Abstract
Core promoters are critical regions for gene regulation in higher eukaryotes. However, the boundaries of promoter regions, the relative rates of initiation at the transcription start sites (TSSs) distributed within them, and the functional significance of promoter architecture remain poorly understood. We produced a high-resolution map of promoters active in the Drosophila melanogaster embryo by integrating data from three independent and complementary methods: 21 million cap analysis of gene expression (CAGE) tags, 1.2 million RNA ligase mediated rapid amplification of cDNA ends (RLM-RACE) reads, and 50,000 cap-trapped expressed sequence tags (ESTs). We defined 12,454 promoters of 8037 genes. Our analysis indicates that, due to non-promoter-associated RNA background signal, previous studies have likely overestimated the number of promoter-associated CAGE clusters by fivefold. We show that TSS distributions form a complex continuum of shapes, and that promoters active in the embryo and adult have highly similar shapes in 95% of cases. This suggests that these distributions are generally determined by static elements such as local DNA sequence and are not modulated by dynamic signals such as histone modifications. Transcription factor binding motifs are differentially enriched as a function of promoter shape, and peaked promoter shape is correlated with both temporal and spatial regulation of gene expression. Our results contribute to the emerging view that core promoters are functionally diverse and control patterning of gene expression in Drosophila and mammals.
The Drosophila melanogaster embryo is an important model system used to study transcriptional regulation of gene expression during development (for review, see Biggin and Tjian 2001). Much recent work has focused on the genome-wide identification and characterization of binding sites for sequence-specific transcription factors in Drosophila, other model animals, and human (The ENCODE Project Consortium 2007; Li et al. 2008; MacArthur et al. 2009). For a global understanding of how transcription factors and other chromatin proteins and their bound genomic regions interact with core promoter regions (for review, see Juven-Gershon and Kadonaga 2010) to regulate transcription, it is necessary to discover and characterize promoter regions comprehensively.
Transcription start sites (TSSs) were first defined by primer extension studies (Qu et al. 1983). Subsequently, improved approaches such as rapid amplification of cDNA ends (RACE) (Frohman et al. 1988) and cap-trapped 5′ expressed sequence tag (EST) sequencing (Carninci et al. 1996) were developed. More recently, cap analysis of gene expression (CAGE), a high-throughput method for promoter discovery, has been used in mouse and human to characterize capped transcript ends (Shiraki et al. 2003; Carninci et al. 2006). In Drosophila, TSSs have been defined on a modest scale by sequencing and annotation of 5′ ESTs generated from cap-trapped cDNA clones (Misra et al. 2002; Stapleton et al. 2002). The current reference annotation of the D. melanogaster genome sequence uses these data, and 5′ ESTs from non-cap-trapped cDNA libraries (Rubin et al. 2000), to define 5′ transcript ends (Drysdale 2008). While these ESTs have been useful for annotating 5′ ends of genes, there are insufficient numbers to identify core promoters of lowly expressed transcripts or to determine the distributions of TSSs within most core promoter regions.
Analysis of 5′ EST clusters and surrounding genomic sequences identified 10 sequence motifs within core promoter regions of D. melanogaster representing binding sites for factors involved in the initiation of transcription (Ohler et al. 2002). Subsequent analysis of FlyBase 5′ transcript ends (Misra et al. 2002; Drysdale 2008) revealed an additional five sequence motifs in promoter regions (FitzGerald et al. 2006). In these studies, promoters were modeled as discrete points rather than as local distributions of TSSs. Yasuhara et al. (2005), using 5′ RLM-RACE to study a small set of transcripts, showed that Drosophila promoters are characterized either by a broad region of distributed TSSs, which they described as “slippery promoters,” or by a single TSS defining a discrete promoter. These findings are consistent with analysis of CAGE data that define “peaked” and “broad” promoter classes in the mouse and human genomes (Carninci et al. 2006). Recent analysis of cap-trapped and non-cap-trapped 5′ ESTs has determined that promoters characterized by a broad distribution of TSSs are also common in Drosophila (Rach et al. 2009). In both mammals and Drosophila, peaked and broad promoters differ in the enrichment of core promoter sequence motifs and are associated with different spatial patterns of activation (Carninci et al. 2006; Rach et al. 2009). Because peaked and broad promoters are distinct, a complete understanding of gene regulation depends on characterizing and classifying these regulatory elements in greater detail.
Two recent genome-wide studies contribute to the characterization of promoters in Drosophila. Ni et al. (2010) describe a new high-throughput method, named PEAT, for paired-end sequencing to map capped 5′ transcript ends and define 5699 clusters of sequence tags in Drosophila embryo, many of which correspond to core promoters. Nechaev et al. (2010) report on high-throughput RNA-sequencing of short nuclear RNAs associated with paused RNA polymerase II (Pol II) in Drosophila embryo-derived S2 cells and find that these RNAs are specifically associated with many core promoters. These reports constitute significant advances, but neither attempted the comprehensive characterization of Drosophila promoters.
As part of the modENCODE project (Celniker et al. 2009), we used two independent methods, CAGE and 5′ RLM-RACE, to map and validate TSS distributions within promoter regions of long capped transcripts expressed at significant levels, either maternally or zygotically, in the developing D. melanogaster embryo. These methods are complementary in two ways. First, like 5′ EST sequencing, CAGE randomly samples capped 5′ transcript ends in proportion to expression level, whereas 5′ RLM-RACE targets capped 5′ ends of specific transcripts and has greater sensitivity for lowly expressed transcripts. Second, CAGE and RACE recover the cap structure at 5′ transcript ends using very different strategies. Both methods were adapted for next-generation sequencing platforms, and we produced large data sets that sample many promoter regions with redundancy sufficient to classify promoters by their TSS distributions. We integrated CAGE, RACE, and EST data to identify and characterize promoters. We used an entropy-based score to show that TSS distributions form a complex continuum of shapes, and we used the score to classify promoters as peaked or broad. We then performed RACE on a subset of the same transcripts in an adult RNA sample and found that promoters that are active in both stages have very similar TSS distributions. This suggests that promoter shape is determined by static features such as local DNA sequence. We showed that peaked promoters are strongly and significantly associated with genes that have restricted temporal and spatial expression patterns. Our integrative analysis suggests that the numbers of active promoters in mammals determined from CAGE data have been overestimated by fivefold. Finally, the genome-wide annotation of promoter architecture described here provides a resource for future studies of the regulation of transcription by factors bound to core promoters and their interactions with cis-regulatory modules and the Pol II complex.
Results
Cap-trapped 5′ ESTs reveal peaked and broad TSS distributions within promoters
In an initial assessment of the distributions of TSSs within active promoters in the D. melanogaster embryo, we analyzed 66,169 previously described embryonic cap-trapped 5′ ESTs (Stapleton et al. 2002), known as RIKEN embryo (RE) ESTs, including 3035 clones represented by full-insert cDNA sequences, to the reference genome sequence (Release 5, http://www.fruitfly.org). Because these ESTs are long sequences (average length 453 nt), >92% (61,429) map uniquely to the genome (Supplemental Table 1).
We associated 50,415 ESTs with 5771 FlyBase r5.12 (FB5.12) gene models (Drysdale 2008). Approximately three-fourths of ESTs associated with annotations share their first splice site with the first splice site of the associated transcript. This agreement is expected because many of these ESTs were used in producing FlyBase annotations (Misra et al. 2002). The median number of associated ESTs per gene is four. Consistent with previous descriptions of TSS distributions within promoters in Drosophila (Yasuhara et al. 2005) and mammals (Carninci et al. 2006), we find that TSS distributions span a range of shapes from peaked to broad (Supplemental Fig. 1). Only 565 genes have a sufficient number of ESTs (20 or more, as shown below) for the distribution of TSSs within their promoters to be classified as peaked or broad. We therefore generated additional TSS data to characterize additional promoters.
Massively parallel mapping of TSSs in the Drosophila embryo using CAGE
To map TSSs of long capped transcripts efficiently in a massively parallel manner, we performed CAGE on total RNA from a 0- to 24-h collection of D. melanogaster embryos. This sample represents the entire period of embryogenesis; it contains maternally expressed RNAs loaded into the oocyte and zygotically expressed RNAs including those expressed in differentiated cell types and tissues arising during embryonic development. We constructed a CAGE library modified for sequencing on the Illumina GAI platform and generated 42 million 27-nt sequence tags.
Alignment of short sequence reads to a complex genome sequence is a challenging problem. We used ELAND (Illumina) to align the CAGE tags to the reference genome sequence, allowing up to two mismatches. This resulted in unique map locations for 23 million tags and multiple map locations for 6 million tags. However, ELAND does not take into consideration the sequence quality of reads, nor does it provide a rigorous estimate of the significance of alignments. To improve the mapping of CAGE data, we mapped tags to the genome using StatMap (Methods), an alignment program built on statistical modeling principles, to assign alignment probabilities to each tag. We identified 26 million tags with significant alignments, excluding reads that aligned to transposable elements. Based on poly(A)+ RNA-seq analysis, 80% of these tags map to genes that are expressed in the 0- to 24-h embryo sample as defined by reads per kilobase per million (RPKM) > 1.
The StatMap alignments of CAGE tags are consistent with transcription start sites of long capped transcripts: 80% of tags map within the 5′ untranslated region (UTR) of a transcript (Fig. 1A; Supplemental Data File 1). Furthermore, poly(A)+ RNA-seq expression levels and CAGE-tag counts are correlated within first exons (Spearman's ρ ∼ 0.47), which include both the first exons of annotated 5′ UTRs and initial coding exons of transcripts without annotated 5′ UTRs, in a log-linear fashion (r ∼ 0.48). The next largest fraction (17%) of tags is distributed throughout protein-coding genes on the transcribed strand, consistent with the existence of an RNA background signal in the CAGE assay that is not associated with TSSs of long capped transcripts. Such peaks have been detected in previous studies (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Ni et al. 2010) and are thought to correspond to RNA processing sites, where a transcript has been cleaved and recapped in the cytoplasm (Schoenberg and Maquat 2009). To account for this, we modeled the stranded signal throughout genes as a mixture of signal originating from promoter regions and background. We modeled the background as a mixture of signal linearly proportional to transcript expression level as measured by stranded RNA-seq of total RNA (Methods) and uniform unstranded signal, which we treated as random noise in a manner similar to Balwierz et al. (2009). All CAGE tags that could be explained by our model as RNA background or random noise (18% of mapped tags) were removed from subsequent analysis (Supplemental Methods), resulting in a set of 21 million filtered aligned CAGE tags.
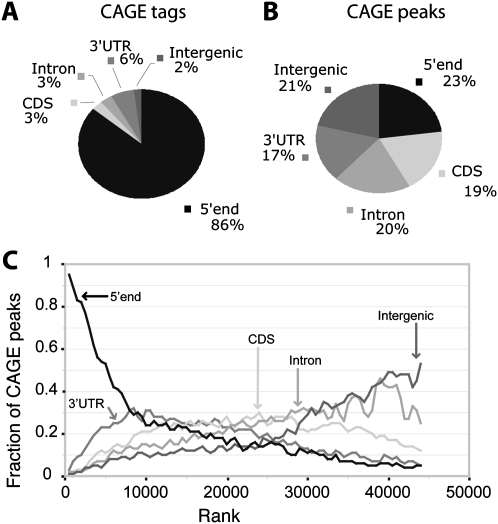
Figure 1.
Intersection of CAGE data with gene annotations. (A) The fractions of total CAGE tags that overlap annotated features. (B) The fractions of CAGE peaks that overlap annotated features. (C) CAGE peaks are ordered by tag count from highest to lowest. For bins of 1000 CAGE peaks, the fractions of peaks that overlap five classes of annotated features are plotted. The CAGE peaks toward the top of the rank list primarily overlap 5′ UTRs, while peaks at the bottom of the rank list tend to be intergenic. At the bottom of the rank list, the fractions of overlap approach expectation as computed by the GSC statistics package.
To understand the impact of our filtering procedure, we grouped the unfiltered and filtered CAGE signals into “CAGE peaks” by iterative hierarchical clustering. The minimum inter-peak distance was 50 bp; closer peaks were merged. Clustering of all 26 million unfiltered aligned tags resulted in 143,000 peaks, of which 57% were low-signal peaks with fewer than 15 tags and only 10% mapped near the 5′ end of a transcript. In contrast, clustering of the 21 million filtered aligned tags resulted in 45,000 statistically significant peaks (Fig. 1B), of which 41% were low-signal peaks with fewer than 15 tags and 23% map near the 5′ end of a transcript. The filtered alignments resulted in a twofold increase in the specificity of CAGE peaks for 5′ transcript ends without imposing an arbitrary threshold on tag counts. It removed 39,000 weak intronic peaks and 9000 peaks in coding sequence, many of them very strong and hence not filterable by thresholding alone. These filtered intronic and coding CAGE peaks likely correspond to uncapped background and low-frequency RNA processing sites. In order to target our study of CAGE data toward promoters, and not RNA processing sites, we used the filtered set for all subsequent analysis. The CAGE peak with the largest tag count maps to CG9184 and contains 326,403 tags, of which 170,000 are aligned to a single base pair, indicating that the dynamic range of the assay is at least 1 × 105.
To interpret the CAGE peaks, we determined their intersections with gene annotations (FB5.12) on the same strand (Methods; Fig. 1). There is a strong correspondence between the assigned annotations and tag counts per peak. Of the 1000 strongest CAGE peaks, 95% overlap a first exon. In contrast, only 5% of the 1000 weakest peaks overlap an annotated first exon, whereas 53% are intergenic. In all, 7073 CAGE peaks (17%) overlap a 5′ UTR (43% of annotated 5′ UTRs), and another 2190 CAGE peaks (5%) map within 100 bp of a 5′ transcript end on the same strand. These peaks together account for 86% of the filtered aligned CAGE tags (Fig. 1A,B) and are likely to represent promoter regions. In addition, 19% of CAGE peaks overlap protein-coding exons (3% of filtered tags), 20% overlap introns (3% of filtered tags), and 21% map in intergenic regions at least 100 bp from a transcript (2% of filtered tags). Finally, 17% of CAGE peaks overlap 3′ UTRs (6% of filtered tags), accounting for 52% of all 3′ UTRs (for this overlap, P-value < 1 × 10−16 as computed using the genome structural correction [GSC]; Bickel et al. 2011; Methods). These peaks are unlikely to represent promoter regions (see below).
Surprisingly, more than 90,000 CAGE tags mapped to the mitochondrial genome in 33 peaks that include the 5′ ends of nearly every transcription unit (Supplemental Fig. 2; Torres et al. 2009). We are not aware of any evidence that mitochondrial transcripts are capped, and there is evidence to the contrary in other animals (Grohmann et al. 1978). We observed a similar mapping of human CAGE tags (The ENCODE Project Consortium, unpublished data on cell lines K562 and GM12878 at http://genome-test.cse.ucsc.edu/cgi-bin/hgTrackUi?db=hg18&g=wgEncodeRikenCage) produced in the ENCODE project (The ENCODE Project Consortium 2007) to human mitochondrial transcripts. We also found that 1.4 million CAGE tags aligned to the Drosophila rDNA repeat (Supplemental Fig. 2; Tautz et al. 1988). The rRNA genes are transcribed by RNA polymerase I into a single, long pre-rRNA transcript that is processed into the mature rRNAs, so these CAGE peaks do not correspond to TSSs. These and previous results indicate that some CAGE peaks do not correspond to Pol II promoters. Thus, in this study, we do not consider CAGE evidence alone sufficient to define a promoter region.
Directed mapping of TSSs in the Drosophila embryo using 5′ RLM-RACE
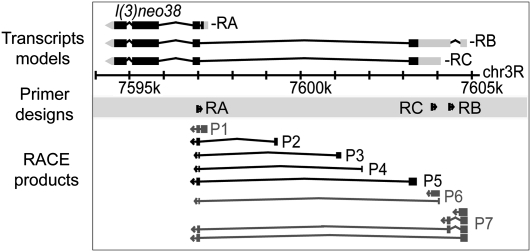
In an approach complementary to and independent of CAGE, we performed 8727 targeted 5′ RLM-RACE experiments on the same 0–24-h embryo total RNA sample used for CAGE, to characterize TSS distributions within promoters of embryonic transcripts of 7742 genes. We produced 2.1 million RACE reads on the 454 Life Sciences (Roche) platform, of which 1.2 million were oriented, trimmed, mapped to the genome, and associated with a transcript. Compared to the 61,429 mapped RE ESTs, this is a 20-fold increase in the number of embryonic long cap-trapped 5′-end reads. The average length of trimmed mapped reads was 154 nt. Of trimmed mapped reads, 29% were spliced, with 2% covering more than one splice junction. A total of 8418 transcripts of 7546 genes (96% of the transcripts targeted) was associated with at least one RACE read, and on average each transcript was associated with 143 RACE reads. A single RACE experiment can sample multiple promoters. For example, three RACE experiments targeting different transcripts of l(3)neo38 detected seven promoters (Fig. 2). Using a threshold of three reads, we identified 698 new promoters detected only by RACE.
Figure 2.
RLM-RACE analysis of the l(3)neo38 gene. RACE primers were designed to target three transcript isoforms of the gene. Three promoters (P1, P6, P7) correspond to annotated start sites for the –RA, –RB, and –RC isoforms, respectively. Four promoters (P2–P5) are new.
Defining promoter regions
We devised an iterative hierarchical clustering procedure to group tags into promoter regions and applied it to the RE EST, CAGE, and RACE data sets independently. Then, we integrated these clusters to produce consensus clusters based on the tags from all three data sets (Fig. 3). We identified 12,454 promoters and associated 11,672 with 8037 genes (Methods). This corresponds to an average of 1.4 promoters per gene: one promoter for each of 5849 genes, two for each of 1403 genes, and three or more promoters for each of 786 genes.
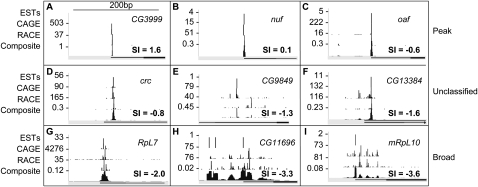
Figure 3.
Integration of RE EST, CAGE, and RACE data and classification of promoter shape. TSS distributions within nine promoter regions are ordered by increasing shape index (SI): (A–C) peaked promoters, (D–F) unclassified promoters, and (G–I) broad promoters. For each promoter, the RE EST, CAGE, RACE, and composite TSS distributions are shown. SI values of the composite distributions and gene associations are indicated.
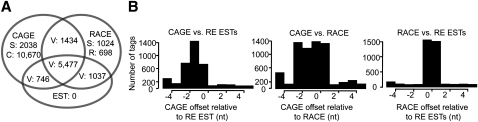
We grouped the promoters based on evidentiary support into three categories: validated (V), supported (S), and RACE-only (R). The validated set (8694 promoters) is defined by two or more data types, the supported set (3062 promoters) by either a CAGE peak or at least three RACE reads overlapping a 5′ UTR, and the RACE-only set (698 promoters) by three or more RACE reads with no support from an overlapping 5′ UTR. Within the validated set, 7657 promoters have CAGE peaks, 7948 have RACE data, 7260 have RE ESTs, and 5477 have all three data types (Fig. 4A). We discovered 2075 new promoters: 1257 have CAGE peaks, 1272 have RACE data, 566 have RE ESTs, and 163 have all three data types (Supplemental Data Files 2 and 3).
Figure 4.
Comparison of promoter regions and TSS distributions determined by RE EST, CAGE, and RACE data. (A) The numbers of clusters in overlapping subsets of CAGE peaks, RACE clusters, and RE EST clusters are indicated. Validated promoters (V) are defined by at least two of the three assays; supported promoters (S) are defined by one assay only but overlap an annotated promoter or 5′ UTR; unsupported CAGE-only (C) and RACE-only (R) clusters do not overlap annotated promoters or 5′ UTRs. (B) The relative offsets of TSS locations by pairwise comparisons of the three assays. The mean pairwise offset is 1.7 nt.
We intersected our set of 12,454 promoters with the recently published set of embryonic 5′ capped transcript end clusters produced using PEAT (Ni et al. 2010). Ni and colleagues report 5699 clusters: 4054 overlap or are within 25 bp of a TSS or 5′ UTR, 88 are in introns, 197 are intergenic, and finally 1360 are in coding exons or 3′ UTRs. We note that their analysis did not distinguish between coding exons and 3′ UTRs and that clusters mapping antisense to genes were included in the intergenic category. Our promoter set overlaps 76% of all the PEAT clusters and 92% of the 4054 TSS and 5′ UTR-associated clusters. Of the clusters unique to the PEAT data, 68% are in coding exons or 3′ UTRs. The authors attributed these clusters, as do we, to re-capping of transcript fragments.
We examined the remaining 10,670 CAGE peaks not supported or validated in our analysis in order to estimate the number of additional promoters in these data. At a threshold of 50 tags (2.5 tags per million aligned tags), CAGE peaks are about as likely to map to 5′ UTRs as to coding exons or introns, and are nearly as likely to map to intergenic regions (Fig. 1C). If we consider CAGE peaks overlapping 5′ UTRs, RACE clusters or RE EST clusters to be promoters, and all the rest to be false discoveries (unlikely since RACE has not been performed on all CAGE peaks), then CAGE peaks in the neighborhood of this threshold have a false discovery rate (FDR) of 25%. Above this threshold, there are 2268 unsupported CAGE peaks in intergenic or intronic regions. To determine whether these CAGE-only peaks represent promoters of unannotated or incompletely annotated transcripts, we intersected these peaks with transcribed regions detected by RNA-seq analysis of a time course of embryonic development with a sequencing depth of 930 million reads (Graveley et al. 2011). We found 196 CAGE peaks (9% of unsupported intergenic and intronic peaks) within 100 bp of the 5′ end of a transcribed region, and these are likely to represent unannotated promoters (Supplemental Data File 4); 12 of these CAGE peaks map to the 5′ ends of newly discovered primary transcripts of microRNA genes (Graveley et al. 2011). Hence, the majority of unsupported CAGE peaks are likely associated with other phenomena, and not with bona fide transcription initiation sites.
At each called promoter, each assay produced a slightly different distribution of tags. Even within technical replicates of the same assay, there is sampling variance, and between two distinct assays, there are assay-specific effects. To understand the TSS distribution in our validated promoter set, we studied the distributions of mapped tags in each of the assays at each promoter. We have achieved “single base-pair resolution” if all three assays appear to be drawn from the same underlying multinomial distribution. However, cross-correlation analysis revealed a tendency of each assay to provide tag distributions that are “shifted” by 1 or 2 bp from each other assay (Fig. 4B). For the 3406 validated promoters with more than one tag from each assay, 99% show a shift of at least 1 bp for at least one pair of assays. We estimate that 5% of CAGE tags have untemplated 5′ dG residues (see Carninci et al. 2006), which does not explain the apparent shifts.
Our approach has generated an average resolution, estimated by cross-correlation, of 1.7 bp (Fig. 4B). To represent TSS distributions within promoters as accurately as possible, we modeled this uncertainty in an assay-agnostic fashion. We estimated the resolution and smoothed the TSS distribution for each assay with a window size given by our estimate. We combined the smoothed distributions across the three assays to obtain variance-normalized consensus probability density functions (PDFs) that are the input to the following downstream analyses.
Defining promoter shapes
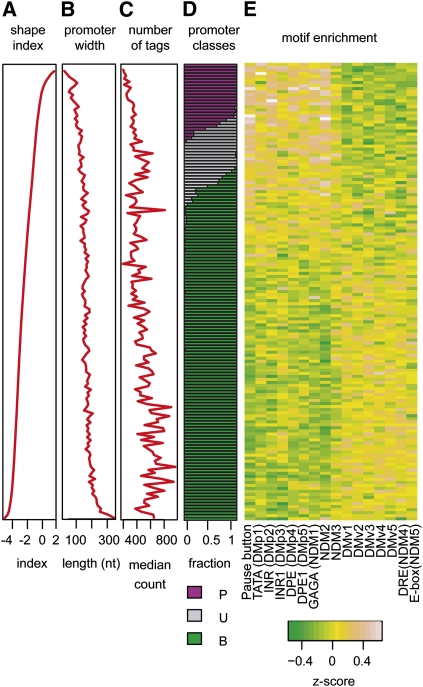
To characterize the tag distribution within each promoter region, we calculated a shape index (SI) based on the observed number of tags at each position (Methods). The SI is analogous to the thermodynamic entropy of a system and quantifies the number of states occupied by the system (the tag heights and locations) and the total possible states (the entire promoter region). The SI is distributed continuously (Fig. 5A) and is correlated with promoter width (Fig. 5B). While promoter width is bi-modally distributed (Supplemental Fig. 3), we find that SI is a better metric because unlike promoter width, it is insensitive to rare outlier tags discovered as the depth of sampling increases. There are 1351 promoters with widths >30 bp where 75% of transcription initiation events occur within 2 bp of the dominant TSS (Fig. 3C). Conversely, 90 promoters with widths <30 bp have TSS usage preferences distributed throughout a broad region. The continuous nature of the SI distribution necessarily makes classification of promoters into discrete classes somewhat arbitrary. However, to study general trends in the data, we classified promoters with SI > −1 as “peaked” and promoters with SI ≤ −1 as “broad” (Methods). Of the 12,454 promoters in the annotated set (V, S, and R), we classify 2337 as peaked, 6607 as broad, and 3456 as unclassified due to low tag count (2487) or class-instability (982) as determined by subsampling from the existing TSS distribution (Methods). A real-valued shape index has considerable advantage over fixed classifications such as those employed in previous studies, since it can be used to rank promoter shapes from most peaked to most broad. We use this property below to study the differences in expression patterns.
Figure 5.
Promoter architecture of the Drosophila embryo. Promoters are ordered by shape index, and each row corresponds to the average of a bin of 50 promoters. Shape index (A), promoter width (B), and number of tags per promoter (C) are plotted. (D) Promoter classification into peaked (P, purple), unclassified (U, gray), and broad (B, green) are indicated. (E) Core promoter motifs are differentially enriched between peaked and broad promoters.
Due to the observed offsets among the CAGE, RACE, and EST data (see above), our method of smoothing can produce an artificially broad composite TSS distribution. If we had classified all promoters that are peaked in the individual assays as peaked in the composite, then as many as 35% of promoters would be classified as peaked. Furthermore, using the previously published classification rule of Ni et al. (2010), we find that 25% of promoters are classified as “Narrow Peak,” 20% as “Broad Peak,” and the remainder as “Weak Peak”—generally, what we classified as broad. These numbers are similar to those reported by Ni et al. (2010) (26% Narrow Peak and 16% Broad Peak), but differ from those reported by Rach et al. (2009) (80% Peak, 20% Broad). Rach et al. (2009) used available EST data only, while Ni et al. (2010) and we used next-generation sequencing assays. Previous classification approaches do not provide a real-valued score for promoter shape, and hence are not as useful as the shape index. Our Supplemental Data Files allow re-analysis of our data to classify promoters according to different criteria. We also provide a movie that displays several hundred promoters in succession and ordered by their shape index that illustrates the range of TSS distributions (Supplementary Data File 5).
There is a median of 64 tags per validated peaked promoter and 182 tags per validated broad promoter. We determined that 20 tags are required, on average, to confidently infer promoter class (Supplemental Methods). Deeper coverage (promoters supported by at least 100 tags) does not, on average, lead to wider called promoters (r ∼ 0.1, ρ ∼ 0.1). This suggests that transcription in broad promoter regions is initiated probabilistically within a well-defined region. TSS distributions within broad promoters are not uniform; the probability of initiation is often complex with multiple peaks and troughs (Fig. 3H). The median width of broad promoters, 162 nt, is approximately the length of DNA in one nucleosome.
TSS distributions are stable for promoters active in both embryos and adults
To determine whether TSS distributions within promoters change during development, we performed 1920 5′ RLM-RACE experiments targeting 1681 genes using total RNA from a mixed-sex, mixed-age collection of adult flies. We generated 296,547 sequence reads and mapped and associated 262,530 with transcripts. From these data we defined 2128 promoters, including 1921 that are also in our embryonic set. In order to determine the stability of promoter shape between adults and embryos, we performed a cross-correlation analysis, as above, treating the adult RACE data as though they were a replicate of the embryonic RACE data. The estimated resolution is 0.15 bp (Supplemental Fig. 4), with 96% of promoters showing maximal cross-correlation at a shift of 0 bp. The median Pearson correlation for these promoters is r ∼ 0.85. This is in contrast to the integrative analysis of CAGE, RACE, and RE ESTs, in which 99% of validated promoters show a shift of one or more base pairs in relative TSS distribution between the assays. Hence, TSS distributions are strikingly stable for promoters active in both embryos and adults. In addition, promoters classified in embryos as peaked or broad retain their peaked or broad classification in adults with 95% identity. Finally, we discovered 185 promoters in adults that we did not observe in the embryo. Of these, 70% are found more than 100 bp from an embryonic promoter, CAGE peak, or RE EST, indicating that these constitute adult-specific promoters.
Core promoter motifs are differentially enriched in peaked and broad promoters
To determine how sequence composition varies with promoter shape, we examined nucleotide content and the occurrences of 15 core promoter motifs (Ohler et al. 2002; FitzGerald et al. 2006) and the pause button (PB) motif associated with Pol II stalling (Hendrix et al. 2008) in validated promoters with at least 100 TSS tags (Fig. 5; Supplemental Table 2). In contrast to mammalian promoters in which CG di-nucleotides are enriched in broad promoters, mono- and dinucleotide contents were similar in peaked and broad Drosophila promoters. The five positionally enriched core promoter motifs, corresponding to TATA, Inr, and DPE elements, were enriched in peaked promoters, consistent with previous reports (Rach et al. 2009; Ni et al. 2010). In addition, the GAGA and PB motifs were enriched in peaked promoters. Four core promoter motifs were overrepresented in broad promoters: the enrichments of Ohler 6 and Ohler 7 were previously reported (Rach et al. 2009), and the enrichments of NDM1 and DMv1 are new. Five remaining motifs lacked significant differential enrichment between peaked and broad promoters.
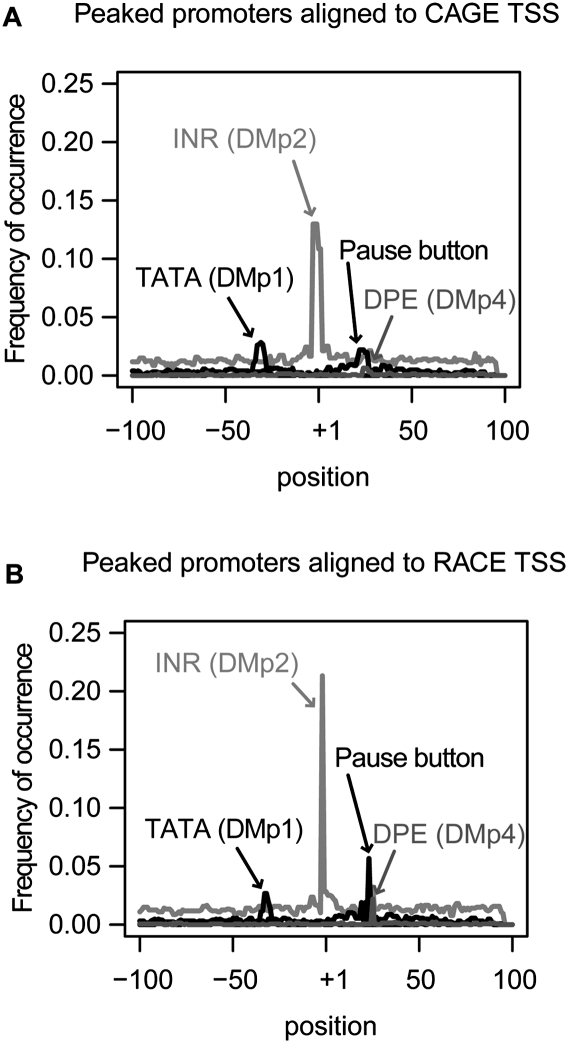
Positional enrichments of core promoter motifs were determined by computing the frequency of occurrence in a 200-bp window centered on the dominant TSS within each promoter region (Supplemental Fig. 5). The TATA-box (Lifton et al. 1978) occurred within 5 bp of position −32 in 16% of peaked promoters and 4% of broad promoters. The INR element (Smale and Baltimore 1989) occurred within 5 bp of position +1 in 70% of peaked promoters and 35% of broad promoters. The DPE element (Burke and Kadonaga 1996) occurred within 5 bp of position +26 in 5% of peaked promoters and 1.5% of broad promoters. Notably, the PB motif was positionally enriched, occurring within 5 bp of position +24 in 19% of peaked promoters and 7.8% of broad promoters. Thus, these motifs are enriched at the expected positions relative to the dominant TSS in peaked promoters, and they are also detected at the same location but at reduced rates relative to the dominant TSS peak in broad promoters (Supplemental Fig. 5).
To assess differences in the CAGE and RACE assays, we studied the locations of the three most positionally enriched motifs relative to CAGE and RACE peaks in peaked promoters (Fig. 6). The average distance between RACE peaks and the TATA-box motif is −32 bp, while for CAGE peaks this distance is −30 bp. Similarly, the INR motif appears precisely at RACE peaks but is shifted by +1 bp from CAGE peaks. Finally, the DPE motif maps at +25 bp relative to RACE peaks and at +24 relative to CAGE peaks. In each case, the average location of the motif relative to RACE peaks is more consistent with published studies (e.g., FitzGerald et al. 2006) and is more sharply delineated than the average location relative to CAGE peaks.
Figure 6.
Comparison of the CAGE and RACE assays by motif analysis in peaked promoters. Motif occurrence frequencies of positionally enriched motifs are plotted. The most abundant TSS within a promoter was used to define position +1. (A) Motif positions in peaked promoters relative to the most abundant TSS defined by CAGE. (B) Motif positions in peaked promoters relative to the most abundant TSS defined by RACE.
Peaked promoters are associated with restricted gene expression patterns
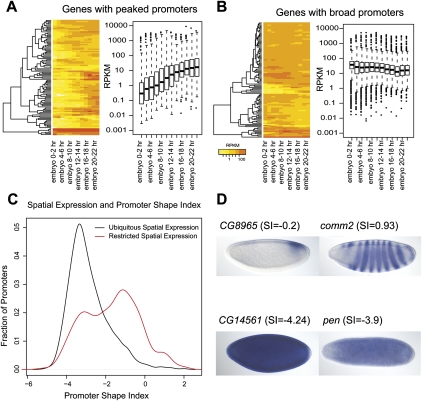
Using poly(A)+ RNA-seq data from 12 2-h windows throughout embryonic development (Graveley et al. 2011), we found that the 100 genes with the broadest promoters (lowest SI) were 2.4-fold more likely than the 100 genes with the most peaked promoters (highest SI) to have a constitutive temporal expression pattern (Fig. 7A). Of the genes with the broadest promoters, 46% were constitutively expressed across the entire 24-h period of embryonic development. Conversely, only 19% of the genes with the most peaked promoters were constitutively expressed, and 56% were expressed during no more than half the period of embryonic development (six of 12 windows).
Figure 7.
Correlation of temporal and spatial gene expression patterns with peaked and broad promoters. (A) Temporal expression profiles of 100 genes whose promoters have the highest SI scores (peaked promoters) are highly variable across a time course of embryonic development, with reads per kilobase per million (RPKM) values fluctuating between <1 (yellow) and >100 (red). The average RPKM value among these genes with peaked promoter is 0.3 at the 0–2-h time point and gradually increases to 10 at the 22–24-h time point. Expression profiles of genes with peaked promoters were also highly variable in the time course, ranging over an order of magnitude between the first and third quartiles (box plots). (B) Temporal expression profiles of 100 genes whose promoters have the lowest SI scores (broad promoters). The average RPKM is 60 across all time points. The first and third quartile RPKMs of genes with broad promoters were within one order of magnitude of the average RPKM, or between 10 and 80 across all time points. (C) Distribution of the shape index (SI) for spatially restricted genes (red) and ubiquitously expressed genes (black). (D) Representative embryonic gene expression patterns in whole-mount embryos, stages 4–5, restricted (upper two panels) and ubiquitous (lower two panels).
We examined the spatial expression patterns of 5750 genes associated with a single, classified embryonic promoter and with documented whole-mount embryonic in situ expression data (Tomancak et al. 2007). Genes with restricted spatial expression patterns tend to have a peaked promoter (mean SI = −0.7), while genes with ubiquitous spatial expression tend to have a broad promoter (mean SI = −3) (Fig. 7B). The majority of genes with peaked promoters, 344 of 401 (85%), were expressed in a spatially restricted pattern; the remainder had ubiquitous expression. In contrast, the majority of genes with broad promoters, 1238 of 1893 (65%), were expressed ubiquitously; the remainder had spatially restricted expression patterns (χ2 test, P-value < 1 × 10−16). Exemplary cases are shown in Figure 7C.
Characterization of CAGE peaks within 3′ UTRs
There are 10,670 CAGE peaks identified by more than 50 tags that do not overlap mapped RACE reads, RE ESTs, or annotated 5′ UTRs. Of these, 4153 (39% of these peaks, accounting for 1.1 million CAGE tags) overlap an annotated 3′ UTR. Such peaks have been reported previously in mammals (Carninci et al. 2006).
Neither the TATA nor the INR motif is positionally enriched in 3′ UTR CAGE peaks, but the PB and DPE motifs are sharply and twofold enriched at position −10 bp from the dominant CAGE-tag position (Supplemental Fig. 6). Surprisingly, we find that 18% of 3′ UTR, CAGE peaks have a PB motif at position −10 bp. In our promoter set, both motifs are positionally enriched 26 bp downstream from the dominant TSS. There is no significant difference in motif enrichment between the peaked and broad classes for CAGE peaks in 3′ UTRs. Hence, 3′ UTR, CAGE peaks are associated with positional signals, but differ substantially from known promoters in the locations of those signals.
Of the 7639 genes with a CAGE peak overlapping a 5′ UTR, 80% also have a peak overlapping the 3′ UTR. The strength of these reciprocal 3′ peaks correlates weakly, but not very linearly, with the strength of the 5′ peak (r ∼ 0.14, ρ ∼ 0.36), and the 3′ peak includes on average 25% as many CAGE tags (179 tags). Thus, there is a prevalent and strong CAGE signal on the sense strand within the 3′ UTRs of protein-coding transcripts. We identified RE ESTs overlapping 14 such CAGE peaks and performed full-insert sequencing to show that the cDNA clones overlap the 3′ UTR of the corresponding mRNA transcripts and terminate in poly(A) tails (Supplemental Results). Therefore, the ESTs do not represent unannotated promoters of downstream genes.
A recent study of short capped nuclear RNAs (<100 nt) in Drosophila embryo-derived S2 cells showed that virtually all such RNAs colocalized specifically with 7400 known promoters (Nechaev et al. 2010). The authors successfully characterized these short RNAs as byproducts of Pol II stalling, and, importantly, observed no 3′-UTR signal (K Adelman, pers. comm.) in contrast to our total RNA CAGE data. We conducted a brief re-analysis of these short RNA-seq data and confirmed this observation: there is no signal in a 3′ UTR except at loci where the 3′ UTR overlaps a 5′ UTR on the same strand (data not shown). Thus, these data support our conclusion that CAGE peaks in 3′ UTRs are unlikely to represent novel sites of transcription initiation. We conclude that CAGE peaks in 3′ UTRs are likely to be associated with transcript degradation products that might be recapped by a recently described cytoplasmic capping complex (Otsuka et al. 2009). Thus, the CAGE peaks within 3′ UTRs appear to represent 5′ ends of cytoplasmic transcript fragments, and not independent promoters.
Discussion
Genome-wide analysis of core promoter architecture in D. melanogaster has been limited by the availability of TSS data. Previous studies have relied on 5′ ESTs generated from large-insert cDNA libraries, including libraries constructed using methods that do not trap the 5′ cap structure (Ohler et al. 2002; FitzGerald et al. 2006; Rach et al. 2009). The recently reported PEAT clusters of Ni et al. (2010) include 4054 promoters, but only the mode of each TSS distribution is reported. We mapped large numbers of TSS tags in the developing Drosophila embryo using two independent methods: CAGE and 5′ RLM-RACE. Comparison of TSS distributions within core promoters as determined by integrative analysis of CAGE, RACE, and cap-trapped 5′ ESTs shows that these methods are consistent and cross-validating in defining promoters and determining their TSS distributions. We report 12,454 embryonic promoters and their TSS distributions (Supplemental Data File 4), providing the first well-documented, genome-wide map of Drosophila promoter architecture. As we continue to generate data on Drosophila promoters in the modENCODE project, we will maintain updated, public versions of the data files on the Berkeley Drosophila Genome Project website (http://www.fruitfly.org).
Unlike previous analysis of genome-wide TSS data, our statistical analysis recognized that the CAGE assay has a biochemical background and modeled this background to assess confidence. This had a major impact on our conclusions. We identified 143,000 CAGE peaks by clustering unfiltered CAGE data, whereas using our RNA-seq-based filtering approach to enrich for CAGE peaks associated with transcription initiation events we find only 45,000 significant CAGE peaks. As has been previously reported, CAGE tags identify a diverse population of RNA elements. We find that these include the 5′ ends of capped transcripts; 5′ ends of some uncapped transcripts including mitochondrial transcripts and rRNAs, which are very abundant in total RNA; and 5′ ends of transcript fragments that tend to be associated with 3′ UTRs. Some of these must result from <100% efficiency in the cap-trapping protocol (see Schoenberg and Maquat 2009). Our analysis of CAGE peaks in 3′ UTRs revealed little or no evidence for a class of long capped RNAs that initiate within 3′ UTRs and instead is consistent with recapping of transcript fragments. After integrative analysis with RACE, RE-ESTs, and gene annotations, we identify 20,365 CAGE peaks corresponding to annotated and putative new promoters. Thus, because our filtering and integrative analysis retained only 14% of CAGE peaks (accounting for 80% of tags), we conclude that previous analyses of CAGE data are likely to have overestimated the number of promoters in mammals by at least fivefold.
The concordance between our integrated promoters and the PEAT clusters recently reported by Ni et al. (2010) is strong near annotated promoters and weaker in other regions. Ni and colleagues used the peak-caller F-seq (Boyle et al. 2008), which was designed for analysis of DNase I hypersensitive site data and masks tags outside of dense clusters. In contrast, we systematically quantified and controlled for background signal using stranded RNA-seq data. We found that, just as in our analysis of CAGE data, a surprising number of PEAT reads (55,000 reads) map to the mitochondrial genome sequence. These clusters may be due to imperfect cap selection, but this phenomenon has been detected by three different methods (5′ RE ESTs, CAGE, and PEAT) and merits further investigation.
Our high-throughput approach to RACE using pooling and the 454 sequencing platform enabled us to target at least one promoter of an unprecedented 7238 genes or 77% of 9409 genes expressed (RPKM > 1) in the 0–24-h embryo sample. The scale of these RACE data has resulted in the characterization of 1722 promoters that were not detected by CAGE or RE ESTs. It is not yet clear why some promoters are detected by RACE but not by CAGE. The intuitive answer, that these genes tend to be expressed at low levels, does not appear to be the case. The set of 5′ transcript ends detected solely by RACE is expressed, on average, at about the same level as the set detected by both CAGE and RACE. Although confounding, this certainly underlines the need for the application of multiple, independent experimental methods to the discovery and validation of promoters.
The precise boundary between peaked and broad promoters in the continuum of our shape index is largely a subjective decision. However, our simple classification allowed us to demonstrate compelling biological correlates of promoter shape. Four classes of promoters have been defined in mammals (Carninci et al. 2006), and we initially used near-identical criteria to define four promoter classes in Drosophila. However, we observed that similar core promoter motifs were enriched between the “peaked” and “broad-with single-peak” classes, and that genes with “broad” and “multimodal” (a broad-with-multiple-peaks class defined in mammals) promoters had similar associations with constitutive gene expression profiles in the developmental time-course data. We found that the strongest discrimination was between just two classes, peaked and broad. Two such classes were defined previously in Drosophila using different criteria (Rach et al. 2009), but that study defined more peaked (81%) than broad (19%) promoters because it was based on low-coverage EST data.
Promoter shape has biological significance. First, core promoter sequence motifs are differentially enriched in the peaked and broad classes. Second, genes with peaked promoters have a marked and highly significant tendency to be expressed in spatially and temporally restricted patterns, and genes with broad promoters do not. Previous studies indicated these tendencies in mammals (Carninci et al. 2006) and Drosophila (Hendrix et al. 2008; Rach et al. 2009; Ni et al. 2010), but the statistical significance of the correlations we report is much higher. Thus, peaked and broad promoters are differentially regulated by mechanisms to be elucidated in future studies.
The CAGE, RACE, and EST data used to define our promoter set were produced from rapidly developing embryos that contain many different cell types and tissues. Thus, it is possible that in some cases mixed peaked and broad signals result from a superposition of peaked and broad promoters. This is unlikely, because these complex promoter shapes are observed in mammalian cell lines (Carninci et al. 2006). There may be subtle tissue-specific differences in TSS distributions within promoters, and this is an important area for future research.
Beyond identifying and classifying promoters, at the finer scale of TSS usage within promoter regions, the CAGE, RACE, and EST data are somewhat discordant. CAGE and RACE are independent methods, and there are many reasons why they might not produce identical results. The approaches described here represent the best available methods in current use for genome-wide TSS mapping. Integrative analysis indicates that we have achieved a resolution of 1.7 nt, near single-nucleotide resolution. We find that in peaked promoters, RACE is better correlated than CAGE with the published location preferences of the position-specific core promoter motifs, but this result may be due to the methods used to determine these published preferences rather than to an advantage of RACE. Hence, our promoter annotations are agnostic with respect to the relative accuracy or precision of CAGE, RACE, and 5′ ESTs. As additional CAGE peaks are validated using RACE or other approaches, it may become clear that one method is fundamentally more informative than the other, in which case a re-analysis of these data may sharpen the resolution we report here. The causes of these offsets and computational methods for coping with them are subjects for future study. Sources of bias may include PCR variance in CAGE and RACE, and sequence-specific preferences of T4 DNA ligase (Romaniuk et al. 1982) in RACE.
Promoter shape was highly similar between embryos and adults for promoters active in both developmental stages; 95% of promoters retain their peaked or broad classifications. We posit that the remaining 5% are due to stochastic noise in RACE data. This result indicates that TSS distributions are innate aspects of promoters, rather than dynamically controlled transcriptional modes. This is consistent with the finding of Frith et al. (2008) that TSS distributions in mammalian promoters, as determined by CAGE, can be predicted from the local DNA sequence. Thus, it may be that the TSS distribution of a promoter can be entirely characterized by assaying at a single biological sample in which the promoter is active.
Finally, the phenomenon of peaked and broad promoter architectures appears to be conserved in Drosophila and mammals. Peaked promoters are associated with position-specific motifs and spatially restricted gene expression in both. Here, we have shown that peaked promoters are also strongly associated with temporally restricted gene expression in the developing Drosophila embryo. Although CpG islands are not found in Drosophila, the broad class of promoters in Drosophila shares features in common with CpG-island promoters in mammals. Both account for a majority of promoters in their genomes, both are characterized by a broad distribution of TSSs, and both are associated with constitutive gene expression. These promoter classes may have a common origin in evolution with the mammalian lineage acquiring the CpG island as a derived feature. Thus, promoter shape appears to represent a fundamental aspect of gene regulation in animals.
Methods
EST analysis
Previously described cap-trapped 5′ RE ESTs (Stapleton et al. 2002) were reanalyzed to ensure accurate vector trimming and genomic alignment. The vector sequence at the junction in EST reads was identified using cross_match (http://www.phrap.org) and aligned to the genome using sim4 (Florea et al. 1998) centered on a region surrounding the BLAST HSP (Altschul et al. 1997). We associated an EST with a gene if an EST alignment shared genomic coordinates with either the start or stop codon, or the start or end coordinate of any exon.
RNA preparation
Total RNA was prepared from a 0–24-h collection of embryos and a collection of adults of the D. melanogaster strain of genotype y1; cn1 bw1 sp1, the same strain used to produce the reference genome sequence (Adams et al. 2000). RNA was produced using the RNeasy procedure (QIAGEN); this method reduces the representation of RNA shorter than 200 nt.
Poly(A)+ RNA-seq analysis
A poly(A)+ RNA-seq library was constructed from the 0–24-h embryo total RNA sample (10 μg) using the mRNA-Seq Sample Prep Kit (Illumina). The library was used to produce paired-end sequences of 76 nt each on the Illumina GAII platform. Sequence reads were aligned to the Release 5 reference genome sequence using TopHat (Trapnell et al. 2009), allowing up to two mismatches per read and including multiply mapped reads. We mapped 13 million reads. The FB5.12 annotation and the alignments were used to compute expression values as reads per kilobase of exon per million reads (RPKM) for all nonredundant initial exons in the annotation. These RNA-seq data have been submitted to the NCBI Sequence Read Archive (SRX015869).
Total RNA-seq analysis
Strand-specific total RNA-seq data produced using the SOLiD platform (Applied Biosystems) from 12 embryo samples in 2-h windows spanning embryonic development are described in Graveley et al. (2011). We aligned these reads using StatMap; the first 27 nt of each 50-nt read were aligned to the reference genome sequence, so that alignments would have similar biases as the alignments of CAGE tags. The alignment results from the 12 samples were combined into a single set of alignments and used to model the background of CAGE tags as described in Supplemental Methods.
CAGE
CAGE was performed on the 0–24-h embryo total RNA sample as described in Valen et al. (2009) with adaptations for the Illumina GA I sequence analyzer. A detailed protocol is provided in Supplemental Methods. The CAGE tags have been submitted to the NCBI Sequence Read Archive (SRX015329).
CAGE data analysis
A total of 41,804,261 (99.2%) of the 42,132,348 CAGE tags were trimmed to remove the 5′ adapter sequence ACACAGCAG; reads that did not match exactly to this sequence were not used in subsequent analysis. CAGE library construction can result in the addition of untemplated dG residues to CAGE products at the position of the TSS (Carninci et al. 2006). These residues were not explicitly trimmed, but were instead modeled as a random process during alignment to the genome sequence. CAGE tags were aligned using StatMap, as described in Supplemental Methods, with the command-line options -p -10 -m 2.
We modeled the stranded CAGE signal as a mixture of signal originating from promoters and background signal, as described in Supplemental Methods. Mapped filtered CAGE tags were clustered to define CAGE peaks as described in Supplemental Methods.
RACE
We performed 5′ RNA ligase mediated rapid amplification of cDNA ends (RACE) using the FirstChoice RLM-RACE procedure (Ambion) with modifications indicated below. A detailed RACE and sequencing protocol is provided in Supplemental Methods.
In the 0–24-h embryo total RNA sample, we targeted all FB5.12 transcript models that overlap 5′ ESTs from the RE (Stapleton et al. 2002) and LD (Rubin et al. 2000) cDNA libraries, both constructed from mixed-stage embryos. We added transcripts of genes expressed in the embryo based on whole-mount RNA in situ hybridization (Tomancak et al. 2007) and literature surveys. From this set of transcripts, we designed nested primer pairs for 5′ RLM-RACE. Primers matching the annealing temperature, sequence composition, and offset from the annotated 5′ transcript end specified by the manufacturer's protocol were designed using Primer3 (Rozen and Skaletsky 2000). To reduce redundancy, transcripts that share an initial exon with another transcript already selected were not included. Pairs of nested transcript-specific oligonucleotide primers were designed within 150 to 250 bp of each annotated 5′ transcript end unless the sequence composition prevented design of a suitable primer in this range. The set contains 8570 distinct primer pairs representing 7742 genes.
Primer sets were used to perform individual RACE reactions without multiplexing. The number of PCR cycles per round of nested PCR was reduced from 40 to 20 to preserve a diversity of product lengths. In 1453 cases in which no detectable product was obtained, five additional PCR cycles were added to the second round of nested PCR. RACE products were quantified and sized before being combined into molar-normalized pools of 1440 to 2787 reactions. The pooled products were sequenced on the 454 Life Sciences (Roche) platform using the manufacturer's library construction and sequencing protocols.
On the adult total RNA sample, we used 1920 of the primer pairs from the embryo RACE experiments and identical protocols to target FB5.12 transcript models known to be expressed in the adult based on overlap with RH (RIKEN Head) ESTs (Stapleton et al. 2002).
RACE data have been submitted to the NCBI Sequence Read Archive (SRA008141).
RACE data analysis
RACE reads were oriented, trimmed, aligned to the reference genome, and associated with transcripts as described in Supplemental Methods.
Defining promoters
For promoters identified by exactly one data type (supported or RACE-only promoters), we take the TSS distribution to be the empirical distribution of mapped tags. Whenever CAGE peaks, RACE clusters, or RE ESTs overlapped, we assigned the clusters to a validated promoter region. Within each promoter region we modeled the distribution of tags within each of the three assays as a multinomial distribution, with each bin corresponding to a single base pair. We then modeled the joint distributions of the mapped tag counts at each position as originating from an underlying “consensus” binomial, as described in Supplemental Methods. We defined the resolution of a validated promoter to be the maximum pairwise offset between the tag distributions of two assays. The output of this analysis is the set of consensus TSS PDFs for our promoters.
Promoter classification
The shape index of the TSS distribution within a promoter is defined as:
where p is the probability of observing a TSS at base position i within the promoter, and L is the set of base positions that have at least one TSS tag. Promoter regions with shape index score >−1 were classified as peaked (P); all others were classified as broad (B). Classifications were subject to statistical testing, as described in Supplemental Methods, to filter out ambiguous results; all ambiguous promoters were relabeled as unclassified (U).
Intersection with gene annotations
Annotation features from one gene can overlap those of another gene, so we adopted a progressive strategy for associating peaks with annotations. We first associated peaks with 5′ UTRs, then with regions within 100 bp of a 5′ transcript end (5′ end), followed by 3′ UTRs, introns, protein-coding exons, and finally other annotations (e.g., pseudogenes and regions within 100 bp of a 3′ end). The remaining peaks are classified as intergenic.
Motif analysis
Known promoter motifs were mapped using 16 position-specific scoring matrices (PSSMs) (Ohler et al. 2002; FitzGerald et al. 2006; Hendrix et al. 2008). Motifs were modeled by these PSSMs and were counted if their scores exceeded the 99th percentile score derived from sampling a background set of sequences with matching nucleotide content. Additional details are given in Supplemental Methods.
P-values and overlap analysis
P-values and associated analyses on the overlap of two sets of genomic annotations were computed using the Genome Structural Correction (GSC) statistical package available from the ENCODE Consortium (Bickel et al. 2011; http://www.encodestatistics.org). P-values have been Bonferroni-corrected by the total number of tests performed during this study. This step is highly conservative, but as in any study where a large number of tests are performed during exploratory data analysis, it is essential to prevent the reporting of spurious associations.
Acknowledgments
We thank the members of the modENCODE Drosophila Trancriptome Group for helpful discussion. We thank Elaine Mardis and the Washington University Genome Sequencing Center for Roche 454 FLX library construction and sequencing services and Leath Tonkin and the QB3 Sequencing Laboratory at the University of California, Berkeley for support and access to a Roche 454 FLX Titanium instrument. We thank Joel Rozowsky for useful discussion of CAGE data and Ann Hammonds for critical reading of the manuscript. This work was funded by an award from the National Human Genome Research Institute modENCODE Project (U01 HG004271) to S.E.C. under Department of Energy contract no. DE-AC02-05CH11231.
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession nos. SRX015329, SRA008141, and SRX015869.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112466.110.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Carninci P, Daub CO, Kawai J, Hayashizaki Y, Van Belle W, Beisel C, van Nimwegen E 2009. Methods for analyzing deep sequencing expression data: Constructing the human and mouse promoterome with deepCAGE data. Genome Biol 10: R79 doi: 10.1186/gb-2009-10-7-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel PJ, Brown JB, Boley N, Huang H, Zhang N 2011. Non parametric methods for genomic inference. Ann Appl Stat (in press) [Google Scholar]
- Biggin MD, Tjian R 2001. Transcriptional regulation in Drosophila: The post-genome challenge. Funct Integr Genomics 1: 223–234 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS 2008. F-Seq: A feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10: 711–724 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. 1996. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327–336 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R 2008. FlyBase: A database for the Drosophila research community. Methods Mol Biol 420: 45–59 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C 2006. Comparative genomics of Drosophila and human core promoters. Genome Biol 7: R53 doi: 10.1186/gb-2006-7-7-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8: 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A 2008. A code for transcription initiation in mammalian genomes. Genome Res 18: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR 1988. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature (in press). doi: 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Amairic F, Crews S, Attardi G 1978. Failure to detect “cap” structures in mitochondrial DNA-coded poly(A)-containing RNA from HeLa cells. Nucleic Acids Res 5: 637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS 2008. Promoter elements associated with RNA Pol II stalling in the Drosophila embryo. Proc Natl Acad Sci 105: 7762–7767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Goldberg ML, Karp RW, Hogness DS 1978. The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harb Symp Quant Biol 42: 1047–1051 [DOI] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80 doi: 10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol 3: RESEARCH0083 doi: 10.1186/gb-2002-3-12-research0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J 2010. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods 7: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3: RESEARCH0087 doi: 10.1186/gb-2002-3-12-research0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu HL, Michot B, Bachellerie JP 1983. Improved methods for structure probing in large RNAs: A rapid ‘heterologous’ sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5′ terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res 11: 5903–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach EA, Yuan HY, Majoros WH, Tomancak P, Ohler U 2009. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol 10: R73 doi: 10.1186/gb-2009-10-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk E, McLaughlin LW, Neilson T, Romaniuk PJ 1982. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur J Biochem 125: 639–643 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ 2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: Methods in molecular biology (ed. Krawetz S, Misener S), pp. 365–386 Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA 2000. A Drosophila complementary DNA resource. Science 287: 2222–2224 [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE 2009. Re-capping the message. Trends Biochem Sci 34: 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. 2003. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci 100: 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Baltimore D 1989. The “initiator” as a transcription control element. Cell 57: 103–113 [DOI] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, et al. 2002. The Drosophila Gene Collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res 12: 1294–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Hancock JM, Webb DA, Tautz C, Dover GA 1988. Complete sequences of the rRNA genes of Drosophila melanogaster. Mol Biol Evol 5: 366–376 [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol 8: R145 doi: 10.1186/gb-2007-8-7-r145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres TT, Dolezal M, Schlotterer C, Ottenwalder B 2009. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucleic Acids Res 37: 7509–7518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. 2009. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res 19: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara JC, DeCrease CH, Wakimoto BT 2005. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci 102: 10958–10963 [DOI] [PMC free article] [PubMed] [Google Scholar]