Abstract
RA-inducible gene I (RIG-I/DDX58) has been shown to activate IFN-β promoter stimulator 1 (IPS-1) on recognizing cytoplasmic viral RNAs. It is unclear how RIG-I functions within the IFN and/or RA signaling process in acute myeloid leukemia (AML) cells, however, where obvious RIG-I induction is observed. Here, we show that the RIG-I induction functionally contributes to IFN-α plus RA-triggered growth inhibition of AML cells. Interestingly, although RIG-I induction itself is under the regulation of STAT1, a major IFN intracellular signal mediator, under circumstances in which it does not stimulate IPS-1, it conversely augments STAT1 activation to induce IFN-stimulatory gene expression and inhibit leukemia cell proliferation. Thus, our results unveil a previously undescribed RIG-I activity in regulating the cellular proliferation of leukemia cells via STAT1, which is independent of its classic role of sensing viral invasion to trigger type I IFN transcription.
IFNs consist of mainly two classes of related cytokines, type I IFNs and type II IFN, whose generation and secretion are stimulated by a variety of inflammatory or physiological regulatory cues (1, 2). When being induced by inflammatory cues or given at pharmacological dosages, IFNs act effectively to trigger two types of basic biological responses within the host eukaryotic cells: initiation of an antiviral status and activation of the regulatory pathways that inhibit cellular proliferation (3). Mechanistic studies have shown that the ligation of type I IFNs or type II IFN to their cognate receptors results in the activation of highly overlapped downstream signal transduction pathways centered on the phosphorylation modifications of STAT members, among which the phosphorylation of STAT1 at its tyrosine 701 site represents a shared crucial event (3). Once being phosphorylated, STAT1 molecules tend to form homodimers or heterotrimers with STAT2 and IFN regulatory factor 9 (IRF9), which, in turn, enable the complexes to translocate into the nucleus to enhance the transcription of numerous IFN stimulatory genes (ISGs) by binding to the specific elements, such as the IFN-stimulated response element (ISRE) or IFN-γ activated site (GAS) on their promoters (4). Worthy of mentioning, in line with the well-documented intensive cross-talk between IFN and RA signaling (5), all-trans retinoic acid (ATRA)-induced cell cycle arrest and differentiation of acute myeloid leukemia (AML) cells also involve the activation of STAT1 (6–8). As the direct target genes of STAT1 transcriptional activity, ISGs constitute the major effector molecules both in antiviral innate immunity by restricting viral RNA propagation (9, 10) and in cellular proliferation regulation by inducing cell death and cell cycle arrest (11, 12).
Recent studies have established RA-inducible gene I (RIG-I; also named DDX58 for DEAD box polypeptide 58) as a crucial sentry molecule to trigger the innate immunity responses against viral propagation (13). Mechanistically, on sensing the cytoplasmic invasion of viral RNA entities, the latent RIG-I molecules will be activated to initiate a robust generation of type I IFN mainly through binding and activating an adaptor protein, IFN-β promoter stimulator 1 (IPS-1; also named MAVS, VISA, or Cardif) (14–17). However, RIG-I was also identified as one gene whose expression was highly up-regulated during ATRA-induced granulocytic differentiation and growth inhibition of acute promyelocytic leukemia (APL) cell line NB4 cells (18). Moreover, our previous work based on the phenotypic analyses of a Rig-I−/− mouse model has unveiled an essential role of RIG-I in regulating myelopoiesis in vivo, at least partially through promoting the myeloid expression of a classic ISG, IFN consensus sequence-binding protein (Icsbp) (19). In this study, we provide the further and relevant mechanistic insight that by positively regulating STAT1 activation in a manner independent of IPS-1 signaling, IFN- and/or RA-induced RIG-I induction exerts a critical feedback effect to augment STAT1 activation and the induction of ISGs, including ICSBP, thus fulfilling the efficacy of IFN and/or RA signaling to curb the cellular proliferation of AML.
Results
RIG-I Induction Functionally Contributes to IFN Plus ATRA-Induced Growth Inhibition of AML Cells.
To test whether RIG-I induction functionally contributes to the growth inhibition of AML cells, we exploited the U937 AML cell line (M5 subtype) whose proliferation in vitro is inhibited by ATRA and IFN (7, 20), both of which are well-documented RIG-I inducers in numerous cell types (19, 21). We noticed that although IFN-α alone but not ATRA alone triggered obvious RIG-I induction in both mRNA and protein levels in U937 cells, the addition of ATRA apparently produced a potent synergistic effect with IFN-α to trigger RIG-I induction and cell growth inhibition (Fig. S1 A–C). Conversely, Rig-I expression was not obviously altered when inducing U937 with phorbol 12-myristate 13-acetate to the monocytic differentiation (Fig. S1D). Taken together, these data indicate that if RIG-I induction is not absolutely required for the growth inhibition of AML cells, it may at least enhance their growth inhibition.
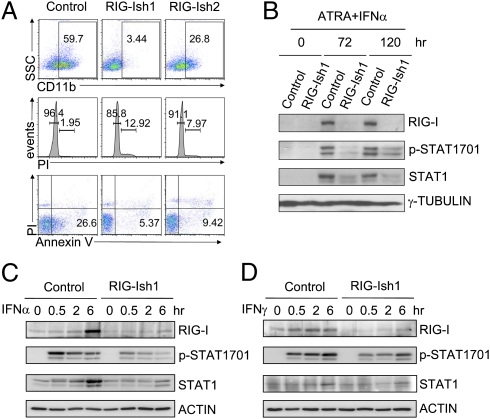
We then established two stably transfected U937 cell lines (RIG-Ish1 and RIG-Ish2) that expressed tetracycline free (Tc−)-inducible shRNA against RIG-I (Fig. S2). As shown in Fig. 1A, IFN-α plus ATRA-induced myeloid differentiation, cell cycle arrest, and cell death of U937 cells were obviously lessened by preventing RIG-I induction with siRNAs, indicating a functional contribution of RIG-I induction to the IFN plus RA signaling-activated program that inhibits cellular proliferation of AML cells.
Fig. 1.
ATRA/IFNα-induced growth inhibition and STAT1 activation in U937 leukemia cells depend on RIG-I induction. (A) U937/control, U937/RIG-Ish1, or U937/RIG-Ish2 cells were treated with 1 μM ATRA plus 1,000 U/mL IFN-α. Ninety-six hours after treatment, CD11b level, cell cycle status, and Annexin V-propidium iodide (PI) staining signals were measured by flow cytometry. (B) Tyrosine 701-phosphorylated and total levels of STAT1 were examined by Western blotting assay at 0, 72, and 120 h after treatment was begun. U937/control and U937/RIG-Ish1 cells were treated with 1,000 U/mL IFN-α (C) or 1,000 U/mL IFN-γ (D) for less than 6 h, as indicated. The tyrosine 701-phosphorylated and total levels of STAT1 were determined by Western blotting assays.
IFN Plus ATRA-Induced RIG-I Expression Augments STAT1 Activation in AML Cells.
Previous studies demonstrate that the key IFN intracellular signaling mediator Stat1 acts as an oncorepressor in numerous tumor types (4). Relevant to this, it has been shown that analogous to the essential role of STAT1 activation in IFN intracellular signaling, the up-regulation and phosphorylation of STAT1 mediate the ATRA signaling-induced differentiation and growth inhibition of U937 cells (6, 7), probably via promoting the expression of IFN and/or the numerous intracellular IFN signaling components (22, 23). We then asked whether a functional relationship exists between RIG-I induction and STAT1 activation. As shown in Fig. S3 A and B, RIG-I induction by IFN-α was almost abrogated in the STAT1-deficient human fibrosarcoma cell line U3A and in Stat1−/− mouse bone marrow (BM) myeloid cells, confirming that RIG-I is a classic ISG to be regulated by STAT1 activation. Conversely, the tyrosine 701-phosphorylated and total levels of STAT1 were also significantly reduced by RIG-I knockdown for 72–120 h after ATRA plus IFN-α treatment (Fig. 1B), suggesting a converse regulatory role of RIG-I induction on STAT1 activation. To delineate further whether RIG-I has a regulatory effect on the phosphorylation modification of STAT1 independent of the accompanied alterations in the protein level, we examined the phosphorylation modification of STAT1 in the control and RIG-I knockdown (RIG-Ish1) U937 cells shortly after IFN stimulation (Fig. 1 C and D). The results unveiled a direct promoting role of RIG-I induction on STAT1 phosphorylation at the 701 site before the alterations occurring to STAT1 protein level (within 2 h poststimulation). Thus, a functional contribution of RIG-I induction to promote STAT1 activation is indicated.
Robust ISG Induction and Stat1 Activation in Myeloid Cells Require Rig-I Induction.
We have previously reported that Rig-I deficiency in mice results in the development of myeloproliferative disorder with aging, which is at least partly ascribed to the reduced expression of Icsbp in Rig-I−/− myeloid cells (19). Because Icsbp belongs to the classic ISGs stimulated by either IFN-α or IFN-γ (24, 25), we then examined whether the expressions of other ISGs might also be affected by Rig-I deficiency. By comparing the mRNA expression profile of Rig-I−/− granulocytes with that of Rig-I+/+ granulocytes, we found that two additional classic ISGs, Klf4 and Trail (26, 27), also appeared in the top-ranked genes whose expression was greatly reduced by Rig-I deficiency in BM granulocyte progenitors or mature granulocytes (Fig. 2A). This trio-ISG expression reduction in Rig-I−/− myeloid cells thus provided confirming data for RIG-I involvement within the IFN intracellular signaling pathway.
Fig. 2.
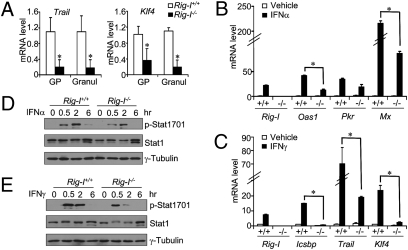
Rig-I deficiency attenuates ISG production and Stat1 activation. (A) Relative mRNA levels of Trail and Klf4 within the granulocytic progenitors (GP) or mature granulocytes (Granul) from Rig-I+/+ or Rig-I−/− mice were determined by the real-time PCR assay (n = 3, mean ± SD; *P < 0.05). Primary Rig-I+/+ or Rig-I−/− BM myeloid cells were stimulated by 1,000 U/mL IFN-α (B) or 30 ng/mL IFN-γ (C) for 12 h. Relative mRNA levels of various ISGs were measured by real-time PCR assay (n = 3, mean ± SD; *P < 0.05). The tyrosine 701-phosphorylated and total levels of Stat1 were examined by Western blotting assay on the Rig-I+/+ or Rig-I−/− BM myeloid cells at the indicated time points after being stimulated by 1,000 U/mL IFN-α (D) or 30 ng/mL IFN-γ (E).
To address this directly under sterile conditions, we stimulated the freshly isolated Rig-I+/+ or Rig-I−/− BM myeloid cells with high concentrations of IFN-α or IFN-γ and subsequently measured the mRNA induction of numerous ISGs with a real-time PCR assay. As mentioned above (28, 29), Rig-I itself was an authentic ISG, and its mRNA level in normal myeloid cells was significantly elevated by either IFN-α or IFN-γ stimulation (Fig. 2 B and C). Interestingly, the inductions of Oas1 and Mx at 6 h after IFN-α stimulation seemed highly dependent on an earlier induction of RIG-I at 2 h poststimulation (Fig. S4). In line with this, for five of six representative IFN-α– or IFN-γ–stimulated ISGs as indicated in Fig. 2 B and C, the inductions of their mRNA levels at 12 h were significantly impaired in Rig-I−/− myeloid cells, indicating a crucial role of Rig-I induction to augment ISG induction.
Next, we applied a Western blotting assay to measure Stat1 phosphorylation levels in Rig-I+/+ or Rig-I−/− myeloid cells following IFN stimulation. In line with what was found in the U937 AML model (Fig. 1B), the tyrosine phosphorylation of Stat1 at the 701 site was significantly reduced under the circumstance of Rig-I deficiency after either IFN-α or IFN-γ stimulation (Fig. 2 D and E). Moreover, the total Stat1 protein level experienced a transient reduction after IFN-γ signaling but not after IFN-α signaling in Rig-I−/− myeloid cells (Fig. 2E), indicating a regulatory role of Rig-I on both Stat1 phosphorylation and protein levels.
RIG-I Induction Alone Triggers STAT1 Activation in a Manner Independent of IPS-1.
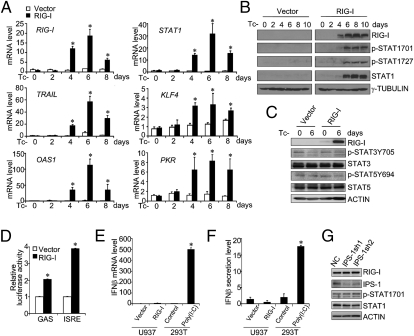
The canonical IFN signaling-induced STAT1 activation occurs through the activation of JAKs and is subject to the modulating effects of waves of molecular events with either positive or negative regulatory activity (3). To test directly how RIG-I induction contributes to STAT1 activation in leukemia cells, we integrated a Tc−-inducible Flag-tagged RIG-I–expressing system in U937 cells to replace the IFN plus RA-triggered induction (Fig. S2). As shown in Fig. 3 A and B, at Tc−-induced day 4, significant increases in both RIG-I mRNA and protein levels were observed, reaching a peak point around Tc−-induced day 6. Coincidentally, increases in the mRNA levels of numerous ISGs, such as STAT1, TRAIL, KLF4, OAS1, and PKR, were also observed (Fig. 3A). Most interestingly, even in the absence of viral infection or foreign IFN and/or RA stimulation, enhanced STAT1 phosphorylations at both the 701 site and the 727 site (but not further activation of STAT3 and STAT5) were evident following this sole RIG-I induction (Fig. 3 B and C). Furthermore, RIG-I induction in U937 cells led to the activation of a GAS or an ISRE element-coupled luciferase reporter gene (Fig. 3D), confirming a functional consequence of RIG-I up-regulation–induced STAT1 activation in U937 cells even without a foreign IFN or ATRA stimulation.
Fig. 3.
RIG-I induction triggers ISG production and STAT1 activation in U937 cells independent of IPS-1. (A and C) U937/vector and U937/RIG-I cells were incubated in the Tc− medium for the different days as indicated. (A) Relative mRNA levels of RIG-I, STAT1, and other ISGs, including TRAIL, KLF4, OAS1, and PKR, were determined by real-time PCR assays (n = 3, mean ± SD; *P < 0.05). The total and phosphorylated levels of STAT1 (B) or those of STAT3 and STAT5 (C) were examined by Western blotting analyses. (D) GAS- or ISRE-coupled luciferase reporter plasmid was transfected into U937/vector or U937/RIG-I cells at day 6 of culture in Tc− medium. The relative luciferase activities of each group were expressed as mean ± SD (n = 3, mean ± SD; *P < 0.05). U937/vector and U937/RIG-I were measured by real-time PCR assays for IFN-β mRNA levels (E) or by ELISA for IFN-β protein levels in the culture supernatant (F) at day 6 of culture in Tc− medium. IFN-β mRNA or protein induction levels in the 293T cells transfected with 1 μg/mL Poly(I:C)[poly(rI):poly(rC)] were measured as the positive control. (G) Two knockdown plasmids specific for IPS-1 (IPS-1sh1 and IPS-1sh2) and one negative control plasmid (NC) were separately transduced into U937/RIG-I cells at day 2 of tetracycline withdrawal. Cells were cultured in the Tc− medium for another 4 d before protein extracts were collected for Western blotting analysis.
It could be argued that even without prior priming by the cognate RNA ligands, a high concentration of RIG-I molecules might activate IPS-1 via a noncanonical mechanism, which would, in turn, activate STAT1 phosphorylation through completing an IFN induction and IFN signaling circuit, because with the in vitro transient-transfection assay, we noted that the deletion of the caspase recruitment domain (CARD) that binds IPS-1 made RIG-I unable to activate STAT1 (Fig. S5). To test this possibility, we monitored the endogenous IFN-β mRNA levels and IFN-β secretion in the case of the sole RIG-I induction in U937 cells. We found that RIG-I induction neither raised the intracellular IFN-β mRNA level nor the IFN-β protein level within the culture supernatant (Fig. 3 E and F), thus excluding the feasible existence of an autocrine IFN signaling circuit created by the sole RIG-I induction. To examine directly whether IPS-1 activation was ever involved in STAT1 activation, we further transfected two IPS-1 shRNA-expressing plasmids into the RIG-I–induced U937 cells (Materials and Methods) and found that IPS-1 knockdown did not prevent STAT1 phosphorylation and up-regulation at all in the RIG-I–induced U937 cells (Fig. 3G), verifying that the prompted activation of STAT1 by the sole RIG-I induction is achieved via a mechanism independent of IPS-1 activation.
IFN-induced JAK1, JAK2, and TYK2 activations are direct upstream molecular events that trigger STAT1 phosphorylation at the 701 site. We then examined whether RIG-I induction ever needs to stimulate JAKs for activating STAT1. Quite unexpectedly, none of these three JAK members was found to be activated in the RIG-I–induced U937 cells as compared with the U937/control cells, whereas JAK1 and TYK2 were obviously stimulated by IFN-α in the U937 cells (Fig. S6 A and B). Moreover, the administration of a phosphatase inhibitor, sodium orthovanadate (OV) (30), failed to rescue the hypophosphorylated status of Stat1 in Rig-I−/− myeloid cells after IFN-γ signaling (Fig. S7), refuting the possibility that Rig-I promotes Stat1 activation through negatively regulating the numerous phosphatase activities that act on the phosphorylated Stat1. Taken together, these results indicate the existence of a so far unidentified noncanonical mechanism through which RIG-I induction promotes the activation of STAT1.
RIG-I Induction Alone Inhibits the Proliferation of AML Cells both in Vitro and in Vivo.
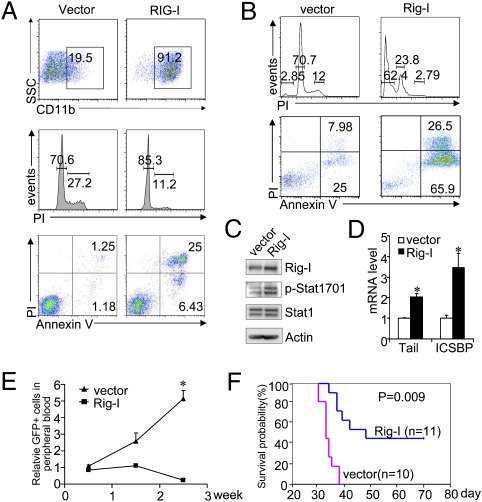
STAT1 activation has been shown to convey a strong oncorepressing activity in numerous tumor models (31). In particular, essential roles of STAT1 activation in ATRA-induced cell growth inhibition of APL cells and the IFN-induced clinical response of patients with chronic myeloid leukemia have been well documented (32, 33). Because the sole RIG-I induction triggers significant STAT1 activation in U937 cells, we then asked whether RIG-I induction might exert a measurable inhibitory effect on AML cell proliferation. Intriguingly, as a partial mimicry of the IFN plus ATRA-induced antiproliferation effect, RIG-I induction alone indeed triggered obvious myeloid differentiation of U937 cells, accompanied by cell cycle arrest and cell death, pointing to RIG-I as an important downstream effector molecule of the IFN or RA signaling cascade (Fig. 4A). To test for a possible regulatory role of RIG-I induction on the proliferation of APL cells in which RIG-I cDNA was originally cloned (18), we isolated APL blast-enriched primary BM cells from a mouse APL model and infected them with a retroviral vector harboring Rig-I cDNA (coupled with GFP expression). In accordance with what was observed in the U937 cell model, the transduction of Rig-I promoted primary APL cells to undergo cell cycle arrest and cell death in vitro (Fig. 4B), in parallel with Stat1 activation and ISG induction (Fig. 4 C and D). Moreover, the enforced Rig-I overexpression into c-Kit+ APL progenitors (GFP+ cells) greatly inhibited the reestablishment of APL after being transplanted into the syngeneic recipients, as compared with the empty vector-transduced APL group (Fig. 4 E and F), demonstrating a capacity of Rig-I induction to inhibit the in vivo proliferation of APL leukemia-initiating cells (LICs).
Fig. 4.
RIG-I induction alone inhibits proliferation of AML cells both in vitro and in vivo. (A) Flow cytometric assays show that RIG-I induction triggers U937 cells to express CD11b and to undergo cell cycle arrest and cell death. (B) Flow cytometric assays show that Rig-I transduction triggers APL cells to undergo cell cycle arrest and cell death. Rig-I transduction induces Stat1 activation (C) and ISG expression (D) in mouse APL cells, as measured by Western blotting and real-time PCR assays (n = 3, mean ± SD; *P < 0.05). (E) Rig-I transduction inhibits the in vivo proliferation of the inoculated GFP+ c-Kit+ APL BM cells (n = 3, mean ± SD; *P < 0.05). The percentage of GFP+ cells in mouse peripheral blood was monitored by flow cytometry at the indicated time points. (F) Survival curves of the normal recipients receiving either empty vector- or Rig-I–transduced c-Kit+ APL BM cells.
RIG-I Induction Triggers ISG Production and Growth Inhibition at Least Partially Through STAT1.
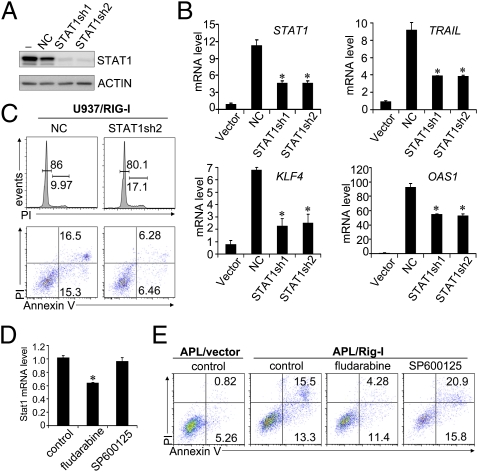
It is reasonable to postulate that RIG-I induction will initiate numerous molecular events other than simply promoting STAT1 activation. Finally, we tested whether RIG-I induction-triggered ISG production and leukemia cell growth inhibition are at least partly dependent on the augmenting role of RIG-I induction on STAT1 activation. As expected, the transduction of these two shRNA-expression plasmids into the RIG-I–induced U937 cells significantly reduced not only the induction of STAT1 mRNA but also those of TRAIL, KLF4, and OAS1 mRNAs (Fig. 5B). In line with numerous previous reports indicating that ISGs, such as TRAIL and KLF4, are involved in regulating tumor cell proliferation (11, 12), RIG-I–induced cell cycle arrest and cell death were inhibited by STAT1 knockdown (Fig. 5C). Likewise, the treatment of Rig-I–transduced c-Kit+ mouse APL cells by fludarabine, a Stat1-specific inhibitor (34), down-regulated the Stat1 mRNA level (Fig. 5D) and, accordingly, Rig-I induction-triggered cell death, both of which were not observed when the APL cells were treated with JNK inhibitor SP600125 (Fig. 5E). Collectively, these results demonstrate that the RIG-I induction-triggered STAT1 activation at least partly contributes to ISG production and growth inhibition of AML cells.
Fig. 5.
RIG-I induction triggers ISG production and growth inhibition of AML cells partly through its promoting role on STAT1 activation. (A) Two shRNA plasmids specific for STAT1 knockdown (STAT1sh1 and STAT1sh2) and one plasmid containing a negative control (NC) sequence were individually transduced into 293T cells, together with STAT1-expressing plasmid. Forty-eight hours later, the cell lysates were collected for STAT1 protein analyses by Western blotting assay. (B and C) Two STAT1 shRNA plasmids and one control plasmid were individually transduced into U937/RIG-I cells at day 2 after tetracycline withdrawal and then cultured in the Tc− medium for another 4 d. (B) Relative mRNA levels of STAT1 and ISGs as indicated were determined by real-time PCR assays (n = 3, mean ± SD; *P < 0.05). (C) Cell cycle measurement by propidium iodide (PI) staining or cell death analysis by Annexin V-PI staining was performed by flow cytometry. (D and E) RIG-I–transduced mouse APL BM cells were treated with 100 μM fludarabine (Stat1 inhibitor) or 5 μM SP600125 (JNK inhibitor) for 48 h. Cells were then measured by real-time PCR for Stat1 mRNA level (n = 3, mean ± SD; *P < 0.05) (D) or analyzed by flow cytometry for cell survival (E).
Discussion
As previously documented, on sensing and binding viral RNA entities, the ubiquitously expressed cytoplasmic RIG-I molecules, presumably at basic levels, are recruited into IPS-1 signalosome to trigger type I IFN production, which, in turn, initiates the innate immunity response (14). Nevertheless, because RIG-I expression itself is highly inducible by IFN or RA signaling (Fig. S2 B and C) (21, 28, 29), we argue that RIG-I might also possess a relatively independent regulatory role on the ensuing IFN intracellular signaling process. In line with this assumption, we have previously demonstrated an important physiological role of RIG-I in negatively regulating myelopoiesis, partly through maintaining the myeloid expression of a classic ISG, Icsbp (19). In light of the findings that IFN signaling or RA signaling in AML cells elevates STAT1 protein level and enhances its phosphorylation to induce cell cycle arrest and differentiation (6, 7), we have conducted the further experiments to show that RIG-I has an intrinsic activity to regulate the cellular proliferation of AML cells through augmenting STAT1 activation (Fig. S8).
Actually, previous studies have already shown that in the absence of viral infection, RIG-I overexpression alone in the mammalian cell line BEAS-2B results in the induction of STAT1 mRNA (28) and that IFN-γ–stimulated expression of IFN regulatory factor 7 is dependent on RIG-I (29). In this present study with the Tc−-inducible expression of RIG-I in the U937 cell line, the dual effects of RIG-I not only on elevating the STAT1 mRNA level but also on enhancing STAT1 phosphorylation, at least at the 701 site, were clearly demonstrated. As shown in the immediate phase after IFN treatment of RIG-I knockdown U937 cells, STAT1 phosphorylation at tyrosine 701 had already been significantly reduced before the STAT1 protein level was ever decreased (Fig. 2 B and C), indicating that the influence of RIG-I on STAT1 701 phosphorylation is independent of its regulation on STAT1 protein level.
Intriguingly, RIG-I induction promotes STAT1 activation in an IPS-1 activation-independent manner (Fig. S8), although its N-terminal CARDs, which harbors the capacity to tether IPS-1, is required for exerting this activity. Quite unexpectedly, RIG-I induction triggers STAT1 activation without further enhancing the phosphorylation of any of the three JAK family members, requiring further study to define precisely how RIG-I induces STAT1 activation downstream of IFN/IFN receptor ligation and what additional noncanonical STAT1 activation regulatory pathways, as influenced by RIG-I activity, might account for this regulatory process. For example, we noted that although the direct mechanism is not understood, in a similar manner to RIG-I, PKC-ε, a serine/threonine kinase, has been shown to promote STAT1 tyrosine phosphorylation without stimulating JAK activation (35). We also cannot exclude the possibility that RIG-I might exert this STAT1 regulation through numerous other accessory regulators, such as suppressor of cytokine signaling family members (36).
As its name implies, RIG-I cDNA was isolated in the biological setting of ATRA-induced granulocytic differentiation of AML cells. However, the functional role of RIG-I induction in AMLs has not been clarified before. In this study, we provide substantial evidence indicating that RIG-I induction participates in promoting STAT1 activation, downstream induction of critical ISGs, and, finally, cellular growth inhibition in the IFN- and/or RA-induced leukemia differentiation model. The inoculation experiments using APL progenitor cells suggest that RIG-I induction alone exerts a significant inhibiting effect on the proliferation of APL LICs in vivo (Fig. 4), pointing to RIG-I induction as a potential therapeutical element for improving treatment of AML.
Materials and Methods
Mice, Cell Culture, and Reagents.
Rig-I−/− 129S3 mice, Stat1−/− 129S6 mice, and normal FVB/NJ mice were bred in the animal facility of Shanghai Jiao-Tong University. The hMRP8-PML/RAR-α–harboring APL cells were passaged in vivo among syngeneic FVB/NJ mice. 293T or U937 cells were grown in DMEM or RPMI-1640 supplemented with 10% (vol/vol) FBS (GibcoBRL). IFN-α and IFN-γ were purchased from R&D Systems, ATRA and SP600125 were purchased from Sigma–Aldrich, and fludarabine was obtained from Bayer. Human and mouse RIG-I antibodies were provided by Abmart Inc. Primary antibodies against IPS-1, STAT1, p-STAT1Y701, p-STAT1S727, STAT3, p-STAT3Y705, STAT5, and p-STAT5Y694 were purchased from Cell Signaling. Actin and γ-tubulin antibodies were purchased from Sigma–Aldrich.
Generation of RIG-I cDNA- or RIG-I shRNA-Inducible U937 Cell Lines.
To generate empty vector or RIG-I cDNA stably transfected U937T cell lines, 1 × 107 U937T cells that constitutively express tetracycline-controlled transactivator were electroporated with pTRE2hyg empty vector or pTRE2hyg–RIG-I plasmid using a Gene-Pulser II (Bio-Rad) and selected with 0.5 mg/mL hygromycin B, 0.5 μg/mL puromycin, and 1 μg/mL tetracycline (Merck–Calbiochem) for 4–6 wk. The U937 cell lines that express inducible RIG-Ish1, RIG-Ish2, or control sequences (listed in Table S1) were generated in a similar manner. RIG-I shRNAs flanked by miR30a sequences were inserted into the 3′-UTR region downstream of an EGFP-encoding region in pTRE2hyg plasmid.
RNAi.
RNAi sequences for STAT1 and IPS-1 were expressed by miR30a-shRNA vector P201 (Open Biosystems). The specific target sequences and negative control used for interference are listed in Table S1.
Real-Time PCR and Western Blotting.
The real-time PCR reactions were performed using an ABI7900 PCR machine (Applied Biosystems) as previously described (19). The primers used are listed in Table S2. For Western blotting assay, the whole-cell lysates were run on SDS/PAGE and transferred to PVDF membranes (GE-Healthcare). Detection was performed using an ECL detection kit (Millipore).
Flow Cytometry.
Myeloid differentiation was monitored using PE-Cy5–labeled anti-CD11b antibody (Beckman Coulter), and the nuclear DNA content was determined by propidium iodide staining. For cell death assay, Annexin V-positive cells were measured using an Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences). The flow data were collected with a BD FACSCalibur machine (Becton Dickinson) and analyzed by FlowJo software (Tree Star Inc.). For the sorting experiment, stained cells were sorted out with a MoFlo high-speed cell sorter (Beckman Coulter).
Retroviral Transduction of APL Cells and Transplantation.
MigR1 retroviral vector expressing mouse Rig-I cDNA was prepared as described (19). The infection was performed on the c-Kit+ primary mouse APL BM cells that were cultured in MyeloCult M5300 (StemCell Technologies) supplemented with 6 ng/mL IL-3, 10 ng/mL IL-6, and 50 ng/mL stem cell factor (R&D Systems). The sorted GFP+ cells were then injected i.v. into syngeneic recipients of FVB mice for in vivo experiments.
Statistical Analysis.
Kaplan–Meier survival analysis (SPSS 10.0; SPSS, Inc.) was used to compare the survival probabilities of different groups. The Student's t test was used to compare the difference between two different groups. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Abmart Inc. for making RIG-I antibodies. This work was supported by Shanghai Municipal Committee of Science and Technology Grants 06dj14002 and 09XD1403000, National Scientific Foundation of China Grant 30871108, Chinese National Key Basic Research Project 973 Grants 2007CB947800 and 2010CB945600, and E-Institutes of Shanghai Municipal Education Commission Grant E03003.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019059108/-/DCSupplemental.
References
- 1.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 2.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: Implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 4.Levy DE, Darnell JE., Jr. Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 5.Chelbi-Alix MK, Pelicano L. Retinoic acid and interferon signaling cross talk in normal and RA-resistant APL cells. Leukemia. 1999;13:1167–1174. doi: 10.1038/sj.leu.2401469. [DOI] [PubMed] [Google Scholar]
- 6.Dimberg A, Karlberg I, Nilsson K, Oberg F. Ser727/Tyr701-phosphorylated Stat1 is required for the regulation of c-Myc, cyclins, and p27Kip1 associated with ATRA-induced G0/G1 arrest of U-937 cells. Blood. 2003;102:254–261. doi: 10.1182/blood-2002-10-3149. [DOI] [PubMed] [Google Scholar]
- 7.Dimberg A, Nilsson K, Oberg F. Phosphorylation-deficient Stat1 inhibits retinoic acid-induced differentiation and cell cycle arrest in U-937 monoblasts. Blood. 2000;96:2870–2878. [PubMed] [Google Scholar]
- 8.Gianni M, et al. Stat1 is induced and activated by all-trans retinoic acid in acute promyelocytic leukemia cells. Blood. 1997;89:1001–1012. [PubMed] [Google Scholar]
- 9.Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.Altucci L, et al. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7:680–686. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, et al. Krüppel-like factor 4 (gut-enriched Krüppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 15.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 16.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 17.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Liu TX, et al. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood. 2000;96:1496–1504. [PubMed] [Google Scholar]
- 19.Zhang NN, et al. RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proc Natl Acad Sci USA. 2008;105:10553–10558. doi: 10.1073/pnas.0804895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedo A, Van Weyenbergh J, Rouillard D, Bauvois B. Synergistic effect of prolactin on IFN-gamma-mediated growth arrest in human monoblastic cells: Correlation with the up-regulation of IFN-gamma receptor gene expression. Immunol Lett. 1996;53:125–130. doi: 10.1016/s0165-2478(96)02622-3. [DOI] [PubMed] [Google Scholar]
- 21.Imaizumi T, et al. Interferon-gamma induces retinoic acid-inducible gene-I in endothelial cells. Endothelium. 2004;11:169–173. doi: 10.1080/10623320490512156. [DOI] [PubMed] [Google Scholar]
- 22.Kolla V, Lindner DJ, Xiao W, Borden EC, Kalvakolanu DV. Modulation of interferon (IFN)-inducible gene expression by retinoic acid. Up-regulation of STAT1 protein in IFN-unresponsive cells. J Biol Chem. 1996;271:10508–10514. doi: 10.1074/jbc.271.18.10508. [DOI] [PubMed] [Google Scholar]
- 23.Luo XM, Ross AC. Physiological and receptor-selective retinoids modulate interferon gamma signaling by increasing the expression, nuclear localization, and functional activity of interferon regulatory factor-1. J Biol Chem. 2005;280:36228–36236. doi: 10.1074/jbc.M505749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egwuagu CE, et al. Interferon-gamma induces regression of epithelial cell carcinoma: Critical roles of IRF-1 and ICSBP transcription factors. Oncogene. 2006;25:3670–3679. doi: 10.1038/sj.onc.1209402. [DOI] [PubMed] [Google Scholar]
- 25.Politis AD, Ozato K, Coligan JE, Vogel SN. Regulation of IFN-gamma-induced nuclear expression of IFN consensus sequence binding protein in murine peritoneal macrophages. J Immunol. 1994;152:2270–2278. [PubMed] [Google Scholar]
- 26.Tecchio C, et al. IFNalpha-stimulated neutrophils and monocytes release a soluble form of TNF-related apoptosis-inducing ligand (TRAIL/Apo-2 ligand) displaying apoptotic activity on leukemic cells. Blood. 2004;103:3837–3844. doi: 10.1182/blood-2003-08-2806. [DOI] [PubMed] [Google Scholar]
- 27.Feinberg MW, et al. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–38258. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 28.Imaizumi T, et al. Involvement of retinoic acid-inducible gene-I in the IFN-gamma/STAT1 signalling pathway in BEAS-2B cells. Eur Respir J. 2005;25:1077–1083. doi: 10.1183/09031936.05.00102104. [DOI] [PubMed] [Google Scholar]
- 29.Imaizumi T, et al. Retinoic acid-inducible gene-I (RIG-I) is induced by IFN-gamma in human mesangial cells in culture: Possible involvement of RIG-I in the inflammation in lupus nephritis. Lupus. 2010;19:830–836. doi: 10.1177/0961203309360540. [DOI] [PubMed] [Google Scholar]
- 30.Baron M, Davignon JL. Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol. 2008;181:5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 31.Sternberg DW, Gilliland DG. The role of signal transducer and activator of transcription factors in leukemogenesis. J Clin Oncol. 2004;22:361–371. doi: 10.1200/JCO.2004.10.124. [DOI] [PubMed] [Google Scholar]
- 32.Landolfo S, et al. Chronic myeloid leukemia cells resistant to interferon-alpha lack STAT1 expression. Hematol J. 2000;1:7–14. doi: 10.1038/sj.thj.6200004. [DOI] [PubMed] [Google Scholar]
- 33.Pelicano L, Brumpt C, Pitha PM, Chelbi-Alix MK. Retinoic acid resistance in NB4 APL cells is associated with lack of interferon alpha synthesis Stat1 and p48 induction. Oncogene. 1999;18:3944–3953. doi: 10.1038/sj.onc.1202802. [DOI] [PubMed] [Google Scholar]
- 34.Frank DA, Mahajan S, Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- 35.Ivaska J, Bosca L, Parker PJ. PKCepsilon is a permissive link in integrin-dependent IFN-gamma signalling that facilitates JAK phosphorylation of STAT1. Nat Cell Biol. 2003;5:363–369. doi: 10.1038/ncb957. [DOI] [PubMed] [Google Scholar]
- 36.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.