Abstract
Bifunctional DNA alkylating agents form a diverse assortment of covalent DNA interstrand crosslinked (ICL) structures that are potent cytotoxins. Since it is implausible that cells could possess distinct DNA repair systems for each individual ICL, it is believed that common structural and dynamic features of ICL damage are recognized, rather than specific structural characteristics of each cross-linking agent. Investigation of the structural and dynamic properties of ICLs that might be important for recognition has been complicated by heterogeneous incorporation of these lesions into DNA. To address this problem we have synthesized and characterized several homogenous ICL-DNAs containing site–specific staggered N4-cytosine-ethyl-N4-cytosine crosslinks. Staggered crosslinks were introduced in two ways: in a manner that preserves the overall structure of B-form duplex DNA, and in a manner that highly distorts the DNA structure, with the goal of understanding how structural and dynamic properties of diverse ICL duplexes might flag these sites for repair. Measurements of base pair opening dynamics in the B-form ICL duplex by 1H NMR linewidth or imino proton solvent exchange showed that the guanine base opposite to the crosslinked cytosine opened at least an order of magnitude more slowly than when in a control matched normal duplex. To a lesser degree, the B-form ICL also induced a decrease in base pair opening dynamics that extended from the site of the crosslink to adjacent base pairs. In contrast, the non-B-form ICL showed extensive conformational dynamics at the site of the cross link, which extended over the entire DNA sequence. Since DNA duplexes containing the B-form and non-B-form ICL crosslinks have both been shown to be incised when incubated in mammalian whole cell extracts, while a matched normal duplex is not, we conclude that intrinsic DNA dynamics is not a requirement for specific damage incision of these ICLs. Instead, we propose a general model where destabilized ICL-duplexes serve to energetically facilitate binding of DNA repair factors that must induce bubbles or other distortions in the duplex. However, the essential requirement for incision is an immobile Y-junction where the repair factors are stably bound at the site of the ICL, and the two DNA strands are unpaired.
Covalent interstrand DNA cross links (ICLs) between complementary stands of the DNA double helix constitute a major and particularly cytotoxic class of DNA damage (1). As these crosslinks affect both DNA strands, the redundant information normally safeguarded by the DNA double helix is jeopardized, and cellular repair processes can induce double strand breaks and other potentially deleterious lesions in the process of removing the lesion (2, 3, 4). These highly toxic ICLs primarily disrupt rapidly dividing cells as they pose physical obstacles to strand separation at the replication fork and to transcriptional bubble formation. Consequently, structurally diverse agents such as psoralen, mitomycin C, cisplatin, and nitrogen mustards that induce ICLs have become important chemotherapeutics and frontline weapons against many types of cancer (1, 2, 3).
ICL repair in mammalian cells proceeds via several overlapping pathways, and the precise repair mechanism depends upon the nature of the crosslink, and the stage of the cell cycle when the damage is discovered (2, 5, 6). Prominent among these pathways is replication-dependent repair (S phase), where the ICL poses a physical barrier to the progressing replication fork, and stalled replication complexes lead to the recruitment of repair machinery that initiates repair via ICL-induced double-strand breaks (5). Similarly, ICLs encountered at or near actively transcribed regions of the genome during Go or G1 stages of the cell cycle can prevent transcriptional bubble formation and stall transcriptional machinery (2, 3, 7). Such transcription-coupled repair (TCR) processes involve recruitment of nucleotide excision repair (NER) factors to the stalled complex (8, 9, 10, 11). Finally, ICL detection in Go or G1 can also occur in unperturbed DNA in a process known as the global genome repair (GGR) pathway of NER, which occurs in the absence of transcription (7, 10, 11, 12). The GGR pathway has only been completely worked out in prokaryotes, and involves NER-dependent incision events on a single DNA strand on both the 5′ and 3′ sides of the ICL resulting in “unhooking” of the crosslink. However, previous studies using mammalian cell extracts showed that NER only leads to dual incisions on the 5′ side of the ICL that do not result in crosslink unhooking (Figure 1) (13).
Figure 1.
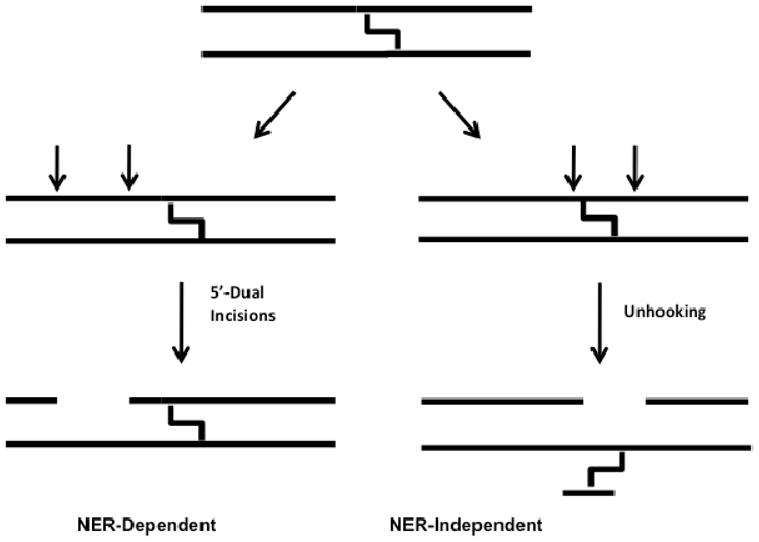
Schematic illustrating NER-dependent and -independent processing of ICLs observed in mammalian cell extracts (32). Interstrand crosslinks prevent mammalian NER enzymes from making incisions surrounding the crosslink. Instead, unknown repair enzyme activities are responsible for unhooking the two strands.
Recently there has been increased interest in the process of ICL detection using model ICL-containing duplexes that serve as defined substrates for DNA repair factors present in mammalian cells(Fig. 1) (11, 14, 15). A model ICL that has been very useful in ex vivo repair studies is one in which the exocyclic N4 nitrogen atoms of staggered cytosine bases on opposite strands of the DNA duplex are connected by an ethyl covalent linker (Fig. 2). This defined crosslink can be site-specifically incorporated using a convertible nucleoside approach and has been extensively characterized using structural and biochemical methodology (2, 16, 17, 18). Due to the inherent anti-parallel nature of the DNA duplex, two different orientations of the staggered CC ICL can be generated, one in which the crosslink is placed in a 5′-CG-3′ sequence and one in which the crosslink is placed in a 5′-GC-3′ sequence. Because of the relative geometries of the staggered cytosine bases in these two forms, only the 5′-CG-3′ orientation allows an ethyl linker geometry that optimally positions the N4 atoms of the linked cytosine bases to maintain normal Watson-Crick hydrogen bonding contacts with their cross strand partner guanine, thereby maintaining the B-form DNA structure (Fig. 2b). 5′-CG-3′ ICL-DNA has been structurally characterized in different sequence contexts using x-ray crystallography, solution state NMR spectroscopy, and atomic force microscopy, and found to adopt a structure that is essentially identical to canonical B-form DNA (16, 18, 19). In contrast, the non-optimal 5′-GC-3′ ICL-DNA form has been found to have a highly distorted duplex structure (16).
Figure 2.
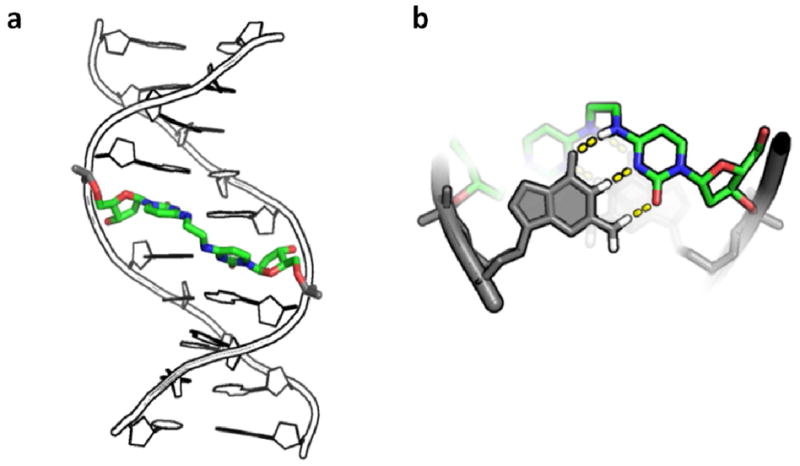
Structure of DNA duplex containing the N4C-ethyl-N4C crosslink. (a) Crystallographic model of duplex DNA (pdb accession name 2OKS) containing a central staggered 5′–CG-3′ N4C-ethyl-N4C interstrand crosslink in the major groove. (b) Structural detail of crosslinked cytosines looking down the DNA helical axis. Dashed lines denote the Watson-Crick hydrogen bonding groups of cytosine that are free to pair with an opposing guanine base shown in grey. For reference, the DNA major and minor grooves are located at the top and bottom of the structure, respectively.
When these staggered CC ICL duplexes were exposed to mammalian cell extracts it was observed that the disordered 5′-GC-3′ construct and the B form 5′-CG-3′ ICL-DNA were incised in two distinct ways (14, 20), but a matched normal DNA was not incised. The first set of incisions occurred on a single strand, 5′ to the ICL and produced a set of oligonucleotides 24–32 nucleotides in length (Fig. 1), similar to those previously reported by Sancar and coworkers (13). The second set of apparent incisions appeared on both the 5′ and 3′ sides of the ICL and led to unhooking of the ICL (Fig. 1) 2. Through the use of extracts derived from cells that were deficient in NER enzymes, it was established that the dual 5′ incisions require NER, while the highly-specific 5′ and 3′ incision activity that leads to unhooking is novel, and not attributable to known DNA repair pathways (14). The disordered 5′-GC-3′ ICL was unhooked with an initial rate that was 6-fold grater than that of the B-form 5′-CG-3′ ICL-DNA, suggesting that DNA distortion plays a role in detection, however the remarkable aspect of both NER-dependent and independent incision events is that the normal matched DNA duplex is never detectably incised (14). Thus, subtle differences in the structure and/or dynamic properties of the B form ICL and the normal duplex must give rise to incision specificity.
While the gross structural distortions provided by the 5′-GC-3′ ICL could lead to its preferred incision (14, 16), the detection of specific incision events in the structurally undistorted 5′-CG-3′ ICL duplex demonstrates that structural distortions can not be the sole determining factor guiding incision. Since structural methods only yield information on the time-averaged conformation of a macromolecule, they are insensitive to short-lived high-energy states that may be present and critical to biological recognition (21, 22, 23, 24). Here we use NMR imino proton exchange measurements to probe the dynamics of B-form and non-B-form duplexes containing staggered CC ICLs. We find that the intrinsic dynamic properties of these ICL-DNAs do not provide an adequate explanation for their specific endonucleolytic incision by DNA repair factors in mammalian cell extracts. The findings suggest that the covalent nature of the two linked DNA strands is the essential determining feature for incision of both ICL’s, and that the differences in the dynamic and structural properties of the two ICL duplexes plays a lesser role. A model for recognition and incision is proposed based on these findings.
Material and Methods
Synthesis of DNA and Sample Preparation
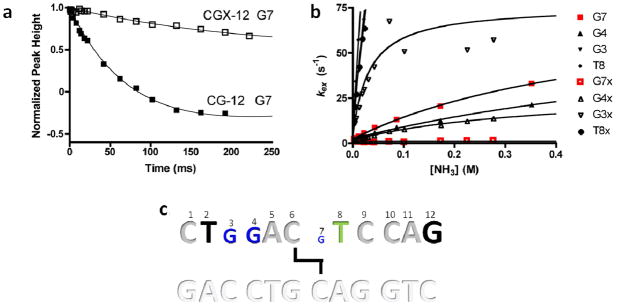
Six palindromic DNA sequences were used in this work (Fig. 3). Three sequences contained a central ethyl linkage between the N4 positions of C6/7 on opposite strands and were synthesized as previously described (17). Non-crosslinked DNA was purchased from Integrated DNA Technologies. The self-complementary DNA oligos were purified by strong anion exchange chromatography HPLC (Dionex PA-100) and desalted using reversed phase HPLC (Aqua column, Phenomenex). The desalted DNA was dried and dissolved in 10 mM sodium phosphate 75 mM sodium chloride, 0.05% sodium azide, and 10 % deuterium oxide for investigation by NMR. Assignment experiments on the DNA were conducted at pH 7.0 to minimize imino exchange rates, but the pH was elevated to 9.2 for the imino exchange rate measurements to ensure significant quantities of added ammonia would be in the active NH3 form.
Figure 3.
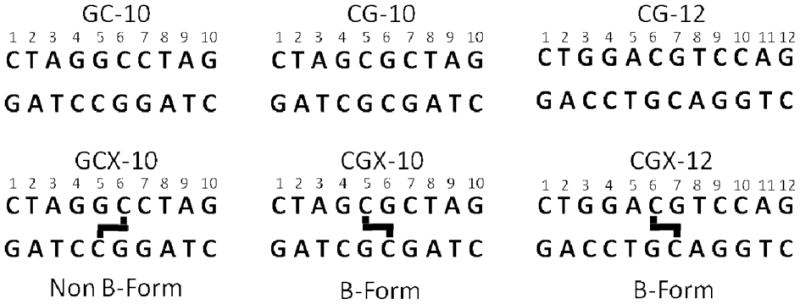
The six palindromic DNA sequences used in this study (top strand 5′ to 3′). Thick black lines indicate the location of ethyl linkages between staggered cytosine exocyclic N4 nitrogens.
Measurement of Imino Exchange Rates
All NMR experiments were preformed on Varian Invoa systems operating at 600 or 800 MHz proton Larmor frequency. Imino exchange rates were measured using water inversion followed by subsequent observation of the imino protons after a variable mixing time as has been widely described elsewhere (25). Water inversion was accomplished with a ISNOB3 pulse shape of approximately 300 μs duration (26) selectively applied to the water resonance. A variable delay time ranging from 500 μs to 10 s was used to measure exchange rates prior to detection of the imino proton signal with a refocused jump-return sequence. Unavoidable imperfections in water selective inversion pulses lead to significant radiation dampening, particularly on high Q-factor cryogenic probes and at longer exchange delay times. To prevent this phenomenological water relaxation rate from contaminating inversion recovery measurements, step gradients were employed during the mixing time (300 usec 12 G/cm gradient immediately after the water inversion pulse and a 0.3 G/cm gradient for the remainder of the mixing delay), and exchange time points were limited to less than 250 ms such that radiation dampening effects were negligible.
An eight second delay was inserted prior to each transient to allow the system to return to equilibrium. The imino region of the unapodized spectra at each exchange time was extracted, baseline corrected, and the peaks fitted as Lorentzian line-shapes using in-house MATLAB and python scripts (see Fig. S1). Linewidth and peak positions were optimized and fixed to those values measured in the fully relaxed (10 s delay) spectrum, and the peak intensities were the only free floating parameters in fitting the other delay times. The extracted peak heights I(t) at each delay time were fit to the following equation:
| (1) |
where t is variable delay time after water inversion and prior to detection, kex is the exchange rate, and R1w and R1i are the longitudinal relaxation rates of water and the imino protons, respectively, and E is the water inversion efficiency (25). Values for R1w were obtained prior to each exchange rate measurement using an adiabatic water inversion pulse and a spin-echo imaging element to minimize the effects of radiation dampening.
Measurement of Base-Pair Opening Rates
Base pair opening rates were obtained by fitting the observed imino exchange rates as a function of ammonia catalyst (at pH 9.2) to the following equation:
| (2) |
where kex is the observed imino exchange rate from eq 1, kop is the rate of base pair opening, kb the pseudo first order collision constant of ammonia (~5 × 108 M−1s−1 under these conditions), [NH3] is the concentration of ammonia under experimental pH and temperature, ko the intrinsic rate of imino proton exchange in the absence of added catalyst, and kcl is the rate of base pair closing.
In some cases the imino proton exchange rates in the absence of exchange catalyst (ko) were too rapid for direct measurement using magnetization transfer methods and were instead obtained by linewidth measurements (lw, eq 3), where R2dip is the intrinsic dipolar spin-spin contribution to observed
| (3) |
relaxation rate, which was estimated from the narrowest imino linewidth of the control or crosslinked sequence minus the effects imino proton exchange as determined by eq 1.
Results
Previous work has established the structural similarity between a normal DNA duplex and a sequence matched CGX ICL duplex, but failed to identify unique structural characteristics of the CGX ICL-DNA that might lead to its recognition by DNA repair factors (16, 18, 19). Because different dynamic properties could be manifested in DNAs that have similar time-averaged structures, we synthesized the family of normal and crosslinked DNA duplexes shown in Figure 3, and measured their dynamic properties using NMR imino proton exchange methods. These duplexes have reduced spectral complexity because of palindromic symmetry. Thus, every imino proton shares an identical chemical environment, and has an identical NMR chemical shift as its cognate pair on the other side of the palindromic axis of symmetry.
General Features of ICL-DNA
In order to assign each imino resonance, a standard 1H-1H NOESY experiment was performed on each DNA construct in Figure 3 (see also Fig. S2). Unambiguous assignments were obtained for all imino protons using this method (Fig. 4), except for those in terminal base pairs, and for T2 of the structurally perturbed GCX-10 duplex, which lacked the standard correlations in the imino region. After eight-days in NMR buffer containing 7.5 mM ammonia catalyst, we also found that the N4-ethyl-N4 crosslink in the GCX-10 duplex was extensively hydrolytically deaminated. Deamination resulted in a uracil on one strand (i.e. a GU mismatch), and a C-N4-ethylamine modification on the opposite strand (see Fig. S3). The slow deamination process had no effect on the NMR imino exchange experiments, which were performed on the freshly synthesized duplex over a time period of less than two days over which time no changes in the spectra indicative of degradation were observed. The imino resonances of all bases in the GCX-10 duplex (except for the terminal bases T2 and G10) were assigned by comparison of the proton spectra of the sequence-matched GC-10 and the deaminated GCX-10 sample (see Fig. S3 for details).
Figure 4.
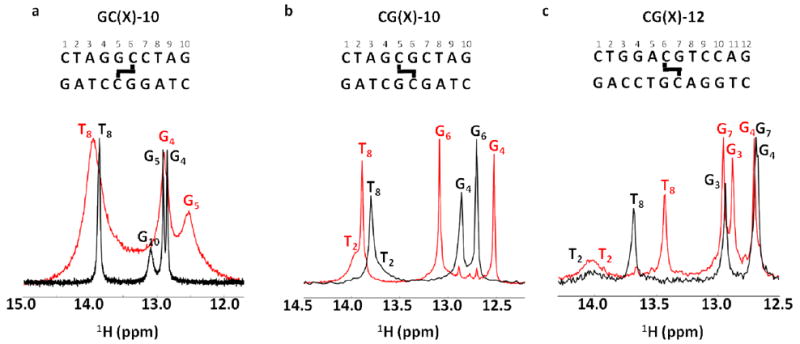
Imino spectra of the six normal and crosslinked DNA duplexes in Figure 2 (GC-10 and CG-10 at 10 °C and CG-12 at 15 °C, pH 7.0 no ammonia exchange catalyst). The imino spectra of the normal duplexes are shown in black, and their cognate crosslinked forms are shown in red. For reference, the DNA sequences and position of the crosslink are shown above each spectrum.
Inspection of the 1H imino proton regions of the 10mer and 12mer normal and ICL-duplexes generally reveals the expected number of imino resonances (Fig. 4). However, end fraying leads to extensive broadening of the imino proton resonances of each terminal base pair such that they are undetectable at 10 or 15 °C. Uniquely, the GCX-10 ICL-DNA shows extensive line broadening of all imino resonances, indicating destabilization of the base pairs and an increased contribution of solvent exchange to the observed linewidths (see Fig. S4). In contrast, the two structurally unperturbing CGX-ICL sequences generally show 1.1 to 1.4 Hz narrower imino proton linewidths relative to their sequence matched controls for every base located more than two nucleotides away from the duplex end indicating decreased exchange contributions to the linewidths. Unique chemical shift perturbations are observed in the CGX-10 and CGX-12 constructs, with the guanine base directly opposite to the crosslink moving 0.3 (G6 of CGX-10) to 0.4 (G7 of CGX-12) ppm downfield compared to their sequenced matched normal controls. The decreased electron density on these protons likely indicates a slightly shorter Watson-Crick hydrogen bond between the crosslinked cytosine and its paired guanine. In contrast, the G5 imino proton directly across from the crosslinked cytosine of the destabilized GCX-10 construct moves 0.4 ppm upfield. The other surrounding bases showed random upfield or downfield changes in their chemical shift as a result of the crosslink. These sensitive measures of environment indicate subtle differences in the time-averaged length of imino hydrogen bond in the ICL-DNAs, or perhaps changes in the local aromatic ring currents owing to subtle changes in base stacking.
Dynamics of the GCX-10 Construct
We investigated the nature of the greatly broadened imino proton linewidths of the GCX-10 DNA relative to its conjugate normal sequence using solvent magnetization transfer measurements (GC-10) or imino proton linewidth measurements (GCX-10) (Fig. S5, Table 1). The indirect linewidth method for measuring imino proton exchange rates of GCX-10 is non-ideal because conformational exchange processes may also contribute to the linewidth. However, this possibility is made unlikely because other resolved carbon bound protons of GCX-10 were not similarly broadened (Fig. S4), indicating that the majority of the observed line broadening arises from chemical exchange with water protons. The exchange rates for the G4, G5 and T8 imino protons of GCX-10 in the absence of added catalyst were 20 to 100-fold larger than the corresponding protons in the matched control duplex indicating significant increases in the base pair opening rates or in the lifetime of the imino proton in the open solvent exposed state (Table 1).
Table 1.
Base pair opening rates of B-form GC-10 and GCX-10 duplexes at 10°Ca
 | ||||
|---|---|---|---|---|
| Base | ko (s−1)b | kslope (s−1 M−1)c | ||
| GC-10 | GCX-10 | GC-10 | GCX-10 | |
| T2 | -d | -d | -d | -d |
| G4 | 1.7 ± 0.2 | 131 ± 11e | 44 ± 4 | 8400 ± 3200e |
| G5 | 2.0 ± 0.2 | 204 ± 22e | 40 ± 4 | -d |
| T8 | 8.7 ± 0.5 | 219 ± 11e | -d | 26400 ± 4000e |
| G10 | 39 ± 4.1 | -d | -d | -d |
The DNA sequence is indicated as well as the crosslinked bases in red. The guanine base G5 opposite to the crosslinked cytosine is indicated in bold.
The observed exchange rate at zero concentration ammonia catalyst (eq 2).
The initial slope of the hyperbolic curve of eq 2, corresponding to the apparent second-order rate constant for general base catalysis of exchange by ammonia, (kop/kcl) × kb.
Exchange broadened beyond detectable limits.
Exchange rates from line width measurements and eq 3. A value for R2dip was taken from the narrowest imino linewidth of the GC-10 spectra minus its ko as determined by imino exchange, 17.9 Hz.
The catalysis of imino exchange by increasing concentrations of ammonia was investigated for both the GC-10 and GCX-10 duplexes (Table 1). Imino proton exchange is a two-step process, with the first step being base pair opening, and the second step, chemical exchange with solvent protons. At low concentrations of ammonia catalyst the chemical exchange step is typically rate limiting, but through the addition of high concentrations of ammonia, the chemical step can be sufficiently accelerated such that base pair opening limits the observed imino proton exchange rate. Such a change in rate-limiting step is experimentally manifested as downward curvature in a plot of kex against catalyst concentration (eq 2). For the GC-10 and GCX-10 DNA sequences, the exchange rates showed a linear response to ammonia catalyst over the entire concentration range investigated (≤ 1.9 mM NH3 base catalyst), indicating that the rate-limiting step for exchange was catalyst-assisted proton abstraction, and not base pair opening. Under these conditions the slope of the observed exchange rate against catalyst concentration (kslope) is equal to (kop/kcl) × kb (eq 2). Since each imino proton may be reasonably assumed to have the same response to ammonia catalyst in the open state (i.e. the kb terms are identical), the ratio of the slopes between the normal and crosslinked duplexes provides a quantitative measure of how much the opening equilibrium (Kop = kop/kcl) is perturbed by the presence of the crosslink. Comparison of the kslope values of the GCX-10 and the normal control DNA establishes that the crosslink causes increases in the base pair opening equilibrium constants of at least several orders of magnitude. The largest increase is observed for the guanine base (G5) that is paired with the crosslinked cytosine, but the effects extend to the neighboring 5′ guanine (G4). The opening equilibrium for the imino proton of T8, which is located three base pairs 3′ to the crosslink, is very large in both the sequence matched normal DNA and the GCX-10 sequence, and so ratios of equilibrium constants cannot be directly compared in this case. However, the k0 of T8 in GCX-10 is nearly 25 fold greater than the matched control, indicating a much greater level of solvent accessibility for the T8 imino proton in GCX-10.
Dynamics of CGX-10
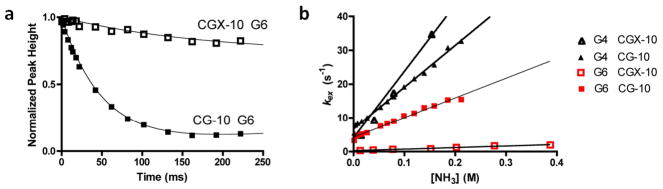
We then investigated the dynamic behavior of the B form CGX-10 construct along with its normal matched duplex using imino proton solvent magnetization transfer measurements (Fig. 5 and Table 2). High-quality magnetization transfer data were obtained for the imino protons of bases G4 and G6, which flank the site of the crosslink (Table 2, kFig. 5). However, the imino proton exchange rate for T8, which is three steps 3′ of the crosslink, was only measureable in the CG-10 sequence in the absence of catalyst (o). Upon the first addition of ammonia, the imino proton of T8 of CG-10 disappears while the corresponding T8 of CGX-10 is only broadened, indicating that the distant crosslink decreases base pair opening at this site. A plot of kex against ammonia catalyst concentration for bases G4 and G6 showed no indication of curvature (Fig. 5), precluding the measurement of the opening and closing rate constants using eq 2. However, as described above for the GC-10 duplexes, a comparison of the initial slopes of these linear responses to catalyst concentration quantifies the effect of the crosslink on the base pair opening equilibrium constants. Using this interpretation, the G6 base that is paired with the crosslinked cytosine shows a 10-fold decrease in its opening equilibrium, while the imino proton of G4 located just 5′ of the crosslink shows a slight increase its opening equilibrium as compared to the normal duplex (Table 2). Thus, unlike the perturbing GCX-10 ICL that caused gross increases in the opening equilibrium constants across the duplex sequence, the nonperturbing CGX-10 crosslink locally decreases the opening equilibrium, and has little effect on nearby base pairs.
Figure 5.
Representative imino-proton exchange by solvent magnetization transfer and exchange catalystsis by ammonia. (a) Representative time courses of solvent magnitzation transfer to imino site of Guanine 6 in CGX-10 and CG-10 duplexes (300 mM [NH3] catalyst concentration). Solid line is the best fit of the data to eq 1. (b) Amonia concentration dependence of immino proton exchange for the indicated residues. Fitted parameters are listed in Table 2.
Table 2.
Base pair opening rates of B-form CG-10 and CGX-10 duplexes at 10°Ca
 | ||||
|---|---|---|---|---|
| Base | ko (s−1)b | kslope (s−1 M−1)c | ||
| CG-10 | CGX-10 | CG-10 | CGX-10 | |
| T2 | 260 ± 100d | 150 ± 30d | -e | -e |
| G4 | 6.7 ± 0.3 | 4.1 ± 3.5 | 124 ± 3.0 | 196 ± 17 |
| G6 | 4.2 ± 0.2 | 0.2 ± 0.1 | 58 ± 2.1 | 4.6 ± 0.3 |
| T8 | 10 ± 4 | 11 ± 6 | -e | -e |
The DNA sequence is indicated as well as the position of the crosslinked base in red. The guanine base opposite to the crosslinked cytosine is indicated in bold. The terminal base pair G10 was not observed due to rapid exchange with water at pH 9.2.
The observed exchange rate at zero concentration ammonia catalyst (see eq 2).
The initial slope of the hyperbolic curve of eq 2, corresponding to the apparent second-order rate constant for general base catalysis of exchange by ammonia kslope = (kop/kcl) × kb.
Determined by linewidth measurement using eq 2 and a R2dip value of 19.0 Hz. derived from the narrowest imino linewidth of the CGX-10 spectra minus its ko as determined by imino exchange experiments.
Exchange broadened beyond detectable limits.
Dynamic Measurements of CG-12 and CGX-12 Duplexes
Because we were unable to achieve conditions where base pair opening was rate-limiting for imino proton exchange using the 10mer duplexes, we turned to investigating a related B form 12mer CG ICL-DNA (Fig. 3), with the dual goals of measuring the opening rates, as well as assessing the effects of these crosslinks in a different sequence context. For the CG-12 and CGX-12 duplexes, opening rates could be measured for bases G3, G4, and G7. However for both duplexes, only lower limit values for the opening rate of T8 were obtained due to its rapid exchange rate (Fig. 6 and Table 3). Comparison of the two data sets indicates that G3 and G7 of CGX-12 open approximately 6 and 80 times slower than the same base pairs in the matched normal DNA, while the ICL induces a lesser 2.5-fold inhibitory effect for G4. The inhibitory effect on the opening rate of G7 is accounted for by its position immediately opposite the crosslink similar to the effect observed in CGX-10. However, G4 and G3 are located two and three base pairs 5′ of the crosslink, respectively, indicating long range stabilizing effects of the crosslink. The long-range effect of the crosslink on the opening rate of G3 likely arises from its unusually rapid opening rate in the normal CG-12 duplex (490 ± 96 s−1, Table 3). This rapid opening rate is atypical for a non-terminal G·C base pair at 15°C, but is consistent with previously reported sequence dependent variances of G·C opening rates, particularly in GC tracts (80–700s−1) (27). Interestingly, the crosslink has no discernable effect on the exchange behavior of T8, which is located two bases 3′ to the crosslink site.
Figure 6.
Representative imino-proton exchange by solvent magnetization transfer and exchange catalystsis by ammonia. (a) Representative time courses of solvent magnitzation transfer to imino site of Guanine 6 in CGX-12 and CG-12 duplexes (240 mM [NH3] catalyst concentration). Solid line is the best fit of the data to eq 1. (b) Amonia concentration dependence of immino proton exchange for the indicated residues. Fitted parameters are listed in Table 3. (c) Summary of the effects of the CG-12 cross-link on imino proton exchange. Relative differences in base pair dynamics in CG-12 and CGX-12 are proportional to letter size. Letters in blue show decreased opening relative to CG-12, bases in green show no change, bases in black are exchanging too rapidly for measurement, and bases in grey to not have imino protons (cytosine and adenine).
Table 3.
Imino proton exchange parameters CG-12 and CGX-12 duplexes at 15°Ca
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| ko (s−1)b | kslope (s−1 M−1)c | kop (s−1)d | kcl (× 108 s−1)e | |||||
| CG-12 | CGX-12 | CG-12 | CGX-12 | CG-12 | CGX-12 | CG-12 | CGX-12 | |
| T2 | 88 ± 60f | 128 ± 79f | -g | -g | -g | -g | -g | -g |
| G3 | 8.2 ± 1.7 | 7.4 ± 0.4 | 2790 ± 140 | 1135 ± 35 | 490 ± 96 | 76 ± 12 | 0.7 ± 0.3 | 0.2 ± 0.1 |
| G4 | 2.2 ± 0.2 | 1.6 ± 0.1 | 96 ± 19 | 94 ± 2.0 | 72 ± 19 | 29 ± 3 | 4.9 ± 1.9 | 1.8 ± 0.4 |
| G7 | 1.9 ± 0.2 | 1.2 ± 0.1 | 181 ± 17 | 0 ± 8h | 80 ± 9 | 1h | 2.8 ± 0.5 | -h |
| T8 | 7.0 ± 1.4 | 12 ± 1 | 2830 ± 115 | 2650 ± 28 | >1000i | >1000i | -i | -i |
The DNA sequence is indicated as well as the position of the crosslink in red. The guanine base opposite to the crosslinked cytosine is indicated in bold. The terminal base pair G12 was not observed at pH 9.2 due to rapid exchange with water.
The observed exchange rate at zero concentration ammonia catalyst (see eq 2).
The initial slope of the hyperbolic curve of eq 2, corresponding to the apparent second-order rate constant for general base catalysis of exchange by ammonia kslope = (kop/kcl) × kb.
The rate of base pair opening derived from fits of the exchange data as a function of catalysts to eq 2.
The rate of base pair closing derived from fits of the exchange data as a function of catalysts to eq 2.
Determined by linewidth measurement using eq 2 and a R2dip value of 21.0 Hz. derived from the narrowest imino linewidth of the CGX-12 spectra minus its ko as determined by imino exchange.
Exchange broadened beyond detectable limits in the presence of ammonia catalyst.
Could not be fit to eq 2 because exchange of this imino proton, opposite the crosslinked cytosine, is not catalyzed by ammonia.
Exchange rate too rapid to fit to eq 2. kop could be established as a lower limit of the true value.
Discussion
Due to the high cytotoxicity and the structural diversity of ICLs, mammalian cells have developed elaborate mechanisms for their repair. The challenge of understanding ICL recognition in vivo is that detection occurs in the context of large amounts of decoy undamaged DNA, and is efficient for ICLs of various structures or chemical compositions. This last property necessitates that common features shared by many ICLs must guide detection. Such features must be limited to either structural distortions of the DNA, altered dynamic characteristics of ICL sites, or the most fundamental fact that the strands are covalently tethered. Because the enzymatic activities in mammalian cell extracts that lead to unhooking of the CC ICL are not yet identified (Fig. 1) (14), the discussion below presents general mechanistic features of ICL recognition that could apply to any enzyme. Furthermore, because our work and the previous ex vivo repair studies use linear duplex ICLs (14), we restrict our discussion to enzymatic detection and strand incision events that occur in the absence of transcription, replication or chromatin suprastructure.
The Merits of Being Flexible
It is straightforward to envision how gross structural distortions to the regular B-form geometry of DNA, similar to those observed in the GCX-10 construct, could be preferentially recognized as damage. This duplex shows significant evidence of dynamic and structural perturbations that extend at least five base pairs from either side of the ICL (14) (Fig. 4a). Such a large and structural flexibility target site could provide a pliable and flexible substrate for repair enzymes that must distort the DNA duplex during the act of binding or catalysis (28). Increased flexibility of the site may lead to a decreased thermodynamic penalty for binding to the site, and thereby increase the probability that a repair enzyme will be productively bound at a damaged site rather than unproductively bound over the vast excess of undamaged sites that are available. In other words, flexibility could provide thermodynamic specificity. Alternatively, or in addition to above effects, the increased affinity for flexible DNA duplexes could be realized at the chemical transition state for the enzyme reaction (in this case DNA strand incision). This specificity at the catalytic step would prevent normal DNA from being incised with the possible deleterious outcome of chromosome rearrangements or mutations. It seems reasonable that both ground state and transition state mechanisms would be selected for over time.
The Price of Being Rigid
The flexibility mechanism described above provides an unsatisfying explanation for the previously observed enzymatic incision of the non-perturbing 5′-CG-3′ CC ICL by mammalian cell extracts (14). Although the rate of 5′-CG-3′ ICL unhooking was 6-fold lower than the distorted 5′-GC-3′ ICL (14), which might be attributed to structural and flexibility differences between these duplexes, both ICLs displayed the same highly-specific NER dependent futile incision events, as well as the NER-independent 5′ and 3′ incision events that led to unhooking (Fig. 1). Since the B form CGX-10 and CGX-12 ICLs studied here resulted in dramatically decreased dynamics at the site of the crosslink, and to a lesser extent nearby base pairs (Fig. 6, Tables 2 and 3), it is difficult to conclude that increased dynamics and gross structural perturbations are a absolute requirement for recognizing such sites. Moreover, the fact that a matched normal sequence is not detectably incised, yet the structurally intact and less dynamic 5′-CG-3′ ICL is recognized reasonably well (14), strongly suggests that the fundamental criterion for NER-dependent or independent strand incision events is the presence of the covalent crosslink, and not the structural or dynamic properties that the crosslink provokes in the DNA. Thus, based on the relative unhooking efficiencies for the B form 5′-CG-3′, and distorted 5′-GC-3′ ICL sequences, the price for being rigid and isostructural with normal DNA is less than an order of magnitude with these ICLs (~1 kcal/mol).
A Model for ICL Recognition
What type of model could account for the specific detection and incision of ICLs that have different structural and dynamic properties yet share the same well-defined crosslink? One model that would satisfactorily account for the increased incision efficiency for the destabilized ICL, yet would still allow for specific incision of a structurally unperturbed and stable ICL, involves a binding step where an enzyme or protein associates with DNA near a crosslink, and then tests for its presence by unpairing of the two DNA strands into a bubble-like structure. Subsequently, migration of the bubble would be prevented by any nearby ICL, and the complex would persist at the site if the enzyme or repair factor possessed specific binding energy for Y-type DNA junctions. This model is general and has the merits of being directly testable if purified components become available.
There is precedent for such a mechanism in the GGR pathway of NER where xeroderma pigmentosum protein C (XPC) binds damaged regions of DNA containing a monoadduct lesion and then induces bubble formation at or near the site of the damage (28). XPC then recruits other processing enzymes to the site of damage to complete excision of the lesion. This strand separation mechanism does not require expenditure of metabolic energy, as thermal fluctuations in the duplex may initiate the process, and specific enzyme binding energy may be used to stabilize the separated duplex strands. This initial step of bubble formation would be more favorable for destabilized duplex regions such those induced by the 5′-GC-3′ ICL, reducing the energetic penalty for enzyme binding and promoting thermodynamic specificity for such sites.
Effects of Structurally Non-Perturbing Crosslinks on Base Pair Opening
Our imino exchange rate measurements of stable crosslinked duplexes also provide new information on lingering unknowns in the mechanism of base pair opening. While imino exchange has long been used to follow base pair opening, the technique is insensitive to both the mechanism and identity of the opening base, as purine or pyrmidine base pair members could equally well swing out of the base stack and allow access of the imino proton to solvent. The identity of the opening base has been investigated in computational work, which found that the purine opening pathway was always preferred over dynamic excursions of its partner pyrimidine base (29). In the present case, the observed slow opening rates of the guanine bases located opposite to the crosslink in the B form CGX-10 and CGX-12 duplexes (1 to 5 s−1), are inconsistent with a purine-only opening model because these guanines are bonded to the crosslinked cytosine via normal Watson-Crick hydrogen bonds, and are not themselves covalently constrained to prevent opening. Although it is formally possible that the N4C-ethyl-N4C cross link situated along the major groove might impinge on the major groove opening pathway of the guanine base, it is not obvious from inspection of the structure, or the pathway for major groove flipping, that the crosslink would constitute a steric impediment (see Figure 2) (30). These data are also inconsistent with a minor groove pathway for exposure of the guanine imino proton, because this pathway is not sterically impeded in the 5′-CG-3′ constructs. Indeed, numerous computational studies have suggested that guanine prefers to open via the major and not minor groove, and our data provide experimental support for these findings (30, 31). In summary, these observations (using duplexes where only the cytosine of a G/C base pair is covalently constrained) are consistent with only a limited number of base opening models: (i) imino proton exchange in normal G/C pairs arises nearly exclusively from cytosine opening, which is completely prevented by the covalent crosslink in our system, (ii) guanine opening through the major groove is the major pathway, but the N4C-ethyl-N4C cross link poses an impenetrable barrier, or (iii) the tether removes essential flexibility in the DNA duplex that is required for low energy barrier opening.
Supplementary Material
Abbreviations
- ICL
Intersrand Cross Link
- NOE
Nuclear Overhouser Effect
- NOESY
Nuclear Overhouser Effect Spectroscopy
- CGX
a DNA construct containing a central staggered 5′-CG-′3 and N4-C-Ethyl-C-N4 crosslink
- GCX
a highly distorted DNA construct containing a central staggered 5′-GC-′3 and N4-C-Ethyl-C-N4 crosslink
- GGR
global genomic repair
- TCR
transcription coupled repair
- NER
nucleotide excision repair
Footnotes
This work was supported by NIH grants GM-056834 (J.T.S) and CA082785 (P.S.M).
Although incisions were reported to occur on either side of the crosslink, it cannot be concluded that both incisions were the result of endonuclease activity. It is possible that an endonuclease cuts on one side of the crosslink and an exonuclease then digests past the crosslink. Previous studies have established that the unhooked crosslink remnant consists only of the N4C-ethyl deoxycytidine and does not have additional nucleotides attached to either side of the deoxycytidine (20), a structure that is consistent with such a mechanism.
Supporting Information Available
A detailed description of the fitting algorithm used to extract peak linewidths and polarization intensities during inversion timecourses. Imino proton regions of 1H-1H NOESY spectra for each duplex. Jump return spectra of imino proton regions of the GC(X)-10 construct used to make assignments of this construct. Jump return spectra containing all proton resonances of the GC(X)-10 constructs downfield of water. Imino proton exchange dependencies of the GC(X)-10 constructs on ammonia catalysts concentrations. Detail of the exchange rate versus catalysis concentration of the fastest exchanging imino protons in the CG(X)-12 duplexes. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hurley LH. DNA and its Associated Processes as Targets for Cancer Therapy. Nature Reviews Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 2.Noll DM, Mason TMG, Miller PS. Formation and Repair of Interstrand Cross-Links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA Interstrand Crosslinks: Molecular Mechanisms and Clinical Relevance. The Lancet Oncology. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 4.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the Roles of Nucleotide Excision Repair and Recombination in the Repair of DNA Interstrand Cross-Links in Mammalian Cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Räschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muniandy PA, Liu J, Majumdar A, Liu S, Seidman MM. DNA Interstrand Crosslink Repair in Mammalian Cells: Step by Step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone MP, Cho YJ, Huang H, Kim HY, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM. Interstrand DNA Cross-Links Induced by a, β-Unsaturated Aldehydes Derived from Lipid Peroxidation and Environmental Sources. Acc Chem Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an Interstrand DNA Cross-Link Initiated by ERCC1-XPF repair/recombination Nuclease. J Biol Chem. 2000;275:26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 9.Fisher LA, Bessho M, Bessho T. Processing of a Psoralen DNA Interstrand Cross-Link by XPF-ERCC1 Complex in Vitro. J Biol Chem. 2008;283:1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 10.Reardon JT, Sancar A. Nucleotide Excision Repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 11.Wood RD. Mammalian Nucleotide Excision Repair Proteins and Interstrand Crosslink Repair. Environ Mol Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muniandy PA, Thapa D, Thazhathveetil AK, Liu S, Seidman MM. Repair of Laser-Localized DNA Interstrand Cross-Links in G1 Phase Mammalian Cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA Interstrand Cross-Links Induce Futile Repair Synthesis in Mammalian Cell Extracts. Mol Cell Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smeaton MB, Hlavin EM, Mason TMG, Noronha AM, Wilds CJ, Miller PS. Distortion-Dependent Unhooking of Interstrand Cross-Links in Mammalian Cell Extracts. Biochemistry (N Y) 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Lao Y, Yang IY, Hecht SS, Moriya M. Replication-Coupled Repair of Crotonaldehyde/Acetaldehyde-Induced Guanine- Guanine Interstrand Cross-Links and their Mutagenicity†. Biochemistry (N Y) 2006;45:12898–12905. doi: 10.1021/bi060792v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noll DM, da Silva MW, Noronha AM, Wilds CJ, Colvin OM, Gamcsik MP, Miller PS. Structure, Flexibility, and Repair of Two Different Orientations of the Same Alkyl Interstrand DNA Cross-Link†. Biochemistry (N Y) 2005;44:6764–6775. doi: 10.1021/bi050014n. [DOI] [PubMed] [Google Scholar]
- 17.Noll DM, Noronha AM, Miller PS. Synthesis and Characterization of DNA Duplexes Containing an N4C– Ethyl– N4C Interstrand Cross-Link. J Am Chem Soc. 2001;123:3405–3411. doi: 10.1021/ja003340t. [DOI] [PubMed] [Google Scholar]
- 18.Noronha AM, Noll DM, Wilds CJ, Miller PS. N4C– Ethyl– N4C Crosslinked DNA: Synthesis and Characterization of Duplexes with Interstrand Cross-Links of Different Orientations†. Biochemistry (N Y) 2002;41:760–771. doi: 10.1021/bi011610u. [DOI] [PubMed] [Google Scholar]
- 19.Swenson MC, Paranawithana SR, Miller PS, Kielkopf CL. Structure of a DNA Repair Substrate Containing an Alkyl Interstrand Cross-Link at 1.65 A Resolution†,‡. Biochemistry (N Y) 2007;46:4545–4553. doi: 10.1021/bi700109r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smeaton MB, Hlavin EM, Noronha AM, Murphy SP, Wilds CJ, Miller PS. Effect of Cross-Link Structure on DNA Interstrand Cross-Link Repair Synthesis. Chem Res Toxicol. 2009;22:1285–1297. doi: 10.1021/tx9000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic Dynamics of an Enzyme Underlies Catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 22.Tang C, Schwieters CD, Clore GM. Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR. Nature. 2007;449:1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 23.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic Capture of an Extrahelical Thymine in the Search for Uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA Binding Shapes the Dynamic Landscape for Enzymatic Recognition of DNA Damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gueron M, Leroy JL. Studies of Base Pair Kinetics by NMR Measurement of Proton Exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 26.Kupce E, Boyd J, Campbell I. Short Selective Pulses for Biochemical Applications. Journal of Magnetic Resonance, Series B. 1995;106:300–303. doi: 10.1006/jmrb.1995.1049. [DOI] [PubMed] [Google Scholar]
- 27.Dornberger U, Leijon M, Fritzsche H. High Base Pair Opening Rates in Tracts of GC Base Pairs. J Biol Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 28.Min JH, Pavletich NP. Recognition of DNA Damage by the Rad4 Nucleotide Excision Repair Protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 29.Priyakumar UD, MacKerell AD., Jr NMR Imino Proton Exchange Experiments on Duplex DNA Primarily Monitor the Opening of Purine Bases. J Am Chem Soc. 2006;128:678–679. doi: 10.1021/ja056445a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banavali NK, MacKerell AD., Jr Free Energy and Structural Pathways of Base Flipping in a DNA GCGC Containing Sequence. J Mol Biol. 2002;319:141–160. doi: 10.1016/S0022-2836(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 31.Giudice E, Várnai P, Lavery R. Base Pair Opening within B-DNA: Free Energy Pathways for GC and AT Pairs from Umbrella Sampling Simulations. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hlavin EM, Smeaton MB, Miller PS. Initiation of DNA Interstrand Cross-Link Repair in Mammalian Cells. Environ Mol Mutagen. 2010;51:604–624. doi: 10.1002/em.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.