Abstract
Understanding how information is encoded and transferred by biochemical networks is of fundamental importance in cellular and systems biology. This requires analysis of the relationships between the stochastic trajectories of the constituent molecular (or submolecular) species that comprise the network. We describe how to identify conditional independences between the trajectories or time courses of groups of species. These are robust network properties that provide important insight into how information is processed. An entire network can then be decomposed exactly into modules on informational grounds. In the context of signalling networks with multiple inputs, the approach identifies the routes and species involved in sequential information processing between input and output modules. An algorithm is developed which allows automated identification of decompositions for large networks and visualization using a tree that encodes the conditional independences. Only stoichiometric information is used and neither simulations nor knowledge of rate parameters are required. A bespoke version of the algorithm for signalling networks identifies the routes of sequential encoding between inputs and outputs, visualized as paths in the tree. Application to the toll-like receptor signalling network reveals that inputs can be informative in ways unanticipated by steady-state analyses, that the information processing structure is not well described as a bow tie, and that encoding for the interferon response is unusually sparse compared with other outputs of this innate immune system.
Keywords: biochemical reaction networks, stochastic kinetics, information, conditional independence, modularization, signalling
1. Introduction
Imagine a record over time of the trajectory of the number of molecules of each type of biochemical species present in a reaction network, together with a record of the time-varying, external signals received by the cell. The network may be large, is not assumed to be in a ‘steady’ or stationary state, and is subject to intrinsic stochasticity. Current understanding in this context of how to analyse the relationships between the various trajectories is very limited. We describe how to identify conditional independence relationships between the trajectories of groups of species. These are robust network properties that provide important insight into how information is encoded and transferred by cellular networks—an area whose importance has been widely recognized [1,2].
Both the external environment and internal state of the cell are inherently dynamic. Signalling and regulatory networks must detect and coordinate responses to both external and internal fluctuations using stochastic biochemistry. The multiple signals arriving at the cell surface are expected to exhibit complicated dynamics and are themselves noisy carriers of information. Advances in understanding information processing within the cell therefore require both a dynamic (see [3]) and a probabilistic approach. Hence the focus here is on the probabilistic relationships between species and module trajectories. There is clear evidence that key components of the cellular signal transduction machinery exhibit involved dynamic behaviour and that different temporal patterns are associated with differential regulation of downstream targets. Spiking and oscillations are exhibited by, for example, cytosolic calcium, cyclic AMP, nuclear NF-κB, and nuclear ERK [4]. Frequency/amplitude modulation has been established for the control of both gene expression and mitochondrial redox state by calcium [5], and for the nuclear translocation of cAMP-dependent protein kinase [6]. Recently, Ashall et al. [7] report that altering the frequency of stimulation by tumour necrosis factor TNFα also results in different patterns of nuclear translocation of NF-κB and of NF-κB-dependent gene expression.
Two related types of question arise from these observations. First, the inferential one concerning what may be learnt from the trajectories of outputs and intermediaries about the dynamics of the input signals and hence about the external state of the cell. This connects with recent work [8,9] that interprets biochemical networks as implementing inference in order to provide a basis for cellular decisions. Second, the question of how external information is encoded by and transferred within biochemical networks exposed to multiple inputs.
The need for a rigorous, dynamic, information-based framework for relating the stochastic trajectories of biochemical networks and for quantifying their information content is thus clear. There has been little previous work in this area (although note [10]). We describe how to identify conditional independences, written At↮Dt Bt, between the trajectories (up to some time t) of groups of biochemical species A, B and D (see §2.2). Such an independence means that the information contained in or encoded by the trajectory Dt makes any information in At irrelevant for Bt (and vice versa). That is, At and Bt contain no mutual information given the trajectory Dt. In this sense, the species in D play the role of informational intermediaries or ‘information carriers’ between A and B within the network. Further discussion of the importance of such conditional independences in the context of signalling networks is given in §2.1. Although probabilistic and information theories have been used to quantify the mutual information content between inputs and outputs for single neurons [11], a small signalling network [12] and simple regulatory transcriptional mechanisms [13,14], the approach adopted here is different. We consider general, potentially large biochemical networks and the sequential information transfer via intermediaries that is central to signal transduction mechanisms. A stochastic process treatment of (bio)chemical networks that focuses on mathematical proof of dynamic independence properties is outside the scope of this paper and is given in Bowsher [15].
In developing our approach to a quantitative theory of information processing by biochemical networks, it has proved fruitful to adopt a modular perspective. This is unsurprising as such networks are increasingly thought of as having a modular structure [16–19], a separable architecture that can be decomposed into units that perform to some extent independently of one another. Furthermore, it is recognized both that understanding such modularity requires a dynamic approach [20] and that rigorous definition and identification of dynamic modularizations is a difficult problem ([21], ch. 3). We show how to exactly decompose or modularize an entire network using informational properties. We view a modularization here as a collection of groups of biochemical species in which, for each module, all information relevant to the trajectory of the module is encoded by the trajectory of the module's overlap (interface) with the rest of the network. With this definition, it turns out that valid conclusions may be drawn by studying the kinetics of each module in isolation from the rest of the system.
As would be expected of a functionally important property of a naturally evolved network, our modularizations and analysis of information processing are robust to (unaffected by) changes in rate parameters. The MIDIA algorithm is developed (§2.3) which allows automated identification of modularizations for large networks, control over the degree of coarse graining, and visualization using a tree that encodes the conditional independences. Only stoichiometric information is used and neither simulations nor knowledge of rate parameters are required. A bespoke version of the algorithm for signalling networks identifies routes of sequential encoding between inputs and outputs, visualized as paths in the tree.
Previous dynamic approaches to module identification adopt various perspectives. Network motifs [22] are an important concept, but the manner in which individual motif function depends on the context within and connections to the rest of the network remains an open question. Graphical, community detection-based methods [18,19] are extended by Saez-Rodriguez et al. [23] to kinetic models of signalling networks—edges are interpreted as retroactivities and ‘inter-modular retroactivity’ is minimized in order to partition species between modules. Some bi-directional, local dynamic influences between modules remain after decomposition. Correlated reaction sets (or ‘Co-sets’; [24]) arise where interest is in a region of the solution space of steady-state fluxes (or reaction rates)—e.g. a Monte Carlo sample from that space [25,26] or the collection of all ‘extreme pathways’ [27]. A Co-set is a group of reactions that have non-zero flux at all points in the solution region under consideration and that thus function together when the system is at steady state—i.e. when species concentrations and reaction rates are constant over time.
Using their stoichiometric reconstruction of the toll-like receptor (TLR) signalling network, Li et al. [28] employ a steady-state flux balance analysis (FBA) to identify signalling pathways for different input–output (I–O) pairs. The objective flux maximized is the flux of the particular reaction regarded as the output of signalling in each case. Although a virtue of this and the other constraint-based methods is that they require knowledge only of the stoichiometric matrix, the steady-state assumption is a strong one that considerably limits their generality. In many settings, signalling and gene regulatory networks are inherently dynamic and non-stationary, as has already been noted. Furthermore, none of the module identification methods discussed allows for stochasticity, which is both an important characteristic of gene expression and signalling dynamics (e.g. [29–33]) and essential for a consideration of how the cell processes noisy information.
2. Results
2.1. Informational encoding, transfer and decomposition—a preview
The approach of the paper relies on two particular properties of the probabilistic relationships between species trajectories in biochemical networks. By the trajectory of a species we mean its dynamic evolution or time course in continuous time. These properties are as follows.
— For a given network, there are usually many ways to partition the network species into non-overlapping groups A, B and D so that the information encoded by the trajectory of D makes any information in the trajectory of A irrelevant for that of B (and vice versa). The species in D thus play the role of informational intermediaries or ‘carriers’. Such species may be identified by establishing the conditional independence between trajectories, At↮Dt Bt.
— Furthermore, the entire network can be decomposed exactly into modules on informational grounds, and the modules arranged in a tree structure that encodes conditional independences between the trajectories of (groups of) modules. The path in the tree from one module to another then identifies the route and species involved in sequential information processing between the two modules.
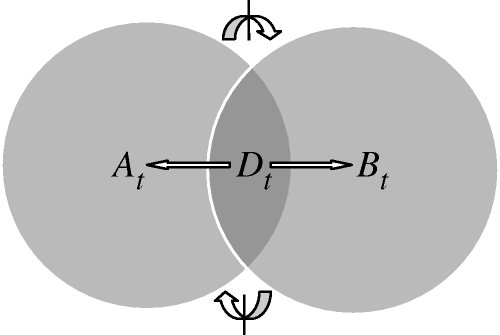
We will denote the state of the molecular network by x(s) = [x1(s), … , xk(s), … , xn(s)], where xk(s) is the number of molecules of type k present at time s. The trajectory Xk,t = (xk(s); 0 ≤ s ≤ t) records the dynamic evolution of the kth species. Figure 1 shows a simple illustration of a network with two modules. The network species are grouped into the three sets A, B and D in such a way that the conditional independence relationship At↮Dt Bt holds. We stress the informational aspect of the relationship—namely that the trajectories At and Bt contain no mutual information given Dt, and the species in D play the role of ‘information carriers’ between A and B within the network. The network is thus said to decompose exactly into the two modules M1 = [A, D] and M2 = [B, D] (shown as circles), with all information relevant to each module's trajectory thus encoded by their region of overlap, D.
Figure 1.
Illustrative example of a network with two modules. Each circle represents a module, i.e. a group of biochemical species. The network species are grouped into three non-overlapping sets A, B and D as shown, in such a way that At↮Dt Bt —i.e. the trajectories (up to any time t) of A and B are independent given the trajectory of D. Hence At and Bt contain no mutual information given Dt, all information transfer between the two modules being conveyed via Dt.
Importantly, our approach is well suited to the analysis of information processing by cell signalling networks. It is commonplace to refer heuristically to information flow within such networks. However, a precise, quantitative understanding of how (and which) internal species of the cell encode information about the trajectories of signalling inputs (ligands) is lacking. Denote by It* the stochastic process by which multiple signalling ligands enter and depart the cell's surroundings, and by O some target output species of the signalling network. We refer to It* as the external input process. By identifying a conditional independence At↮Dt Bt as above, where the trajectory At (resp. Bt) implies that of It* (resp. Ot), we are able to identify encoding species D such that the conditional independence It*↮Dt Ot holds. (Furthermore It*↮Et Ot whenever the species D are contained in E.)
The implication is that, given the trajectory of the species in D, the trajectories It* and Ot contain no mutual information. This may be interpreted in two ways. First, the encoding of information by the trajectory Dt makes the original information in the external input process It* irrelevant for the outputs Ot. Furthermore, this irrelevance holds no matter what other trajectories in addition to those of D are also considered. Second, inferences about the multiple signals received, It*, based on Dt are unaffected by taking the outputs Ot into account. These properties of the trajectory Dt thus capture the essence of signal ‘transduction’ by intermediary species.
In general, the signalling network modularizations we identify are represented using a tree structure that allows us to trace sequential routes of information transfer through the network. Denote the groups of species on the path in the tree between signalling inputs (I*) and outputs (O)—i.e. the relevant ‘edges’ in the tree—by S1, S2, … , SN. Then an important result (see §2.4) will be that It*↮Sj,t Ot for j = 1, … , N, i.e. conditional independence holds for all of the species groups Sj, and moreover (It*, S1t, … , Sj−1,t)↮Sj,t (Sj+1,t, … , SN,t, Ot). The latter property means that (S1, S2, … , SN) is a sequence of encoders, each one of which makes the information in both It* and all earlier encoders in the sequence irrelevant for both all later encoders in the sequence and the outputs Ot. A comparative analysis varying the groups I* and O for a given network then reveals important differences of information processing for the various I–O combinations.
2.1.1. Module kinetics may be studied in isolation
The problem of when it is appropriate to study smaller subsystems without reference to the network in which they are embedded arises frequently in systems biology. We therefore mention the following property of our network decompositions (without further development). It turns out that, given the definition of a decomposition or modularization here (§2.3), valid conclusions may be drawn by studying the kinetics of each module in isolation from the rest of the system. Consider again the two-module network in figure 1 (where it is assumed that conditions 1, 2 and 3 of §2.2 apply). Observing the trajectories of all species in the first module M1 = [A, D] alone, without also observing the trajectories of any species in B, allows many of the rate parameters of the reactions governing the dynamics of M1 to be estimated using maximum likelihood. (We abstract from practical issues of data collection such as discrete sampling.) More specifically, the estimators may be computed for the rate parameters of all reactions that change some species in A and of reactions that change species in D alone. This is a surprising result and one that arises from the ability to decompose the likelihood function along the lines of the conditional independence At↮Dt Bt—see ([15], theorem 4.5). (The only reactions changing M1 whose rate parameters cannot be estimated are those that change both B and D but not A—inevitably there is some loss from not observing B.) These comments extend straightforwardly to decompositions with greater than two modules.
2.2. Identifying dynamic conditional independences
The first stage in identifying conditional independences between trajectories for a given reaction network is to construct its kinetic independence graph or KIG. The nodes in this directed graph correspond to the different biochemical species. The graph is constructed in order to display, for each species, those nodes whose concentration influences the instantaneous, stochastic kinetics of that species (see the electronic supplementary material, SM1.2; [15]). An arrow (directed edge) is therefore drawn from species j to species k whenever j is a reactant in a reaction that either results in the overall production or consumption of molecules of k. In the former case k usually participates in the reaction only as a product whereas in the latter case j and k react together. It is an obvious but important point here that only the concentrations of reactants influence instantaneous reaction rates or ‘intensities’. (Reverse reactions are treated as separate reactions in the formalism.)
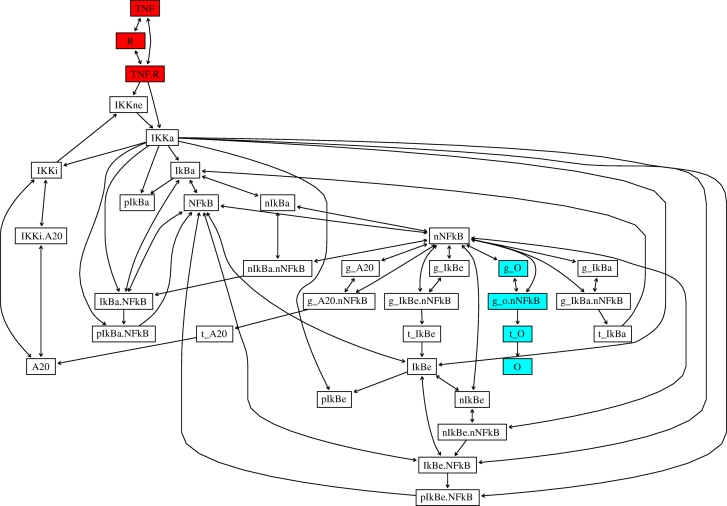
For any collection of species A, it is possible to show that the instantaneous kinetics of A depend only on the current concentration (or ‘levels’) of A itself and those species that are parents of A in the KIG—i.e. those species (outside A) that lie at the tail of an arrow that points to some node in A. Construction of the KIG of a reaction network requires only limited stoichiometric information about the network—often all that is needed is a signed binary, (0, ±1)-version of the stoichiometric matrix identifying, for each reaction, those species that are overall consumed and produced. Importantly, no knowledge of rate parameters is required. Figure 2 shows the KIG for the NFκB signalling network example discussed below.
Figure 2.
Kinetic independence graph of the NFκB signalling network. TNF is the tumour necrosis factor ligand, R is the ligand's receptor, IkBa is IκBα, and IkBe is IκBɛ; IKK has three forms (ne neutral, a active, and i inactive); O denotes a generic output protein; the prefix g denotes gene, t transcript, p phosphorylation and n nuclear location. For ease of viewing, input- and output-associated species are coloured red and blue, respectively. Complete details of the reaction network are given in the electronic supplementary material, SM2.
The following means of identifying conditional independences applies to a general class of stochastic biochemical reaction networks termed (standard) stochastic kinetic models. The class includes, but goes considerably beyond, the class corresponding to the stochastic simulation algorithm of Gillespie [34]. (The SKM class is described formally in the electronic supplementary material, SM1.1.) Let G be the (undirected) graph obtained from the KIG of the reaction network by substituting lines for all arrows. A reaction is said to change A if it causes the level of some species in A to change (no matter which particular species). For a partition [A, D, B] of the species in the network, the conditional independence of trajectories At↮Dt Bt holds whenever two conditions are both satisfied (for mathematical proof, we refer to [15], corollary 4.6 and proposition 4.9):
— A and B are separated by D in the graph G—i.e. every path along lines in G that begins at a node in A and (without revisiting any node) ends at a node in B visits some node in D (condition 1); and
— for any two reactions that change D and result in identical changes to all species in D, either both reactions change A or neither changes A, and either both change B or neither changes B (condition 2).
Additionally, we require that a weak regularity condition, condition 3, holds for the set of reactions that change both A and B, namely that these reactions be ‘identified by consumption of reactants’ (see the electronic supplementary material, SM1.1). Condition 1 is equivalent to there being no species in A that is a reactant in a reaction that results either in the net production or consumption of some species in B (i.e. in a reaction that changes B), and vice versa. When this fails to hold, there is a direct influence of A on the instantaneous kinetics of B, or of B on A (or both). Condition 2, together with the first, ensures the conditional independence of the trajectories up to time t (for all t > 0). A full understanding of the role of this condition is rather involved but the following point is apparent—given the trajectory of D, the possible occurrence of two reactions that change D identically can be distinguished using the trajectory of B if one of them changes B but the other does not; this information is then often relevant for the trajectory of A (contradicting the conditional independence of A and B).
In the mathematical literature, the dynamic conditional independence relationship At↮Dt Bt is usually written as At ⫫ Bt|Dt (and, technically, is a relationship between σ-fields). We prefer the more intuitive notation At↮Dt Bt in this context. Note that the relationship concerns the trajectories up to time t. (There may, owing to a lag effect, be information in At relevant for the trajectory of B beyond time t and up to time u (when Dt is given) but this information will be encoded in Du so that Au↮Du Bu holds at the later time u.)
2.2.1. Illustrative example—NFκB signalling
The exposition of the results and methods of the paper includes, as a running example, a kinetic model of the core mechanisms of NFκB signalling. An important contribution of the work lies in the ability to analyse information processing by large, complex reaction networks (see the TLR example in §2.4.1). However, the smaller NFκB example is useful both in aiding rapid understanding of the techniques and in revealing the often subtle informational properties of biochemical networks.
The network used is based on a state-of-the-art, stochastic dynamic model recently developed using extensive experimental data by Ashall et al. [7]. Essential aspects include the TNF ligand-induced activation of the kinase IKK; subsequent phosphorylation of IκB.NFκB and degradation of pIκB, leading to release of NFκB; and translocation to the nucleus of NFκB. Three negative feedback loops are set up as NFκB itself activates transcription of IκBα, IκBɛ, and A20. The latter inhibits conversion from the inactive to the neutral form of IKK. Complete details of the reaction network (together with species names) may be found in the electronic supplementary material, SM2.
Figure 2 shows the associated KIG. To represent generic outputs of the signalling network, the subgraph gO.nNFκB → tO → O is included (the NFκB-activated expression of protein O). Much more complicated ‘output graphs’ would not alter our conclusions—the important aspect is that the output species are separated from all other species in the KIG by gO.nNFκB (or its equivalent for multiple promoters)—feedback from O is thus ruled out. Throughout we use 𝒱 to denote all species in a given network; the notation\ should be read as ‘excluding’ (and corresponds to set difference).
Let TNFt* = It* be the external ligand or input process. We will show how to use conditions 1 and 2 above to address the following hypotheses or conjectures concerning information processing by the NFκB network (the associated conditional independence At↮Dt Bt is shown in parentheses).
— All information relevant to signalling output is encoded by the trajectory of the promoter occupancy of output genes ([tO, O]t↮gO.nNFκBt [𝒱\{gO.nNFκB,tO,O}]t).
— The encoding of information by the trajectory of the active kinase, IKKat, makes all information about the external input process TNFt* and about ligand binding and unbinding by receptors irrelevant for the rest of the network, including the output (St↮IKKat [𝒱\{S,IKKa}]t, where S = {TNF, R, TNF.R} and TNFt* is implied by St).
Both hypotheses perhaps appear plausible given knowledge of the reaction network—however, only the first is correct. In the case of the first, the graphical separation required by condition 1 is satisfied (figure 2), as is condition 2 since no two reactions cause the same change in gO.nNFκB (see the electronic supplementary material, SM2). In particular, all information about TNFt* relevant to output is encoded in the trajectory of the promoter occupancy of output genes. The second hypothesis is false. Condition 1 fails (owing to the edge TNF.R → IKKne in the KIG)—the trajectory of the neutral form of the kinase, IKKnet, in fact encodes additional information about TNF.Rt. The reason is that the reaction list of the network (see the electronic supplementary material, SM2) means that although the trajectory IKKat implies the history of the activation reaction TNF.R + IKKne → IKKa + TNF.R, it does not imply knowledge of the production and hence trajectory of IKKne. Since activation of IKK involves both TNF.R and IKKne, the trajectory IKKnet provides additional information about the trajectory of receptor occupancy, TNF.Rt.
It is straightforward to verify using conditions 1 and 2 that St↮IKKat, IKKnet [𝒱\{S,IKKa, IKKne}]t, from which we are able to conclude that TNFt*↮IKKat, IKKnet[tO,O]t. That is, all information about the external stimulus relevant for outputs is encoded in the trajectory of the distribution of IKK across its different isoforms (the level of free plus complexed IKKi being implied by the other two forms). This interesting observation demonstrates that thinking in terms of a signal transduction or activation ‘path’ such as TNF.R → IKKa → pIκB.NFκB → NFκB → nNFκB can deliver misleading conclusions about informational properties of the network. Note that A20 does not appear—in particular IKKi.A20t need not be included in the conditioning information because the relevant effect of the negative feedback loop involving A20 is ‘impounded’ in the trajectory IKKnet. The example serves to emphasize that a rigorous, mathematical framework is both indispensable for reasoning correctly about information encoding and transfer, and able to deliver biologically relevant conclusions.
2.3. Identifying exact network decompositions
Using our methods, as was emphasized in §2.1, a given biochemical network can usually be decomposed exactly into modules and the modules arranged in a tree structure encoding conditional independences between the trajectories of groups of modules. This then provides a framework for analysing biochemical information processing and is particularly beneficial for larger networks. A modularization here, {Md}, is a collection of groups of species with the following two properties: (i) all network species are assigned to at least 1 module; and (ii) the trajectory of each module Md is conditionally independent of the trajectory of the other modules, given the trajectory of the overlap of Md with the rest of the network—i.e. Md,t↮Sd,t [∪e≠d Me]t, where the overlap (or interface) Sd = Md ∩ {∪ e≠d Me}, and ∩,∪ denote the intersection and union of sets, respectively. Note that this dynamic conditional independence is equivalent to [Md\Sd]t↮Sd,t [𝒱\Md]t. All information relevant to a module's trajectory is encoded by the module's overlap region with the rest of the network. (It should be noted that this definition of a modularization is considerably stronger than the mathematically abstract one proposed by [15]. Although computationally much more demanding, this definition is the biologically relevant one.)
We have developed a modularization identification by dynamic independence algorithm (MIDIA) based on graphical decomposition methods. It implements automated identification, with the modularization displayed and visualized as a junction tree, 𝒯M. A junction tree is a (connected, acyclic) undirected graph in which the nodes of the graph are the modules themselves. Furthermore, the overlap or intersection of any two modules of the tree, Md ∩ Me (d ≠ e), is contained in every module on the unique path in the tree between Md and Me.
Identifying a network modularization by manual inspection of the KIG is far from straightforward even for the small NFκB signalling network (recall figure 2). With even a modest increase in the size and complexity of the network, a principled and computationally efficient approach to module identification becomes indispensable. Three particular problems must be addressed. First, while using the KIG to find some three-way partitions of the species satisfying condition 1 can be relatively straightforward, it is far from obvious how to assign all network species to modules in a consistent manner so that each module is separated in the KIG from the rest of the network by its interface. Second, it is desirable to have control over the degree of coarse graining of the modularization identified. Third, ensuring that condition 2 is satisfied for every module interface Sd is particularly challenging. The solutions to these problems are, respectively, clique decomposition, clique aggregation and species copying within the junction tree.
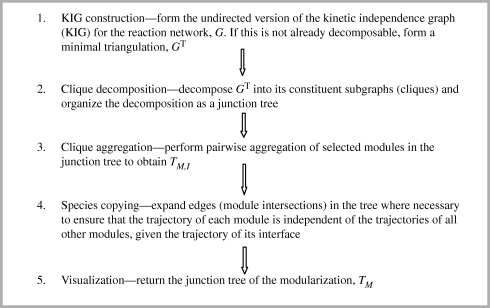
An overview of the various stages of the MIDIA approach is given as a flowchart in figure 3 (see the electronic supplementary material, SM3, for further details). Stages 1 and 2 involve decomposition of the (undirected, minimally triangulated) KIG into a junction tree in which the nodes of the tree are the cliques. Use is then made of results concerning properties of the clique junction tree for a triangulated graph. These properties turn out to be stable when neighbouring modules in the tree are ‘merged’ or aggregated in stage 3. The selection of module pairs for aggregation is automated and allows the degree of coarse graining of the network to be controlled by setting a minimum size for the module residuals, Md\Sd. The resultant junction tree, 𝒯M,I, constitutes a modularization in which the instantaneous kinetics of each module residual are independent of all the network species outside the module. It is an important output in its own right, giving insight about the kinetic structure of the network. Stages 4 and 5 are explained in the following subsection.
Figure 3.
Overview of the stages of the MIDIA algorithm. The algorithm automatically identifies a modularization of the biochemical network on dynamic, informational grounds (see text) and displays it as a junction tree in which the modules are the nodes of the tree. The undirected version of the KIG, G, is formed simply by replacing all arrows with (‘undirected’) lines. In forming GT, enough edges are added (but no more) to make the graph triangulated. The cliques of GT are its maximally complete subgraphs. For these and other concepts in basic graph theory see Cowell et al. [37, ch. 4]. The details of the MIDIA algorithm are explained in the electronic supplementary material, SM3.
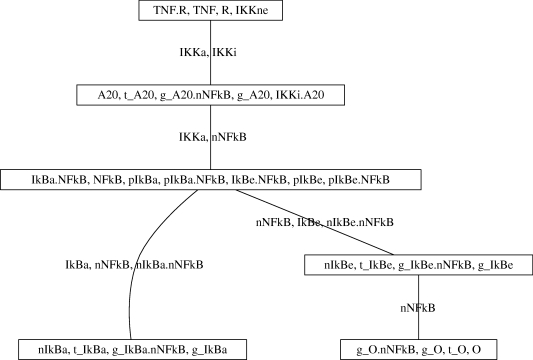
The junction tree, 𝒯M,I, for the NFκB network (§2.2.1) is shown in figure 4. The benefits of a principled modularization in revealing the architecture of the network implicit in the KIG are obvious even for this small network. Working from the top to the bottom of 𝒯M,I, the NFκB network is decomposed into modules associated with: the TNF receptor and IKK; the A20 negative feedback mechanism; cytoplasmic NFκB, IκB and their complexes; nuclear IκBα and its gene expression; nuclear IκBɛ and its gene expression; and finally the signalling outputs. Each edge in the tree contains species determining the instantaneous kinetics of the two adjacent module residuals. The central roles played by IKKa, nuclear NFκB, and nuclear import and export processes are thus made clear.
Figure 4.
Modularization 𝒯M,I for the NFκB signalling network. The modularization is the one returned after stage 3 of the MIDIA algorithm (using a minimum residual size of 4 (except for the ‘root’ module)), and is based on instantaneous kinetics alone (see text). Each module Md is labelled with the corresponding residual (Md\Sd); each edge is labelled with the intersection of the two modules it connects.
We emphasize that for moderately sized or large networks an automated tool for investigating the modular architecture of the network based on its dynamic properties is essential—manual inspection of reaction mechanisms and their raw graphical representations becomes infeasible (consider, for example, the TLR signalling network in §2.4.1). The junction tree in figure 4, obtained from the fully automated algorithm, illustrates MIDIA's efficacy in this regard using the example of a relatively small network for transparency.
2.3.1. Encoding dynamic conditional independences in junction trees
This section explains how to read conditional independences between species trajectories from the junction tree returned by the MIDIA algorithm. As is usual, each edge between adjacent modules in a junction tree is associated with the modules' overlap which we denote for modules d and e by Sde = Md ∩ Me. Consider first the intermediate tree, 𝒯M,I, returned by stage 3. Imagine cutting any one of its edges, say the one between the modules Md and Me. Denote the species present in each of the two subtrees obtained from the cutting procedure by Vde and Ved, respectively. Then an important property is that Vde and Ved are separated in the undirected KIG by the modules' intersection—there is no direct influence of either ‘subtree’ on the instantaneous kinetics of the other. The species copying procedure (stage 4) is designed (i) to preserve this property in the final tree, 𝒯M, thus allowing application of our condition 1 (§2.2) and (ii) to ensure via condition 2 that for the two subtrees obtained by cutting any one of its edges, the conditional independence of trajectories Vde,t↮Sde,t Ved,t holds (see theorem 1 of the electronic supplementary material, SM1.3). It is then possible to show for each module Md in 𝒯M that Md,t↮Sd,t [∪e≠d Me]t (see theorem 2 of the electronic supplementary material, SM1.3). These dynamic conditional independences, of course, constitute the defining property of our modularizations.
In stage 4, the MIDIA algorithm works ‘backwards’ through the junction tree 𝒯M,I, imposing condition 2 on each edge of the tree, Sde, so as to preserve the junction tree property and to leave edges treated in previous steps unchanged. Each edge or intersection of neighbouring modules, Sde, is enlarged by ‘copying’ the appropriate species from a module containing them, Mg say, both to Md (which lies on the unique path between g and e) and to Me. The species are also copied to all other modules on that path. For example, in the case of the NFκB network, the edge {IKKa, nNFκB} in 𝒯M,I is enlarged by including g_A20 also in the third module. (The whole tree 𝒯M returned after stage 5 for the NFκB network is shown in the electronic supplementary material, SM4.) Cutting this edge in 𝒯M and applying theorem 2 then reveals the non-obvious conditional independence St↮[IKKa,nNFκB,g_A20]t [𝒱\{S, M1, M2}]t, where S = {TNF, R, TNF.R} as before, and M1, M2 are the first and second modules (from the top) of 𝒯M. As is the case for the trajectory of the distribution of IKK across its isoforms, the trajectory [IKKa, nNFκB, g_A20]t is thus another encoder with only a few species that makes the information in St irrelevant for the trajectories of all species containing cytoplasmic and nuclear NFκB, for the trajectories of the IκB's (i.e. their protein complexes, and genes and transcripts) and for the trajectories of the output module. This encoder is equivalent to the trajectory of active IKK and nuclear NFκB, both free and bound to the A20 promoter. Space precludes further discussion of the NFκB example here. Sequential information processing is considered in general and in the context of the TLR signalling network example below.
2.4. Identifying paths of sequential information processing in signalling networks
As was discussed earlier in §2.1, our modular approach and the MIDIA algorithm are well suited to analysing information processing by signalling networks, especially large ones. Let MI* be a module identified whose trajectory contains that of an external input process, It*, and let Mo be a module containing the target output O. Consider the (unique) path in the junction tree 𝒯M from MI* to Mo, which we write as MI* − S1M1 − …− SN−1MN−1 − SNMo, N being the number of edges on the path. The sequence of edges (S1, S2, … , SN) reveals the sequential process by which the network encodes and transfers information. Imagine cutting the edge Sj—then applying the result of §2.3.1 on the subtree trajectories implies that It*↮Sj,t (Sj+1,t, … , SN,t, Ot) for j = 1, … , (N − 1), and furthermore implies the main result that (It*, S1t, … , Sj−1,t)↮Sj,t (Sj+1,t, … , SN,t, Ot) for j = 2, … , N. To see this, recall that (using this notation), for example, Sj+1,t = Mj ∩ Mj+1, and therefore the species (Sj+1, … , SN, O) are in the ‘subtree’ Vj,j−1 while (I*, S1, … , Sj−1) is ‘contained in’ the subtree Vj−1,j.
The main result here then, (It*, S1t, … , Sj−1,t)↮Sj,t (Sj+1,t, … , SN,t, Ot), means that (S1, S2, … , SN) is a sequence of encoders, each one of which encodes information that makes both It* and all earlier encoders in the sequence irrelevant for all later encoders in the sequence and for the outputs Ot. Using the junction tree of the modularization identified by MIDIA we are thus able to identify truly sequential routes of information processing by the network. Since identifying where the information resides is a prerequisite for understanding how it is encoded and determining the extent of any information loss involved, this is an important step in the development of a quantitative theory of cellular information processing.
The MIDIA algorithm is tailored to identify the routes of sequential encoding as follows. After performing stages 1 and 2, the modules containing the external input (processes) and outputs are determined, and all edges in the tree that lie on a path between such an input and an output module identified. We call these the I–O paths. In stage 3, clique aggregation is performed according to the following steps: (i) to remove any edges containing either an input or output and (ii) by aggregating as many modules as possible without altering any of the modules on the I–O paths, in order to focus attention exclusively on those paths. Stages 4 and 5 are as usual.
2.4.1. The TLR signalling network
As a proof of principle, the tailored MIDIA algorithm was applied to the stoichiometric reconstruction of the TLR signalling network presented by Li et al. [28]. The TLR network is large, having 752 species and 1034 reactions (after accounting for reaction reversibility; for species names see [28], table S1). There are 16 external input processes. These correspond to the extracellular trajectories of the individual receptor-specific ligands (with the exception of the input process for NOD1 which also includes the cytoplasmic trajectory of the NOD1 ligand since some ligand arrivals are owing to an export reaction from the cytoplasm). We focus on three groups of output species of the network—(AP1-JUN, AP1-JUN-FOS), (ISRE-IRF3, ISRE-IRF7) and vacuolar NADPH-oxidase (in two different phospho-forms). The first two consist of active transcription factor (TF) complexes bound to their DNA sites, while the latter is responsible for reactive oxygen species production.
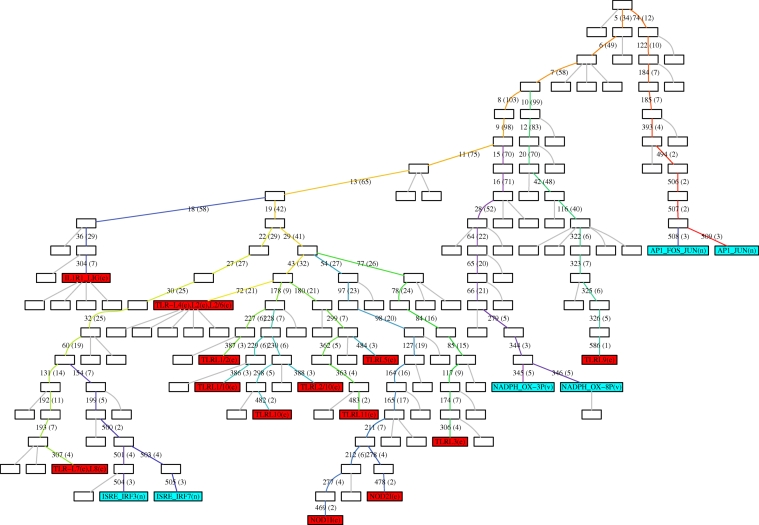
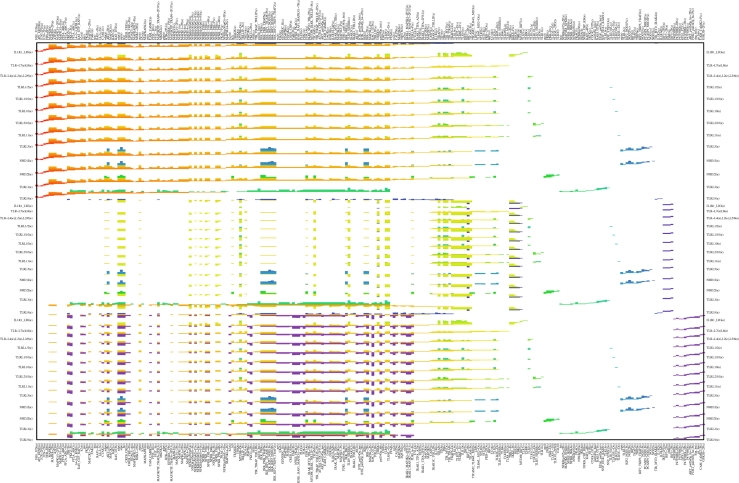
Figure 5 shows the junction tree 𝒯M and I–O paths for the TLR network. In order to investigate further the structure of the I–O paths while explicitly taking the species content of the edges into account, we introduce a visualization technique called the I–O path matrix. The construction of the I–O path matrix is explained in figure 6, which depicts the matrix for all of the inputs in the TLR network and three outputs (one from each output pair). On-screen viewing of the matrix is recommended. Together, the tree 𝒯M and the I–O path matrix are powerful, complementary visual tools for investigating information processing by the network.
Figure 5.
Modularization of the TLR signalling network showing I–O paths. Tree, 𝒯M, returned by the MIDIA algorithm tailored to analyse the I–O paths of the network (see text). Modules containing external input processes are coloured red and those containing outputs blue. Edges on the path between any input and any output module are uniquely numbered (with the number of species in the edge also shown in parentheses)—the colours of edges match the ones used in figure 6. Species names are as in [28], table S1) where L and LIG both denote a receptor ligand, but with the following exceptions: NOD1I(e) is the external input process for the NOD1 receptor, and similarly for NOD2I(e); NADPH-OX-3P(v) corresponds to PHOX-GTP-3P(v) in ([28], table S1), and similarly for NADPH-OX-8P(v).
Figure 6.
Sequential information processing by the TLR network: the I–O path matrix. Each column corresponds to a species (named as in [28], table S1)) and input processes are labelled on the vertical axis. The upper, central and lower regions of the plot correspond to the outputs AP1-JUN, ISRE-IRF3 and NADPH-OX-3P, respectively. Each row in the matrix indicates the species composition of a given edge on an I–O path, with edges colour-coded (as in figure 5). Rows of the matrix are grouped according to the individual I–O combinations and ordered within a group from top to bottom according to the sequence of the edges in the corresponding I–O path in 𝒯M in figure 5.
This is not the place to undertake a lengthy and detailed analysis of information processing by the TLR network. Rather, the utility of our approach is illustrated with a discussion of the insight provided into the following aspects. First, the grouping or clustering of signalling inputs (TLR) is found to be similar on the basis of how information is encoded by the network. Second, as indicated by Kanae & Kitano [35], there is a need to go beyond a ‘bow tie’ conception of the architecture of the network when considering signal transduction. The tree 𝒯M provides such a ‘structural’ view of how the network processes information between various inputs and outputs. We are able to identify the species involved in information encoding for the various outputs (figure 6), the sequential I–O paths and their associated shapes, and those ‘core’ encoding species in common between the I–O paths. The analysis reveals the breadth of encoding species in less peripheral parts of the I–O paths rather than a bottleneck—the information processing structure is not well described as a bow tie. Third, we find that the extent to which information encoding is distributed broadly across many species differs significantly between the ISRE-IRF and the AP1/NADPH-oxidase outputs (the TLRL9 input excluded). Encoding in the case of the ISRE output is much sparser, while the other two outputs share more similar I–O paths (figure 6). Fourth, for a large cluster of 12 TLRs, we identify a surprisingly small group of species responsible for mediating information transfer from the ‘core’ encoding species to the ISRE-IRF outputs. Fifth, we show that in the context of multiple stimuli, inputs can be informative for an output in ways not revealed by steady-state FBA. Each of these aspects is considered in turn below.
The structure of the tree 𝒯M immediately implies a hierarchical clustering of the input processes on the basis of their associated I–O paths—the dendogram or tree for the clustering being derived directly from the structure of 𝒯M in the obvious way. Since the species contents of edges in 𝒯M can overlap to differing degrees, we note that the I–O path matrix in figure 6 strongly supports this hierarchical clustering of inputs using 𝒯M. In particular, the following groups of inputs are seen to possess (within each group) extremely similar sequential paths of information processing to all three output types: (TLR-L1/2, L1/10, L10, L2/10), (TLR-L4, L2, L2/6), (TLR-L5, L11), (NOD1I, NOD2I) and (TLR-L7, L8). At a higher level within the clustering hierarchy, there is a large group of 12 inputs contained in the subtree given by cutting edge 29 in 𝒯M—this group, denoted TLRI\IL1,7:9, contains all but four of the external inputs (the IL1R input and TLR-L7,8,9). TLRL9 stands out as the most exceptional individual input, followed by ILR1L. As can be seen from figure 6, the 12 external inputs comprising TLRI\IL1,7:9 share substantively similar I–O paths.
The breadth of encoding species in less peripheral parts of the I–O paths is apparent, as opposed to any bottleneck or narrow ‘bow-tie centre’. The number of species in the constituent edges of each I–O path in 𝒯M (and also in 𝒯M,I) is roughly ‘diamond-shaped’ going from input to output—i.e. early and late edges have few species while intermediate edges have many. Almost always, the earliest edges in the sequence contain exclusively receptors, ligands and receptor–ligand complexes, while the latest edges contain species closely related to the outputs (the output TFs and binding sites, or the constituents of the NADPH-oxidase complex). The diamond shape might appear to be largely the result of the ‘confluence of multiple paths’ within the tree that could nevertheless retain ‘functionality’ alone—however, this is not the case. The structure of the tree 𝒯M reflects the edge content and hence path content of the undirected KIG (owing to stages 1 and 2 of MIDIA); hence, even when inputs and outputs are considered only pairwise, an edge on an I–O path in 𝒯M,I is a minimal separator of the particular input and output in the undirected KIG in 92 per cent of all cases. In connection with the breadth of encoding species in less peripheral parts of the I–O paths, we note the existence of edges on the path to each output that contain a relatively large number of species and are shared by all or nearly all of the inputs—e.g. edges 7, 8, 9 and 11 for AP1 outputs, edges 11 and 15 for NADPH-OX outputs, and edges 22 and 27 for ISRE-IRF.
Perhaps the most immediately obvious feature of figure 6 is the similarity between I–O paths corresponding to a given output (the TLRL9 input excluded), compared with the clear differences across outputs. In order to focus on these differences, the following discussion is for the group of 12 external inputs TLRI\IL1,7:9 which share very similar I–O paths. The structure of 𝒯M and consideration of the species involvement (see figure 6) strongly indicate that the I–O paths are closely related for the NADPH-OX and AP1 outputs while encoding for the ISRE output is much ‘sparser’ (involves fewer species) and is rather different. For the group TLRI\IL1,7:9, consider for each output the edges on the I–O paths in 𝒯M that are in common between all of the inputs—e.g. the sequence of edges 29, 22, … , 501, 504 in the case of ISRE. (The group of species found on all of the I–O paths for a particular output is identical to the species content of these common edges, except for the inclusion of one additional, identical species in each case.) These common edges contain 57 different species for ISRE-IRF—compared with 137 for NADPH-OX and 165 for AP1—of which 48 are found on the I–O paths for all three outputs and only nine are specific to the ISRE output alone. A further 77 species are found on the I–O paths for both NADPH-OX and AP1 but not for ISRE. There are thus 12 species specific to the NADPH-OX output alone and 40 species specific to AP1 alone.
For the large group of 12 inputs, TLRI\IL1,7:9, we thus identify a ‘core’ of 48 species involved in the three sequences of encoders ending at the respective output modules (and beginning in each case with edge 29). These species, listed in the electronic supplementary material, SM5, are also located in a central region of the tree, in edges 19, 22 and 29 (the latter consisting only of core species). For the ISRE output, the path of sequential information processing involves only these 48 species and (excluding the four that are direct components of the output species themselves) the following: TBK1-P, TBK1, IKKɛ-P, IKKɛ and MYD88_TRAF6_IRF7. (Together with cytoplasmic IRF7-2P and ubiquitin the latter group comprises edge 154.) The small group (TBK1-P, TBK1, IKKɛ-P, IKKɛ, MYD88_TRAF6_IRF7) can thus be seen as responsible for mediating information transfer from the ‘core’ encoding species to the ISRE output. We view encoding for the other two output types as distributed across many more species trajectories (as less sparse), with the species involved for NADPH-OX typically involved also for AP1. Edge 11, for example, contains 75 species and is common to the sequence of encoders for both outputs.
We conclude this section by drawing a number of contrasts with the steady-state FBA of the same network in Li et al. [28]. The FBA analysis there takes a collection of steady-state ‘snapshots’ of the network—in each one, a single input flux is held at a constant non-zero level and the network is assumed to optimize a single output flux. These snapshots are then combined in the manner described in Li et al. [28] to obtain discrete signalling (‘DIOS’) pathways. Groupings of inputs are provided by ([28], fig. 4), but the same individual input may appear in groups associated with different DIOS pathways, depending on the output flux chosen as the objective to be maximized. As has already been emphasized, our analysis makes no steady state or stationarity assumption. Nor is it restricted to the separate analysis of particular I–O combinations. Rather, the signalling system is exposed to any number of the possible time-varying input processes and all output trajectories are determined simultaneously by the network biochemistry, as in the living cell. The individual I–O paths in figures 5 and 6 are system properties that hold simultaneously—indeed, 𝒯M may be used to analyse paths of sequential information processing between a group of inputs (e.g. the group TLRI\IL1,7:9) and a group of outputs, as has just been described.
In steady state, Li et al. [28] report that certain individual inputs are unable to affect certain outputs—e.g. no input affects the ISRE-IRF7 output, NOD1 and NOD2 do not affect any of our outputs, and TLRL3 does not affect AP1 outputs. Computational analysis of the KIG for the network reveals by contrast that there is a directed path in the KIG (i.e. one following arrows), not involving ‘currency’ species, from all individual inputs to all outputs. Such paths correspond to chains of direct kinetic influence from input to output (each ‘link’ or edge of which will be operative at any point in time provided no reactant level is zero for the reaction giving rise to it). Thus, for example, the NODI1 and NODI2 input processes can be informative for all output trajectories, including the ISRE-IRF3 and -IRF7 outputs. One way in which this can occur (revealed by interrogation of the KIG) is that activation of both NOD inputs raises IKKα,β,γ-P levels, which consumes substrates of IKKε-P (such as NFκB(p50/p65)), thus reducing sequestration of IKKε-P and consequently increasing phosphorylation of IRF3 and (in the presence of MYD88_TRAF6_IRF7) of IRF7. The encoders in the rows of figure 6 corresponding to NOD1I and NOD2I render these inputs uninformative (e.g. IKKα,β,γ is seen to be an encoding species in the case of the IRF3 output). Similarly, TLRL3 can be informative for the AP1 outputs. Thus in the context of multiple stimuli, inputs can be informative for an output in ways not revealed by steady-state FBA.
3. Discussion
Understanding how information is encoded and transferred by the biochemical reaction networks inside cells requires analysis of the relationships between the stochastic trajectories of their constituent molecular (or submolecular) species. We have emphasized the importance in this context of conditional independences between the trajectories or time courses of groups of species, represented by At↮Dt Bt. Such an independence means that the information encoded by the trajectory Dt makes any information in At irrelevant for the trajectory Bt (and vice versa). That is, At and Bt contain no mutual information given the trajectory Dt, and the species in D play the role of informational intermediaries or ‘information carriers’ between A and B within the network.
We describe how to identify such conditional independences between trajectories using a pair of conditions that are straightforward to check in practice. We have demonstrated, it seems for the first time, that entire networks (including very large ones) can be decomposed systematically into modules so that all information relevant to a particular module's trajectory is encoded by the trajectory of those species in the region where the module overlaps with the rest of the network. In the context of signalling networks with multiple inputs, the approach identifies the routes and species involved in sequential information processing between input and output modules. The dynamic conditional independences, modularizations and I–O paths we identify are themselves robust features of a given network in three senses emphasized recently by Gunawardena [36]—they are invariant to changes in all rate parameter values (cm> 0), in the initial condition (x0), and in the ‘dynamical’ or reaction intensity functions (gm{·}, eqn (1) of the electronic supplementary material, SM1.1). Since the cellular networks under study are naturally evolved, the absence of any need to fine-tune rate parameters to obtain these properties is consistent with their functional importance.
Furthermore, the methods of analysis presented apply to a rather general form of stochastic dynamics for biochemical networks that includes, but goes considerably beyond, the form employed in a Gillespie [34] simulation algorithm or a variant thereof (for a more technical discussion, see the electronic supplementary material, SM1.1). In particular, each conditional reaction intensity may depend on the entire past trajectory of its reactants (e.g. on the time elapsed since the last jump in some reactant levels) and also on the time evolution of variables taken to be deterministic such as temperature, light level and time itself. The class of stochastic dynamics used can thus accommodate, for example, general memory effects (owing perhaps to inhomogeneity arising from imperfect mixing), non-exponential waiting times, and deterministic circadian or environmental effects.
The MIDIA algorithm is developed which allows automated identification of exact decompositions for large networks, control over the degree of coarse graining, and visualization using a junction tree that encodes the conditional independences. The algorithm relies on construction of the KIG for the network and uses only stoichiometric information. No knowledge of rate parameters or use of simulations are required. A bespoke version designed for signalling networks identifies the process of sequential encoding and visualizes its structure using the collection of I–O paths in the junction tree. Together with the I–O path matrix, this provides a powerful, visual tool for investigating information processing by signalling networks.
The stochastic, dynamic signals received by cells are impounded in the stochastic trajectories of the intermediate species of signalling networks and ultimately in the trajectories of their targets. The use of dynamic conditional independences thus allows us to identify those species trajectories which fully encode the relevant information and to trace the sequential process of information transfer through the network. We have demonstrated using a small NFκB signalling network that a rigorous framework is indispensable for reasoning correctly about informational properties.
Application of the techniques to the TLR network establishes both their utility for studying the properties of cell signalling and their computational feasibility for large reaction networks. Joint inspection of the junction tree and the I–O path matrix enables a comparative analysis of different I–O combinations. We find that encoding in the case of the interferon response outputs is much sparser, while the AP1 and NADPH-oxidase outputs share more similar I–O paths. For a large cluster of 12 TLRs, we identify a surprisingly small group of species responsible for mediating information transfer from the ‘core’ encoding species to the interferon response outputs. Overall, the information processing structure is not well described as a bow tie owing to the breadth of encoding species away from the periphery. Our analysis reveals that, in the context of multiple stimuli, inputs can be informative in ways unanticipated by steady-state analyses.
Stoichiometric approaches to network analysis have had a large impact on systems biology, owing in part to the modest knowledge required about reaction parameters. The dynamic conditional independence-based techniques introduced here provide a modular approach for analysing information encoding and transfer by biochemical networks, while also requiring only stoichiometric information. Importantly, the approach fully incorporates both stochastic and non-steady-state dynamics. We believe it constitutes a significant step in the development of a quantitative theory of cellular information processing.
Acknowledgments
The author is grateful to the anonymous referees for helpful comments on the manuscript and for the research environment provided by the Cambridge Statistics Initiative, University of Cambridge, UK. The research was jointly funded by the EPSRC and MRC (United Kingdom). The author declares that he has no conflict of interest.
References
- 1.Barkai N., Shilo B.-Z. 2007. Variability and robustness in biomolecular systems. Mol. Cell 28, 755–760 10.1016/j.molcel.2007.11.013 (doi:10.1016/j.molcel.2007.11.013) [DOI] [PubMed] [Google Scholar]
- 2.Nurse P. 2008. Life, logic and information. Nature 425, 424–426 10.1038/454424a (doi:10.1038/454424a) [DOI] [PubMed] [Google Scholar]
- 3.Samoilov M., Arkin A., Ross J. 2002. Signal processing by simple chemical systems. J. Phys. Chem. A 106, 10 205–10 221 10.1021/jp025846z (doi:10.1021/jp025846z) [DOI] [Google Scholar]
- 4.Shankaran H., Ippolito D. L., Chrisler W. B., Resat H., Bollinger N., Opresko L. K., Wiley H. S. 2009. Rapid and sustained nuclear–cytoplasmic ERK oscillations induced by epidermal growth factor. Mol. Syst. Biol. 5, 332. 10.1038/msb.2009.90 (doi:10.1038/msb.2009.90) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berridge M. J., Bootman M. D., Roderick H. L. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529 10.1038/nrm1155 (doi:10.1038/nrm1155) [DOI] [PubMed] [Google Scholar]
- 6.Dyachok O., Isakov Y., Sågetorp J., Tengholm A. 2006. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting β-cells. Nature 439, 349–352 10.1038/nature04410 (doi:10.1038/nature04410) [DOI] [PubMed] [Google Scholar]
- 7.Ashall L., et al. 2009. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324, 242–246 10.1126/science.1164860 (doi:10.1126/science.1164860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libby E., Perkins T. J., Swain P. S. 2007. Noisy information processing through transcriptional regulation. Proc. Natl Acad. Sci. USA 104, 7151–7156 10.1073/pnas.0608963104 (doi:10.1073/pnas.0608963104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins T. J., Swain P. S. 2009. Strategies for cellular decision-making. Mol. Syst. Biol. 5, 326. 10.1038/msb.2009.83 (doi:10.1038/msb.2009.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tostevin F., Ten Wolde P. R. 2009. Mutual information between input and output trajectories of biochemical networks. Phys. Rev. Lett. 102, 218101. 10.1103/PhysRevLett.102.218101 (doi:10.1103/PhysRevLett.102.218101) [DOI] [PubMed] [Google Scholar]
- 11.Rieke F., Warland D., de Ruyter van Steveninck R., Bialek W. 1997. Spikes: exploring the neural code. Cambridge, MA: MIT Press [Google Scholar]
- 12.Mehta P., Goyal S., Long T., Bassler B. L., Wingreen N. S. 2009. Information processing and signal integration in bacterial quorum sensing. Mol. Syst. Biol. 5, 325. 10.1038/msb.2009.79 (doi:10.1038/msb.2009.79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tkacik G., Callan C. G., Jr, Bialek W. 2008. Information flow and optimization in transcriptional regulation. Proc. Natl Acad. Sci. USA 105, 12 265–12 270 10.1073/pnas.0806077105 (doi:10.1073/pnas.0806077105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walczak A. M., Mugler A., Wiggins C. H. 2009. A stochastic spectral analysis of transcriptional regulatory cascades. Proc. Natl Acad. Sci. USA 106, 6529–6534 10.1073/pnas.0811999106 (doi:10.1073/pnas.0811999106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowsher C. G. 2010. Stochastic kinetic models: dynamic independence, modularity and graphs. Ann. Statist. 38, 2242–2281 10.1214/09-AOS779 (doi:10.1214/09-AOS779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong H., Mason S., Barabási A.-L., Oltvai Z. 2001. Lethality and centrality in protein networks. Nature 411, 41–42 10.1038/35075138 (doi:10.1038/35075138) [DOI] [PubMed] [Google Scholar]
- 17.Ravasz E., Somera A. L., Mongru D. A., Oltvai Z. N., Barabási A.-L. 2002. Hierarchical organization of modularity in metabolic networks. Science 297, 1551–1555 10.1126/science.1073374 (doi:10.1126/science.1073374) [DOI] [PubMed] [Google Scholar]
- 18.Guimerà R., Amaral L. A. N. 2005. Functional cartography of complex metabolic networks. Nature 433, 895–900 10.1038/nature03288 (doi:10.1038/nature03288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashtan N., Alon U. 2005. Spontaneous evolution of modularity and network motifs. Proc. Natl Acad. Sci. USA 102, 13 773–13 778 10.1073/pnas.0503610102 (doi:10.1073/pnas.0503610102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander R. P., Kim P. M., Emonet T., Gerstein M. B. 2009. Understanding modularity in molecular networks requires dynamics. Sci. Signal. 2, pe44. 10.1126/scisignal.281pe44 (doi:10.1126/scisignal.281pe44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szallasi Z., Periwal V., Stelling J. 2006. On modules and modularity. In System modeling in cellular biology, ch. 3 Cambridge, MA: MIT Press [Google Scholar]
- 22.Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U. 2002. Network motifs: simple building blocks of complex networks. Science 298, 824–827 10.1126/science.298.5594.824 (doi:10.1126/science.298.5594.824) [DOI] [PubMed] [Google Scholar]
- 23.Saez-Rodriguez J., Gayer S., Ginkel M., Gilles E. D. 2008. Automatic decomposition of kinetic models of signaling networks minimizing the retroactivity among modules. Bioinformatics 24, 213–219 10.1093/bioinformatics/btn289 (doi:10.1093/bioinformatics/btn289) [DOI] [PubMed] [Google Scholar]
- 24.Papin J. A., Reed J. L., Palsson B. Ø. 2004. Hierarchical thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem. Sci. 29, 641–647 10.1016/j.tibs.2004.10.001 (doi:10.1016/j.tibs.2004.10.001) [DOI] [PubMed] [Google Scholar]
- 25.Price N. D., Schellenberger J., Palsson B. Ø. 2004. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys. J. 87, 2172–2186 10.1529/biophysj.104.043000 (doi:10.1529/biophysj.104.043000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiele I., Price N. D., Vo T. D., Palsson B. Ø. 2005. Candidate metabolic network states in human mitochondria. J. Biol. Chem. 280, 11 683–11 695 10.1074/jbc.M409072200 (doi:10.1074/jbc.M409072200) [DOI] [PubMed] [Google Scholar]
- 27.Papin J. A., Price N. D., Palsson B. Ø. 2002. Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome Res. 12, 1889–1900 10.1101/gr.327702 (doi:10.1101/gr.327702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F., Thiele I., Jamshidi N., Palsson B. Ø. 2009. Identification of potential pathway mediation targets in toll-like receptor signaling. PLoS Comput. Biol. 5, e1000292. 10.1371/journal.pcbi.1000292 (doi:10.1371/journal.pcbi.1000292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kærn M., Elston T. C., Blake W. J., Collins J. J. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451–464 10.1038/nrg1615 (doi:10.1038/nrg1615) [DOI] [PubMed] [Google Scholar]
- 30.Shahrezaei V., Swain P. S. 2008. The stochastic nature of biochemical networks. Curr. Opin. Biotechnol. 19, 369–374 10.1016/j.copbio.2008.06.011 (doi:10.1016/j.copbio.2008.06.011) [DOI] [PubMed] [Google Scholar]
- 31.Skupin A., Kettenmann H., Winkler U., Wartenberg M., Sauer H., Tovey S. C., Taylor C. W., Falcke M. 2008. How does intracellular Ca2+ oscillate: by chance or by the clock? Biophys. J. 94, 2404–2411 10.1529/biophysj.107.119495 (doi:10.1529/biophysj.107.119495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. 2009. Regulated fluctuations in Nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149. 10.1371/journal.pbio.1000149 (doi:10.1371/journal.pbio.1000149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson D. J. 2009. Stochastic modelling for quantitative description of heterogeneous biological systems. Nat. Rev. Genet. 10, 122–133 10.1038/nrg2509 (doi:10.1038/nrg2509) [DOI] [PubMed] [Google Scholar]
- 34.Gillespie D. T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 10.1021/j100540a008 (doi:10.1021/j100540a008) [DOI] [Google Scholar]
- 35.Kanae O., Kitano H. 2006. A comprehensive map of the toll-like receptor signaling network. Mol. Syst. Biol. 2, 2006.0015 10.1038/msb4100057 (doi:10.1038/msb4100057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunawardena J. 2009. Models in systems biology: the parameter problem and the meanings of robustness. In Elements of computational systems biology (eds Lodhi H. M., Muggleton S. H.), pp. 21–48 New York, NY: Wiley [Google Scholar]
- 37.Cowell R. G., Dawid A. P., Lauritzen S. L., Spiegelhalter D. J. 2007. Probabilistic networks and expert systems. New York, NY: Springer [Google Scholar]