
| Mode | Origin | Literature | Human skin |
Engineered skin |
||
|---|---|---|---|---|---|---|
| Epidermis | Dermis | Epidermis | Dermis | |||
| Amide A (N–H stretching) | Peptide, protein | 3300 | 3298 | 3298 | 3292 | 3292 |
| Amide B (N–H stretching) | Peptide, protein | 3080 | 3078 | 3078 | 3080 | 3082 |
| C–H stretching of alkenes | Phospholipid, glycolipid, fatty acid, ceramide, triglyceride | 3010 | 3010 | 3010 | 3012 | 3012 |
| Asymmetric CH3 stretching | Phospholipid, glycolipid, fatty acid, ceramide, triglyceride | 2957 | 2960 | 2960 | 2962 | 2958 |
| Asymmetric CH2 stretching | Phospholipid, glycolipid, fatty acid, ceramide, triglyceride | 2920 | 2920 | 2920 | 2916 | 2918 |
| Symmetric CH3 stretching | Phospholipid, glycolipid, fatty acid, ceramide, triglyceride | 2872 | 2868 | 2868 | 2874 | 2874 |
| Symmetric CH2 stretching | Phospholipid, glycolipid, fatty acid, ceramide, triglyceride | 2851 | 2850 | 2850 | 2850 | 2850 |
| CO stretch | Phospholipid | 1738 | 1740 | 1740 | 1738 | 1738 |
| Amide I (80% CO stretch) | Peptide, protein, ceramide | 1655 | 1654 | 1654 | 1664 | 1664 |
| Amide II (60% N–H in plane bend, 40% C–N stretch) | Peptide, protein, ceramide | 1545 | 1548 | 1548 | 1552 | 1550 |
| CH2 scissoring | Phospholipid, glycolipid, fatty acid, triglyceride | 1468 | 1468 | 1468 | 1454 | 1460 |
| CO stretch of COO– | Fatty acid | ~1395 | 1400 | 1400 | 1402 | 1392 |
| CH3 symmetric bend | Phospholipid, fatty acid, triglyceride | 1378 | 1384 | 1384 | 1392 | 1390 |
| CH2 wagging | Phospholipid, fatty acid, triglyceride | 1343–1370 | 1344 | 1338 | 1344 | 1336 |
| Amide III (40% C–N stretch, 30% N–H in plane bend, 20% methyl–C stretch) | Collagen | 1283 | 1284 | 1284 | 1284 | 1284 |
| Asymmetric PO2–stretch | Phosphate salt, nucleic acid, DNA, RNA | ~1245 | 1242 | 1240 | 1240 | 1238 |
| Amide III (40% C–N stretch, 30% N–H in plane bend, 20% methyl–C stretch) | Collagen | 1236 | 1236 | 1234 | 1236 | 1236 |
| Asymmetric PO2–stretch | Phosphate salt, nucleic acid, DNA, RNA | 1223 | 1224 | 1226 | 1224 | 1226 |
| Amide III (40% C–N stretch, 30% N–H in plane bend, 20% methyl–C stretch) | Collagen | 1204 | 1204 | 1204 | 1204 | 1204 |
| Ester C–O asymmetric stretch | Ester | 1170 | 1170 | 1168 | 1160 | 1170 |
| Symmetric PO2–stretch | Phosphate salt, nucleic acid, DNA, RNA | ~1080 | 1082 | 1082 | 1082 | 1082 |
| C–OP stretch | Phosphate ester | 1047 | 1056 | 1058 | 1034 | 1030 |
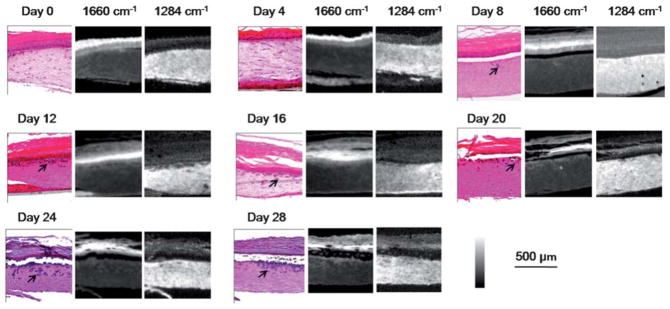
We engineered skin samples with malignant melanocytes as described in the methods section. Samples were removed and analyzed every fourth day for a total time period of 28 days. Two consecutive 5 μm tissue sections were used for H&E staining and FT-IR imaging, respectively. We observed that tumors are limited to small cell clusters at the beginning of the experiment, but grow larger and deeper into the tissue over the time (Fig. 2). Tumors can be tracked in absorbance images and classifiers for the same could be developed following previous methods in automating histological recognition.41 In this case, however, we can readily identify the tumor by differential absorption and are more interested in following the evolution of the tumor and tissue over time.
Fig. 2.
Melanoma progression in 28 days. Tumors (arrows) grow larger and invade into the dermis over time, as shown by both H&E-stains followed by histological recognition and IR spectroscopic imaging. IR absorbance images at 1660 cm−1 and 1284 cm−1 highlight epidermal and dermal regions, respectively, indicating different chemical compositions between these two regions and their facile delineation by simple spectral metrics.
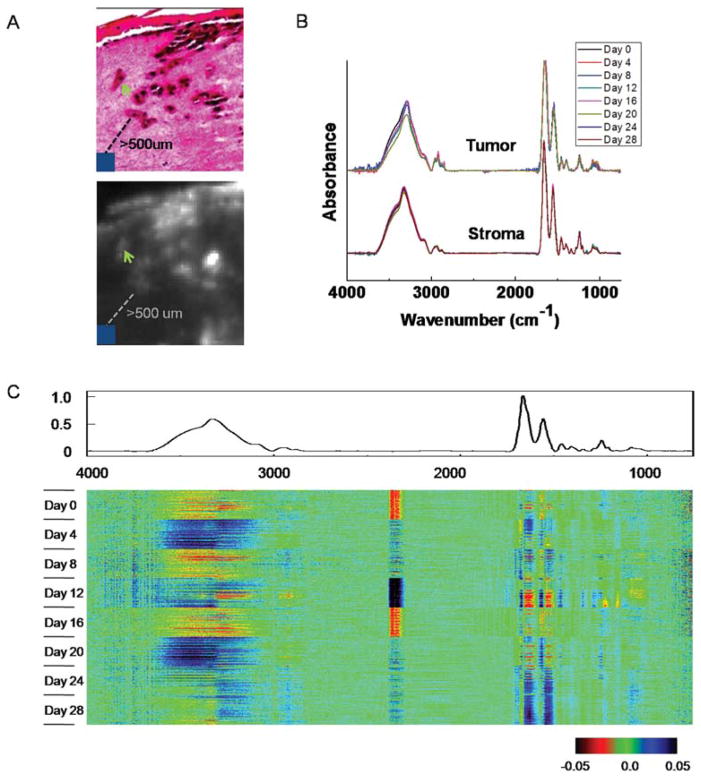
To determine the stability of these engineered structures for spectroscopic analyses, we examined the temporal variation of spectra for “normal stroma” or dermal regions at least 500 μm away from tumors. 100 representative pixels from normal stroma were extracted from each of the eight samples (Fig. 3A, 3B). We employed a recently proposed visualization and statistical analysis tool, termed Comprehensive Data Map (CDM),46 to analyze spectral variation among these pixels (Fig. 3C). Briefly, CDM provides a convenient method of graphically displaying data so as to convey the view of signal and variance and identify patterns and trends in complex datasets. The data are mean centered using an average for the entire data set and the average spectrum is shown above the data. The central data block consists of rows of spectra, with each row having a particular set of data associated with it. The display is the difference from the average. The spectral regions at which differences occur are apparent from the color display in the figure. It appears that differences are primarily concentrated at large absorbance areas (amide I, amide II and amide A peaks). While there are also differences at characteristic spectral regions, such as the fingerprint region among some of the samples, these differences are quite small, even after we changed the way CDM presents the differences by normalizing them to the magnitude of the average spectrum (data not shown). In addition, these differences do not appear to have any particular patterns. These results indicate that the skin samples are quite consistent for our infrared spectroscopic imaging analysis, which primarily utilizes the information from those characteristic spectral regions. A measure of the biological noise, hence, is the standard deviation in time.
Fig. 3.
Temporal analysis of tumor and normal stroma. (A) Spectra of 10 pixels of tumors (green arrow) and 10 × 10 pixels of normal stroma that are at least 500 μm away from tumors (blue square) are extracted for temporal analysis. (B) Comparison of average spectra from tumor and normal stroma regions. (C) CDM visualization of spectra of normal stroma. The average spectrum is shown above the data and the central data block displays the difference of each pixel spectrum from the average spectrum.
Next, we examined the spectral variation of tumor pixels during the course of melanoma invasion starting from day 8 (Fig. 3A, 3B). No changes were observed that could be related to the time course of invasion, which is also confirmed by CDM analysis (data not shown). The inability of IR spectroscopy to capture these changes could arise from insufficient sensitivity of the technique or experimental limitations, for example in the signal to noise ratio.47 We believe, however, that the consistency in spectra likely arises from the biological basis of these cells in culture. The cells likely obtain their proliferative capacity prior to culture and are not modified considerably by the culturing environment or process. It is likely that melanocytes in engineered skin have previously experienced interactions with both epidermal and dermal cells in tissue and likely do not transform in the new milieu of the engineered tissue. In either case, there is no apparent signature in the tumor cells that correlate with tumor progression. To understand transformations associated with malignancy, hence, a comparative analysis of benign and malignant cells is desirable but beyond the scope of the manuscript here. Studies have previously reported melanoma analysis using IR spectroscopy,26,48–53 but, to our knowledge, have not examined the spectral properties as a function of tumor progression.
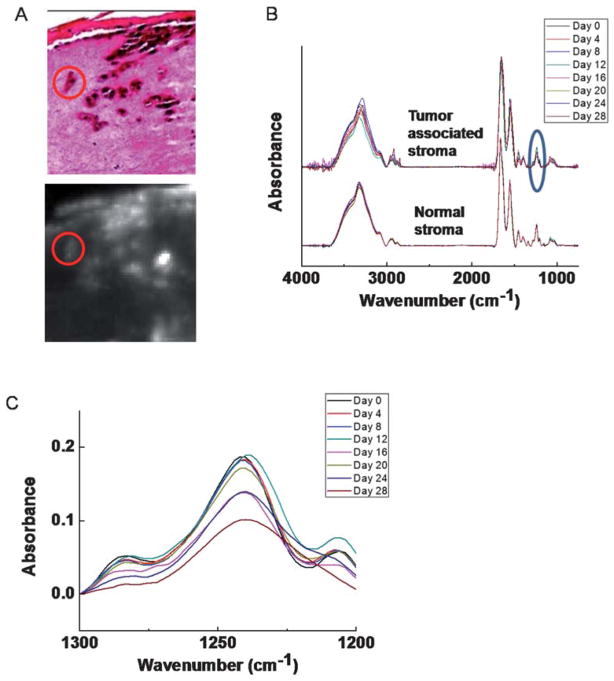
While changes in tumor cells are one aspect of progression, interactions between the tumor and its microenvironment is another important and rapidly emerging aspect of understanding tumor evolution.54,55 Here, we hypothesize that cells that are in close contact with tumor may possibly undergo spectral changes that reflect the underlying change of biomolecular expression. Since it is difficult to postulate every change a priori, a holistic method of measuring changes is likely useful. Hence, we employed FT-IR imaging here to probe the stromal regions surrounding tumors at different time points (Fig. 4A). Two stromal regions–one closely associated with the tumor in the vicinity of 10–100 μm and the other at least 500 μm away–are identified as tumor associated stroma and normal stroma respectively. Average spectra from these two regions were obtained and compared (Fig. 4B). One of the most significant variations over the time occurs at the fingerprint region, especially from 1200–1300 cm−1 (Fig. 4C). This region is closely associated with the proteins of the connective tissue and extracellular matrix. Hence, the changes likely indicate a matrix remodeling rather than large scale changes in stromal cells. Further, the changes appear to start between day 12 and day 16, corresponding to the histological observation that tumor invasion becomes apparent around day 12. Therefore, the spectra from regions next to tumor appear to have undergone a transition due to the tumor development, suggesting the possibility of heterotypic interactions and transformations in engineered tissue. It is notable that the biochemical changes do not appear to precede the histological changes. Hence, the methods we have used do not appear to be sensitive to biochemical changes preceding apparent morphological changes, if such are present.
Fig. 4.
Temporal behavior of normal and tumor-associated stroma. (A) Spectral regions of tumor (red circle) and tumor associated stroma are extracted for analysis of temporal changes. (B) Spectral analysis of normal and tumor-associated stroma demonstrates likely differences between the two, especially including significant changes in the 1200–1300 cm−1 (blue circle) region. (C) Spectral detail of the 1200–1300 cm−1 region indicates that the biochemical changes likely occur around day 16.
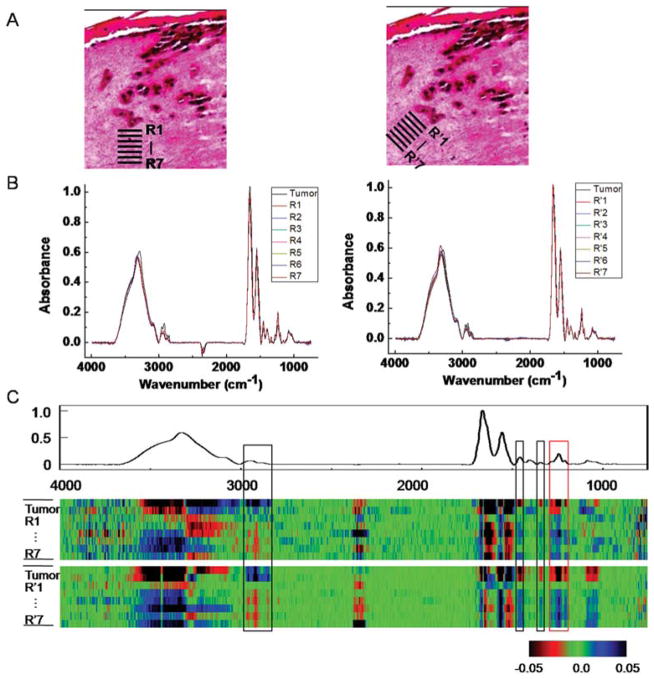
This change in stromal characteristics is also known as “field effect” for many other tumors.56,57 While field effect changes are histologically hard to differentiate, the chemical change is seemingly well-suited to analysis by FT-IR imaging. To validate that we are indeed observing the field effect, we sought to examine the spatial dependence of stromal transformation. Regions of interest close to the tumor and from progressively increasing distance were analyzed. Seven spatial regions adjacent to the tumor (Fig. 5A, indicated by R1 through R7) are chosen and average spectra of these seven regions of interest (ROIs) are compared to the average spectrum from the tumor (Fig. 5B). While average spectra are useful, visualization of changes and the relative importance of changes are difficult to appreciate. To facilitate the identification of specific spectral regions that undergo transformations, we employed the CDM tool again to show the variation among these ROIs (Fig. 5C). The difference of each spectrum in a few regions from the mean spectra, including a prominent peak from 1200–1300 cm−1 clearly shows a trend of transition from tumor to distal stromal regions. Same conclusion can be drawn from the analysis of ROI regions (R′1–R′7) obtained from a 45° angle.
Fig. 5.
Analysis of stromal transformation around a tumor. (A) Spectra at tumor-neighboring dermal region at different depths and angles (R1–R7, R′1–R′7) are extracted. (B) Analysis of ROIs from the tumor and 7 peritumoral regions at both angles shows spectral differences. (C) CDM suggests a transition pattern of spectra from tumor to distal stromal regions at a few spectral peaks (highlighted by boxes in the picture), including a prominent peak 1200–1300 cm−1 (red box). A sample from day 20 is used for the analysis.
The map indicates several regions corresponding to the transformation. The first region is between 2800–3000 cm−1, which arises from various C–H stretching vibrational modes (Fig. 5C). The most interesting changes, however, are those in fingerprint region, including peaks in the 1430–1480 cm−1 range −1 attributed to arising from CH2 scissoring modes, 1330–1360 cm CH2 wagging modes, and the protein specific tri-peaks at 1200–1300 cm−1 that also differ significantly at the surrounding stromal regions over the course of tumor invasion as we discussed above (Table 1). This tri-peak spectral region is often confused with features from nucleic acids, which are located in the similar region. Here, the lack of nuclear density and abundance of collagen in the extracellular matrix indicate that these features likely arise from collagen’s C–N stretching and in-plane bending modes.45 The regions close to the tumor appear to have adopted certain level of similarity to the neighboring tumors, while this similarity diminishes when the region is further away. While this may indicate a mesenchymal-to-epithelial type of transformation, we caution that the interpretation of results is made difficult by the lack of specific molecular features indicative of such a transformation. What is apparent is that the effect extends over a distance of 50–100 μm. Hence, spectroscopic imaging presents an attractive means to determining holistic chemical changes,58 which can then be further probed for molecular details by other modalities.
Similar results are observed in samples with other tumors in which the tumor is large enough to be identified, namely from days 20, 24 and 28. We especially focused on the collagen-specific region (1200–1300 cm−1) as it offers an interpretation of the field effect for our measurement. Similar to above studies, spectra from tumor and 7 neighboring regions are plotted (Fig. 6A). Spectra from regions next to tumor are distinct from either tumor or stroma, and appear as a transition between tumor and benign stroma in the distal region, suggesting the possibility of heterotypic interactions and transformations in engineered tissue. CDM was also used to visualize the variance of the spectra, giving the same 4 regions of transition we saw earlier (Fig. 6B). The trend demonstrated is consistent with the biological understanding of collagen, the major component of extracellular matrix, being biochemically remodeled59 and eventually occupied by invading tumor cells.60 As a result, the spectral contributions from collagen cross-linking diminish closer to the tumor and likely diminish over the time prior to tumor invasion. The most interesting observation here is that this type of transition appears to occur over a region much larger than the dimension of a single tumor cell, for example. Further, this chemical change appears to have begun before changes that are apparent in traditional histological methods, which might be critical for early tumor detection and treatment. The effect seems to be limited to 50–100 μm close to the tumor. We must emphasize that this is a molecular measure of the “field effect” as opposed to prior studies using histological,57 optical61 or specific gene/protein62 measure of the effect. Reconciling the various data on model systems, such as the ones proposed here, would likely prove beneficial. Finally, the measurements here are still 2D-histological analyses of a 3D tumor. Methods to study tumor growth in 3D using non-invasive imaging techniques will likely prove useful. Heterotypic interactions between malignant and benign tissues are an important component of tumor progression. By identifying and localizing this aspect in the model system, detailed molecular studies of these interactions can now be performed. Stromal-epithelial interactions present an emerging and promising approach to control tumor growth. Hence, the combination of model disease systems in culture and holistic methods of analysis, such as FT-IR spectroscopic imaging, can provide valuable research and clinical information. This study is a step towards that direction by presenting a framework for analysis and evidence for stromal involvement in tumors.
Fig. 6.
Chemical analysis of melanomastromal interactions. Regions of interest are extracted from tumors and from progressively increasing distance in stroma (R1–R7). Two duplicated samples from day 20, 24 and 28 are used. (A) The spectral region of 1200–1300 cm−1, corresponding to collagen specific peaks, is presented to show the magnitude of changes. (B) CDM is used to visualize the differences among spectra. Regions of transition are highlighted in squares, including the collagen region (red square).
Conclusions
We present here the rationale and a preliminary analysis of a melanoma model in engineered skin using FT-IR spectroscopic imaging. The technique allowed for rapid visualization of skin constituents and tumors induced within without the use of stains or dyes. We then conducted numerical analysis to demonstrate that the tumor cells demonstrated stable spectral profiles over time. Using comparative spectral profiles, we determined that the stroma was remodeled around a tumor in the 50–100 μm vicinity. The study presents a novel tool for various applications. It provides the cancer research community with a powerful analytical tool to characterize the extent of tumor influence, presents the spectroscopy community access to model systems for studying cancer progression, provides a platform for the chemometrics community to develop better tools and algorithms in cancer research by providing longitudinal samples and, finally, the tissue engineering community in measure holistic chemical changes during the development and maturation of samples.
Acknowledgments
We gratefully acknowledge the financial support from the Department of Defense and Susan G. Komen for the Cure. The project described was supported by Award Number R01CA138882 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Melanoma Study Group of the Mayo Clinic Cancer Center. Mayo Clin Proc. 2007;82:364–380. [Google Scholar]
- 2.Thompson JF, Scolyer RA, Kefford RF. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- 3.James WD, Berger TG, Elston D. Andrews’ Diseases of the Skin. 9. Philadelphia: 2000. pp. 881–889. [Google Scholar]
- 4.Elder DE. Clin Cancer Res. 2006;12:2308s–2311s. doi: 10.1158/1078-0432.CCR-05-2504. [DOI] [PubMed] [Google Scholar]
- 5.Hsu MY, Meier F, Herlyn M. Differentiation. 2002;70:522–536. doi: 10.1046/j.1432-0436.2002.700906.x. [DOI] [PubMed] [Google Scholar]
- 6.Ruiter D, Bogenrieder T, Elder D, Herlyn M. Lancet Oncol. 2002;3:35–43. doi: 10.1016/s1470-2045(01)00620-9. [DOI] [PubMed] [Google Scholar]
- 7.Lanza R, Langer R, Vacanti JP. Principles of Tissue Engineering. Elsevier; New York: 2007. [Google Scholar]
- 8.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 9.Debnath J, Brugge JS. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 10.Sokolov K, Galvan J, Myakov A, Lacy A, Lotan R, Richards-Kortum R. J Biomed Opt. 2002;7:148. doi: 10.1117/1.1427052. [DOI] [PubMed] [Google Scholar]
- 11.Shevchenko RV, James SL, James SE. J R Soc Interface. 2010;7:229–258. doi: 10.1098/rsif.2009.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabolinski ML, Alvarez O, Auletta M, Mulder G, Parenteau NL. Biomaterials. 1996;17:311–320. doi: 10.1016/0142-9612(96)85569-4. [DOI] [PubMed] [Google Scholar]
- 13.Jeffs AR, Glover AC, Slobbe LJ, Wang L, He SJ, Hazlett JA, Awasthi A, Woolley AG, Marshall ES, Joseph WR, Print CG, Baguley BC, Eccles MR. PLoS One. 2009;4:e8461. doi: 10.1371/journal.pone.0008461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Kempen LC, Ruiter D, van Muijen GN, Coussens LM. Eur J Cell Biol. 2003;82:539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- 15.Committee on Revealing Chemistry through Advanced Chemical Imaging & National Research Council of the National Academies. Visualizing Chemistry: The Progress and Promise of Advanced Chemical Imaging. National Academies Press; Washington, D.C: 2007. [PubMed] [Google Scholar]
- 16.Mendelsohn R, Diem M. Biochim Biophys Acta, Biomembr. 2006;1758:813. [Google Scholar]
- 17.Lewis EN, Treado PJ, Reeder RC, Story GM, Dowrey AE, Marcott C, Levin IW. Anal Chem. 1995;67:3377–3381. doi: 10.1021/ac00115a003. [DOI] [PubMed] [Google Scholar]
- 18.Levin IW, Bhargava R. Annu Rev Phys Chem. 2005;56:429–474. doi: 10.1146/annurev.physchem.56.092503.141205. [DOI] [PubMed] [Google Scholar]
- 19.Krafft C, Sergo V. Spectroscopy. 2006;20:195–218. [Google Scholar]
- 20.Zhang GJ, Moore DJ, Flach CR, Mendelsohn R. Anal Bioanal Chem. 2007;387:1591–1599. doi: 10.1007/s00216-006-0852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendelsohn R, Rerek ME, Moore DJ. Phys Chem Chem Phys. 2000;2:4651–4657. [Google Scholar]
- 22.Garidel P. Phys Chem Chem Phys. 2002;4:5671–5677. [Google Scholar]
- 23.Garidel P. Phys Chem Chem Phys. 2003;5:2673–2679. [Google Scholar]
- 24.Chan KLA, Kazarian SG. J Biomed Opt. 2007;12:044010. doi: 10.1117/1.2754686. [DOI] [PubMed] [Google Scholar]
- 25.Schultz CP, Mantsch HH. Cell Mol Biol. 1998;44:203–210. [PubMed] [Google Scholar]
- 26.Tfayli A, Piot O, Durlach A, Bernard P, Manfait M. Biochim Biophys Acta, Gen Subj. 2005;1724:262–269. doi: 10.1016/j.bbagen.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 27.Hammody Z, Sahu RK, Mordechai S, Cagnano E, Argov S. Sci World J. 2005;5:173–182. doi: 10.1100/tsw.2005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammody Z, Agrov S, Sahu RK, Cagnano E, Moreh R, Mordechai S. Analyst. 2008;133:372–378. doi: 10.1039/b712040k. [DOI] [PubMed] [Google Scholar]
- 29.Ly E, Piot O, Wolthuis R, Durlach A, Bernard P, Manfait M. Analyst. 2008;133:197–205. doi: 10.1039/b715924b. [DOI] [PubMed] [Google Scholar]
- 30.Mendelsohn R, Chen HC, Rerek ME, Moore DJ. J Biomed Opt. 2003;8:185–190. doi: 10.1117/1.1560645. [DOI] [PubMed] [Google Scholar]
- 31.Mendelsohn R, Flach CR, Moore DJ. Biochim Biophys Acta, Biomembr. 2006;1758:923–933. doi: 10.1016/j.bbamem.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Morganti F, Bramanti E, Solaro R, Benedetti E, Chiellini E, Nannipieri E, Narducci P, Krauser SF, Samour CM. J Bioact Compat Polym. 1999;14:162–177. [Google Scholar]
- 33.Dary CC, Blancato JN, Saleh MA. Regul Toxicol Pharmacol. 2001;34:234–248. doi: 10.1006/rtph.2001.1462. [DOI] [PubMed] [Google Scholar]
- 34.Cott M, Dumas P, Besnard M, Tchoreloff P, Walter P. J Controlled Release. 2004;97:269–281. doi: 10.1016/j.jconrel.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Andanson JM, Chan KLA, Kazarian SG. Appl Spectrosc. 2009;63:512–517. doi: 10.1366/000370209788347011. [DOI] [PubMed] [Google Scholar]
- 36.Andanson JM, Hadgraft J, Kazarian SG. J Biomed Opt. 2009;14:034011. doi: 10.1117/1.3130324. [DOI] [PubMed] [Google Scholar]
- 37.Tetteh J, Mader KT, Andanson JM, McAuley WJ, Lane ME, Hadgraft J, Kazarian SG, Mitchell JC. Anal Chim Acta. 2009;642:246–256. doi: 10.1016/j.aca.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Bhargava R, Levin IW. Appl Spectrosc. 2004;58:995–1000. doi: 10.1366/0003702041655412. [DOI] [PubMed] [Google Scholar]
- 39.Baker MJ, Gazi E, Brown MD, Shanks JH, Clarke NW, Gardner P. J Biophotonics. 2009;2:104–113. doi: 10.1002/jbio.200810062. [DOI] [PubMed] [Google Scholar]
- 40.Paget S. Lancet. 1889;1:571–573. [Google Scholar]
- 41.Fernandez DC, Bhargava R, Hewitt SM, Levin IW. Nat Biotechnol. 2005;23:469–474. doi: 10.1038/nbt1080. [DOI] [PubMed] [Google Scholar]
- 42.Muhart M, McFalls S, Kirsner RS, Elgart GW, Kerdel F, Sabolinski ML, Hardin-Young J, Eaglstein WH. Arch Dermatol. 1999;135:913–918. doi: 10.1001/archderm.135.8.913. [DOI] [PubMed] [Google Scholar]
- 43.Mantsch HH, Chapman D. Infrared Spectroscopy of Biomolecules. Wiley–Liss; New York: 1995. [Google Scholar]
- 44.Gunzler H, Heise HM. IR-Spektroskopie. Eine Einfuhrung. VCH; Weinheim: 1996. [Google Scholar]
- 45.Jackson M, Choo LP, Watson PH, Halliday WC, Mantsch HH. Biochim Biophys Acta. 1995;1270:1–6. doi: 10.1016/0925-4439(94)00056-v. [DOI] [PubMed] [Google Scholar]
- 46.Reddy RK, Walsh MJ, Bhargava R. Appl Spectrosc. 2010 doi: 10.1366/11-06568. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhargava R. Anal Bioanal Chem. 2007;389:1155–1169. doi: 10.1007/s00216-007-1511-9. [DOI] [PubMed] [Google Scholar]
- 48.Lasch P, Naumann D. Cell Mol Biology. 1998;44:189–202. [PubMed] [Google Scholar]
- 49.Buzug TM, Schumann S, Pfaffmann L, Reinhold U, Ruhlmann J. Conf Proc IEEE Eng Med Biol Soc. 2006;1:2766–9. doi: 10.1109/IEMBS.2006.259895. [DOI] [PubMed] [Google Scholar]
- 50.Chew SF, Wood BR, Kanaan C, Browning J, MacGregor D, Davis ID, Cebon J, Tait BD, McNaughton D. Tissue Antigens. 2007;69:252–258. doi: 10.1111/j.1399-0039.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 51.Ly E, Piot O, Durlach A, Bernard P, Manfait M. Analyst. 2009;134:1208–1214. doi: 10.1039/b820998g. [DOI] [PubMed] [Google Scholar]
- 52.Cruz GAS, Bertotti J, Marin J, Gonzalez SJ, Gossio S, Alvarez D, Roth BMC, Menendez P, Pereira MD, Albero M, Cubau L, Orellano P, Liberman SL. Appl Radiat Isot. 2009;67:S54–S58. doi: 10.1016/j.apradiso.2009.03.093. [DOI] [PubMed] [Google Scholar]
- 53.Acerbo AS, Miller LM. Analyst. 2009;134:1669–1674. doi: 10.1039/b823234b. [DOI] [PubMed] [Google Scholar]
- 54.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 55.Tlsty TD, Coussens LM. Annu Rev Pathol: Mech Dis. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 56.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS. Science. 1996;274:2057–2059. doi: 10.1126/science.274.5295.2057. [DOI] [PubMed] [Google Scholar]
- 57.Barcellos-Hoff MH, Ravani SA. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- 58.Ellis DI, Goodacre R. Analyst. 2006;131:875–885. doi: 10.1039/b602376m. [DOI] [PubMed] [Google Scholar]
- 59.Li L, Dragulev B, Zigrino P, Mauch C, Fox JW. Int J Cancer. 2009;125:1796–1804. doi: 10.1002/ijc.24463. [DOI] [PubMed] [Google Scholar]
- 60.Angeli F, Koumakis G, Chen MC, Kumar S, Delinassios JG. Tumor Biol. 2009;30:109–120. doi: 10.1159/000218708. [DOI] [PubMed] [Google Scholar]
- 61.Keller MD, Kanter EM, Lieber CA, Majumder SK, Hutchings J, Ellis DL, Beaven RB, Stone N, Mahadevan-Jansen A. Disease Markers. 2008;25:323–337. doi: 10.1155/2008/230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang X, Shigematsu H, Bekele BN, Roth JA, Minna JD, Hong WK, Gazdar AF, Wistuba II. Cancer Res. 2005;65:7568–7572. doi: 10.1158/0008-5472.CAN-05-1705. [DOI] [PubMed] [Google Scholar]