Abstract
Mutations in HFE cause the most common form of hereditary hemochromatosis (HH). We previously showed that liver-specific, transgenic overexpression of murine Hfe stimulates production of the iron regulatory hormone hepcidin. Here, we developed several additional transgenic mouse strains to further interrogate the structural basis of HFE function in the pathophysiology of HH. We hypothesized that the small, cytoplasmic domain of HFE might be necessary for HFE-mediated induction of hepcidin. We demonstrate that, like the full-length protein, overexpression of Hfe proteins lacking the cytoplasmic domain leads to hepcidin induction, iron deficiency and a hypochromic, microcytic anemia. However, high-level expression of a liver-specific Hfe transgene carrying the mouse equivalent of the common HFE C282Y human disease-causing mutation (murine C294Y) did not cause iron deficiency. Furthermore, hepcidin induction by transgenes encoding both WT Hfe and Hfe lacking its cytoplasmic domain is greatly attenuated in the absence of hemojuvelin (Hjv). Our observations indicate that the extracellular and transmembrane domains of Hfe are sufficient, and Hjv is essential, for Hfe-mediated induction of hepcidin expression.
Introduction
The common iron overload disease hereditary hemochromatosis (HH) is caused by a chronic increase in dietary iron absorption. A unifying model has emerged to explain the pathogenesis of genetically distinct forms of HH.1 Aberrant iron homeostasis results from interruption of a regulatory axis involving the hormone hepcidin, predominantly produced by hepatocytes, and the iron exporter ferroportin, present in duodenal enterocytes and macrophages. Hepcidin binds to ferroportin to trigger its internalization and lysosomal degradation.2 In this way, circulating hepcidin controls both intestinal iron absorption and the release of iron from macrophages into the plasma. There are 3 general classes of genetic defects known to cause HH. First, recessive mutations in the gene encoding hepcidin (HAMP) prevent the production of functional peptide.3 Second, some dominant mutations in ferroportin (encoded by SLC40A1) render it resistant to hepcidin regulation.4,5 Third, recessive mutations in the genes encoding HFE, transferrin receptor-2 (TFR2) or hemojuvelin (HFE2 or HJV), alter hepcidin expression by the hepatocyte.6–8
Most hemochromatosis patients are homozygous for a C282Y mutation in HFE (equivalent to C294Y in mouse Hfe), an atypical major histocompatibility complex (MHC) class I-like molecule9 that heterodimerizes with β2-microglobulin10 and associates with the major transferrin receptor (TFR1).11,12 HFE is similar to MHC class I proteins in that it contains a signal sequence, 3 sequential immunoglobulin superfamily α-domains, and a transmembrane domain followed by a 19 amino acid, cytoplasmic domain. The cytoplasmic domain of human HFE has 2 conserved, potential phosphorylation sites at 335Ser and 342Tyr. Although it has been shown that the cytoplasmic domains of Class I MHC proteins are not necessary for transmitting T-cell activation signals,13 and overexpression of HFE proteins containing either a S335M or Y342C mutation decreased ferritin levels in HEK293 cells,14 we hypothesized that these 2 phosphorylation sites might be involved in HFE-mediated hepcidin induction in vivo. Cytoplasmic motifs similar to the YVLA (342Tyr) motif in HFE function as endocytic-sorting motifs in other transmembrane proteins, including TFR1.15,16 Recently, Gao and colleagues reported that a truncated protein containing only the α3 and cytoplasmic domains of HFE was able to regulate hepcidin expression in cultured cells.17 Taken together, these data suggested that the cytoplasmic tail might play a role in HFE function or trafficking.
HFE acts in the liver where it contributes to regulation of hepcidin expression. Mice lacking Hfe only in hepatocytes display a hemochromatosis-like phenotype.18 HFE may also function in other cells involved in iron recycling, such as Kupffer cells,19 but its roles have not been fully characterized in cell types other than hepatocytes. We showed that Tfr1 sequesters Hfe, preventing it from participating in the induction of hepcidin expression.20 Hfe also interacts with Tfr2, and Hfe/Tfr2 complex formation is favored over Hfe/Tfr1 interaction when high levels of diferric transferrin (Fe2-Tf) are present, as is true in iron overload.21 Based on these observations, we and others have hypothesized that competition between Tfr1 and Tfr2 for Hfe binding allows hepcidin expression to be regulated in response to the amount of circulating Fe2-Tf.17,20,21 However, other work suggests that Hfe and Tfr2 may not function in the same signaling pathway, as humans and mice with mutations in both HFE and TFR2 have a clinically more severe phenotype than individuals with either mutation alone.22,23
HJV is mutated in patients with a severe, early onset form of HH. HJV acts as a bone morphogenetic protein (BMP) co-receptor, binding BMP ligands in association with Type I BMP receptors and triggering a sons of mothers against decapentaplegic (SMAD)–dependent signaling cascade that leads to increased hepcidin expression.24 Accordingly, disease-associated mutations in HJV result in decreased BMP signaling and decreased hepcidin expression.24 Mice lacking Hjv produce very little hepcidin mRNA.25,26 Furthermore, hepatocyte-specific inactivation of Smad4 or germ line deletion of Bmp6, which encodes a BMP ligand that responds to iron status, leads to a failure of hepcidin expression and marked iron overload.27–29 Although Bmp6 is up-regulated in response to iron overload in Hfe−/− animals, there is no stimulation of SMAD signaling and hepcidin levels are not increased.30,31 Thus, it appears that Hfe may function, at least in part, through the BMP-SMAD signaling pathway, but the implied functional dependence on Hjv has not been demonstrated experimentally.
To further interrogate the role of the Hfe in vivo, we generated Hfe transgenic mice that express Hfe lacking its short cytoplasmic domain and Hfe carrying a C294Y missense mutation. Transgenic lines were bred to animals lacking either endogenous Hfe or Hjv. Our results indicate that the extracellular and transmembrane domains of Hfe are sufficient for Hfe-mediated regulation of hepcidin in this overexpression model. However, loss of Hjv greatly attenuates Hfe-mediated hepcidin regulation.
Methods
Generation of transgenic mice expressing modified Hfe under control of the transthyretin (TTR) promoter
A full-length Hfe transgene (HfeWT) under the control of the transthyretin (TTR) promoter (pPJS095) was developed previously.20 The C294Y mouse mutation (equivalent to the human C282Y disease-causing mutation) was introduced into vector pPJS095 with the QuickChange site-directed mutatgenesis kit using primers mC294YF and mC294YR (HfeC294Y). A truncated 1020bp mouse Hfe cDNA lacking the final 19 amino acids (HfeΔCD) was amplified using the primers HFEtgRnotail and HFEtgFOR from a cDNA template contained in pcDNA3.1, and subcloned into pCR2.1-TOPO. The truncated 1032bp mouse Hfe cDNA with a cMyc epitope replacing the final 15 amino acids (HfeΔCD−MYC) of the protein was amplified using the primers HFEmycR and HFEtgFOR from a cDNA template contained in pcDNA3.1, and subcloned into pCR2.1-TOPO. In this second model 4 additional basic amino acids (RKRK) immediately after the transmembrane domain were left in place to increase the likelihood that the protein would localize to the cell membrane.32 Both fragments were liberated with SpeI and EcoRV. The 3′ overhangs were filled with Klenow enzyme, and the resulting sequence ligated into a blunted StuI site in the pTTR1exV3 vector (kind gift of Terry Van Dyke, University of North Carolina at Chapel Hill School of Medicine). Correct orientation and sequence were confirmed by sequencing analysis. The excised HindIII fragments were purified from the vectors by electroelution and microinjected into C57BL/6NCrl oocyte pronuclei at the Children's Hospital Boston Center for Molecular Developmental Hematopoiesis and then bred onto the C57BL6/J background. These mice were bred with an Hfe−/− strain of the same genetic background to generate Hfe−/− animals carrying the integrated TTR-Hfe transgene. Mice expressing the full-length Hfe or HfeΔCD−MYC truncated transgene on a C57BL6/J background were bred to Hjv−/− animals on a 129SvEv/Tac background (N3 generation final). All animals were heterozygous for each Hfe transgene.
Oligonucleotide primers
Oligonucleotide primers employed in the generation of transgenic mice, for PCR genotyping and for quantitative PCR analysis are listed in supplemental Table 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Animal care and analysis
All genetically modified mice were born and housed in the barrier facility at the Children's Hospital Boston and handled according to Institutional Animal Care and Use Committee–approved protocols. Animals were maintained on the Prolab RMH 3000 diet (Lab Diet, 380ppm iron). The facility employs a constant dark-night light cycle, and all animals were provided both water and food ad libitum. Due to differences in iron metabolism between male and female animals, only females were analyzed. All animals were euthanized and analyzed at 8 weeks of age.
PCR genotyping
Puregene DNA isolation (Gentra Systems) or DNeasy Blood & Tissue kits (QIAGEN) were used to prepare genomic DNA from tail snips. Transgenic TTR-Hfe mice were genotyped by polymerase chain reaction (PCR) with primers PS-133 and PS-134 to yield a 450bp band. HFENeo, HFEKO, Map3, and Map4 were also used to genotype mice by PCR, yielding a 230bp wild-type (WT) band and a 200bp Hfe knockout (KO) allele. Hjv−/− mice were genotyped by PCR using primers FH07, FH08, FH03 and FH04 yielding a 500bp WT band and a 250bp KO band. Tfr2Y245X/Y245X mice were genotyped by PCR using primers TFR2 Y245XF and TFR2 Y245XR yielding a 814bp WT band and a 922bp KO band (supplemental Table 3).
Immunohistochemisty
Liver tissue samples were fixed in 10% buffered formalin for 24 hours and then embedded in paraffin. Formalin-fixed paraffin embedded tissue sections were mounted on microscope slides. Immunohistochemical staining was optimized using a Ventana Discovery XT automated immunohistochemistry slide processing platform according the manufacturer's instructions (Ventana Medical Systems). After the Closed Loop Assay Development (CLAD) protocol (Ventana Medical Systems), c-Myc immunoreactivity (rabbit anti-cMyc, Abcam) was optimized using the OmniMap DAB anti–Rabbit (HRP) detection kit (Ventana Medical Systems). Standard quality control procedures were undertaken to optimize antigen retrieval, primary antibody dilution, secondary antibody detection and other factors for both signal and noise. The Children's Hospital Boston Pathology Research Core Laboratory performed immunohistochemical procedures. Brightfield images were acquired using 40×/0.75 or 20×/0.5 objective lenses and 2× camera adaptor (final magnification of 80× or 40×, respectively) of a BX50 microscope with DP25 Digital Camera employing DP2-BSW Version 2.1 software (Olympus).
Blood and tissue iron analysis
Whole blood for complete blood counts was collected retro-orbitally into EDTA-coated microtainer tubes (Becton Dickinson) from animals anesthetized with Avertin. Samples were analyzed on an Avida 120 analyzer (Bayer) by the Clinical Core Laboratories located at Children's Hospital Boston. Whole blood for other purposes was collected by retro-orbital bleeding into serum separator tubes (Becton Dickinson), and serum was prepared according to the manufacturer's instructions. Serum iron values were determined with the Serum Iron/UIBC kit (Thermo Fisher) according to the manufacturer's instructions. Liver and spleen tissues were collected and tissue non-heme iron concentrations were determined as described previously.33
Cell culture
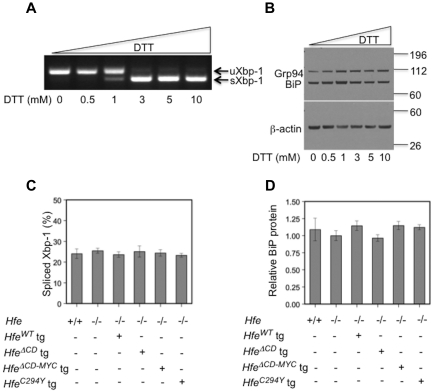
Mouse embryonic fibroblast (MEF) cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 1% l-glutamine (Cellgro), 10% heat-inactivated fetal bovine serum (Hyclone) and 1% penicillin/streptomycin (Cellgro). Cultured cells were grown at 37°C in 5% CO2. For ER stress control experiments, MEF cells were seeded and grown to near confluence in 10-mm cell culture dishes, and transferred to fresh media and treated with increasing concentrations (0-10 mM) of dithiothreitol (DTT; Sigma) for 6 hours.
RNA extraction, RT-PCR, semiquantitative and quantitative PCR
Total liver RNA was isolated from flash-frozen tissue with TRIzol (Invitrogen) or from cells using RNeasy mini kit (QIAGEN). Total RNA was treated with DNase I (Roche) to remove contaminating genomic DNA. cDNA was synthesized from the resulting RNA using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's protocol. Real-time quantification of hepcidin (Hamp) and β-actin,20 and Id1 and Bmp634 mRNA transcript levels was performed as described previously. Semiquantitative reverse transcription (RT)–PCR was employed to measure unspliced (205 bp) or spliced (179 bp) Xbp-1 mRNA forms in harvested MEF cells and liver samples. PCR products from 50 ng of starting cDNA were separated on 2% agarose gels and liver samples were quantified with ImageJ 1.42q software (NIH). All primers are listed in supplemental Table 3.
Immunoblot analysis
Tissues or cells were manually lysed in modified RIPA buffer (50mM Tris pH 7.5, 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with Complete Mini, EDTA-free protease inhibitor cocktail tablets (Roche). Cell debris was removed by centrifugation. 50, 100, or 150 μg of total liver tissue protein or 25 μg cell lysate protein was diluted in 2X Laemmli buffer (0.2 M DTT final), boiled and electrophoresed on precast 10% Criterion polyacrylamide (Bio-Rad) or 4%-12% NuPage (Invitrogen) gels. Proteins were transferred onto nitrocellulose membranes and immunoblot analysis was performed using rabbit anti–mouse Hfeα3 (1:250, a gift from Alain Townsend, University of Oxford Weatherall Institute of Molecular Medicine), rabbit anti-cMyc (1:1000, Abcam), rabbit anti–mouse Tfr2 (1:1000, Alpha Diagnostic), mouse anti–human TFR1 (1:500, Zymed), mouse anti–rat KDEL [10C3] (1:1000, Abcam) or rabbit anti–β-actin (1:1000, Cell Signaling). Blots were then incubated with anti–rabbit or anti–mouse (1:5000) secondary antibody conjugated to horseradish peroxidase and then subjected to chemiluminescence (Amersham, ECL) per the manufacturer's directions. Between immunoblot analyses, blots were stripped with ReBlot Strong (Chemicon). BiP protein expression relative to β-actin protein was quantified using ImageJ 1.42q software (NIH).
Results
The Hfe cytoplasmic domain is not essential for induction of hepcidin expression
Earlier studies suggested that the cytoplasmic domain might be important for HFE function and/or trafficking. We previously demonstrated that hepatocyte-specific expression of WT Hfe cDNA under the control of the transthyretin (TTR) promoter (HfeWT) prevented iron overload in Hfe−/− animals, and, in fact, resulted in iron deficiency attributable to increased hepcidin expression.20 To assess whether the cytoplasmic domain of Hfe is necessary for Hfe-mediated induction of hepcidin expression, we performed similar experiments employing truncated Hfe transgenes.
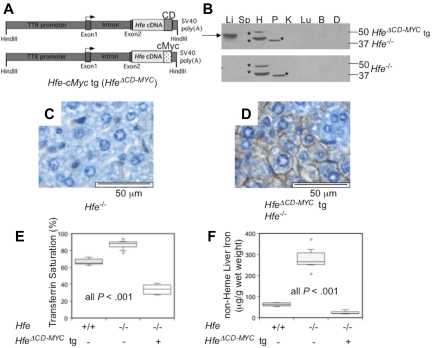
First, we constructed a TTR-Hfe transgene that encodes a protein lacking the entire 19-amino acid, C-terminal cytoplasmic domain, leaving the extracellular and transmembrane portions intact (HfeΔCD; supplemental Figure 1A). Unfortunately, it was not possible to detect endogenous Hfe with available antibodies, and we were concerned that we would not be able to visualize a truncated protein product. To circumvent this problem, we created a second truncated transgene (HfeΔCD−MYC) in which the cytoplasmic domain of Hfe was replaced with a c-Myc epitope tag (Figure 1A). Mouse lines carrying each transgene were bred to Hfe−/− animals of the same genetic background and analyzed for perturbations in iron homeostasis. We found that animals carrying the HfeΔCD−MYC transgene expressed high levels of chimeric protein in the liver (Figure 1B) and modest amounts in the pancreatic islets of Langerhans (supplemental Figure 2A), consistent with the known activity of the TTR promoter.35 In hepatocytes, the HfeΔCD−MYC chimeric protein localized to the basolateral membrane (Figure 1C-D). This is consistent with previous observations that functional wild-type HFE protein is localized to the cell surface.10
Figure 1.
Phenotypic analysis of mice expressing a truncated hepatocyte-specific Hfe-cMyc transgene (HfeΔCD−MYC tg). Schematic drawing of the HfeΔCD−MYC tg (A) substituting a c-Myc epitope (cMyc) for the Hfe cytoplasmic domain (CD). (B) Hfe-cMyc protein in the liver (Li), spleen (Sp), heart (H), pancreas (P), kidney (K), lung (Lu), brain (B), and duodenum (D) of 8 week-old Hfe−/− HfeΔCD−MYC tg animals (top panel) and Hfe−/− animals (bottom panel) analyzed by Western blot. Hfe-cMyc (arrow) and non-specific bands (*) are noted. Liver immunohistochemistry for cMyc epitope in Hfe−/− (C) and Hfe−/− HfeΔCD−MYC (D) animals (magnification 80X). Box plots depicting the (E) serum transferrin saturation (%), and (F) non-heme liver iron (μg/g wet weight). The bar within the box represents the median, while the top and bottom of the box are the 75th and 25th percentiles, respectively. The top and bottom whiskers depict the 90th and 10th percentiles, respectively. Data points outside of the 10th and 90th percentiles are drawn as circles. WT (n = 7), Hfe−/− (n = 12) and Hfe−/− HfeΔCD−MYC (n = 6) are depicted. P values were calculated with Microsoft Excel 2008 Version 12.2.6 (Student t test).
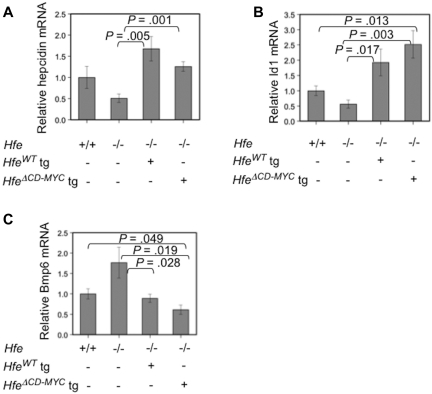
We compared the phenotypes of Hfe−/− animals with and without liver-specific expression of C-terminally truncated Hfe transgenes. We found that both truncated Hfe transgenes overcorrected the Hfe−/− iron loading phenotype, and caused iron deficiency (supplemental Figure 1B-C, Figure 1E-F). compared with Hfe−/− animals, this was associated with a normalization of hematologic parameters in HfeΔCD (supplemental Table 1) and a hypochromic, microcytic anemia in HfeΔCD−MYC mice (Table 1). Specifically, Hgb, MCV, and MCH were increased significantly in Hfe−/− compared with WT animals and these parameters decreased upon expression of HfeΔCD (supplemental Table 1). The results were even more dramatic in HfeΔCD−MYC expressing mice; Hgb, MCV, MCH, and CHr were decreased significantly compared not only to Hfe−/− but also WT animals (Table 1). These results are similar to our previous observations in Hfe−/− mice expressing a wild-type Hfe transgene,20 and comparable with mice with liver-specific transgenic expression of hepcidin itself.36,37 Compared with Hfe−/− animals, Hfe−/− mice expressing the HfeΔCD transgene demonstrated a trend toward increased hepcidin mRNA (data not shown). Hfe−/− mice carrying either the previously described HfeWT20 or HfeΔCD−MYC transgenes expressed significantly more hepcidin mRNA than Hfe−/− mice (Figure 2A), providing an explanation for the more severe anemia observed in these animals (Table 1 and Schmidt et al20).
Table 1.
Hematologic features of Hfe−/− HfeΔCD−MYC transgenic animals
| Genotype | n | Hgb (g/dL) | Hct (%) | MCV (fl) | MCH (pg) | RDW (%) | Retic (%) | Chr (pg) |
|---|---|---|---|---|---|---|---|---|
| WT | 7 | 14.8 ± 0.2 | 48.2 ± 0.6 | 50.1 ± 0.6 | 15.4 ± 0.2 | 13.0 ± 0.2 | 3.2 ± 0.1 | 15.7 ± 0.1 |
| Hfe−/− | 12 | 15.4 ± 0.1* | 48.6 ± 1.2 | 53.9 ± 0.5† | 17.2 ± 0.4§ | 12.6 ± 0.2 | 3.3 ± 0.2 | 16.9 ± 0.1† |
| Hfe−/−HfeΔCD−MYC tg | 6 | 12.6 ± 0.3†¶ | 43.7 ± 2.6 | 41.4 ± 1.1†¶ | 12.1 ± 0.4†¶ | 20.8 ± 0.6†¶ | 4.4 ± 0.1§‡ | 12.9 ± 0.4†¶ |
The red blood cell parameters hemoglobin (Hgb), hematocrit (Hct), mean cell volume (MCV), mean cell hemoglobin (MCH), red cell distribution width (RDW), reticulocyte count (Retic), and reticulocyte mean cell hemoglobin (CHr) were measured in 8-week-old female wild-type (WT), Hfe−/−, and Hfe−/− HfeΔCD−MYC tg mice. Data are presented as mean ± SEM. P values were calculated by the Student t test using Microsoft Excel 2008 Version 12.2.6.
P < .05 versus WT.
P < .01 versus WT.
P < .001 versus WT.
P < .01 versus Hfe−/−.
P < .001 versus Hfe−/−.
Figure 2.
Analysis of BMP signaling in Hfe−/− mice expressing hepatocyte-specific Hfe transgenes. Total mRNA was harvested from wild-type (WT), Hfe−/−, Hfe−/− Hfe-full length transgene (HfeWT) or Hfe−/− Hfe-cMyc transgenic (HfeΔCD−MYC tg) livers (n = 5 for each genotype) and hepcidin (Hamp; A), Id1 (B), and Bmp6 (C) mRNA was assessed by quantitative real-time PCR, normalized to β-actin (Actb), and then expressed relative to the WT value whose mean was defined as 1.0. Ratios are expressed ± SEM.
Signaling through the Bmp pathway is decreased relative to liver non-heme iron content in Hfe−/− mice.30,31 Considering the inappropriately low hepcidin mRNA levels and elevated non-heme liver iron concentration in Hfe−/− mice, and inappropriately high hepcidin mRNA levels and depressed non-heme liver iron concentration in all of the transgenic animals, we predicted that Id1 mRNA, a transcriptional target of Bmp signaling,28,29 would be decreased in Hfe−/− animals and elevated in both HfeWT and HfeΔCD−MYC transgenic mice. Accordingly, we found that overexpression of either of these Hfe transgenes increased Id1 mRNA expression (Figure 2B). As was observed with hepcidin induction, this increase reached significance for both the HfeWT and HfeΔCD−MYC-transgenic lines compared with Hfe−/− alone. Conversely, it has been shown that Bmp6 mRNA levels are increased in response to increased iron stores resulting from either increased dietary iron or loss of endogenous Hfe.30,34 Correspondingly, we observed an increase in Bmp6 mRNA levels in Hfe−/− mice (Figure 2C). Furthermore, mice expressing either of these Hfe transgenes had significantly decreased Bmp6 mRNA expression compared with Hfe−/− animals.
Overexpression of Hfe or Hfe mutant proteins does not initiate an unfolded protein or ER stress response
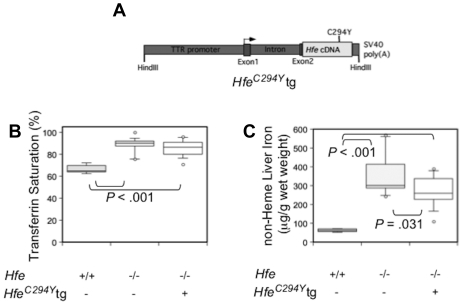
It is possible that overexpressed Hfe might lead to hepcidin induction by a mechanism distinct from that of endogenous Hfe. It was recently reported that hepcidin can be induced through the unfolded protein response (UPR) and endoplasmic reticulum stress response.38,39 The common human HFE C282Y (murine C294Y) disease-causing mutation alters the secondary structure of HFE, rendering it unable to reach the cell surface,10 and induces UPR and ER stress in cell culture systems.40 To test the hypothesis that it might have a similar effect in vivo, we created TTR-Hfe C294Y transgenic animals (HfeC294Y, Figure 3A) and analyzed parameters of iron metabolism. We bred HfeC294Y transgenic animals to Hfe−/− mice of the same genetic background, and compared Hfe−/− mice carrying the HfeC294Y transgene to Hfe−/− littermates. In contrast to Hfe−/− mice expressing the HfeWT transgene,20 we found that HfeC294Y animals did not have decreased transferrin saturation, and had similar liver iron overload compared with Hfe−/− animals (Figure 3B-C). In agreement with these findings, hepcidin levels in Hfe−/− HfeC294Y animals were not significantly elevated compared with Hfe−/− littermates (data not shown) and complete blood counts revealed no significant changes (supplemental Table 1).
Figure 3.
Phenotypic analysis of mice expressing a mutated (C294Y) hepatocyte-specific Hfe transgene (HfeC294Y tg). Schematic drawing of the HfeC294Y tg construct (A). (B-C) Box plots depicting the measurement of serum transferrin saturation (%; B), and non-heme liver iron (μg/g wet weight; C). WT (n = 7), Hfe−/− (n = 13) and Hfe−/− HfeC294Y tg (n = 16), are depicted in box plots as in Figure 1.
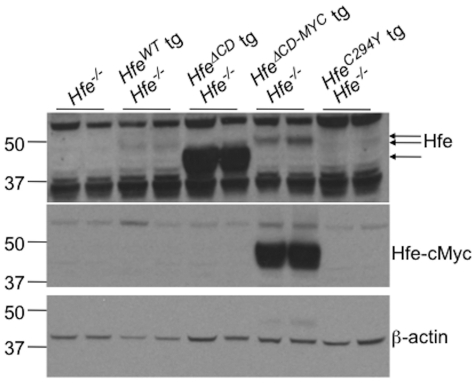
To assess whether differences observed with each of the transgenes was related to overexpression, and hence the potential to induce the UPR or ER stress response, we measured total liver Hfe mRNA by quantitative RT-PCR. We found that Hfe mRNA was increased in all transgenic lines compared with both WT and Hfe−/− mice but that there were no significant differences in Hfe expression between the HfeΔCD and HfeΔCD−MYC or the HfeWT and HfeC294Y strains (supplemental Figure 3). Although it was not possible to detect endogenous Hfe, we were able to compare transgene protein expression levels (Figure 4 top panel). HfeWT, HfeΔCD−MYC or HfeC294Y animals expressed similar amounts of Hfe protein; however, the HfeΔCD protein product was greatly overexpressed. We do not yet understand why HfeΔCD transgenic animals express such a high level of Hfe protein relative to mRNA expression. Expression of the Hfe-cMyc chimeric protein was also confirmed by blotting with an anti-cMyc antibody (Figure 4 middle panel).
Figure 4.
Analysis of Hfe transgene expression. Liver protein lysates were analyzed for Hfe (top panel) and Hfe-cMyc protein (middle panel) in 8-week-old wild-type (WT), Hfe−/−, Hfe−/− HfeC294Y, Hfe−/− Hfe-truncated transgenic (HfeΔCD tg), or Hfe−/− Hfe-cMyc transgenic (HfeΔCD−MYC tg) animals by Western blot. Equivalent loading of liver lysates was confirmed by immunoblot analysis for β-actin (bottom panel).
Having established that transgene-expressed protein levels were comparable in most of the lines, we evaluated the possibility that 1 or more of the variant Hfe proteins induced hepcidin through a cellular stress response. Appearance of the spliced form of the transcription factor Xbp-1 has been shown to be an indicator of ER stress.41 Treatment of mouse embryonic fibroblast (MEF) cells with increasing concentrations of dithiothrietol (DTT) induced ER stress and led to the appearance of an alternatively spliced form of Xbp-1 (Figure 5A). BiP (Grp78), an ER chaperone protein that is a key regulator of the UPR response and is induced by ER stress,39,42 was also increased by treatment with DTT (Figure 5B). In vivo, overexpression of the HfeWT, HfeΔCD, HfeΔCD−MYC or HfeC294Y –transgenes, however, did not lead to an increase in either Xbp-1 mRNA splicing or BiP protein expression (Figure 5C-D). Most significantly, in contrast to previous in vitro observations,40 gross overexpression of the HfeΔCD protein product, or overexpression of the C294Y Hfe missense mutation in vivo, does not cause measurable ER stress in mouse livers. We conclude that the elevated hepcidin level in these mice is not caused by ER stress.
Figure 5.
Hfe transgenes do not induce endoplasmic reticulum (ER) stress or the unfolded protein response (UPR). Dithiothreitol (DTT) induction (A-B) of the endoplasmic reticulum (ER) stress response in mouse embryonic fibroblast (MEF) cells. (A) Semiquantitative RT-PCR for unspliced (205 bp, uXbp-1) or spliced (179 bp, sXbp-1) Xbp-1 mRNA forms after treatment with increasing concentrations of DTT that induces ER stress. Total cell lysates (B) were analyzed for the KDEL motif-containing proteins Grp94 (Hsp90b1) and BiP (Hspa5) in MEF cells by Western blot. Equivalent loading of liver lysates was confirmed by immunoblot analysis for β-actin. ER stress in transgenic mouse livers was evaluated by measuring Xbp-1 mRNA splicing (C) using semiquantitative RT-PCR (n = 3, results presented as percentage spliced) and protein expression of BiP (D, n = 3), an ER-resident chaperone, relative to β-actin. Mean protein expression for Hfe−/− mice was set as 1.0 and all other data were expressed in relation to this. (D-E) All comparisons are statistically not significant.
Hfe transgene-induced hepcidin expression occurs in the absence of detectable Tfr2
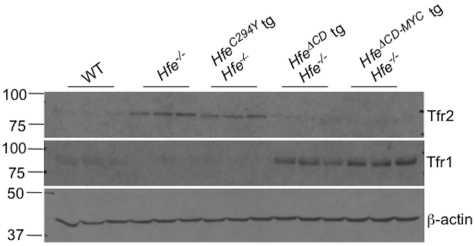
Exposure to elevated Fe2-Tf is associated with stabilization of TFR2 protein in both cell culture and animal models.20,43,44 Accordingly, both Hfe−/− and Hfe−/− HfeC294Y-transgenic animals, which have increased transferrin saturation, have increased hepatic Tfr2 expression (Figure 6 top panel). In contrast, Tfr1 was essentially undetectable by immunoblot in Hfe−/− and Hfe−/− HfeC294Y-transgenic animals, consistent with an appropriate response to hepatocellular iron overload (Figure 6 middle panel). Conversely, as expected in animals with decreased transferrin saturation and decreased non-heme liver iron concentration, both truncated Hfe transgenic lines had decreased Tfr2 and increased Tfr1 protein in the liver (Figure 6 top 2 panels). Increased Tfr1 and greatly decreased Tfr2 protein expression was also observed in animals overexpressing the HfeWT transgene.20 Previous work demonstrated that the interaction between Hfe and Tfr2 is promoted by exposure to Fe2-Tf21 and this complex was postulated to lead to hepcidin-induction. However, here we show that hepatocyte-specific expression of Hfe can induce hepcidin even in the absence of detectable levels of Tfr2 protein. This observation is concordant with other data in human patients and mouse models suggesting that TFR2 may not be essential for HFE-mediated induction of hepcidin.22,23 Our data indicated that an overexpressed, truncated form of Hfe lacking its cytoplasmic domain is sufficient for Hfe-dependent hepcidin induction in vivo.
Figure 6.
Tfr1 and Tfr2 protein expression in Hfe−/−Hfe-transgenic animals. Liver protein lysates were analyzed for Tfr2 (top panel) or Tfr1 protein (middle panel) in 8-week-old wild-type (WT), Hfe−/−, Hfe−/− HfeC294Y, Hfe−/− Hfe-truncated transgenic (HfeΔCD tg), or Hfe−/− Hfe-cMyc transgenic (HfeΔCD−MYC tg) animals by Western blot. Equivalent loading of liver lysates was confirmed by immunoblot analysis using anti–β-actin antibody (bottom panel).
Loss of Hjv greatly attenuates Hfe-mediated hepcidin induction
Homozygous mutations in HJV cause a more severe form of HH than do homozygous HFE mutations and are associated with markedly decreased hepcidin production.45 Accordingly, Hjv−/− mice develop more severe iron overload and produce much less hepcidin than do Hfe−/− mice.25,26,46,47 It is not yet known whether direct physical or indirect functional interactions between HJV and HFE are necessary to induce hepcidin. Considering the strong effect of wild-type and truncated Hfe transgenes in modulating hepcidin expression, we asked whether transgenic expression of Hfe could overcome the loss of Hjv in vivo.
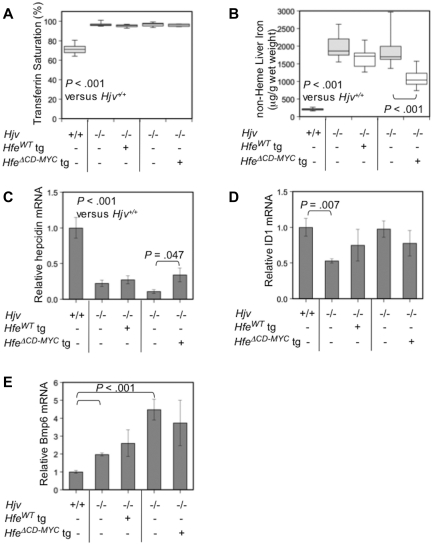
We bred animals expressing HfeWT and HfeΔCD−MYC transgenes to Hjv−/− mice and compared transgenic Hjv−/− animals to their Hjv−/− littermates. Neither Hfe transgene was able to prevent the greatly increased transferrin saturation and high liver iron concentration that are hallmarks of the Hjv−/− phenotype25 (Figure 7A-B). However, both Hfe transgenes were associated with decreased non-heme liver iron and this reduction was statistically significant for the HfeΔCD−MYC transgene. As expected, hematologic parameters were normal in all strains (supplemental Table 2).
Figure 7.
Loss of Hjv prevents expression of the Hfe-transgenic phenotype. Box plots depicting (A) serum transferrin saturation (%), and (B) non-heme liver iron (μg/g wet weight) in WT or Hjv−/− animals expressing either a full-length (HfeWT) or HfeΔCD−MYC tg. WT (n = 5), Hjv−/− (n = 8), Hjv−/− HfeWT transgene (n = 6), Hjv−/− (n = 9), Hjv−/− HfeΔCD−MYC tg (n = 8) are shown. Liver hepcidin (Hamp; C), Id1 (D), and Bmp6 (E) mRNA was analyzed and results were depicted as in Figure 2.
We anticipated that, similar to Hjv−/− mice, Hfe-transgenic animals lacking Hjv would express inappropriately low levels of hepcidin compared with their hepatic iron burden. We found that hepcidin expression in Hjv−/− animals carrying either of the Hfe transgenes was similar to that of Hjv−/− littermates, and greatly decreased compared with 129SvEv/Tac WT animals (Figure 7C). As expected in animals with iron overload and increased transferrin saturation, Tfr2 protein levels were increased, and Tfr1 was greatly diminished in the liver (supplemental Figure 5).
To examine signaling through the Bmp pathway in these Hjv−/− models, we determined Id1 mRNA levels (Figure 7D) and found a trend toward a decrease in all animals lacking endogenous Hjv. Conversely, in agreement with the elevated iron stores of Hjv−/− animals, Bmp6 mRNA expression (Figure 7E) increased in all Hjv−/− or Hjv−/− Hfe-trangenic animals in comparison to WT mice. Hfe cDNA was greatly overexpressed in all Hjv−/− Hfe-transgenic animals (data not shown). These results indicate that Hfe-mediated induction of hepcidin is highly dependent upon Hjv and that absence of either protein is associated with diminished Bmp pathway-dependent hepcidin induction.
Discussion
Hfe is required for the proper regulation of hepcidin production46–48 but its role in this process is not well understood. We sought to determine which portions of Hfe are necessary for induction of hepcidin expression, and to determine the functional relationship between Hfe and Hjv, in vivo. Two mouse models were developed expressing a truncated form of Hfe lacking its cytoplasmic C-terminus. A third mouse model was produced in which Hfe is predicted to be unable to fold properly. We demonstrated that Hfe lacking its cytoplasmic domain is sufficient for induction of hepcidin expression in hepatocytes. Our results recapitulate earlier observations showing that the C294Y mutation results in a severe loss of function. Our data indicate that hepcidin expression is not a consequence of ER stress induced by overexpression of a structurally normal or abnormal protein. However, Hfe-induced hepcidin expression is minimal in the absence of endogenous Hjv.
Each of our models has provided insight into the structural requirements for Hfe function, but our understanding is still incomplete in several ways. First, prior studies demonstrated that the α3 domain of Hfe is required to interact with Tfr2.43 Gao et al expressed a chimeric protein containing the HFE α3 and cytoplasmic domains fused to the transmembrane domain of HLA-B7 in HepG2 cells and showed that it was necessary and sufficient for Fe2-Tf-dependent induction of transcription from a hepcidin promoter construct.17 However, our in vivo results are distinctly different. Liver-specific expression of a truncated Hfe molecule lacking its 19 amino acid cytoplasmic domain or of a chimeric Hfe molecule substituting a cMyc epitope for the final 15 amino acids of the Hfe protein complemented Hfe deficiency. Furthermore, expression of the HfeΔCD−MYC transgene was associated with an increase in hepcidin transcription, iron deficiency and a hypochromic, microcytic anemia. We do not yet have an explanation for the differences between the in vitro results and our in vivo results, although it is possible that components of the Tf2-Fe sensing apparatus were not present in the cultured cells.
We also observed that HfeΔCD−MYC-transgenic mice had a more severe phenotype than HfeΔCD expressing animals, consistent with their greater liver hepcidin production. This was not due to higher expression of the HfeΔCD−MYC transgene. In fact, the HfeΔCD protein product was expressed at higher levels than the Hfe-cMyc chimeric protein. It is possible that HfeΔCD truncated protein associates less well with β-2-microglobulin than the HfeΔCD−MYC protein. Alternatively, it has been shown that basic amino acids immediately after the transmembrane domain of membrane proteins are necessary for proper insertion and topology.32 Thus, it is possible that loss of these amino acids in the HfeΔCD truncated protein leads to improper or decreased membrane insertion causing a diminution in protein activity at the cell membrane.
Overexpression of mutant HFE in cell culture triggers an unfolded protein response (UPR)40 and ER stress can induce hepcidin production.38,39 Using 2 separate methods we were unable to detect ER stress in any of the HFE overexpressing animals, including animals overexpressing an ortholog of the common C282Y human missense mutation that has been shown to induce ER stress in cell culture.40 It is possible that there is acute stress early in the development of the mice, but that cellular adaptations to chronic overexpression of proteins occur over time.
HJV is crucial for hepcidin production and hepcidin mRNA is nearly absent when HJV is absent.7,25,26 It is not yet clear, however, whether other known hepcidin regulators, including HFE and TFR2, act in concert with, or independently of HJV. HJV appears to be a primary regulator of hepcidin expression, whereas HFE and TFR2 seem to have secondary, modulatory roles. We observed that expression of Hfe transgenes in mice lacking endogenous Hjv does not rescue hepcidin or Id1 mRNA expression, indicating that HFE is not likely to function downstream of HJV in a signaling pathway. Nonetheless, non-heme liver iron levels do decrease in Hfe transgenic, Hjv−/− animals, suggesting the possibility that Hfe signaling is parallel to, rather than in series with, Hjv-dependent BMP signaling. Alternatively, it is possible that another protein plays a key role. Neogenin, a DCC (deleted in colorectal cancer) family member, interacts with Hjv49,50 and loss of endogenous neogenin expression leads to decreased cell-surface Hjv and iron overload due to decreased BMP-mediated hepcidin signaling.51 Although Lee and colleagues51 postulate that neogenin functions to stabilize Hjv on the cell surface, leading to increased hepcidin expression and decreased iron uptake, it is possible that overexpression of Hfe could cause BMP-mediated hepcidin induction through neogenin rather than Hjv. Finally, it is possible that HFE may also function to affect iron homeostasis independently of hepcidin. Overexpression of HFE in cell culture has been shown to inhibit iron export in colonic HT29 cells52 and inhibit iron uptake via down-regulation of Zip14 in HepG2 cells.53 HFE may also function in a non hepcidin-dependent manner in Kupffer cells of the recycling compartment.19 In any case, the observation that mice lacking Hjv or Hfe have inappropriately low hepcidin and Id1 mRNA expression relative to non-heme liver iron stores argues that Hjv- and Hfe-dependent signaling are, at a minimum, partially convergent on the BMP signal transduction cascade, if only indirectly.
In our studies, phenotypic parameters and hepcidin mRNA expression were measured at a single time point in mature animals. It is possible that overexpression of Hfe can induce minimal BMP-mediated hepcidin expression in the absence of the BMP co-receptor Hjv, and this might be sufficient over the lifetime of the Hfe transgenic animals to account for the observed influence on liver iron loading. Measurement of hepcidin over a longer time-course in animals with either genetically induced iron-overload or anemia may help to elucidate how Hfe-mediated hepcidin expression is modulated at various stages of development.
Based on these mouse models and the in vitro work by Gao, et al,17 we propose that the Hfe α3 domain is involved in Hfe-mediated hepcidin induction. The α3 extracellular domain has been shown to interact with Tfr2,43 but the requirement for this interaction in hepcidin induction is still uncertain. Furthermore, the inability of transgenically expressed Hfe to complement the iron overload in Hjv knockout animals demonstrates that Hfe function is at least in part dependent on BMP/SMAD signaling facilitated by Hjv. Finally, our transgenic animal models offer another useful tool to help elucidate the specific roles of various iron sensing and signaling proteins. Future work will be necessary to better understand the link between members of the TF-Fe2 sensing machinery, including HFE and TFR2, and elements of the BMP signal-transducing pathway such as BMP6 and HJV.
Supplementary Material
Acknowledgments
We thank Terry Van Dyke for the pTTR1exV3 vector, Alain Townsend for the anti–mouse HFEα3 serum, Robert Fleming for HfeY245X/Y245X animals, Yuko Fujiwara and the Children's Hospital Boston Center for Molecular Developmental Hematopoiesis for pronuclei microinjections (NIH P30 DK49216-14), Tom Bartnikas for technical advice and helpful suggestions, and members of the Andrews and Fleming laboratories for helpful discussions.
This work was supported by NIH R01 DK53813 (N.C.A.), NIH R01 DK080011 (M.D.F.), and NIH K01 DK074410 (P.J.S.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.J.S. conceived and designed the murine experiments, analyzed the data, and wrote the manuscript; and N.C.A. and M.D.F designed research and assisted in writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul J. Schmidt, PhD, Department of Pathology, Children's Hospital Boston, Enders Research Bldg, Rm 1122, 320 Longwood Ave, Boston, MA 02115; e-mail: pschmidt@enders.tch.harvard.edu.
References
- 1.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112(2):219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 3.Roetto A, Papanikolaou G, Politou M, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33(1):21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- 4.De Domenico I, Ward DM, Nemeth E, et al. The molecular basis of ferroportin-linkedhemochromatosis. Proc Natl Acad Sci U S A. 2005;102(25):8955–8960. doi: 10.1073/pnas.0503804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schimanski LM, Drakesmith H, Merryweather-Clarke AT, et al. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood. 2005;105(10):4096–4102. doi: 10.1182/blood-2004-11-4502. [DOI] [PubMed] [Google Scholar]
- 6.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105(4):1803–1806. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 7.Papanikolaou G, Samuels ME, Ludwig EH, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36(1):77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 8.Bridle KR, Frazer DM, Wilkins SJ, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361(9358):669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 9.Feder JN, Gnirke A, Thomas W, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 10.Feder JN, Tsuchihashi Z, Irrinki A, et al. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem. 1997;272(22):14025–14028. doi: 10.1074/jbc.272.22.14025. [DOI] [PubMed] [Google Scholar]
- 11.Feder JN, Penny DM, Irrinki A, et al. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci U S A. 1998;95(4):1472–1477. doi: 10.1073/pnas.95.4.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West AP, Jr., Giannetti AM, Herr AB, et al. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J Mol Biol. 2001;313(2):385–397. doi: 10.1006/jmbi.2001.5048. [DOI] [PubMed] [Google Scholar]
- 13.Gur H, Geppert TD, Wacholtz MC, Lipsky PE. The cytoplasmic and the transmembrane domains are not sufficient for class I MHC signal transduction. Cell Immunol. 1999;191(2):105–116. doi: 10.1006/cimm.1998.1417. [DOI] [PubMed] [Google Scholar]
- 14.Roy CN, Carlson EJ, Anderson EL, et al. Interactions of the ectodomain of HFE with the transferrin receptor are critical for iron homeostasis in cells. FEBS Lett. 2000;484(3):271–274. doi: 10.1016/s0014-5793(00)02173-6. [DOI] [PubMed] [Google Scholar]
- 15.Lizee G, Basha G, Jefferies WA. Tails of wonder: endocytic-sorting motifs key for exogenous antigen presentation. Trends Immunol. 2005;26(3):141–149. doi: 10.1016/j.it.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Jing SQ, Spencer T, Miller K, Hopkins C, Trowbridge IS. Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J Cell Biol. 1990;110(2):283–294. doi: 10.1083/jcb.110.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Chen J, Kramer M, Tsukamoto H, Zhang AS, Enns CA. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009;9(3):217–227. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vujic Spasic M, Kiss J, Herrmann T, et al. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7(2):173–178. doi: 10.1016/j.cmet.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Garuti C, Tian Y, Montosi G, et al. Hepcidin expression does not rescue the iron-poor phenotype of Kupffer cells in Hfe-null mice after liver transplantation. Gastroenterology. 2010;139(1):315–322. e311. doi: 10.1053/j.gastro.2010.03.043. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7(3):205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281(39):28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- 22.Pietrangelo A, Caleffi A, Henrion J, et al. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128(2):470–479. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 23.Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined deletion of Hfe and transferrin receptor 2 in mice leads to marked dysregulation of hepcidin and iron overload. Hepatology. 2009;50(6):1992–2000. doi: 10.1002/hep.23198. [DOI] [PubMed] [Google Scholar]
- 24.Babitt JL, Huang FW, Wrighting DM, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38(5):531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 25.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115(8):2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115(8):2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang RH, Li C, Xu X, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2(6):399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41(4):478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 29.Andriopoulos B, Jr., Corradini E, Xia Y, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41(4):482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corradini E, Garuti C, Montosi G, et al. Bone morphogenetic protein signaling is impaired in an Hfe knockout mouse model of hemochromatosis. Gastroenterology. 2009;137(4):1489–1497. doi: 10.1053/j.gastro.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kautz L, Meynard D, Besson-Fournier C, et al. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114(12):2515–2520. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- 32.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174(4):671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 33.Torrance JD, Bothwell TH. Tissue iron stores. In: Cook JD, editor. Methods in Hematology. Vol. 1. New York: Churchill Livingstone Press; 1980. pp. 104–109. [Google Scholar]
- 34.Kautz L, Meynard D, Monnier A, et al. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112(4):1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 35.Westermark GT, Westermark P. Transthyretin and amyloid in the islets of Langerhans in type-2 diabetes. Exp Diabetes Res. 2008;2008:429274. doi: 10.1155/2008/429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicolas G, Bennoun M, Porteu A, et al. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci U S A. 2002;99(7):4596–4601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy CN, Mak HH, Akpan I, Losyev G, Zurakowski D, Andrews NC. Hepcidin antimicrobial peptide transgenic mice exhibit features of the anemia of inflammation. Blood. 2007;109(9):4038–4044. doi: 10.1182/blood-2006-10-051755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vecchi C, Montosi G, Zhang K, et al. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325(5942):877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira SJ, Pinto JP, Picarote G, et al. ER stress-inducible factor CHOP affects the expression of hepcidin by modulating C/EBPalpha activity. PLoS ONE. 2009;4(8):e6618. doi: 10.1371/journal.pone.0006618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Almeida SF, Fleming JV, Azevedo JE, Carmo-Fonseca M, de Sousa M. Stimulation of an unfolded protein response impairs MHC class I expression. J Immunol. 2007;178(6):3612–3619. doi: 10.4049/jimmunol.178.6.3612. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279(25):25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Chloupkova M, Gao J, Chapman-Arvedson TL, Enns CA. HFE Modulates Transferrin Receptor 2 Levels in Hepatoma Cells via Interactions That Differ from Transferrin Receptor 1-HFE Interactions. J Biol Chem. 2007;282(51):36862–36870. doi: 10.1074/jbc.M706720200. [DOI] [PubMed] [Google Scholar]
- 44.Johnson MB, Chen J, Murchison N, Green FA, Enns CA. Transferrin receptor 2: evidence for ligand-induced stabilization and redirection to a recycling pathway. Mol Biol Cell. 2007;18(3):743–754. doi: 10.1091/mbc.E06-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papanikolaou G, Tzilianos M, Christakis JI, et al. Hepcidin in iron overload disorders. Blood. 2005;105(10):4103–4105. doi: 10.1182/blood-2004-12-4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muckenthaler M, Roy CN, Custodio AO, et al. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34(1):102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- 47.Nicolas G, Viatte L, Lou DQ, et al. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet. 2003;34(1):97–101. doi: 10.1038/ng1150. [DOI] [PubMed] [Google Scholar]
- 48.Vujic Spasic MV, Kiss J, Herrmann T, et al. Physiologic systemic iron metabolism in mice deficient for duodenal Hfe. Blood. 2007;109(10):4511–4517. doi: 10.1182/blood-2006-07-036186. [DOI] [PubMed] [Google Scholar]
- 49.Zhang AS, West AP, Jr., Wyman AE, Bjorkman PJ, Enns CA. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J Biol Chem. 2005;280(40):33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- 50.Yang F, West AP, Jr., Allendorph GP, Choe S, Bjorkman PJ. Neogenin interacts with hemojuvelin through its two membrane-proximal fibronectin type III domains. Biochemistry. 2008;47(14):4237–4245. doi: 10.1021/bi800036h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DH, Zhou LJ, Zhou Z, et al. Neogenin inhibits HJV secretion and regulates BMP induced hepcidin expression and iron homeostasis. Blood. 2010;115(15):3136–3145. doi: 10.1182/blood-2009-11-251199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies PS, Enns CA. Expression of the hereditary hemochromatosis protein HFE increases ferritin levels by inhibiting iron export in HT29 cells. J Biol Chem. 2004;279(24):25085–25092. doi: 10.1074/jbc.M400537200. [DOI] [PubMed] [Google Scholar]
- 53.Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem. 2008;283(31):21462–21468. doi: 10.1074/jbc.M803150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.