Abstract
HIV-1 latency is considered the last hurdle toward viral eradication in the presence of antiretroviral therapy. Studies of viral latency in vivo are complicated by the low frequency of latently infected cells found in HIV-1 patients. To be able to study the signaling pathways and viral determinants of latency and reactivation, we have developed a novel method that generates high numbers of latently HIV-1 infected cells, which are derived from human primary CD4+ T lymphocytes. This method allows for the study of different aspects of HIV-1 latency, such as the transcription factors needed for viral reactivation and the signaling pathways involved. In this review, we describe in detail an experimental protocol for the generation of HIV-1 latency using human primary CD4+ T cells. We also present the salient points of other latency models in the field, along with key findings arising from each model.
1. Introduction
Shortly after the introduction of antiretroviral therapy (ART), several groups were able to recover replication competent virus from patients’ cells even when viremia had been suppressed to undetectable levels (1–3). This observation led to the hypothesis that HIV-1 can establish a latent infection in the presence of ART. Eliminating the pool of HIV-1 latently infected cells is likely the next hurdle towards viral eradication in the presence of antiretroviral therapy (ART). In vivo analysis of the latent pool reveals that the main HIV-1 reservoir resides in resting, CD4+ memory T cells (1, 2, 4). Among the different subtypes of memory cells, central memory T cells (TCM) are thought to be an important HIV-1 reservoir (5, 6). However, despite the low frequency of latently infected cells in vivo (about 1 in 106 resting CD4+ T cells) (4), their longevity and their being impervious to antiretroviral treatment represent serious challenges toward eradication. It has been estimated that the half-life of a latently infected cell is about 44 months, and the time to viral eradication via ART has been predicted to be longer than 60 years (7, 8).
Understanding the molecular mechanisms governing HIV-1 latency in vivo is complicated by the low levels of latently infected cells and also by the lack of known phenotypic markers that can distinguish latently infected from uninfected cells. During the last decade, improvements in polymerase chain reaction (PCR)- based techniques detecting integrated HIV-1 and our increasing knowledge of phenotypes and functions of various immune cell subsets have allowed us to learn a great deal about the characteristics of the latent viral reservoir.
1.1 Phenotype of CD4+ T cells from the latent reservoir
Among CD4+ T cells, resting memory CD4+ T cells are the predominant subset that can harbor latent HIV-1 (4, 7). In the past few years, extensive characterization of memory T cells has led to the conclusion that the pool of memory CD4+ T cells (and also CD8+ T cells) is subdivided into subsets with different homing capacity and effector function (for a detailed review, see (9)). The two main subsets are central memory T cells (TCM) and effector memory T cells (TEM). TCM and TEM cells are easily identifiable by the expression of different surface receptors, including homing receptors. TCM are recognized by the dual expression of the chemokine receptor, CCR7, and the co-stimulatory molecule, CD27. On the other hand, differentiation into TEM is concomitant with loss of expression of CCR7 and CD27 (9). Other markers that distinguish TEM from TCM are specific to different subsets of TEM. For example, Th1 cells express CCR5, IL-12βR and intracellular IFNγ whereas Th2 express CrTH2 and intracellular IL-4. TCM do not express any of the above-mentioned five markers that characterize Th1 or Th2. TCM and TEM share expression of CD45RO, while naïve cells express CD45RA. A summary of markers that characterize T cell subsets can be found in Table I.
Table I.
Phenotypic characterization of T cell subsets
| Marker | Function | Naïve | TCM/NP | TEM | |
|---|---|---|---|---|---|
| TH1 | TH2 | ||||
| CD45RA | Phosphatase | + | − | − | − |
| CD45RO | Phosphatase | − | + | + | + |
| CCR7 (CD197) | Chemokine receptor | + | + | − | − |
| L-selectin (CD62L) | Homing to lymph nodes | + | + | − | − |
| CD27 | Co-activation | + | + | − | − |
| CCR5 | Chemokine receptor | − | − | + | − |
| IL-12Rβ | Cytokine receptor | − | − | + | − |
| CrTH2 (CD294) | Prostaglandin D2 receptor | − | − | − | + |
| IFNγ | Cytokine | − | − | + | − |
| IL-4 | Cytokine | − | − | − | + |
| IL-7Rα (CD127) | Homeostatic proliferation | + | + | − | − |
Recently, Chomont et al. reported that TCM cells isolated from blood, lymph nodes and gut from aviremic HIV-1 patients constitute the main reservoir for latent HIV-1 (6). They also found that HIV-1 establishes a significant degree of latency in transitional memory cells (TTM), but not in TEM, naïve T, or terminally differentiated T cells (6). In view of the fact that all of the above CD4+ T cell subsets are permissive to HIV-infection, their different abilities to harbor latent proviruses are striking and could potentially result from differences in the repertoires of transcriptional regulators.
1.2. The resting state of latently infected cells
Another important characteristic of the cells in the latent reservoir in vivo is their resting state. The resting state is characterized by the lack of activation markers as well as by the absence of proliferation. Latently infected cells are found in G0, with low levels of DNA and RNA synthesis (10). However, whether and how entry into G0 is a requirement for the establishment of latency, and whether transition from G0 to G1 is required for viral reactivation remain unanswered questions.
1.3 Epigenetic modifications and latency
Several epigenetic marks have been associated with the existence of HIV-1 latent infections (for reviews, see (11, 12)). For example, histone hypoacetylation has been correlated with transcriptional repression in several models of latency using cell lines (13–16). Direct evidence for the role of histone deacetylases (HDAC) in the maintenance of HIV-1 latency was obtained when specific HDAC inhibitors reactivated HIV-1 in resting cells from aviremic patients (17, 18).
Recently, it has been shown that CpG methylation of the HIV-1 5′ LTR is another important epigenetic mechanism contributing to the control of HIV-1 latency (19, 20). Analysis of the HIV-1 5′ LTR in the latent reservoir from aviremic patients showed a high degree of methylation (20). Data obtained from Blazkova et al. suggested that CpG methylation of the LTR DNA was not required for viruses to establish latency (20). However, over time, methylation of such sequences led to a “locked” latent state that was more difficult to reactivate than the unmethylated one (20).
2. Overview of existing methods to study latency
In the past two decades several in vitro models for HIV-1 latency have been developed. However, no single experimental system of HIV-1 latency is thought to represent a complete recapitulation of the biologic state of the latent cell reservoir in vivo. This is primarily due to two reasons. First, the latent or persistent HIV reservoir in vivo is likely to reside in multiple cell types. Second, as we review in the following paragraphs, the molecular mechanisms that lead to the establishment of a latent infection are likely to be multiple as well.
Rational design of drugs to target HIV latency is not possible at the moment, because we do not have precise knowledge of all the cellular factors and activation pathways that impact viral transcription. A second obstacle to rational therapy design lies in the notion that while the desired compound should trigger viral reactivation, it should induce minimal to no cellular activation. Presently, we do not have the depth of knowledge that allows us to dissociate viral from cellular activation. Therefore, compounds that fulfill these characteristics must be found through an unbiased discovery process. Drug screening has already produced candidate compounds that appear to fulfill these requirements in select model systems (21, 22).
No single cell model of latency is likely to be able to capture the broad spectrum of stimuli that can directly or indirectly induce transcription of the HIV-1 LTR. In addition, we do not know currently which activation pathways are most relevant to viral reactivation in vivo. Although studies in transformed cell line models have helped to reveal candidate mechanisms in the control of HIV latency, caution must be applied in extrapolating such findings to the actual state of viral latency found in resting cells in vivo.
2.1 Chronically infected cell lines
The generation of chronically infected cell lines, such as U1 (23), ACH2 (24), JΔK (25) and J-Lat (26) has contributed valuable insights into the many possible pathways and mechanisms that can lead to a latent HIV-1 infection.
The U1 model consists of a chronically infected clone from the parent promonocyte cell line, U937 (23). U1 cells contain two HIV-1 pro-viruses, which are non-replicating (27). HIV-1 expression can be induced in U1 cells by incubation with certain cytokines, including granulocyte/macrophage colony-stimulating factor (23); or with PMA (28). In U1 cells, latency was found to be associated with a suboptimal level of Tat protein activity. HIV-1 expression in U1 cells can be rescued by addition of exogenous Tat (29). The suboptimal activity of Tat was caused by mutations in both copies of the tat gene (one tat allele lacked an initiation codon, while the other contained an H-to-L mutation at amino acid residue 13 (H13L) (30).
The ACH2 model is based in the generation of a chronically infected T-cell clone from the parental cell line, A3.01 contains one latent provirus (24). In ACH2 cells, TNF-α is able to reactivate HIV-1 from latency (24). In this case, a mutation in the transactivation responsive region (TAR) impaired the transactivation function of Tat (31).
Early work with the ACH2 and U1 cell lines also facilitated studies aimed at understanding how the state of the chromatin impacts HIV-1 transcription. For example, Verdin et al. characterized, for the first time, the existence of nucleosome-occupied and nucleosome-free regions within the HIV-1 LTR using ACH2 and U1 cells (32).
JΔK is a chronically infected cell line derived from the parental Jurkat cell line (25). The HIV-1 strain in these cells contains a deletion in the long terminal repeat (LTR) NFκB binding sites. In JΔK cells, HIV-1 expression can be restored by incubation with PMA, sodium butyrate, or hexamethylene bisacetamide in an NFκB-independent fashion (25).
The J-Lat model is based on the generation of clonal cells lines derived from Jurkat infected with an HIV-1-based retroviral vector containing the Tat and GFP open reading frames both under the control of the HIV-1 promoter in the 5′ long terminal repeat (LTR) (26). These clonal cell lines showed low or no detectable GFP expression in the absence of stimulation. Treatment of the cells with phorbol esters or TNF-α reactivated GFP expression (26). In this model, latency was attributed to the integration of HIV-1 in or in close proximity to heterochromatic regions (26). These studies were pivotal in our early understanding of the influence of integration sites on viral gene expression. Several more recent studies have addressed how the directionality of integration, with respect to cellular transcriptional units in close proximity, influences viral gene expression and latency (33, 34).
Duverger et al. used a GFP-expressing virus to develop a Jurkat model in which a new infection is established with each experiment (35). Therefore, this model does not rely on chronically infected cells or stably integrated proviruses. Duverger’s infection model re-creates a true infection system where multiple integrations sites are permitted, presumably in an unbiased manner. Using this model, the authors explore the concept of “silent integration”, a theoretical pathway leading to latency. The silent integration model proposes that the virus must remain transcriptionally silent throughout the early phase of infection. According to this model, the onset of viral gene expression at any time during the infection cycle will preclude the establishment of a latent infection. Duverger et al. provide compelling evidence that this may indeed constitute a major pathway leading to latency in Jurkat cells (35). Whether silent integration is a requirement for establishment of latency in vivo remains unknown.
2.2 Animal models
Due to the low frequency of latent HIV-1 infections in humans, an animal model for the study of latency would be a promising alternative to in vivo studies. Brooks et al. developed a model that uses human fetal thymus and liver tissues implanted into severe-combined immunodeficient mice (Thy/Liv SCID-hu) (36). In this model, latency is generated at a high frequency during thymopoiesis. The latently infected cells thus generated are resting, mature, naïve CD4+ T cells. The virus in naïve T cells remains latent until cells become activated through T-cell receptor engagement, an event that promotes vigorous viral reactivation (36). This model allows for the study of latency in naïve CD4+ T cells. Early studies showed that HIV-1 sequences could be detected in naïve CD4+ T cells isolated from infected individuals, although at lower levels than those found in the memory subset (37). Furthermore, at least a fraction of the detected viruses present in naïve cells were latent, as evidenced by their ability to reactivate ex vivo (37). A more recent study has shown that the contribution of naïve cells to the HIV-1 reservoir is about 1.9%, while the contributions of TCM and TTM were estimated to be about 51.7 and 34.3%, respectively (6).
2.3 Primary cell cultures
Our growing knowledge of the immunological cell subsets in vivo, their functions and their roles in HIV-1 infection, has prompted the development of model systems that utilize primary cells (22, 38–42).
In the model developed by Saleh and colleges, resting CD4+ T cells can efficiently be infected after incubation with one the CCR7 ligands, CCL19 or CCL21 (38). It is noteworthy that incubation with CCL19 or CCL21 did not induce appreciable activation or proliferation, yet it rendered the cells susceptible to HIV-1 infection. Strong activation signals applied to the cells several days after infection were able to stimulate virus production from the latently infected cells. An important contribution of this ex vivo model is the concept that a viral latent state can be achieved through direct infection of a quiescent, resting memory CD4 T cell (38). This is unique as other models utilize activated cells as targets for infection, which are then allowed to return to a resting state via IL-2 incubation (40, 41).
Yang et al. developed a novel model of latency that involves lentiviral transduction of primary CD4+ T cells with a Bcl-2 cDNA to increase cell survival in vitro (22). These cells were then activated with anti-CD3/CD28 antibodies and IL-2, and then virus infected. When cells infected in this fashion were allowed to return to a quiescent state, via incubation in the absence of cytokines, a population of latently infected cells was clearly identifiable. Viral reactivation was achieved with a variety of stimuli, which induced signaling through a shared PKC/NFκB pathway. It is unclear whether ectopic expression of Bcl-2 in this system may introduce artifactual effects in terms of cellular activation, differentiation that, in turn, may affect viral latency. However, preliminary characterization of the signaling pathways that trigger viral reactivation showed that latently infected cells respond to reactivation stimuli in concordance with observations made in previous ex vivo and in vitro models (22). The phenotype of the latently infected cells in the model by Yang et al., was highly consistent with that of TEM cells, as evidenced by the high levels of CD45RO and low levels of CCR7 present on these cells (22). The utility of this model was demonstrated by the investigators’ ability to screen a small library of random drug-like molecules, which resulted in the discovery of an exciting anti-latency compound (22).
Our laboratory has recently developed an ex vivo paradigm of viral latency that utilizes primary T cells (40). The main difference with other primary cell models resides in the isolation of naïve CD4+ T cells to near purity. Naïve cells are then activated and induced to differentiate into a TCM-like phenotype known as non-polarized cells (NP) (40, 43). Infection with HIV-1 is performed while the cells are in an activated state, and viral latency is strongly favored by the natural progression of activated cells to a quiescent, memory-like state (40). This method is described in detail in Section 3, below.
In the system developed by Tyagi and Karn, primary CD4 cells are isolated in bulk from peripheral blood, stimulated with αCD3+αCD28 antibodies in the presence of rIL-2, and then infected with VSV-G-pseudotyped, HIV-1 vectors that express GFP or mCherry reporters. The HIV vectors lack env and contain either wild-type or attenuated Tat (H13L mutation). After 2 days, productively infected cells, expressing the reporter gene, are purified by flow cytometry sorting and placed into culture in the presence of αCD3+αCD28 plus rIL-2. The infected cell culture is expanded over the course of 4 – 6 weeks. Then, the antibody (αCD3+αCD28)-coated beads are removed and the cells are co-cultured on H80 feeder cell monolayers in the presence of rIL-2, as previously described (42). After approximately 6 weeks, 70 – 90% of the infected cells harbor latent virus that no longer express the reporter gene (41). At this point, the latently infected CD4 cell population has a small resting cell morphology, a phenotype resembling that of TCM cells, and contains ~65% BrdU+ or Ki67+ cells. Virus can be reactivated in the majority of cells, following secondary stimulation with immobilized αCD3+αCD28 antibodies. Analysis by ChIP has demonstrated that TCR activation results in increased histone acetylation and recruitment of NFκB to the LTR promoter region of integrated HIV-1. Exposure to TNFα alone does not reactive viral transcription, due to limiting levels of P-TEFb in primary T cells.
3. Description of the method
3.1 Overview
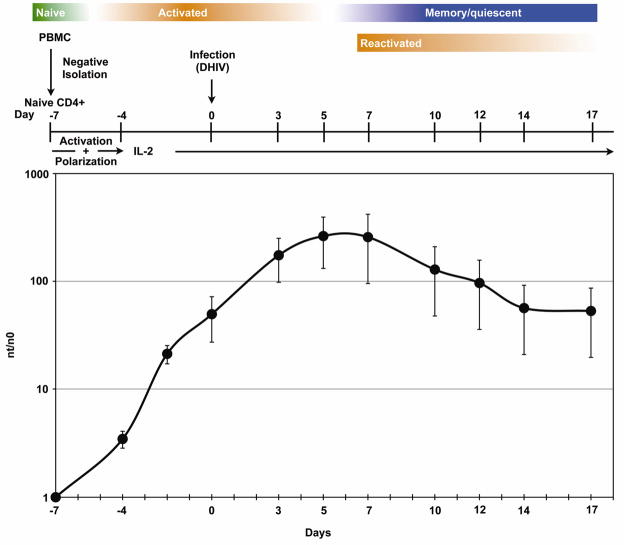
The steps for the generation of latently infected NP cells are graphically depicted in Fig. 1. Mononuclear cells are isolated from peripheral blood from healthy, anonymous donors using Ficoll gradient centrifugation. Naïve CD4+ T cells are then purified using magnetic columns, and these cells are then polyclonally activated using antibodies against CD3 and CD28 (Day −7), in the presence of TGF-β, αIL-12 and αIL-4 antibodies, which induce the generation of non-polarized cells (NP) (43). Under these conditions, vigorous cell activation and proliferation are promoted, and more than 95% of cells develop the phenotypic characteristics of NP cells (40, 43). A detailed discussion about the similarities between NP cells and TCM is presented below in section 3.2.
Figure 1. Protocol for the generation of memory T cells ex vivo.
Colored bars illustrate the differentiation, activation state of the cells. Graph depicts cellular proliferation over time, expressed as nt (cell number at the time of measurement) over n0 (initial cell number). Activation refers to the incubation in the presence of anti-CD3 and anti-CD8 antibodies, and polarization refers to the choice among non-polarizing conditions, Th1- or Th2-polarizing conditions (see text for details).
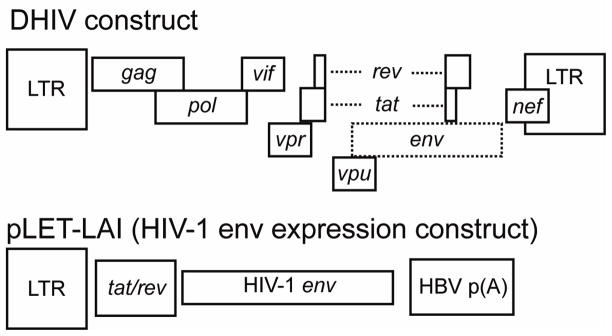
Cells are infected during the exponential proliferation phase (Day 0; see Fig. 1). The virus we utilize is the reference strain HIV-1NL4-3, which was rendered replication-deficient via inactivation of the envelope glycoprotein gene. In the virus production stage, in 293T cells, HIV-1 env is provided in trans via co-transfection of the appropriate expression vector. This virus, deemed defective HIV or “DHIV”, is competent for entry, reverse transcription, integration, and viral gene expression for all genes except for env, but not for the production of infectious progeny (Fig. 2). Therefore, DHIV is unable to spread beyond the first infection event, and, therefore, is only able to cause cell death during the first and only round of infection. An additional advantage of using a defective virus for latency studies is that the appearance of intracellular p24Gag upon cellular stimulation is an unequivocal sign of viral reactivation and is not due to viral spread from cell to cell.
Figure 2. Plasmids used for the generation of defective HIV-1 virions by co-transfection.
Boxes represent open reading frames and select cis acting sequences. The envelope gene in the proviral construct named “DHIV” was inactivated by mutation. Only viral sequences are shown and bacterial plasmid sequences are omitted.
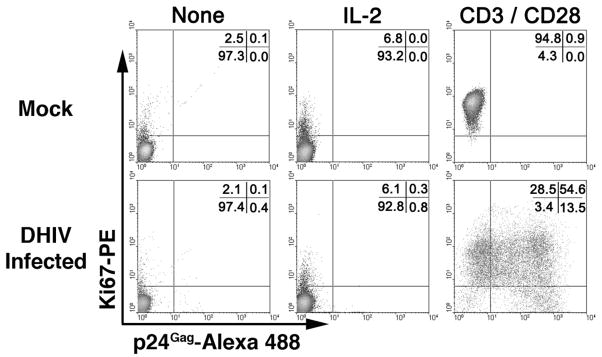
Infected cells are kept in vitro for an additional 7-day period or longer (up to three weeks after infection) in the presence of rIL-2. As shown in Fig. 1, cells cease to proliferate between day 5 and 7 post-infection. To reactivate potential latent viruses, cells are treated with antibodies against CD3 and CD28 at 7 days post infection (reactivation). Three days post-reactivation, cells were stained for p24Gag and the Ki67 antigen (Fig. 3). Ki67 is a known nuclear marker present during all active phases of the cell cycle (G1, S, G2 and mitosis), but is absent in resting (G0) cells (44). As shown in Fig. 3 left panels, when cells were cultured in the absence of IL-2 during the time of reactivation (day 7 to day 10), approximately 97% of the cells become quiescent (Ki67−). The use of recombinant IL-2 (rIL-2) in these cultures does not significantly increase the proliferation rate when compared with cultures in the absence of rIL-2 (Fig. 3). Stimulation of the cells with antibodies against CD3 and CD28 (αCD3/αCD28) induced entry into the cell cycle (Ki67+) in the vast majority of the cells (Fig. 3, right panels). In the absence of reactivation stimulus (“none” or “IL-2”), little viral gene expression is detected as evidenced by intracellular p24 positivity. However, stimulation of the cells with αCD3/αCD28 led to efficient reactivation of latent viruses. These results indicate that the presence of rIL-2 in the culture does not modify the resting state of the latently infected cells and that addition of IL-2 alone does not induce viral reactivation in NP cells.
Figure 3. Central memory T cells harbor a latent HIV-1 reservoir that is vigorously reactivated by anti-CD3 plus anti-CD28 stimulation, but not by IL-2 stimulation.
The effect of stimulation on cell proliferation is measured via Ki67 nuclear antigen staining, and the effect on viral reactivation is measured via intracellular p24 stain.
3.2 Non-polarized T cells as the in vitro equivalent of TCM
Sad and Mosmann in 1994 described, for the first time, the existence of a certain T helper precursor cell type (Thp) which, upon antigenic stimulation, secreted IL-2 but none of the Th1 or Th2-specific cytokines (45). Generation of these uncommitted Thp cells was accomplished when mouse CD4+ T cells were antigen-stimulated in the presence of a combination of TGF-β and anti-IFN-γ (45). Thp cells had the ability to further differentiate into either Th1 or Th2 phenotypes when re-stimulated in the presence of specific cytokine combination (45). Therefore, the existence of a bi-potential precursor that could differentiate into Th1 ad Th2-like cells was demonstrated.
In 1999 Sallusto et al. introduced the terminology of central memory (TCM) and effector memory (TEM) to define T cell subsets with different phenotypes and effector functions (46). TCM express the chemokine receptor 7 (CCR7); the homing receptor, L-selectin (CD62L); and the IL-7 receptor (IL-7R/CD127). Due to the presence of L-selectin and CCR7, TCM can home to secondary lymphoid organs (47). In addition, upon antigenic stimulation, TCM cells produce IL-2 but not IFN-γ or IL-4, and have enhanced responsiveness (i.e., a higher degree of proliferation) when compared with naïve T cells. TEM, on the other hand, do not express CCR7, CD62L or IL-7R and are, instead, positive for markers that are defining of either Th1 or Th2 phenotypes. The two cell types are developmentally related as stimulation of TCM results in their differentiation to TEM (46).
The term NP was created to denote an ex vivo derived T cell subset with an intermediate level of differentiation between naïve and effector cells, devoid of expression of effector cytokines that define Th1 and Th2 polarized cells, but with the potential for differentiation into Th1 or Th2 (48). Non-polarized cells (NP) are considered the in vitro equivalents of TCM (43) and are probably identical or closely related to the Thp cells described by Sad and Mosmann (45).
Furthermore, mouse NP cells generated in vitro from a transgenic BALB/c mouse carrying an I-Ed-restricted TCR specific for an influenza hemagglutinin peptide, when adoptively transferred into syngeneic normal mice, co-expressed CCR7 and CD62L and had lymph node homing capacity (48). Iezzi et al. also showed that NP cells had lower activation thresholds and faster activation kinetics when compared to naïve CD4+ T cells, not only in in vitro experiments but also in vivo after adoptive transfer (48).
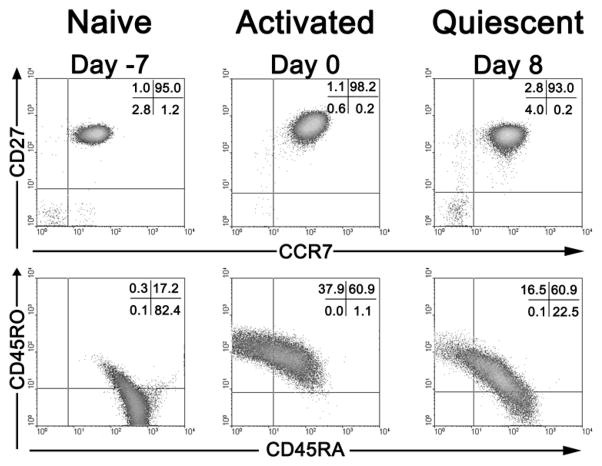
One notable difference between NP cells and fresh TCM is the re-emergence in NP cells of the naïve cell marker, CD45RA, which is then co-expressed with the memory cell marker, CD45RO, (see lower right panels in Fig. 4). The significance of this in vitro phenomenon, also reported in another primary cell model (22), is unknown at present.
Figure 4. Phenotypic characterization of naïve, activated and quiescent cells.
Upper panels show CD27 and CCR7 expression and lower panels show CD45RO and CD45RA. See text for details.
The inability of NP cells to produce IFN-γ or IL-4 after antigen re-stimulation was associated with hypoacetylation of the corresponding promoters (43). This hypoacetylation status is also found in freshly isolated TCM (43). Finally, the authors showed that NP cells can differentiate to Th1 or Th2 when activated in the presence of the differentiation cytokines, IFN-γ and IL-4, respectively (43). Therefore, NP cells obtained in vitro via differentiation of naïve cells closely parallel TCM found in vivo, both in terms of phenotype and function.
3.3. Detailed protocols
3.2.1. Virus generation
A replication-deficient proviral clone that has a small deletion in env (DHIV; Fig. 2) was previously described (49). To generate the virus, 293FT cells were transfected using a CaPO4 transfection protocol, using the following plasmids: 25 μg of DHIV plasmid and 5 μg of pLET-LAI (encoding a X4 envelope (50)). For transfection, 293FT cells were grown in 10cm dishes at 80% confluency. One hour prior to transfection, the culture medium was replaced with 20 ml of DMEM (supplemented with 10% FBS and L-glutamine in the absence of antibiotics). At the time of transfection, plasmids were diluted with ddH2O up to 900 μl. After dilution of the plasmids, 100 μl of 2.5M CaCl2 and 1 ml of 2×HBS (pH 7.05) were added, tubes were vortexed and incubated for 1 min. at room temperature. To increase transfection levels, 20 μl of 100mM chloroquine was added. After approximately 18h, the transfection medium was replaced with 20ml of fresh medium (DMEM + 10 % FBS and L-glutamine in the absence of antibiotics) and kept for an additional 36 hours. Supernatants were then collected and pre-cleared by centrifugation at 2,000 rpm for 10 min. After centrifugation, the supernatant was filtered thorough a 0.4 μm filter, aliquoted, and frozen at −80°C. A small fraction of the frozen supernatant was used to quantify the amount of p24 by enzyme-linked immunosorbent assay (ELISA; ZeptoMetrix, Buffalo, NY).
3.2.3. Isolation of naïve CD4+ T cells (Day −7)
Peripheral blood mononuclear cells are obtained from unidentified, healthy donors by density centrifugation. 25 ml of whole, heparinized blood is overlaid on 20ml of Ficoll-Paque™ PLUS (GE Healthcare Bio-Sciences) and centrifuged at 1,400 rpm (405 g) during 30 min. in a Sorvall® Super T21 using an ST-H750 rotor. After centrifugation, the buffy coat is transferred to a clean tube and washed twice with PBS plus 2mM EDTA to remove platelets. Naïve CD4+ T cells are isolated from the PBMCs by magnetic cell sorting by either of the two following methods:
Naïve CD4+ T cells are isolated by MACS microbead-negative sorting using the human naïve T-cell isolation kit (Miltenyi Biotec, Auburn, CA). Cells are sorted using the “possel_s” program in an AutoMACS Separator (Miltenyi Biotec). Using this program instead the “deplete” program increases the purity of the negative. Cells are collected from the outlet port, “neg1”, in warm, complete medium consisting of RPMI supplemented with 10% pooled human serum (Innovative Research) and Pen/Strep.
Alternatively, naïve CD4+ T cells can be isolated using the EasySep® Negative Selection Human Naïve CD4+ T cell Enrichment Cocktail (StemCell Technologies Inc, Vancouver, Canada). To improve the purity of the isolated population, the kit was modified by adding an anti-CD25 antibody to the negative selection cocktail. Cells were subjected to two rounds of magnetic separation.
Either of the above methods typically yields a population with the phenotype CD4+CD45RA+CD45RO−CCR7+CD62L+CD27+ with purity levels equal or higher than 95% (Fig 4, Day −7 panels).
3.2.4. Activation of naïve CD4+ T cells (Day −7)
After Isolation, cells were activated in non-polarizing (NP) conditions using a variation of a previously described method (43) as follows. 5×105 cells were stimulated with 5×105 beads coated with αCD3 and αCD28 antibodies (Dynabeads CD3/CD28 T cell Expander, Dynal/Invitrogen) in 1 ml of complete medium, in the presence of 10 ng/ml of TGF-β1, 2 μg/ml of anti-Human IL-12 and 1μg/ml of anti-Human IL-4 (All of them from Peprotech, Rocky Hill, NJ). To ensure homogenous activation of the cells, 100μl of the cell mixture, together with cytokines and beads were plated in a 96-well, round bottom plate (BD Falcon, Bedford, MA) and incubated for 3 days at 37 °C.
After activation, cells were resuspended to remove clumps. Dynabeads were removed using a Magnetic Particle Concentrator (Dynal MPC®-L, Invitrogen). Activated T cells were re-plated at 1×106 cells/ml in complete medium with 30 IU/ml of rIL-2 (AIDS Research and Reference Reagent Program) for 4 days at 37 °C. Medium and rIL-2 were subsequently replaced every 2 days.
3.2.5. Infection of activated T cells and generation of latently infected cells (Day 0)
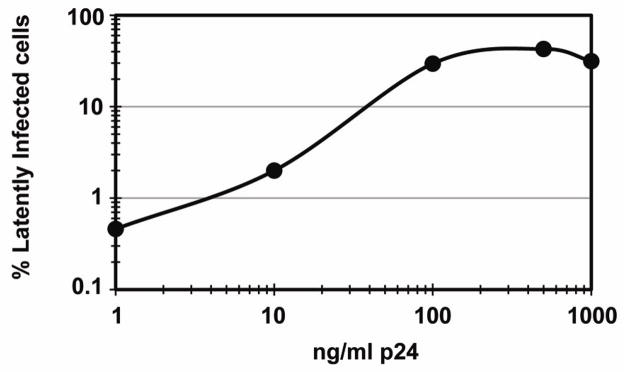
Activated T cells (7 days after stimulation with αCD3/CD28) were infected by spinoculation of 1×106 cells in 1 ml of virus-containing supernatant. Spinoculations were performed in a Sorvall® Super T21 using an ST-H750 rotor at 2,900 rpm (1,741×g) during 2h at 37 °C. To ensure a maximum frequency of infected cells, we exposed cells to 500 ng/ml of p24 (Fig. 5). Infection with higher amounts of p24 did not enhance the production of latently infected cells.
Figure 5. Correlation between percentage of latently infected cells and the size of the virus inoculum.
Percent latently infected cells was calculated via stimulation of latent populations via anti-CD3 plus anti-CD28 antibodies followed by intracellular p24 stain at day 3 post infection. Virus inoculum size was measured via p24 quantitation by ELISA.
After infection, the virus-containing supernatant was removed and cells kept in culture at 1×106 cells/ml in complete medium with 30 IU/ml of rIL-2 at 37 °C. At days 3, 5 and 7 post infection, media and rIL-2 were replaced and cells were kept at a density of 1×106 cells/ml. It is important to note that the efficiency of infection and the subsequent generation of latency are maximized when cells continue to proliferate during the first three days post- infection (days 0–3, Fig. 1; unpublished results).
4. Concluding remarks
We here present a novel method of generating latently infected, resting TCM. This model has been used to analyze the main signaling pathways that lead to viral reactivation after TCR engagement (40) and to begin characterize the methylation status of the viral promoter in primary cells (19). This paradigm is suitable for a variety of analyses. For example, we propose to utilize ex vivo latently infected TCM cells to dissect the cellular events controlling the establishment of latency and the onset of reactivation. Specifically, we propose to examine how various modes of cellular activation (i.e., in response to TCR engagement, or cytokine stimulation) may affect latency. We envision that this model will also help us to understand how homeostatic proliferation (i.e., as induced by IL-7 or IL-15) and other microenvironmental cues affect the size and behavior of the latent reservoir. Finally, it will also be possible to investigate the virological determinants of latency, such as the potential roles of the HIV-1 accessory genes in latency induction and reactivation, and the contribution of various LTR cis-acting elements to the transcriptional state of the virus in primary memory cells.
Viral tropism has been shown to be strongly linked to the latent reservoir. Specifically, it has been shown that the genotype of viruses in the latent reservoir predicts R5 tropism rather than X4 (51). When cells differentiate into TCM (in vivo) or NP (ex vivo) they are known to naturally downregulate CCR5, but they maintain CXCR4. This phenomenon has limited the scope of our experiments because, as one would expect, R5 tropic viruses infect NP cells inefficiently (Bosque and Planelles, unpublished observations). Therefore, only X4 or dual tropism viruses can be used in our primary cell model. Two possible explanations may satisfy the above conundrum. First, CCR5 could be expressed at certain times in the life of a central memory cell in vivo, providing R5-tropic HIV-1 with a window of time for infection leading to latency. We have observed that R5 viruses do infect TCM cells although with very low efficiency (2% compared with 40–60% infection by X4 viruses, using the same amounts of p24 in the inocula; unpublished data). This leads to the second possibility we would like to suggest. We do know that the size of the reservoir is small. Therefore, one may speculate that the inefficient infection of TCM by CCR5 viruses may be a relevant factor determining the relatively small size of the latent pool. This issue should be examined in the near future.
The existence of, perhaps yet another reservoir, residing in the TTM subset has recently been proposed (6). It is plausible that TTM may exhibit differences with respect to TCM in terms of the types of stimuli that will induce cellular activation and/or viral reactivation. Therefore, when considering the design or discovery of anti-latency drugs, one should take into consideration the potential effects of drugs on both cell types.
Acknowledgments
A.B. is supported by an AmFAR Mathilde Krim postdoctoral fellowship. This work was supported by NIH grant AI087508 to V.P. We are grateful to Dr. Celsa Spina for helpful comments and suggestions on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Science. 1997;278:1295–300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Science. 1997;278:1291–5. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Proc Natl Acad Sci U S A. 1997;94:13193–7. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Nature. 1997;387:183–8. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. J Virol. 2004;78:1160–8. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Nat Med. 1999;5:512–7. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 8.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Nat Med. 2003;9:727–8. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 9.Sallusto F, Geginat J, Lanzavecchia A. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 10.Richman DP. J Cell Biol. 1980;85:459–65. doi: 10.1083/jcb.85.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams SA, Greene WC. Cytokine. 2007;39:63–74. doi: 10.1016/j.cyto.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. Nat Rev Microbiol. 2009;7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 13.Van Lint C, Emiliani S, Ott M, Verdin E. EMBO J. 1996;15:1112–20. [PMC free article] [PubMed] [Google Scholar]
- 14.He G, Margolis DM. Mol Cell Biol. 2002;22:2965–73. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. Embo J. 2006;25:139–49. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyagi M, Karn J. EMBO J. 2007;26:4985–95. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. AIDS Res Hum Retroviruses. 2009;25:207–12. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Archin NM, Keedy KS, Espeseth A, Dang H, Hazuda DJ, Margolis DM. AIDS. 2009;23:1799–806. doi: 10.1097/QAD.0b013e32832ec1dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. PLoS Pathog. 2009;5:e1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J, Hirsch I. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang HC, Shen L, Siliciano RF, Pomerantz JL. Proc Natl Acad Sci U S A. 2009;106:6321–6. doi: 10.1073/pnas.0809536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang HC, Xing S, Shan L, O’Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. J Clin Invest. 2009 doi: 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Science. 1987;238:800–2. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 24.Folks TM, Clouse KA, Justement J, Rabson A, Duh E, Kehrl JH, Fauci AS. Proc Natl Acad Sci U S A. 1989;86:2365–8. doi: 10.1073/pnas.86.7.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antoni BA, Rabson AB, Kinter A, Bodkin M, Poli G. Virology. 1994;202:684–94. doi: 10.1006/viro.1994.1390. [DOI] [PubMed] [Google Scholar]
- 26.Jordan A, Bisgrove D, Verdin E. Embo J. 2003;22:1868–77. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pomerantz RJ, Trono D, Feinberg MB, Baltimore D. Cell. 1990;61:1271–6. doi: 10.1016/0092-8674(90)90691-7. [DOI] [PubMed] [Google Scholar]
- 28.Folks TM, Justement J, Kinter A, Schnittman S, Orenstein J, Poli G, Fauci AS. J Immunol. 1988;140:1117–22. [PubMed] [Google Scholar]
- 29.Cannon P, Kim SH, Ulich C, Kim S. J Virol. 1994;68:1993–7. doi: 10.1128/jvi.68.3.1993-1997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emiliani S, Fischle W, Ott M, Van Lint C, Amella CA, Verdin E. J Virol. 1998;72:1666–70. doi: 10.1128/jvi.72.2.1666-1670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emiliani S, Van Lint C, Fischle W, Paras P, Jr, Ott M, Brady J, Verdin E. Proc Natl Acad Sci U S A. 1996;93:6377–81. doi: 10.1073/pnas.93.13.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdin E, Paras P, Jr, Van Lint C. EMBO J. 1993;12:3249–59. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenasi T, Contreras X, Peterlin BM. Cell Host Microbe. 2008;4:123–33. doi: 10.1016/j.chom.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, Lin YB, An W, Xu J, Yang HC, O’Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF. Cell Host Microbe. 2008;4:134–46. doi: 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O. J Virol. 2009;83:3078–93. doi: 10.1128/JVI.02058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks DG, Kitchen SG, Kitchen CM, Scripture-Adams DD, Zack JA. Nat Med. 2001;7:459–64. doi: 10.1038/86531. [DOI] [PubMed] [Google Scholar]
- 37.Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. J Virol. 1999;73:6430–5. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR. Blood. 2007 doi: 10.1182/blood-2007-06-097907. [DOI] [PubMed] [Google Scholar]
- 39.Marini A, Harper JM, Romerio F. J Immunol. 2008;181:7713–20. doi: 10.4049/jimmunol.181.11.7713. [DOI] [PubMed] [Google Scholar]
- 40.Bosque A, Planelles V. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyagi M, Pearson RJ, Karn J. J Virol. 84:6425–37. doi: 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu GK, Lee K, Ji J, Braciale V, Baron S, Cloyd MW. Virology. 2006;355:127–37. doi: 10.1016/j.virol.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 44.Scholzen T, Gerdes J. J Cell Physiol. 2000;182:311–22. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 45.Sad S, Mosmann TR. J Immunol. 1994;153:3514–22. [PubMed] [Google Scholar]
- 46.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 47.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 48.Iezzi G, Scheidegger D, Lanzavecchia A. J Exp Med. 2001;193:987–93. doi: 10.1084/jem.193.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen JL, DeHart JL, Zimmerman ES, Ardon O, Kim B, Jacquot G, Benichou S, Planelles V. PLoS Pathog. 2006;2:e127. doi: 10.1371/journal.ppat.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Challita-Eid PM, Klimatcheva E, Day BT, Evans T, Dreyer K, Rimel BJ, Rosenblatt JD, Planelles V. AIDS Res Hum Retroviruses. 1998;14:1617–24. doi: 10.1089/aid.1998.14.1617. [DOI] [PubMed] [Google Scholar]
- 51.Pierson T, Hoffman TL, Blankson J, Finzi D, Chadwick K, Margolick JB, Buck C, Siliciano JD, Doms RW, Siliciano RF. J Virol. 2000;74:7824–33. doi: 10.1128/jvi.74.17.7824-7833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]