Abstract
We present an integrative machine learning method, incRNA, for whole-genome identification of noncoding RNAs (ncRNAs). It combines a large amount of expression data, RNA secondary-structure stability, and evolutionary conservation at the protein and nucleic-acid level. Using the incRNA model and data from the modENCODE consortium, we are able to separate known C. elegans ncRNAs from coding sequences and other genomic elements with a high level of accuracy (97% AUC on an independent validation set), and find more than 7000 novel ncRNA candidates, among which more than 1000 are located in the intergenic regions of C. elegans genome. Based on the validation set, we estimate that 91% of the approximately 7000 novel ncRNA candidates are true positives. We then analyze 15 novel ncRNA candidates by RT-PCR, detecting the expression for 14. In addition, we characterize the properties of all the novel ncRNA candidates and find that they have distinct expression patterns across developmental stages and tend to use novel RNA structural families. We also find that they are often targeted by specific transcription factors (∼59% of intergenic novel ncRNA candidates). Overall, our study identifies many new potential ncRNAs in C. elegans and provides a method that can be adapted to other organisms.
The massive amounts of data from tiling arrays and high-throughput sequencing (Margulies et al. 2005; Shendure et al. 2005; Fejes-Toth et al. 2009) have driven the discovery of novel transcripts. Many of these transcripts are functional without being translated into proteins and hence are called noncoding RNAs (ncRNAs). ncRNAs include many well-known RNA types such as rRNA, tRNA, and snoRNA, as well as small RNAs such as miRNA, siRNA, and piRNA. They also refer to more recently discovered RNA types, such as promoter-associated short RNAs (PASRs) (Fejes-Toth et al. 2009), whose function has not been well studied. Many small ncRNAs, including miRNAs and siRNAs, contribute to the complexity of regulatory networks in eukaryotes.
Before large-scale experimental data became available, genome-wide identification of ncRNAs had relied on computational approaches. For certain types of ncRNAs, specific databases and prediction methods are available, such as tRNA-SE and GtRNAdb for tRNA (Lowe and Eddy 1997; Chan and Lowe 2009), snoscan and snoRNABase for snoRNA (Schattner et al. 2005; Lestrade and Weber 2006), and miRBase for miRNA (Griffiths-Jones et al. 2008). For the more general task of identifying all ncRNAs from a genome, one common approach is based on comparative genomic analysis. For example, the methods QRNA, DDBRNA, and MSARI make use of the conservation of RNA secondary structures in identifying ncRNAs (Rivas and Eddy 2001; di Bernardo et al. 2003; Coventry et al. 2004). Newly developed methods using this approach have been applied to human (Pedersen et al. 2006; Washietl et al. 2007) and Caenorhabditis elegans (Missal et al. 2006). A large amount of conserved secondary structures, identified from multiple genomes by the so-called Infernal method, are organized into structural families in Rfam (Gardner et al. 2009), which can be used to search for their structural homologs in other genomes using the Infernal software package (Nawrocki et al. 2009).
It has been shown that different types of data provide different kinds of information to the ncRNA identification process, and computational methods could perform better by integrating multiple data types. For example, when secondary structure stability and conservation information are combined using machine learning techniques, integrative methods such as RNAz and Dynalign/SVM demonstrate improved accuracy in identifying ncRNAs in various prokaryotic and eukaryotic genomes (Washietl et al. 2005, 2007; Torarinsson et al. 2006; Uzilov et al. 2006; Torarinsson et al. 2008).
With the advent of high-throughput technologies, it is now also possible to experimentally survey novel transcriptomes to find ncRNAs (He et al. 2007). Other genome-wide experimental data could also potentially provide additional information about ncRNAs. For example, in recent studies, chromatin signatures have been used to identify large conserved ncRNAs (lincRNAs) in mammals (Guttman et al. 2009), and profiles of small RNA reads have been used to find novel ncRNAs in the Drosophila genome (Jung et al. 2010).
In this article, we describe a comprehensive model, incRNA (integrated ncRNA finder), which integrates sequence, structure, and expression data. We gathered large-scale expression data sets from the modENCODE consortium (Celniker et al. 2009; Gerstein et al. 2010), which were obtained not only by using both tiling arrays and deep sequencing but also from different tissues and developmental stages of C. elegans. In addition, our model includes a carefully selected set of other features useful for separating ncRNAs from other genomic elements. We demonstrate that our integrative method significantly improves the accuracy of ncRNA identification compared with some previous methods. Importantly, we show that no single feature could achieve the top performance of the features when combined, which strongly suggests their complementary nature and the advantage of integrative approaches.
We describe how we used incRNA to predict 7237 novel ncRNA candidates (1678 of them are intergenic ncRNA candidates) and experimentally validated the expression of a random sample. We also characterize the novel ncRNA candidates by their genomic locations, structural properties, expression patterns, binding sites of RNA polymerase II (Pol II), and 22 transcription factors. Finally, we summarize these results in a master table to facilitate future validation and functional characterization of the candidates.
Results
Single features and feature pairs only partially separate different types of annotated genomic elements
We gathered a large compendium of sequence, structure, and expression features of C. elegans that could potentially identify ncRNAs. The expression features were expression signals produced by small RNA-seq, poly-A+ RNA-seq, total RNA tiling arrays, and poly-A+ tiling arrays in different developmental stages and conditions. The sequence and structure features include GC content, genome-wide predictions of DNA and protein conservation, as well as secondary structure stability and conservation. We expected that in general, functional ncRNAs would have some expression signals and stronger GC content and DNA conservation. We also expected ncRNAs to be distinguishable from coding transcripts by having, on average, stronger small RNA-seq signals, lower poly-A+ signals, and stronger secondary structures but lower levels of predicted protein-level conservation.
Since functional ncRNAs are likely to be conserved and since RNA secondary structure prediction is more reliable in conserved regions, we only considered regions with a high conservation score (15% of C. elegans genome) from the pairwise alignment between C. elegans and Caenorhabditis briggsae. Two hundred nineteen known ncRNAs from WormBase were covered by the conserved regions, including long (rRNA), small (miRNA), and medium-sized (tRNA, snoRNA, etc.) ncRNAs. In order to predict local RNA secondary structure, we divided the pairwise alignment into small bins (each bin has 150 aligned columns; see details in Methods). Altogether we generated 439,815 bins, which cover 29,655,415 bases of the plus and minus strands. We then annotated each bin as coding sequence (CDS), untranslated region (UTR), ncRNA, or intergenic if it overlapped with the corresponding annotations from WormBase. These annotated bins were used to construct a gold-standard set (see definition in Supplemental Methods and numbers in Supplemental Fig. 3, upper panel) for further analysis. The local expression, structural, and conservation properties of each genomic element (e.g., ncRNA or CDS) can be reflected by the bins within it.
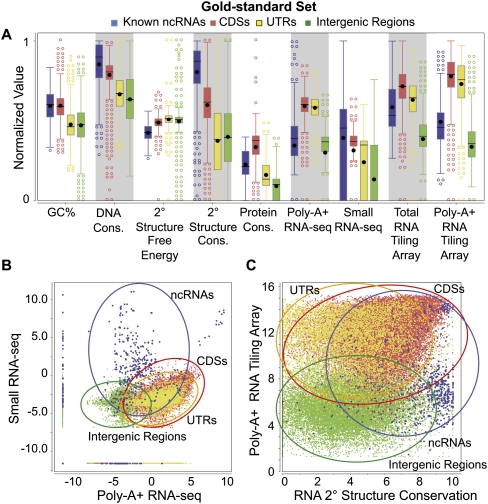
We found that different features were useful in identifying different classes of genomic elements (Fig. 1A). For example, known ncRNAs have, in general, higher signals from small RNA-seq experiments than do the other three classes of gnomic elements. However, there are also ncRNAs with very low signals, as well as CDSs with signals even stronger than those of most ncRNAs. In fact, no single feature could distinguish all known ncRNAs from other genomic elements (Fig. 1A). The four classes of genomic elements became more separated by considering pairs of features (Fig. 1B,C); yet in all cases, some ncRNAs are still indistinguishable from the other classes.
Figure 1.
Distributions of nine genomic feature values. The distributions of values of the nine features are shown for the gold-standard set (for the definition of the gold-standard set, see Supplemental Methods) of the four types of genomic elements: known ncRNAs, coding sequences (CDSs), untranslated regions (UTRs), and intergenic regions. The values of each expression feature are the maximum of the corresponding values from all the expression data sets of the same type. (A) Box plots of individual features (normalized values). (B) Two-dimensional scatter-plot of the maximum small RNA-seq signal against the maximum poly-A+ RNA-seq signal. (C) Two-dimensional scatter-plot of the maximum poly-A+ RNA tiling array signal against the predicted secondary structure conservation. Expression values in B and C are the log-transformed normalized read counts (DCPM, depth of coverage per million reads).
ncRNAs are clearly separated from other genomic elements by machine learning methods using the integrated features
Since the different features capture different kinds of information about genomic elements, we perceived that our ability to identify ncRNAs would be maximized if we integrated all the features. A systematic way to perform this integration is to employ machine-learning methods to model the subtle interactions between the features in annotated data. The learned statistical models can then be used to identify novel ncRNA candidates from the unannotated genomic regions.
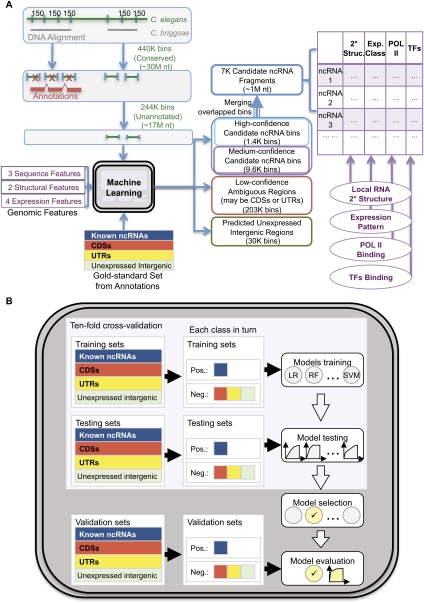
We implemented a machine learning module for this purpose and integrated it into the pipeline of incRNA with additional modules for preprocessing data and characterizing our predictions (Fig. 2A). The annotated bins from the gold-standard set were used to train machine learning models and predict ncRNAs from the unannotated bins. We took the bins predicted with higher confidence and merged them with adjacent bins that could belong to the same ncRNAs into genomic regions we call candidate ncRNA fragments. We then characterized them using various types of information. The details will be given in the coming sections.
Figure 2.
A flowchart of incRNA (integrated ncRNA finder) for predicting and characterizing novel ncRNA candidates in C. elegans. (A) We looked for ncRNAs from conserved regions from the genome alignment between C. elegans and C. briggsae and divided them into small bins. Annotated bins from the gold-standard set were used to build a machine learning model based on nine expression, sequence, and structural features. The model was then used to score each unannotated bin by its likelihood of belonging to four genomic element classes (ncRNA, CDS, UTR, and unexpressed intergenic region). Adjacent bins predicted to be novel ncRNA candidates with high or medium confidence were merged into candidate ncRNA fragments, which were further characterized by their predicted RNA secondary structures, expression patterns, and the binding signals of Pol II and different transcription factors. (B) We used an unbiased procedure to build and evaluate our machine model. Multiple models were trained and tested using cross-validation, and the one with the highest cross-validation accuracy was evaluated using an independent validation set. For details, see Methods and Supplemental Methods.
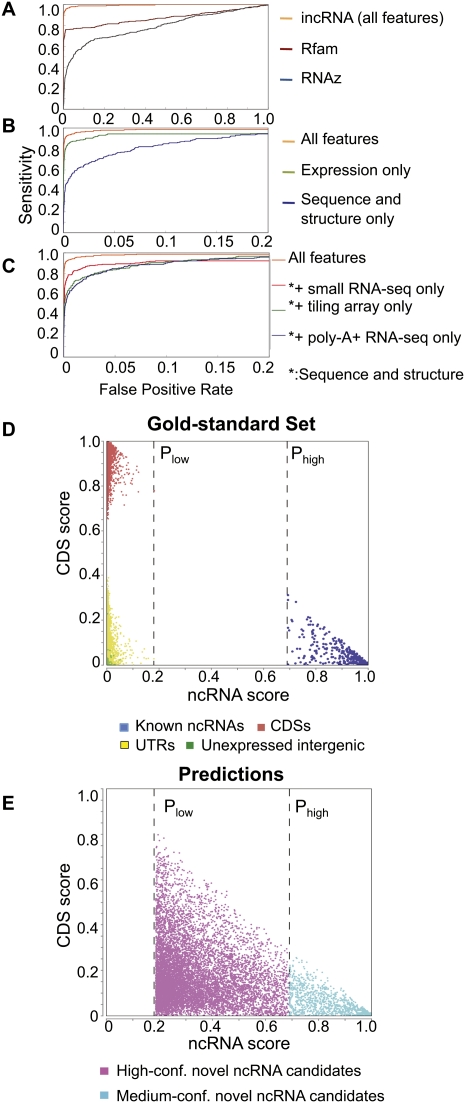
To ensure an unbiased evaluation of the effectiveness of our machine learning model, we used a rigorous procedure that involves a cross-validation step for choosing the best model from those produced by a chosen set of learning methods, and a final evaluation step using an independent validation set not involved in model learning (Fig. 2B; see Supplemental Methods). The procedure recommended Random Forest as the best learning method. When instructed to separate ncRNAs from the other genomic elements in the independent evaluation set, Random Forest showed high accuracy both in absolute terms and in comparison with Rfam/Infernal (Gardner et al. 2009) and RNAz (Washietl et al. 2005), two methods that identify sequences with potential RNA secondary structures (Fig. 3A, Supplemental Fig. 1b). The area under the receiver-operator characteristic curve (AUC) for our predictions was 97%, which indicates that our method was able to identify most of the ncRNAs in the validation set before making any false predictions. We have also separated the ncRNA examples in the independent validation set into four subsets according to their sequence identity and GC content values (lower/higher than median for the two features). The resulting AUC values of the best case (high identity, high GC) and the worst case (high identity, low GC) differ by only 0.01, which shows the robustness of our method (Supplemental Table 1b).
Figure 3.
Prediction performance of incRNA. (A) Comparison between the performance of incRNA and two previously published methods, Rfam/Infernal and RNAz. (B) Comparison between the performance of our method using all features, only expression data (tiling array and RNA-seq features), and sequence and structural features only (GC%, DNA conservation, RNA secondary structure, and protein conservation). (C) Comparison between the performance of our method using all features, and sequence and structural features in addition to only small RNA-seq, tiling array, or poly-A+ RNA-seq data. (D) Predicted ncRNA scores and CDS scores of annotated genomic elements (gold-standard set) assigned by our full model. All known ncRNA bins have ncRNA scores of at least Phigh and all other genomic elements (bins) have ncRNA scores of, at most, Plow. (E) Unannotated bins with ncRNA scores of at least Phigh form our high-confidence candidate ncRNA bins, while bins with ncRNA scores between Plow and Phigh form our medium-confidence candidate ncRNA bins. All predictions are applied on the pairwise alignments of C. elegans and C. briggsae.
We noticed that while Rfam/Infernal could very accurately identify about 80% of known ncRNAs in the gold-standard set, it was unable to distinguish the remaining 20% from the other genomic elements (Fig. 3A), which suggests potential undiscovered ncRNA structural families. The comparison with Rfam/Infernal and RNAz also suggests that information about RNA secondary structure alone is not sufficient to separate all ncRNAs from other genomic elements. This is possibly related to a previous finding that local RNA structures can also occur in coding regions (Katz and Burge 2003). We also need to note that Rfam/Infernal and RNAz could also pick up structured regulatory elements in UTRs, since these two programs aim to detect structured RNA sequences instead of ncRNA genes only. The comparison has some caveats because the specificities of Rfam/Infernal and RNAz will not be high by definition (UTRs containing no known ncRNA genes are negatives in our training set).
To ensure the robustness of our analysis, we repeated the machine learning procedure using various definitions of negative sets and various combinations of genomic element types (see Supplemental Methods). Our prediction accuracy for ncRNAs was very stable in the different cases (Supplemental Fig. 1a; Supplemental Table 1A). Interestingly, we observe that some ncRNA types form distinct clusters in the feature space (Supplemental Fig. 2). For example, rRNAs have a unique signature of high conservation and high expression values in both poly-A+ and small RNA-seq experiments; miRNAs have high small RNA-seq signals but relatively low poly-A+ RNA-seq signals; snoRNAs have small ranges of medium expression values in both types of RNA-seq experiments; and tRNAs form several distinct clusters, with different RNA-seq signals and levels of conservation. Integrating different types of data could thus not only distinguish ncRNAs from other classes of genomic elements but also potentially differentiate between different ncRNA types. Prediction at this granularity is currently limited by the small number of examples of certain types of ncRNAs.
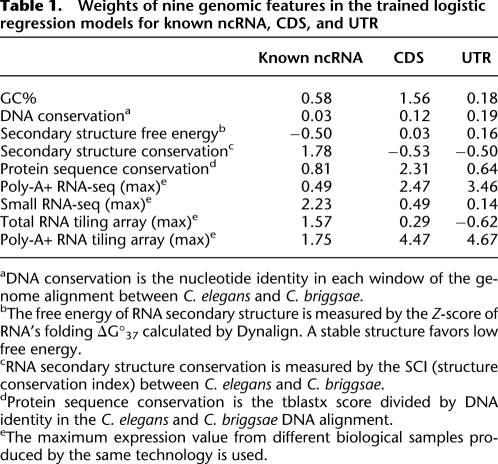
Table 1.
Weights of nine genomic features in the trained logistic regression models for known ncRNA, CDS, and UTR
aDNA conservation is the nucleotide identity in each window of the genome alignment between C. elegans and C. briggsae.
bThe free energy of RNA secondary structure is measured by the Z-score of RNA's folding ΔG°37 calculated by Dynalign. A stable structure favors low free energy.
cRNA secondary structure conservation is measured by the SCI (structure conservation index) between C. elegans and C. briggsae.
dProtein sequence conservation is the tblastx score divided by DNA identity in the C. elegans and C. briggsae DNA alignment.
eThe maximum expression value from different biological samples produced by the same technology is used.
Top prediction performance requires the integration of all features
We next studied the relative importance of the different features by checking the resulting accuracy of our machine learning procedures when we used only a subset of features. We found that the expression features were better at identifying ncRNAs than structural- and sequence-based information (Fig. 3B), yet combining both types of features gives an additional 5%–10% precision at a wide range of false-positive rates. It is thus useful to include both types of data in identifying ncRNAs.
As a way to reduce the cost of experiments, it is interesting to see if it is sufficient to only include the expression data from a single type of experiment. We found that no single type of expression data could completely substitute for the others (Fig. 3C), suggesting that each type of experiment is able to identify some unique ncRNAs.
We also used the weights of the different features in the learned Logistic Regression model as a second indicator of the importance of the features (Table 1). Consistent with the above analysis, we find that while small RNA-seq and RNA secondary structure conservation have the heaviest absolute weights in the ncRNA class, some other features, such as poly-A+ RNA-seq and tiling array data, are also heavily weighted in other classes, indicating their different roles in separating ncRNAs from other genomic element classes.
Using incRNA to predict novel ncRNA candidates
Having verified the predictive power of incRNA on the gold-standard set, we then extended our predictions to all the bins. For each (annotated and unannotated) bin, our model gives an “ncRNA score” that indicates the likelihood that the bin lies in an ncRNA gene. It also gives a CDS score, a UTR score, and an intergenic region score in similar ways. From the bins in the gold-standard set, we find that our model separated known ncRNAs from the other bins by a large score margin (Fig. 3D). These results stand in sharp contrast to the unsatisfactory differentiations made by individual features and feature pairs (Fig. 1), and provide more proof of the robustness of our method.
In particular, we found that all known ncRNA bins had predicted ncRNA scores of at least 0.69, while all the other elements had scores of 0.18 at most (Fig. 3D). We called these values Phigh and Plow, respectively, and used them as thresholds for defining novel ncRNA candidates. Specifically, we defined each unannotated bin with an ncRNA score of Phigh or higher as a high-confidence candidate ncRNA bin and each unannotated bin with a ncRNA score between Plow and Phigh as a medium-confidence candidate ncRNA bin (Fig. 3E, see also Fig. 2A). Altogether, the two sets contain 10,994 bins (covering 1,045,795 bases in total), among which 1413 are high-confidence predictions and 9581 are medium-confidence predictions.
To estimate the accuracy of these candidate ncRNA bins, we repeated the above procedure of defining the two thresholds using only the cross-validation set and then examined the ncRNA scores of the bins in the independent validation set. We found that all the bins with ncRNA scores higher than Phigh, and 221 out of 242 bins with ncRNA scores higher than Plow, were known ncRNA bins, corresponding to positive predictive values (PPVs) of 100% and 91%, respectively. On the other hand, since there are a total of 247 known ncRNA bins in the validation set, the two thresholds lead to prediction sensitivities of 66% and 89%, respectively.
The two sets of candidate ncRNA bins thus serve different purposes. The high-confidence set is expected to have few false-positives, while the medium-confidence set provides additional coverage so that the two sets together have relatively few false-negatives. We expect some of the medium-confidence candidates to be novel ncRNAs with properties different from those of known ncRNAs.
We have examined the 9% of the bins included in the medium-confidence set but are not annotated as ncRNAs (the false-positives) and the 11% ncRNAs not included in the medium-confidence set (the false-negatives). Among the false-positives, 58% are CDSs and 42% are UTRs. They are characterized by a particularly weak tiling array and poly-A+ RNA-seq signals. The false-negatives are characterized by particularly strong poly-A+ RNA-seq signals, weak predicted structures, and low sequence identities.
For bins with ncRNA scores below Plow, we further divide them into two sets. The bins with very low ncRNA, CDS, and UTR scores are grouped into a predicted set of unexpressed intergenic regions. The remaining bins form a low confidence set of ambiguous regions, which could be CDS or UTR (see Supplemental Methods; Fig. 2A). We summarize the number of bins in each set in Supplemental Figure 3.
Since different predicted bins may come from the same ncRNA genes, we merged adjacent bins into longer regions. We call them candidate ncRNA fragments, as they represent candidate regions of novel ncRNA but may not cover the full-length genes. Altogether we produced 7237 such fragments from the high-confidence and medium-confidence sets of candidate ncRNA bins. The length distribution of these fragments is comparable to that of full-length known ncRNA transcripts (Supplemental Fig. 4).
Many candidate ncRNA fragments are inside the introns of coding genes, at the antisense strand of exons or close to coding sequences (see Supplemental Methods). Among the 7237 candidate ncRNA fragments, 1678 of them (merged from 2469 bins) are located in strictly defined intergenic regions based on our gold-standard annotations (see Supplemental Methods), which occupy 1223 distinct genomic locations, not considering the strand information. These intergenic candidates are most likely to be novel ncRNAs, as their expression is unlikely to be due to nearby coding genes. On the other hand, the other candidates could also be authentic ncRNAs, as many ncRNAs have been found inside introns (Bartel 2004; Li et al. 2007) and antisense to exons (Mercer et al. 2008).
About 20% of the candidate ncRNA fragments are overlapped with repeats or inverted repeat regions. It is well known that many pi-RNAs are transcribed from transposons and other repeated sequence elements (Klattenhoff and Theurkauf 2008). Recent analysis has also identified telomeric repeat-containing RNA (TERRA) in animals and fungi (Luke and Lingner 2009). We therefore decided to keep these predictions in our set.
We overlapped our intergenic candidate ncRNA fragments with the novel genelets generated by the modENCODE consortium (Gerstein et al. 2010), where all the splice junctions and all the splice leader sites were incorporated in order to map spliced poly-A+ RNA-seq reads. A small fraction of our intergenic candidate ncRNA fragments (44 of the 1678) are overlapped with the genelets. This is expected because most of our novel ncRNA candidates are probably not having poly-A tails. Further experiments are needed to confirm if they are unannotated coding exons or novel ncRNAs.
Experimental validation of predicted novel ncRNA candidates
In addition to computational validation of our predictions, using an independent set of annotated regions not involved in model learning, we also validated a random sample of our candidate ncRNA fragments located inside intergenic regions by means of RT-PCR experiments. To get a better sense of the lengths of these potential noncoding transcripts, we overlapped our candidate ncRNA fragments with the transcriptionally active regions (TARs) obtained from the tiling array data sets of the modENCODE consortium (Gerstein et al. 2010). We call the resulting overlapped regions the candidate ncRNA TARs. As the TARs were defined from expressed regions of at least 100 nucleotides (nt) long, many novel ncRNA candidates from short transcripts are not covered by our TARs. Only 730 of the longer fragments among our 1678 intergenic candidate ncRNA fragments overlap with the TARs in at least one of the 41 tiling array data sets. Among them, 54 overlap in late embryos. We then picked 15 candidate ncRNA TARs (Supplemental Table 2) for RT-PCR validation using total RNA from N2 late embryos (see Methods). We detected clear bands on 2% agarose gel for 14 of the 15 candidate ncRNA TARs, and of those, 13 had significantly greater amplification than the control reactions to which no reverse transcriptase was added (Fig. 4A). All the RT-PCR products were confirmed by direct DNA sequencing (see Methods). The results confirmed that the expression of our predictions (especially repeat-related ncRNA candidates 1 and 3) is not likely due to cross-hybridization in tiling arrays.
Figure 4.
Validation of our novel ncRNA candidates. (A) Fifteen novel ncRNA candidates were tested using RT-PCR. Lanes 1–15 on the left (+RT) correspond to the PCR results of the novel ncRNA candidates, and lane 16 is the positive control, HEX-3. The right lanes (−RT) are the negative controls without reverse transcriptase. The PCR sizes are ∼150 bp in length. Fourteen of the candidates (except lane 15) were detected on the gel, among which 13 (except lane 11) showed a clear enrichment of expression signals in contrast to the negative control. (B) Example of a validated novel ncRNA candidate (ncRNA at lane 7 in A) with support from multiple information sources. The first and second rows (from top) correspond to the ChIP-seq reads from PHA-4 and Pol II, respectively. The heights of signals are normalized by their total mapped reads. The fourth and fifth rows are the log-transformed values of the tiling array in late embryo and its TARs. The bottom two rows are the reads from small RNA sequencing and the reads from poly-A+ RNA sequencing. The last row is the annotated genes from RefSeq.
We further estimated the expression and size of five out of 15 candidate ncRNA TARs (Supplemental Table 2) using the Northern blot analysis. Because the high-throughput data we used are very sensitive, our model is able to predict many lowly expressed ncRNA candidates (Supplemental File 1). The Northern blot method is less sensitive than the RT-PCR analysis, and can only detect three out of five candidates (Supplemental Fig. 5a). We estimated from the Northern blot that two of them are larger than 500 bases. We also checked their genome location and did not find any annotated genes close to them (within ∼2 kb). This proves that our model has the potential to identify novel long ncRNAs. Because of the complicated process in ncRNA (e.g., miRNA precursor), it is hard to estimate the real length of the ncRNA precursor or mature product using tiling array TARs. This is one of the reasons for the low sensitivity of the Northern blot, since the probe we selected may miss the highly expressed part (e.g., mature sequence part of miRNA).
Furthermore, we found that many novel ncRNA candidates are supported by multiple information sources. An example is shown in Figure 4B for one candidate (lane 7 in RT-PCR gel), which is located inside a transcribed region detected by the tiling array in late embryo. No significant expression was detected from Poly-A+ RNA sequencing or small RNA sequencing, indicating that it is unlikely to be a coding gene or a small interfering RNA. From ChIP-seq data, we observe a Pol II binding peak immediately upstream of ncRNA (lane 7) and strong binding signals of PHA-4 across the region. These binding signals suggest potential regulation of ncRNA7 by Pol II and PHA-4. Since PHA-4 is a key factor that regulates the development of the pharynx/foregut during embryogenesis, this candidate ncRNA could potentially play a role in development during the embryonic stage. Two more examples are shown in Supplemental Figure 5b,c.
Characterizing novel ncRNA candidates with RNA secondary structure and DNA conservation
We next studied various properties of the novel ncRNA candidates. We first examined the local structural and expression properties using the candidate ncRNA bins and then analyzed Pol II and transcription factor binding signals using the candidate ncRNA fragments (merged bins), as the positional relationship between binding sites and ncRNAs are better captured by the fragments. All these results are presented in the coming sections.
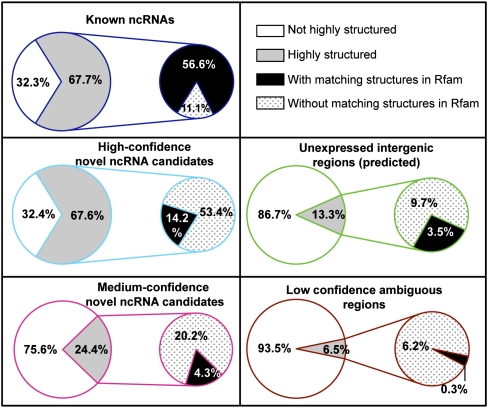
As known ncRNAs generally have stable secondary structures (Fig. 1), we first calculated the potential consensus free energy for both the known ncRNA bins and candidate ncRNA bins with the Dynalign program (Harmanci et al. 2007). We predicted about two-thirds of the known ncRNA bins and high-confidence candidate ncRNA bins to be highly structured (Fig. 5). The fraction is lower for the medium-confidence bins (about one-fourth), yet it is still much higher than the bins predicted to be unexpressed (13%) or ambiguous (6.5%).
Figure 5.
Structural properties of the known ncRNAs and novel ncRNA candidates. The percentages of highly structured ncRNA bins (with predicted Z-score of folding free energy less than the mean by at least one standard deviation) are shown for the known ncRNA bins and for high-confidence and medium-confidence candidate ncRNA bins. Among the highly structured bins, the percentages that overlap with structural homologs of Rfam families are also shown. The same calculations were performed for the bins predicted to be unexpressed intergenic regions and the low-confidence ambiguous regions (bins) for comparison.
We checked if the highly structured novel ncRNA candidates have secondary structures similar to the known ncRNAs. When we used Infernal to predict structural homologs of known families in the C. elegans genome cataloged in Rfam (Gardner et al. 2009), 84% of the highly structured known ncRNA bins (56% of 67.7%) were found to overlap with the Rfam/Infernal predictions (Fig. 5), which is consistent with our earlier observation that Rfam/Infernal could only predict approximately 80% of the known ncRNAs before producing substantial false-positives (Fig. 3A). In contrast, about 80% of the candidate ncRNA bins we predicted to be highly structured did not overlap with the Rfam/Infernal predictions (Fig. 5), which suggests potential novel structural families of ncRNAs. The enrichment of novel structures is also observed for the subset of novel ncRNA candidates inside intergenic regions (Supplemental Fig. 6).
We also compared the characteristics of known ncRNA and novel ncRNA candidates using other genomic features (Supplemental Fig. 7; Supplemental Table 3). The high-confidence candidate ncRNA bins are found to share many common properties with known ncRNA bins, including high conservation between C. elegans and C. briggsae at DNA and RNA level. The conservation of the medium-confidence candidate ncRNA bins is relatively low (Supplemental Fig. 7).
As conservation is an indicator of potential function of ncRNAs, we also used a multiple sequence alignment between five nematode species to double-check the conservation of our predictions. We found that 94% of our candidate ncRNA bins overlap the conserved aligned regions (see Supplemental Methods), which provides extra support for these predictions.
Characterizing novel ncRNA candidates with expression patterns
Next, we compared the expression profiles of the known ncRNA transcripts and novel ncRNA candidates. In particular, we analyzed whether they exhibit developmental stage-specific expression patterns. Since our small RNA-seq data cover the largest number of developmental stages (see Supplemental Methods), in this part of analysis we focus on this type of expression data. Also, since miRNAs form a major class of small RNAs among all known ncRNAs, we separate them from other known ncRNAs to determine whether they have distinct expression patterns as reported by the small RNA-seq experiments.
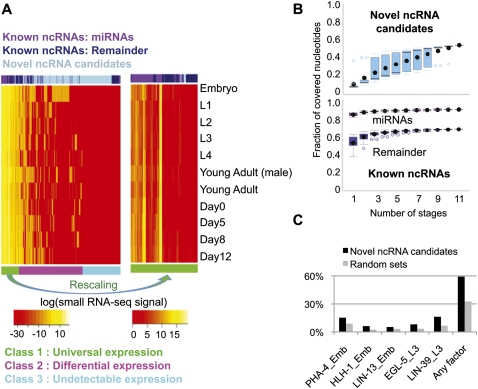
By clustering the bins according to their expression patterns across the developmental stages, we observe that there are three subclasses of ncRNAs (Fig. 6A):
ncRNAs that are universally expressed in all stages. This subclass contains 58% of the known ncRNAs, with a slight enrichment of miRNAs. Some of the ncRNAs in this subclass exhibit fluctuations of expression levels across stages (Fig. 6A, right panel).
ncRNAs that are expressed only in some stages. This subclass contains only 2% of the known ncRNAs, with a significant enrichment of novel ncRNA candidates (P-value from Fisher's exact test less than 1 × 10−15).
ncRNAs with no detectable expression by small RNA-seq in all stages.
Figure 6.
Expression patterns of the novel ncRNA candidates and binding signals of Pol II and 22 transcription factors around their genomic regions. (A) Expression patterns of known miRNA transcripts, other known ncRNA transcripts, and the candidate ncRNA bins based on our small RNA sequencing data at 11 developmental stages. Expression values are the log-transformed normalized read counts (DCPM, depth of coverage per million reads). Three subclasses formed according to the expression patterns are shown in the bottom row in three different colors. All known ncRNA transcripts are shown, while 1000 bins were randomly sampled from a total of 10,994 candidate ncRNA bins for this heat map visualization. The right panel shows a magnified view of class 1, with the colors rescaled to show the fluctuation of expression patterns across the different developmental stages. (B) Saturation plots of expressed known ncRNA transcripts and candidate ncRNA bin in different developmental stages. The fractions of expressed regions (with small RNA-seq signals stronger than the average signal of gold-standard intergenic regions) at the 11 developmental stages are computed using all possible combinations of the stages. The x-axis corresponds to the number of stages considered, and each point at a given number of stages corresponds to a different combination of stages. (C) Fractions of intergenic candidate ncRNA fragments potentially targeted by a selected subset of transcription factors. The total fractions targeted by any of the transcription factors in any of the stages are also shown. Each bar is labeled by the name of the transcription factor followed by the stage at which the binding experiment was performed. The bindings on random genome locations with the same size are also shown. EMB, embryo; YA, young adult.
The numbers of high-confidence and medium-confidence candidate ncRNA bins in the three subclasses are shown in Supplemental Table 4, with the numbers of candidates inside intergenic regions shown separately in the table. In general, the expression levels of novel ncRNA candidates are lower than the universally expressed known ncRNAs, such as rRNAs and tRNAs. The finding that most differentially expressed ncRNAs come from the novel candidates is intriguing. It could suggest that the novel ncRNA candidates play more specialized roles in specific stages.
To gain more insight into how differential expression affects the identification of ncRNAs, we identified the bins with detectable expressions in each combination of developmental stages (see Supplemental Methods). Then for each combination of stages (e.g., L2 + L4 + Young Adult), we computed the fractions of expressed known miRNAs, other known ncRNAs, and candidate ncRNA bins. Finally, we grouped all combinations with the same number of stages (e.g., both L2 + L3 and L4 + Young Adult have two stages) to form a distribution of expressed fractions. The resulting plot, called the saturation plot, is shown in Figure 6B.
We observe that the amount of novel ncRNA candidates with detectable expressions keeps increasing when small RNA-seq data sets of more stages are added, suggesting that for each stage there is a set of novel ncRNA candidates that express only in that stage (Fig. 6B). The same trend is observed when we consider all our expression data sets (Supplemental Fig. 8). In contrast, for known ncRNAs, saturation is reached when only a few stages are considered, regardless of the exact combination of the stages (Fig. 6B).
We notice that in the embryonic stage there is a much larger fraction of expressed novel ncRNA candidates (Fig. 6B). This observation is consistent with a finding in a previous study (Kato et al. 2009) showing that 65.6% of small RNA sequencing reads in embryos could not be mapped to known miRNAs or 21U-RNAs, whereas the percentages of unmapped reads in other hermaphrodite stages were only 27%–38%. Additional experiments in embryo replicas are needed to provide further confirmation.
Characterizing intergenic novel ncRNA candidates with binding sites of Pol II and different transcription factors
As a first step to understanding the underlying mechanisms that caused the observed expression patterns, we studied the potential regulation of the novel ncRNA candidates by Pol II and transcription factors. We obtained the binding signals of Pol II and 22 transcription factors from ChIP-seq experiments produced by the modENCODE consortium (Gerstein et al. 2010). We looked for Pol II binding signals near the starting site (±150 bp) of the intergenic candidate ncRNA fragments (see Methods) and found that about 15% of them had significant Pol II binding signals in at least one of the seven developmental stages (Supplemental Fig. 9). Using previous results from the modENCODE consortium, we also located the binding sites of 22 transcription factors, with a total of 27 experiments across different developmental stages (see Methods). Many of these binding sites are located at potential promoter regions of about 59% of intergenic candidate ncRNA fragments (Fig. 6C; Supplemental Fig. 9). Compared to random genome locations with the same size, the binding of TFs has twofold enrichment for the intergenic candidate ncRNA fragments (P-value < 0.001; Fig. 6C). However, the enrichment or depletion of Pol II binding is not significant (P-value > 0.05; Supplemental Fig. 9). This is consistent with the known ncRNAs, where only certain types of ncRNAs (e.g., miRNA) are transcribed by Pol II. The binding sites of Pol II as found from ChIP-seq data may still be true, although we did not find any enrichment.
Master table of novel ncRNA candidates
We summarize our whole list of novel ncRNA candidates (Supplemental Table 5) predicted from incRNA in a master table (Supplemental File 1), and the subset in intergenic regions in another table (Supplemental File 2). Each candidate is associated with its prediction scores, predicted class, feature values, genomic coordinate and location (such as inside intron or intergenic region), structural properties, expression pattern class, binding signals of Pol II and transcription factors, and the conservation score among the five nematodes. In order to facilitate follow-up studies, we have ranked our predictions into nine levels based on these associated pieces of evidence (see Supplemental Methods). The predictions in higher levels are more likely to be functional ncRNAs.
Discussion
We have shown that none of the individual expression, sequence, and structural features is able to clearly separate known ncRNAs from protein coding sequences and UTRs. Instead, by integrating all the features using a machine learning framework, incRNA, we were able to identify ncRNAs with high accuracy. The learned model is robust, with prediction accuracy above 97% AUC, regardless of the expression thresholds for defining the unexpressed intergenic regions (Supplemental Fig. 1a). The massive experimental data from the modENCODE consortium (Gerstein et al. 2010) contributed significantly to our model.
We have validated our predictions by multiple means. The validation RT-PCR experiments have confirmed that the expression levels of our novel ncRNA candidates are not likely due to the cross-hybridization of the tiling array or a mapping error in RNA-seq. The sequences of the RT-PCR products were verified by direct DNA sequencing, and sizes of the candidate ncRNA transcripts were estimated by Northern blots. Most of our candidate ncRNA bins (94%) were found to be conserved based on a multiple sequence alignment of five nematodes that was not used in model learning. We have also used cross-validation and an independent validation set of known ncRNAs to show that our models are able to identify known ncRNAs that were not included in the model training process. To further identify the subset of our predictions corresponding to functional noncoding genes, more biological functional assays need to be performed in addition to these computational and biochemical validations.
The accuracy of RNA secondary structure predictions (percentage of known base pairs that are correctly predicted) using Dynalign (Harmanci et al. 2007) and RNAz (Washietl et al. 2005) depends heavily on the quality of the genome alignment. Research has shown that the sensitivity of ncRNA sequence identification gets worse as the DNA sequence identity drops below 60% in the alignment (Gorodkin et al. 2009). Among our aligned bins, 26.5% have a sequence identity of less than 60%. However, we still successfully identified most ncRNAs (Fig. 3), because other features involved in the machine learning models are not directly affected by the degree of conservation. We also remark that methods that use local alignments, such as FOLDALIGN (Havgaard et al. 2007), have the potential of making better structure predictions for short RNAs, which may in turn increase the relative contribution of the structural features in the overall ncRNA identification process.
In the C. elegans genome, only a small portion (13%–15%) could be aligned with C. briggsae (Missal et al. 2006) at a quality level sufficient for Dynalign or RNAz to make meaningful predictions. The small portion of aligned genome illustrates a tradeoff between precision and coverage; although the information about secondary structures and conservation could improve prediction accuracy (Fig. 3B), it also excluded a lot of genomic regions that could potentially contain novel ncRNA candidates. In this study our goal is to identify ncRNA candidates with high confidence, and thus we decided to focus on the highly conserved regions with a 73% median DNA identity. For studies that aim at high coverage, one possible alternative approach is to use weakly aligned regions from the multiple alignment of MULTIZ, which provides a better coverage (∼25% of the genome) (Kiontke and Fitch 2005), with the tradeoff of a lower conservation (66% median DNA identity). This approach would generate a more permissive set of ncRNA candidates.
One direct consequence of using a genome alignment is the possibility of having a novel ncRNA only partially covered by the aligned regions. We expect the potential ncRNA fragments we constructed by merging candidate ncRNA bins to be shorter than the full-length ncRNA transcripts. Nevertheless, these fragments form a detailed map of genomic regions that could be used to locate novel ncRNAs at a more refined level.
A significant portion of our novel ncRNA candidates are overlapped with repeats and introns, or are antisense to exons. This is partially due to the compactness of the C. elegans genome. While many interesting ncRNA candidates have been previously found in these kinds of regions, caution should be taken when studying them, as the expression levels of intronic or antisense ncRNA candidates could have been affected by the enclosing genes. In order to facilitate follow-up studies that require a more conservative set of predictions with high precision, we have defined a set of intergenic novel ncRNA candidates, which are far away from annotated coding sequences (see Supplemental Methods).
The tradeoff between precision and coverage is also illustrated by our choice of unannotated regions. In order to minimize the number of false-positives in our predicted set of intergenic ncRNAs, we used a permissive set of genome annotations, such that bins that overlap with any confirmed, unconfirmed, and predicted genes from multiple models were all excluded from our final set of predictions.
Since our machine learning models were trained on known types of ncRNAs, they tend to predict novel ncRNA candidates that exhibit properties similar to these types of annotated ncRNAs. Expressed regions with more distinct properties are likely predicted as CDS-like or UTR-like ambiguous regions. For instance, since the annotated ncRNAs have low poly-A+ RNA signals in general (Fig. 1), this feature was used as a negative indicator of ncRNAs, as reflected by its negative weight in the Logistic Regression model (Table 1). As a result, our novel ncRNA candidates tend to have weak poly-A+ RNA signals. Also, while the majority of known ncRNAs are small- or medium-sized, we also included some non-mRNA-like long ncRNAs (i.e., rRNAs) in our training set. In our predictions, there are indeed fragments of long candidate ncRNA transcripts (>500 nt) (Northern blot result in Supplemental Fig. 5a).
Methods
Machine learning methods
We combined the sequence, structure, and expression data sets and generated nine genomic features for training the machine models and making the predictions. Among the nine genomic features, four of them are expression features, corresponding to the maximum signals of (1) the six poly-A+ RNA-seq experiments, (2) the 11 small RNA-seq experiments, (3) the 29 total RNA tiling arrays, and (4) the 12 poly-A+ RNA tiling arrays. Three of the features are related to sequence information, including GC%, DNA conservation, and predicted protein sequence conservation. The remaining two features are related to RNA structures, namely, predicted secondary structure free energy and predicted secondary structure conservation. The details of the nine gnomic features and machine learning method are described in the Supplemental materials.
RT-PCR and Northern blot confirmation
We used a three-step process to pick candidate ncRNAs for validation. First, we overlapped candidate ncRNA bins with the tiling array TARs defined and optimized by the modENCODE consortium (Agarwal et al. 2010; Gerstein et al. 2010). We then discarded TARs that overlap with any exonic or intronic regions using a permissive set of annotations from WormBase, and those predicted as ncRNAs by Rfam. Finally, we retained only TARs with a positive small RNA-seq read count or with a log2-transformed, normalized total RNA tiling array signal larger than 8 at late embryo. Subsequently, 15 candidates (Supplemental Table 3) were randomly picked from the remaining set for validation.
Total RNA was isolated from N2 late embryos and treated with DNase I from Ambion. Reverse transcription was then performed using random hexamers following the Omniscript RT kit instruction from QIAGEN. Subsequently, primers were designed within the candidate regions and amplified by PCR. The RT-PCR products were purified using the PCR purification kit (QIAGEN). The purified products (10–20 ng DNA) were then sent to the Keck Biotechnology Resource Laboratory (Yale University, New Haven, CT) for direct sequencing. The details of sequences, PCR primers, and products are provided in Supplemental materials.
We then manually picked five ncRNA candidates (out of 15 candidates validated above) with clear signals on the PCR gel to undergo a Northern blot assay. The total RNA was extracted from late embryos and hybridized with labeled probes (see detail in Supplemental materials).
Scoring novel ncRNA candidates with Pol II and transcription factor signals
Pol II and transcription factor binding data from ChIP-seq experiments were scored using the method from the modENCODE consortium (Gerstein et al. 2010) with a default PeakSeq q-value cut-off (Rozowsky et al. 2009) of 0.001. We defined the sets of candidate ncRNA fragments potentially targeted by Pol II and the various transcription factors using a previous target calling method (Zhong et al. 2010). Random locations from the genome with the same size as candidate ncRNA fragments were tested using the same binding peaks. We repeated the random process 20 times, and the average chances of being targeted were plotted in Figure 6C and Supplemental Figure 9.
Acknowledgments
We thank David H. Mathews and Shan Zhao at the University of Rochester for providing the shuffled RNA sequence free energy calculation method. We thank the Yale University Biomedical High Performance Computing Center for its support, which is funded by NIH grant RR19895. We thank Nicholas Carriero from the Computing Center for helping us to map small RNA sequencing reads.
Footnotes
[Supplemental material is available for this article. All data sets, prediction results, and the prediction software are available at http://incrna.gersteinlab.org/.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.110189.110.
References
- Agarwal A, Koppstein D, Rozowsky J, Sboner A, Habegger L, Hillier LW, Sasidharan R, Reinke V, Waterston RH, Gerstein M 2010. Comparison and calibration of transcriptome data from RNA-Seq and tiling arrays. BMC Genomics 11: 383 doi: 10.1186/1471-2164-11-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM 2009. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coventry A, Kleitman DJ, Berger B 2004. MSARI: Multiple sequence alignments for statistical detection of RNA secondary structure. Proc Natl Acad Sci 101: 12102–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Bernardo D, Down T, Hubbard T 2003. ddbRNA: Detection of conserved secondary structures in multiple alignments. Bioinformatics 19: 1606–1611 [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. 2009. Rfam: Updates to the RNA families database. Nucleic Acids Res 37: D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorodkin J, Hofacker IL, Torarinsson E, Yao Z, Havgaard JH, Ruzzo WL 2009. De novo prediction of structured RNAs from genomic sequences. Trends Biotechnol 28: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmanci AO, Sharma G, Mathews DH 2007. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics 8: 130 doi: 10.1186/1471-2105-8-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havgaard JH, Torarinsson E, Gorodkin J 2007. Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput Biol 3: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang J, Liu T, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, et al. 2007. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res 17: 1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS 2010. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics 11: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L, Burge CB 2003. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res 13: 2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K, Fitch DH 2005. The phylogenetic relationships of Caenorhabditis and other rhabditids. WormBook 11: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Tang P, Lin WC 2007. Intronic microRNA: Discovery and biological implications. DNA Cell Biol 26: 195–207 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Lingner J 2009. TERRA: Telomeric repeat-containing RNA. EMBO J 28: 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missal K, Zhu X, Rose D, Deng W, Skogerbo G, Chen R, Stadler PF 2006. Prediction of structured non-coding RNAs in the genomes of the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J Exp Zoolog B Mol Dev Evol 306: 379–392 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D 2006. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol 2: e33 doi: 10.1371/journal.pcbi.0020033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Eddy SR 2001. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2: 8 doi: 10.1186/1471-2105-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB 2009. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–W689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309: 1728–1732 [DOI] [PubMed] [Google Scholar]
- Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J 2006. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res 16: 885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo WL, Gorodkin J 2008. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. Genome Res 18: 242–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzilov AV, Keegan JM, Mathews DH 2006. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics 7: 173 doi: 10.1186/1471-2105-7-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF 2005. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci 102: 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermuller J, Hertel J, Lindemeyer M, Reiche K, Tanzer A, et al. 2007. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res 17: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HYK, Preston E, et al. 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6: e1000848 doi: 10.1371/journal.pgen.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]