Abstract
Histone deacetylase 5 (HDAC5), a class IIa deacetylase, is a prominent regulator of cellular and epigenetic processes that underlie the progression of human disease, ranging from cardiac hypertrophy to cancer. Although it is established that phosphorylation mediates 14–3-3 protein binding and provides the essential link between HDAC5 nucleo-cytoplasmic shuttling and transcriptional repression, thus far only four phospho-acceptor sites have been functionally characterized. Here, using a combinatorial proteomics approach and phosphomutant screening, we present the first evidence that HDAC5 has at least 17 in vivo phosphorylation sites within functional domains, including Ser278 and Ser279 within the nuclear localization signal (NLS), Ser1108 within the nuclear export signal, and Ser755 in deacetylase domain. Global and targeted MS/MS analyses of NLS peptides demonstrated the presence of single (Ser278 and Ser279) and double (Ser278/Ser279) phosphorylations. The double S278/279A mutation showed reduced association with HDAC3, slightly decreased deacetylation activity, and significantly increased cytoplasmic localization compared with wild type HDAC5, whereas the S278A and S1108A phosphomutants were not altered. Live cell imaging revealed a deficiency in nuclear import of S278/279A HDAC5. Phosphomutant stable cell lines confirmed the cellular redistribution of NLS mutants and revealed a more pronounced cytoplasmic localization for the single S279A mutant. Proteomic analysis of immunoisolated S278/279A, S279A, and S259/498A mutants linked altered cellular localization to changes in protein interactions. S278/279A and S279A HDAC5 showed reduced association with the NCoR-HDAC3 nuclear corepressor complex as well as protein kinase D enzymes, which were potentiated in the S259/498A mutant. These results provide the first link between phosphorylation outside the known 14–3-3 sites and downstream changes in protein interactions. Together these studies identify Ser279 as a critical phosphorylation within the NLS involved in the nuclear import of HDAC5, providing a regulatory point in nucleo-cytoplasmic shuttling that may be conserved in other class IIa HDACs—HDAC4 and HDAC9.
Epigenetic regulation of gene expression provides control over important cellular processes without requiring direct DNA sequence modification. Gene activation/repression is accomplished through chromatin remodeling, the dynamic restructuring of nucleosomes, as well as by regulating the activity and localization of transcription factors. Essential to epigenetic regulation is the concerted effort of histone acetyltransferases and deacetylases (HDACs)1 (1). Specifically, HDACs mediate transcriptional repression by removing acetyl groups from DNA-bound histones and often carry out this function as components of larger protein complexes. Based on domain similarity to yeast deacetylases, HDACs are classified as: class I (yRPD3 homologs), class II (yHDA1 homologs), class III (ySIR2 homologs), and class IV (class I and II homology) (2). Class II HDACs are further subdivided into class IIa (HDACs 4, 5, 7, and 9) and class IIb (HDACs 6 and 10) based on primary sequence homology.
Because of their significant roles in human diseases, HDACs have been the subject of intense interest over the last decade. Of these, the class IIa deacetylase HDAC5 has been selectively shown to play critical roles in cardiac disease, epigenetic response to drug stimuli, viral infection, and cancer (3, 4). Intimately linked with these disease processes is the regulation of gene expression indirectly mediated by HDAC5 nucleo-cytoplasmic shuttling, a common regulatory mechanism for all class IIa HDACs (2). This shuttling mechanism may allow for interactions with nonnuclear substrates, and because HDACs evolutionarily predate histone proteins, it is conceivable that cytoplasmic targets remain undiscovered. The deacetylation activity of HDAC5 involves the enzymatic contribution of HDAC3, a class I deacetylase that also can shuttle between the nucleus and cytoplasm (5). HDAC3 binds the C-terminal domain of HDAC5 via transcriptional corepressor complexes containing SMRT/N-CoR (6). HDAC5 also possesses a unique N-terminal region containing transcription factor binding domains, such as for the MEF2 (myocyte enhancer factor-2) family members, and a nuclear localization signal (NLS), as well as a C-terminal nuclear export signal (NES) (7, 8).
Phosphorylation is a central mechanism mediating nucleo-cytoplasmic shuttling and the transcriptional repressor functions of class IIa HDACs (9–11). To date, five phosphorylation sites have been identified on HDAC5. The CaMK-dependent phosphorylation of Ser259 and Ser498 that flank the NLS provide docking sites for 14–3-3 chaperone binding, triggering nuclear export and the negative regulation of repressor activity, a mechanism conserved among class IIa HDACs (12, 13). Two of the additional phosphorylations that have been identified are Ser661, with an uncharacterized function (14), and Thr292 within the NLS, which is phosphorylated by protein kinase C-related kinase (PRK)-2 and PRK1 (15). A recent study showed that Ser279 within the NLS can be in vitro phosphorylated by protein kinase A (PKA) and that in Cos7 cells and cardiomyocytes(NRVMs), PKA-dependent phosphorylation is inhibitory toward HDAC5 nuclear export via disruption of 14–3-3 association (16). Regulation of HDAC5 by phosphorylation is of considerable interest as physiological and pathophysiological processes, such as neuronal activity (17) and cardiac hypertrophy (18), have been shown to converge on specific signal-responsive phosphorylation sites (19). Nevertheless, other as yet undiscovered phosphorylations are thought to be involved in shuttling and transcriptional regulation (19).
Here, we present the first comprehensive phosphorylation study on a class IIa histone deacetylase, HDAC5. We show that HDAC5 is phosphorylated in vivo at 17 sites, suggestive of multiple phosphorylation-dependent mechanisms that dynamically regulate HDAC5. Phosphorylation sites are distributed throughout the protein sequence, including the deacetylation domain, NLS and NES. Functional assessment of distinct phosphorylation sites, revealed decreased deacetylase activity and HDAC3 interaction resulting from serine to alanine mutation within the NLS, leading us to propose a role for Ser278/Ser279 within the NLS in the regulation of HDAC5 localization and function. We demonstrated mutation of these sites lead to predominant localization of HDAC5 to the cytoplasm in concert with changes in protein interactions, including decreased association with the nuclear corepressor complex and absence of PKD enzymes. Targeted mass spectrometry and cellular localization studies of HDAC5-EGFP from stable cell lines expressing the double (S278/279A) and single (S279A) NLS mutants revealed Ser279 phosphorylation as a primary locus that influenced the steady-state distribution of HDAC5.
EXPERIMENTAL PROCEDURES
Reagents
Antibodies used were in-house developed rabbit polyclonal anti-GFP (20), monoclonal anti-GFP (Roche), anti-14–3-3 ε (T-16, Santa Cruz, CA), anti-HDAC3 (Cell Signaling Technology, Inc. Danvers, MA) and Mab414 (Covance, gift from J. Glavy, Stevens Institute). Unless stated otherwise, all reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Construction of EGFP-tagged HDAC5 and HDAC5 Mutants and Cell Lines
The HDAC5 plasmid (gift from E. Seto, Moffitt Cancer Center) (21), was used to obtain the ORF of HDAC5 and EGFP was obtained from pEGFP-N1 (Clontech, Mountain View, CA). A human embryonic kidney (HEK)293 cell line stably expressing EGFP-FLAG-tagged HDAC5 was constructed using the PhoenixTM retrovirus expression system (Orbigen, San Diego, CA). Briefly, pLXSN-HDAC5-EGFP-FLAG was transfected in Phoenix cells using FuGENE (Roche, Indianapolis, IN), cells were grown to 90% confluence, and the resulting retrovirus was used to infect HEK293 cells. Stably expressing cells were selected with 300 mg/L G418 (EMD, Gibbstown, NJ) for 14 days and sorted by FACS (Vantage S.E. with TurboSort II, Becton Dickinson, Franklin Lakes, NJ). For experiments utilizing cells transiently transfected with HDAC5-EGFP or HDAC5 mutants, subconfluent monolayers (∼30%) of HEK293 cells were transfected using FuGENE (Roche, Indianapolis, IN), cultured as above, and harvested 36 h post transfection. See Supplemental Table S1 for primers used.
Immunoaffinity Purification of HDAC5 complexes
WT or mutant HDAC5 and control EGFP were isolated by immunoaffinity purification via the EGFP tag on magnetic beads, as previously described (20, 22). For mass spectrometric analyses of HDAC5 and co-isolated proteins, 1.2–1.5 g cells were used as starting material. Briefly, HEK293 cells were harvested, frozen, and cryogenically lysed in 9 × 2.5 min cycles at 30 Hz using a Retch MM301 Mixer Mill (Retch, Newtown, PA). The frozen cell powder was homogenized in lysis buffer (20 mm HEPES-KOH, pH 7.4, 0.1 m potassium acetate, 2 mm MgCl2, 0.1% Tween-20, 1 μm ZnCl2, 1 μm CaCl2, 0.5% Triton X-100, 250 mm NaCl, 4 μg/ml DNase, 1/100 (v/v) protease and phosphatase inhibitor cocktails (Sigma), subjected to centrifugation at 8000 × g for 10 min at 4 °C. Affinity isolations were performed by incubating cell lysates for 1 h at 4 °C with 8 mg M-270 epoxy magnetic beads (Invitrogen) conjugated to polyclonal anti-GFP antibodies. Proteins were eluted either with 0.5 N NH4OH, 0.5 mm EDTA for 20min at RT or with 50 μl 1× LDS sample buffer containing 50 mm dithiotreitol (Invitrogen) for 10 min at 70 °C. Base-eluted protein complexes were lyophilized overnight, suspended in sample buffer and reduced with dithiotreitol, whereas SDS-eluted complexes were processed immediately.
Deacetylation Assays
The deacetylation activity of isolated HDAC5-complexes was measured using the Fluor-de-Lys® HDAC fluorometric activity assay kit (ENZO, Farmingdale, NY) according to manufacturer's specifications. Briefly, protein complexes (isolated from 0.1 g cells using 1% Triton in the lysis buffer described above) were incubated with 50 μl of 200 μm Fluor-de-Lys substrate for 90 min at 37 °C, in the presence or absence of 2 μm trichostatin A. Fifty microliters of 1× Fluor de Lys developer adjusted to 2 μm trichostatin A was added and incubated for 15 min at 37 °C. Magnetic beads were separated and the supernatant was analyzed using a fluorescence reader (Tecan, Durham, NC) at excitation and emission wavelengths of 379 and 465 nm, respectively. Data was analyzed using MagellanTM software. Background-corrected fluorescence intensity values were normalized by the amount of isolated HDAC5 determined by Western blotting and densitometry analysis (ImageQuant TL; GE Healthcare, Piscataway, NJ).
Microscopy
HEK293 or U2OS cells, transiently transfected with HDAC5-EGFP or mutants, were cultured on glass-bottom dishes, pretreated with poly-d-Lys (Sigma, St. Louis, MO). After 24 h, cells were fixed with 2% paraformaldehyde, washed with d-PBS, incubated with 1 μg/ml DAPI in d-PBS for 15 min, and visualized by confocal microscopy on a Zeiss LSM510 (Zeiss, Dublin, CA) using 63× oil-immersion lens. For live cell imaging, cells were transiently transfected with WT and S278/279A HDAC5-EGFP for 24 h, treated with 80 ng/ml leptomycin B, and visualized on a RS3 Spinning disk (PerkinElmer). HDAC5-EGFP-FLAG stably expressed in HEK293 and U2OS cells were cultured, fixed as above, permeabilized with 0.1% Triton X-100 in d-PBS (PBST) for 15 min and blocked in 2% (w/v) bovine serum albumin, 0.2% (v/v) Tween-20 in d-PBS at RT for 60 min. Incubation with primary antibody was performed at RT for 60 min in blocking buffer. The cells were then washed with PBST and incubated with either goat anti-mouse or goat anti-rabbit secondary antibodies conjugated to Alexa 546 or 633 (Invitrogen). Cells were incubated with 1 μg/ml DAPI in PBS for 15 min and visualized using either 100× oil-immersion lens or 40× water lens when performing quantification of cytoplasmic and nuclear-localized HDAC5-EGFP-FLAG. Quantification of HDAC5 nuclear-cytoplasmic localization (cytoplasmic enrichment) was performed using CellProfiler 2.0 (23). Nuclear HDAC5 was quantified as the integrated intensity of GFP within the nucleus, as defined by DAPI staining. Cytoplasmic HDAC5 was quantified as the integrated intensity of GFP within the whole cell minus nuclear HDAC5. The outer boundary of GFP signal was determined for each cell by the propagation method within CellProfiler (23).
Matrix-Assisted Laser Disorption Ionization (maldi)-MS and Tandem MS Phosphopeptide Analyses
HDAC5 and co-isolated proteins were alkylated, separated on 4%–12% NuPAGE Novex Bis-Tris gels (Invitrogen), and stained with SimplyBlueTM SafeStain (Invitrogen). Proteins were enzymatically digested in-gel with trypsin (Promega, Madison, WI) or Lys-N (Seikagaku Corp, Tokyo, Japan) at 12.5 ng/μl in 50 mm ammonium bicarbonate for overnight at 37 °C. For MALDI MS phosphopeptide analyses, HDAC5 peptides were extracted from gel pieces with 1% formic acid (FA) followed by 1% FA/50% acetonitrile (ACN) for 4 and 2 h at room temperature, respectively. Samples were concentrated by vacuum centrifugation, suspended in 5 μl of 25 mg/ml 2,5-dihydroxybenzoic acid matrix in 50% methanol/20% ACN/0.1% trifluoroacetic acid/1% phosphoric acid (matrix solution), and deposited on the MALDI target. For immobilized metal ion affinity chromatography (IMAC) analyses, peptides were extracted as above, concentrated, and suspended in 50 μl of 250 mm acetic acid/30% ACN (binding buffer). Aliquots (20 μl) of Fe3+ IMAC resin slurry (PhosSelect, Sigma) were prewashed, and incubated with peptides at RT for 45 min, washed with binding buffer, and eluted with 0.4N NH4OH for 5 min at RT. Samples were acidified, lyophilized, suspended in matrix solution, and deposited on the MALDI target. Peptide mass fingerprinting and targeted MS/MS were performed as described (22).
Liquid Chromatography (LC)-MS/MS Phosphopeptide Analyses
Peptides were extracted as above for phosphopeptide analyses, concentrated and suspended in 10 μl 0.1% FA, and analyzed by a linear trap quadrupole (LTQ)-Orbitrap Velos coupled to a Surveyor MS pump and autosampler or LTQ-Orbitrap XL (Thermofisher Scientific, San Jose, CA) coupled to an Ultimate 3000 nanoRSLC system (Dionex Corp., Sunnyvale, CA) running mobile phase A, 0.1% FA in water, and mobile phase B, 0.1% FA in ACN. For LTQ-Orbitrap Velos analysis, peptides were washed onto a nanocapillary column (Magic C18, Michrom Bioresources, Auburn, CA) for 10 min at 7% B at a flowrate of ∼300 nL/min. Peptides were eluted with a 90 min gradient from 7% to 40% B. The mass spectrometer was set to repetitively scan m/z from 350 to 1700 (r = 60,000), followed by data-dependent MS/MS scans on the five most abundant ions. For each data-dependent event, an higher-energy C-trap dissociation (HCD) and ETD MS/MS spectra was recorded with the following parameters: minimum signal of 5000, isolation width of 2.0 (HCD) and 3.0 (ETD), normalized collision energy of 35, and waveform injection, dynamic exclusion, and supplemental activation (ETD) enabled. Fourier transform (FT)-MS full scan AGC target value was 5e5, whereas FT-MS and ion trap MSn AGC were 5e4 and 1e4, respectively. FT-MS full scan and MSn fill time was 100 and 200 ms, respectively, whereas ion trap MSn fill time was 150 ms.
For LTQ-Orbitrap XL collision induced dissociation (CID) analysis, peptides were preconcentrated, desalted online for 5 min at 4 μl/min in 0.5% trifluoroacetic acid/98.5% water/1% ACN using a reverse phase trap column (100 μm × 2.5 cm, Magic C18, 3 μm), and separated on a nanocapillary reverse phase column (75 μm × 25 cm, PepMap C18 1.8 μm, Dionex Corp.) for 90 min using a linear gradient from 4% to 35% B. The mass spectrometer was set to repetitively scan m/z from 350 to 1700 (r = 60,000), followed by data-dependent MS/MS scans on the 10 most abundant ions. For each data-dependent event, a CID MS/MS spectra were recorded with the following parameters: minimum signal of 2000, isolation width of 2.0, normalized collision energy of 33, and waveform injection and dynamic exclusion enabled. FT-MS full scan AGC target value was 1e6, whereas ion trap MSn AGC was 5e3. FT-MS full scan and ion trap MSn fill time was 1000 and 100 ms, respectively. Data-independent analysis of phosphopeptides was performed by CID analysis on an LTQ-Oribtrap XL as described above, except the mass spectrometer acquired spectra as follows: single FT-MS event (r = 7500, m/z = 300 - 600) followed by a maximum of 5 data-independent MS2 events on selected ion species corresponding to known or predicted phosphopeptides. The following m/z values and tryptic peptide sequences from WT and S279A HDAC5 were used for CID-MS2 events: 414.76, RSSPLLR; 454.74, RS*SPLLR/RSS*PLLR; 376.69, S*SPLLR/SS*PLLR; 328.71, SAPLLR; 368.69, S*APLLR (* - phosphosite). All parameters were the same as above, except the number of MSn microscans was 2. All spectra were manually inspected for peptide sequence assignment.
Mass Spectrometric Phosphopeptide Identification Data Analysis
MALDI MS spectra were deisotoped using Xtract RAW (Xcalibur v2.0.7) and searched by XProteo (www.xproteo.com) as described (24). Significant protein hits (d' score > 4) were selected for confirmation by MS/MS (≥2 peptides). RAW files from nLC-ESI experiments were processed by Proteome Discoverer (v1.1). ETD spectra were preprocessed to remove precursor, charge reduced, and neutral loss species. The extracted and filtered CID/HCD/ETD spectra were searched independently using SEQUEST (v1.12) against the human UniProt-SwissProt protein sequence database (release 15.7; 2009/09) plus common contaminants (20,276 sequences), which were automatically reversed and concatenated to the forward sequences. Spectra were searched with the following criteria: full enzyme specificity; two missed cleavages; precursor mass tolerance: 10 ppm; fragment mass tolerance: 0.8 Da (ETD)/50 mmu (HCD); fixed modification of cysteine by carbamidomethylation; max. variable modifications: four; variable modification of methionine oxidation and phosphorylation of serine, threonine, and tyrosine. Using peptide spectrum matches assigned to the reverse database entries, peptide spectrum matches were filtered to <1% false discovery rate using ProteomeDiscoverer false discovery rate filters. For phosphopeptide site localization, tandem mass spectra peak lists corresponding to phosphopeptides identified by SEQUEST were searched by Mascot (ver. 2.3; www.matrixscience.com) with the above parameters, exported to pepxml (Mascot, ver. 2.3), and analyzed by SLoMo (ver. 2) (25) using a fragment error of 400 ppm (CID/ETD) or 12 ppm (HCD). Phosphosites with scores >19 correspond to an false discovery rate of ∼ 1% and were considered unambiguously localized.
Analysis of Protein Interactions Using Mass Spectrometric Spectral Counting
Isolation of HDAC5 complexes and data-dependent LC-CID-MS/MS analyses on an LTQ-Orbitrap XL were performed using HEK293 cells stably expressing wild-type and serine-to-alanine HDAC5 mutants as described above, except RAW files were processed by Bioworks (v3.3.1) and peptide spectrum matches were generated by SEQUEST (v.28, rev12) database searching against the human UniProt-SwissProt protein sequence database (release 15.7; 2009/09) plus common contaminants (20,276 sequences), which were manually reversed and concatenated to the forward sequences. All search parameters were the same as described above except the use of a fragment tolerance of 0.5 Da.
To control of peptide/protein error rates and facilitate comparison of protein identifications, peptide spectrum matches derived from independent immunoisolations of HDAC5 complexes were analyzed by Scaffold 3.0.5 (Proteome Software, Inc., Portland, OR)(26). Peptide and protein identifications from paired HDAC5 wild-type and mutant immunoisolations (n = 2 biological replicates each) were filtered by assigned probability scores, including the requirement of a minimum of 3 unique peptides per protein identified in both biological replicates for either wild-type or mutant isolations. These stringent criteria controlled estimated protein false discovery rate to <1% as assessed by proteins hits to reverse database entries. Proteins identified in isolations from EGFP-expressing cell lines were removed (see Supplemental Table S2). To compare interacting proteins that change between wild-type and mutant HDAC5 conditions, a spectral counting (SC) approach was used. Protein isoforms were considered present only if unique peptides (n ≥ 3) for each isoform were identified. Proteins that were identified only under one condition (wild-type or mutant), but having zero spectral counts in the other, were assigned a value of one to facilitate the calculation of fold change. SC for protein isoforms were the sum of both unique and shared peptides among the isoforms. SC for each protein were normalized to the SC for wild-type HDAC5; biological replicates were averaged and filtered to include only proteins with ≥10 SC in at least one condition. The fold-change (relative to wild type HDAC5) in SC was used as a measure of the approximate relative abundance of interacting proteins (see Tables S4 and S5).
RESULTS AND DISCUSSION
In vivo HDAC5 Interactions With Kinases and a Phosphatase Reflect its Regulation by Phosphorylation-Dependent Mechanisms
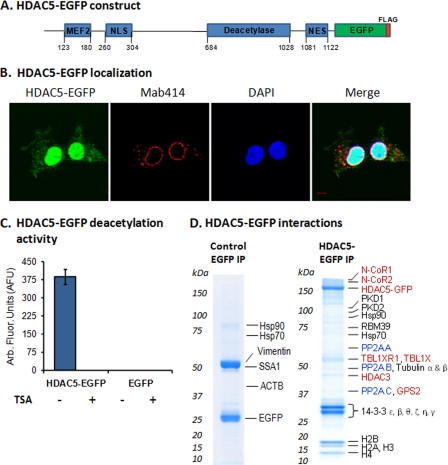
To study HDAC5 interactions and phosphorylation, we used a retrovirus expression system to generate a human embryonic kidney (HEK) 293 cell line stably expressing HDAC5 tagged at the C terminus with EGFP-FLAG (Fig. 1A). In agreement with its known nucleo-cytoplasmic shuttling, HDAC5-EGFP displayed both nuclear and cytoplasmic localization, with a predominant nuclear localization (Fig. 1B). A fluorometric substrate-based deacetylation assay demonstrated that the HDAC5-EGFP complex, isolated via the GFP tag, maintains its enzymatic activity and sensitivity to the HDAC inhibitor trichostatin A (Fig. 1C). It is known that HDAC3 is a primary contributor to the deacetylation activity of HDAC5, and in our experiments, the HDAC3 isolated with HDAC5-EGFP is the endogenous, untagged form (Fig. 1D). Overexpression of HDAC5-EGFP could result in increased binding of HDAC3 and increased deacetylation activity when compared with the endogenous HDAC5. Alternatively, the availability of HDAC3 (or other corepressor complex components) could limit its binding to HDAC5-EGFP and may have a negligible effect on deacetylation activity. The lack of an antibody against HDAC5 that has suitable affinity for immunoaffinity purification precluded us from comparing the activity of the tagged protein with that of the endogenous protein. Nevertheless, our results clearly demonstrate that the EGFP-tagged HDAC5 complex has deacetylation activity that retains its sensitivity to trichostatin A. As a control, we generated a HEK293 cell line stably expressing EGFP-FLAG and showed that the isolated tag did not display deacetylation activity.
Fig. 1.
HDAC5-EGFP has deacetylation activity and associates in vivo with the N-CoR-HDAC3 complex and heterotrimeric PP2A enzyme. A, Schematic of EGFP-FLAG-tagged HDAC5 with functional domains: MEF2, myocyte enhancer factor 2 binding domain; NLS, nuclear localization signal; Deacetylase, deacetylase activity domain; NES, nuclear export signal. B, The HDAC5-EGFP localizes to both the nucleus and cytoplasm in the HEK293 cell line; HDAC5 (green, EGFP), nuclear periphery (red, Mab414 against nuclear pore complex proteins), nucleus (blue, DAPI); 100× oil-immersion lens; Bar, 5 μm. C, HDAC5-EGFP-FLAG has deacetylation activity. Deacetylation activity of HDAC5-EGFP and EGFP, isolated from HEK293 cell lines, was measured (n = 6, AFU±S.D.) using Fluor-de-Lys assay in the absence (-) or presence (+) of trichostatin A. D, HDAC5-EGFP associates with known interacting partners. Immunoaffinity purifications were performed via the EGFP tag, co-isolated proteins were resolved by one-dimensional SDS-PAGE, stained with Coomassie Blue, and analyzed by mass spectrometry. Red, known members of the N-CoR-HDAC3 corepressor complex; Blue, the three subunits of protein phosphatase 2A.
We next assessed the in vivo interactions of HDAC5-EGFP by integrating cryogenic cell lysis, affinity purifications, and mass spectrometry. HDAC5-EGFP was isolated with its known interacting partners, including components of the N-CoR-HDAC3 corepressor complex: N-CoR1, N-CoR2 (SMRT), HDAC3 (6), TBL1X, TBL1XR1 (27), and GPS2 (28) (Fig. 1D, Supplemental Fig. S1, and Supplemental Table S2). Additionally, present in our isolation were protein kinases, PKD1 that interacts with HDAC5 and phosphorylates the 14–3-3 consensus sites (29, 30), PKD2, which phosphorylates HDAC5 in vitro (31), as well as the 14–3-3 isoforms β, ε, θ, ζ, η, and γ that interact with HDAC5 and induce its nuclear export (32). These proteins were not identified in the control isolation of EGFP from the HEK293 cell line stably expressing EGFP-FLAG, indicating that the interactions do not occur via nonspecific association to the tags or the magnetic beads (Fig. 1D and Supplemental Table S3). Overall, these proteins are representative of known nuclear and cytoplasmic functional interactions of HDAC5. Interestingly, we found that HDAC5 associates in vivo with three subunits of protein phosphatase 2A, the catalytic (PP2AC), scaffolding A (PR65), and regulatory B (PR55) subunits, which can form a heterotrimeric holoenzyme complex (33). Although determining which of these interactions are direct or indirect requires further study, these findings are supported by recent reports highlighting the importance of protein phosphatases in regulating deacetylases. In particular, PP2A was showed to play a role in the nucleo-cytoplasmic shuttling of HDAC4 (11) and HDAC7 (34) via dephosphorylation of 14–3-3 binding sites. Other co-isolated proteins included p53, a subset of the nucleosome remodeling and deacetylase (NuRD) complex (HDAC1, RBBP4, RBBP7, and MTA2), and the DNA-dependent protein kinase (PRKDC), illustrated as an interaction network in Supplemental Fig. S1. These proteins may represent indirect associations, as p53 (35) and NuRD components are known to bind HDAC3 (36), and PRKDC is thought to phosphorylate HDAC3 (37). It remains to be determined if these proteins can also interact directly with HDAC5.
In summary, HDAC5-EGFP stably expressed in HEK293 cells is functional, displaying deacetylation activity and associating with proteins known to modulate its localization and transcriptional activity. Furthermore, this targeted proteomic approach allowed us to demonstrate that HDAC5 not only associates in vivo with the PP2A core heterodimer (PP2AD) (34), but also with the PR55 subunit, a component of the heterotrimeric form. Although the PP2AD core structure may exist independently in cells, regulatory B-type subunits can dynamically associate with the core dimer, modulating PP2A substrate specificity and/or localization (33). Interestingly, PR55-containing PP2A trimers have been shown to bind and dephosphorylate the microtubule-associated protein tau, which has been found hyperphosphorylated within neurofibrillary protein aggregates in Alzheimer's disease (38). Although we identified tubulin at background levels in the EGFP control immunoisolation (Supplemental Table S3), an increase in sequence coverage and unique peptides was observed for α- and β-tubulin in isolations of HDAC5-EGFP (Supplemental Table S2). Therefore, the association of HDAC5 with PR55 and tubulin (Fig. 1D) may reflect a role in microtubule regulation. Yet as the cellular contexts that influence the dynamic association of B-type subunits with the core heterodimer are not well-defined, the function of B-type subunits in influencing PP2A-HDAC5 association remains unknown.
HDAC5 is Phosphorylated In Vivo Within the Deacetylation Domain and Nuclear Localization and Export Signals
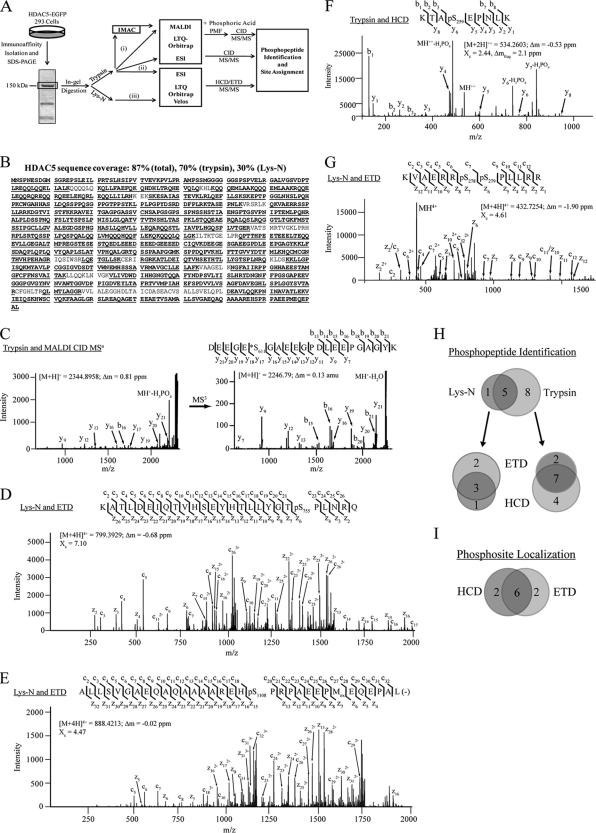
To comprehensively map phosphorylation sites on HDAC5, we employed a combinatorial mass spectrometric approach that integrated several strategies and instrument configurations (Fig. 2A). First, isolated HDAC5-EGFP was enzymatically digested with trypsin or Lys-N. MALDI MS and MS/MS analyses were performed on a MALDI LTQ-Orbitrap XL instrument, with and without prior phosphopeptide enrichment by immobilized metal affinity chromatography (IMAC). Detection of phosphorylated peptides was enhanced by phosphoric acid treatment during deposition on the MALDI target. Peptides were also analyzed using reverse phase nano-liquid chromatography (nLC) coupled online to either an electrospray ionization (ESI) LTQ-Orbitrap XL or a LTQ-Orbitrap Velos instrument. Peptide identification and phosphorylation site determination was facilitated by three peptide fragmentation techniques: (1) collision induced dissociation (CID), (2) higher-energy C-trap dissociation (HCD), and (3) electron transfer dissociation (ETD).
Fig. 2.
Combinatorial mass spectrometric approach allows a comprehensive view of in vivo phosphorylation on HDAC5. A, Workflow for phosphopeptide characterization of HDAC5 using immunoaffinity isolation, in-gel digestion, and complementary mass spectrometric approaches. Proteomics strategies employed include (i) MALDI-IMAC analysis, (ii) ESI-LC CID analysis, and (iii) ESI-LC HCD/ETD analysis. B, 87% sequence coverage of HDAC5, including unmodified and phosphorylated peptides, was obtained. C, MALDI CID MS/MS of tryptic phosphopeptide. Prominent neutral losses of phosphoric acid (H3PO4) and water were observed. MS3 fragmentation of the neutral loss species (M+H+- H3PO4) localized the phosphorylation site (Ser611) as a dehydroalanine, marked with *. (D and E) MS/MS ETD spectra corresponding to multiply charged Lys-N phosphopeptides. Phosphosites (Ser755 and Ser1108) were unambiguously localized, with nearly full sequence coverage of both c and z ion series. F, MS/MS HCD spectrum of tryptic phosphopeptide. Neutral losses of H3PO4 from the precursor and y6 and y7 fragment ions supported the phosphosite at Ser259. G, MS/MS ETD spectrum of a doubly phosphorylated peptide (Ser278 and Ser279) within the nuclear localization signal. For all mass spectra, the precursor m/z, charge state, mass error (Δm), and cross correlation score (Xc) are indicated. H, Venn diagram comparison of phosphopeptides identified from Lys-N and trypsin-digested HDAC5, and each as a function of fragmentation technique (ETD versus HCD). I, Venn diagram of the number of phosphosites unambiguously localized by HCD and ETD. Representative annotated, mass labeled phosphopeptide spectra are included in Supplemental Fig. S2.
Using MALDI and ES ionization, the diverse length and charge states of HDAC5 peptides following trypsin and Lys-N digestions in conjunction with complementary peptide fragmentation resulted in improved protein sequence coverage of 87% (Fig. 2B). The complementarity of peptide fragmentation by CID, HCD, and ETD is illustrated in Fig. 2C–G. For singly charged phosphopeptides, an additional stage of CID fragmentation (MS3) was often performed on neutral loss species to obtain nearly full sequence coverage and site of modification (Fig. 2C, right). Identification of phosphorylation sites by HCD fragmentation benefited from the high mass accuracy of fragment ions, as shown for the doubly charged peptide containing Ser259 (Fig. 2F). Lys-N generates longer peptides with a basicity shifted toward the N terminus, which aids in fragmentation by ETD for multiply charged ions (39) and fragmentation and quantification by CID for singly charged ions (40). The length and presence of internal Arg residues of Lys-N-derived peptides afforded higher charge states (z >3), permitting nearly complete sequencing by ETD (Figs. 2D, 2E, and 2G). Trypsin digestion led to the identification of a greater number of phosphopeptides, whereas Lys-N provided a high degree of confirmation (Fig. 2H). Identification of phosphopeptides from HCD and ETD mass spectra was more robust for trypsin and Lys-N digestions, respectively. Site-specific determination of the phosphorylated residues was aided by a probabilistic algorithm for confidence scoring (SLoMo, pSc, Table I and Supplemental Fig. S2) (25). When comparing HCD and ETD, certain sites were only localized with high confidence when using a single technique, each affording two unique sites (Fig. 2I), including Ser498 and Ser755, respectively. Overall, 17 unique phosphorylation sites were unambiguously localized (score > 19; <1% false positive rate) using CID, HCD, and ETD (Table I). One additional phosphorylation was assigned at Ser225, however, ambiguity in its localization restricted further study.
Table I. Histone deacetylase 5 phosphorylation site mapping by complementary mass spectrometry analysis. The presence (+) or absence (−) of the phosphopeptide identification is indicated for each instrument configuration. n.a. indicates Lys-N-generated peptides that were analyzed only by the LTQ-Orbitrap Velos configuration.
| pSTa | Phosphopeptide sequenceb | pScc | Enzd | Xce | z | MALDI LTQ Orbi XLf NE IM | LC-ESI LTQ Orbi XL CID | LC-ESI LTQ Orbi Velos HCD ETD | ||
|---|---|---|---|---|---|---|---|---|---|---|
| S3 | (-)MNS3PNESDGMSGREPSLEILPR(T) | 103 | T | 4.66 | 3 | − | + | + | + | + |
| S3/S7 | (-)MNS3PNES7DGMSGREPSLEILPR(T) | 55 | T | 3.85 | 3 | − | − | − | − | + |
| S53 | (R)AMoxPSSMoxGGGGGGS53PSPVELR(G) | 61 | T | 3.41 | 2 | − | − | + | + | + |
| S55 | (R)AMoxPSSMoxGGGGGGSPS55PVELR(G) | 38 | T | 2.85 | 2 | − | − | + | + | + |
| S66 | (R)GALVGS66VDPTLR(E) | 49 | T | 3.63 | 2 | − | − | + | − | − |
| S206 | (K)SKEPTPGGLNHS206LPQHPK(C) | 32 | T | 2.64 | 3 | − | − | + | − | − |
| T234 | (P)KCamWGAHHASLDQSSPPQSGPPGT234PPSY(K) | 58 | L | 5.18 | 4 | − | n.a. | n.a. | + | + |
| S259 | (R)KTAS259EPNLK(V) | 90 | T | 2.44 | 2 | + | + | + | + | − |
| S259 | (R)KTAS259EPNL(K) | 53 | L | 2.27 | 2 | + | n.a. | n.a. | + | − |
| S278 | (Q)KVAERRS278SPLLRR(K) | 26 | L | 4.06 | 4 | + | n.a. | n.a. | − | + |
| S278/S279 | (Q)KVAERRS278S279PLLRR(K) | n.c. | L | 4.61 | 4 | − | n.a. | n.a. | − | + |
| S278/S279 | (K)VAERR S278S279PLLR(R) | n.c. | T | 3.94 | 3 | + | + | − | + | − |
| S368 | (R)ALPLDSSPNQFSLYTSPS368LPNISLGLQATVTVTNSHLTASPK(L) | 23 | T | 5.47 | 3 | − | − | + | − | − |
| S498 | (R)TQS498SPLPQSPQALQQLVMQQQHQQFLEK(Q) | 24 | T | 5.69 | 4 | + | + | + | + | + |
| S611 | (K)DEEGES611GAEEGPDLEEPGAGYK(K) | 154 | T | 7.40 | 3 | + | + | + | + | + |
| S611 | (V)KDEEGES611GAEEGPDLEEPGAGY(K) | 155 | L | 6.95 | 3 | + | n.a. | n.a. | + | + |
| S661 | (R)TQS661SPAAPGGMK(S) | 58 | T | 2.78 | 2 | − | + | + | + | − |
| S671 | (R)TQSSPAAPGGMKS671PPDQPVK(H) | 193 | T | 4.52 | 3 | − | + | + | + | + |
| S661/671 | (R)TQS661SPAAPGGMKS671PPDQPVK(H) | 28 | T | 3.89 | 3 | + | + | + | + | + |
| S755 | (R)KATLDEIQTVHSEYHTLLYGTS755PLNRQ(K) | 29 | L | 7.10 | 4 | − | n.a. | n.a. | + | + |
| S755 | (R)KATLDEIQTVHSEYHTLLYGTS755PLNR(Q) | 32 | T | 5.54 | 4 | + | + | + | − | + |
| S1108 | (M)ALLSVGAEQAQAAAAREHS1108PRPAEEPmEQEPAL(-) | 40 | L | 4.47 | 4 | − | n.a. | n.a. | − | + |
| Amb | (H)HASLDQSS225PPQSGPPGTPPSYK(L) | Amb | T | 4.39 | 3 | − | − | − | − | + |
| Amb | (P)KCamWGAHHASLDQSS225PPQSGPPGT234PPSY(K) | Amb | L | 4.64 | 3 | − | n.a. | n.a. | + | + |
a Phosphorylated residue in human HDAC5.
b Phosphopeptide sequences; methionine oxidation (Mox) and cysteine carbamidomethylation (Cam) are specified.
c Phosphosite confidence score were calculated by SLoMo (Bailey, JPR 2009). Scores ≥19 were considered confidently localized, corresponding to an estimated error rate of ≤ 1.3%, while phosphopeptide spectra below this threshold are indicated as “Amb”. n.c. indicates no site localization was required.
d Enzyme, trypsin (T) or Lys-N (L), which generated the phosphopeptide.
e SEQUEST cross-correlation score (Xc) and charge state (z).
f MALDI LTQ-Orbitrap XL analysis was conducted with (IM) or without (NE) IMAC.
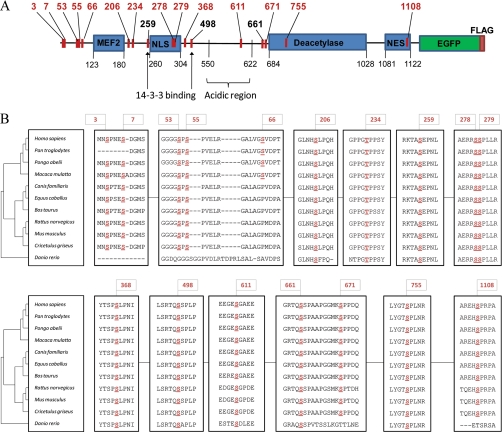
Together, the enrichment of HDAC5-EGFP by immunoaffinity purification and its analysis using a combinatorial mass spectrometric approach provided a comprehensive view of the in vivo phosphorylation status of HDAC5, leading to the unambiguous identification of 17 phosphorylated sites (Fig. 3A), of which only Ser259, Ser279, Ser498 (14–3-3 binding sites) and Ser661 have been previously reported (10, 14, 16, 32). Several phosphorylation sites were clustered at the N terminus of HDAC5 (Ser3, 7, 53, 55, 66), whereas interestingly, others lay within or in close proximity to functional domains critical for protein activity and cellular localization (e.g. Ser206, Thr234, Ser368). We identified phosphorylation sites within the NLS (Fig. 2G), one novel (Ser278) and one recently reported (Ser279) (16), and for the first time, in the NES (Ser1108, Fig. 2E), which may contribute to phosphorylation-dependent nucleo-cytoplasmic shuttling. Phosphorylation within the NLS was observed as either a single Ser278 or double Ser278/279 modification (Table I). HDAC5 was also phosphorylated in the deacetylase domain (Ser755, Fig. 2D), as well as within an acidic region (Ser611, Fig. 2C) comprised of ∼50% Glu/Asp residues. Two additional phosphorylation sites were located immediately N-terminal to the deacetylation domain—the previously identified Ser661 (14) and the novel Ser671 site.
Fig. 3.
In vivo HDAC5 phosphorylation reveals site conservation among class IIa HDACs. A, Location of phosphorylated sites along the HDAC5-EGFP-FLAG sequence. B, Conservation of phosphosites among HDAC5 vertebrate species. NCBI gi numbers used were: Homo sapiens 62750347, Pongo abelii 55730067, Pan troglodytes 114666888, Macaca mulatta 109116100, Bos taurus 194676212, Equus caballus 194216834, Canis familiaris 73965560 (isoform 1), Cricetulus griseus 28627825, Mus musculus 148702151, Rattus norvegicus 109491954, Danio rerio 189517029. The cladogram of vertebrate HDAC5 sequence divergence was generated from NCBI BLASTp by the TreeView program (43).
As the conservation of phosphorylation among species may indicate sites critical for the function of a protein, we performed a comparison of the amino acid sequences for available full-length HDAC5 sequences. A high degree of conservation was observed for the majority of residues (Fig. 3B), with the exception of Ser66 that was only conserved in primates. These sites may subserve potential functions or represent binding sites of HDAC5 that are conserved among vertebrate species. The substantial number of identified phosphorylations not only suggests HDAC5 as an integrator of cellular signaling events, but also that phosphorylation may be critical for protein folding and conformational changes that facilitate interactions with proteins and substrates. As phosphorylation-dependent functions of HDAC5 are implicated in human disease, our study opens new avenues to pursue and better understand the regulation of this enzyme.
The Deacetylation Activity of HDAC5 is Dependent on its Phosphorylation Status
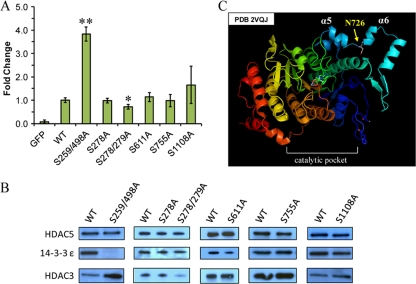
Conservation of residues that were targets of phosphorylation suggested that these sites may confer functional significance. Therefore, we examined individual phosphorylation sites with respect to deacetylation activity using EGFP-tagged HDAC5 Ser to Ala mutants in a pEGFP-N1 vector that allowed for efficient transient transfection and visualization in HEK293. We focused on sites located within functional domains (Fig. 4), generating a double mutant of the 14–3-3 binding sites (S259/498A), single and double mutants within NLS (S278A, S278/279A), and single mutants for sites in the acidic region (S611A), deacetylation domain (S755A) and NES (S1108A). Immunoaffinity purified HDAC5 mutants, wild type HDAC5-EGFP, and EGFP were assessed for enzymatic activity using a substrate-based fluorogenic deacetylation assay (Fig. 4A). Because HDAC3 is co-isolated with HDAC5 (Fig. 1) and known to impart a significant portion of the deacetylation activity measured for HDAC5 (6), we related the activity of the mutant HDAC5 to its association with HDAC3 (Fig. 4B).
Fig. 4.
HDAC5 phosphorylation influences its deacetylation activity and association to HDAC3. A, HEK293 cells were transiently transfected with EGFP, HDAC5-EGFP (wild type), or Ser to Ala HDAC5-EGFP mutants. Following isolations via EGFP, the deacetylation activity was measured using the Fluor-de-Lys assay, and expressed as fold change ± S.D. versus wild type (n ≥ 3). Statistical significance was performed by student's t test comparing wild type to mutant HDAC5; * and ** indicate p = 0.03 and p = 0.0001, respectively. B, Isolated HDAC5 mutants were assessed for their association with HDAC3 and 14–3-3 ε by Western blotting; a quantitative assessment of the relative intensities for the illustrated images was performed by densitometry and shown in Supplemental Fig. S3. C, Crystal structure of the HDAC4 deacetylation domain (PDB 2VQJ) highlighting Asn726, located between α-helix 5 and 6, and which aligns to Ser755 in HDAC5.
The S259/498A HDAC5 mutant exhibited a significant increase in deacetylation activity (3.82 ± 0.31, n = 3, p = 0.0001). This is likely an indirect effect of the loss of interaction with 14–3-3 proteins (Fig. 4B), which results in the nuclear localization of this mutant (8, 32). The nuclear subset of HDAC5 associates with HDAC3 and corepressor proteins required for its activity and recruitment to substrates, as supported by the increased level of HDAC3 associated with the S259/498 HDAC5 mutant (Fig. 4B and Supplemental Fig. S3). Although the S278A HDAC5 mutant within the NLS did not alter the deacetylation activity, the S278/279A mutant led to a slight decrease in the activity (0.72 ± 0.11-fold, n = 3, p = 0.03) and association with HDAC3 compared with wild type. The association with 14–3-3 ε remained constant for both NLS mutants. The S611A mutation did not affect the deacetylation activity or the association with HDAC3. Interestingly, the S755A mutant within the deacetylation domain also did not disrupt the deacetylation activity or the association with HDAC3. Examination of the crystal structure of the HDAC4 deacetylase domain (41) (Fig. 4C), which contains an Asn726 at the conserved site (Ser755) in HDAC5, suggests that Ser755 is located distally from the catalytic pocket. Hence, Ser755 is not likely to interfere significantly with the domain structure or accessibility, but rather may affect the recruitment of substrates to HDAC5. Lastly, S1108A mutation of the NES site showed a slight, but not significant increase in HDAC5 deacetylation activity (1.65 ± 0.80-fold, n = 6, p = 0.08), and only minimally increased HDAC3 association (Fig. 4B and Supplemental Fig. S3). Although several phosphorylation sites did not affect the deacetylation activity of HDAC5 and may be involved in protein folding and/or regulation of interactions, others that prominently affected deacetylation activity could result from indirect mechanisms that are dependent upon HDAC5 localization (e.g. S259/498) or from altering the association with proteins containing deacetylase activity. Interestingly, these novel phosphorylation sites had little effect on 14–3-3 binding, pointing toward additional mechanisms that regulate HDAC5 function.
Phosphorylation within the NLS is required for HDAC5 Nuclear Localization
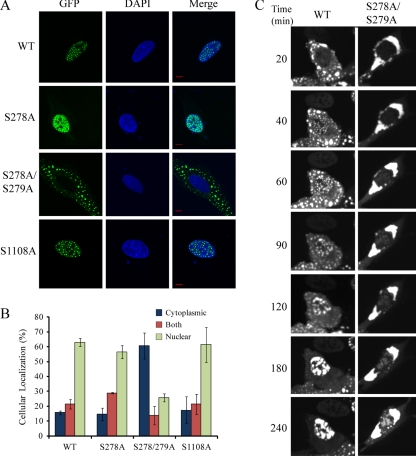
The identification of HDAC5 phosphorylation within the NLS and NES, as well as the reduced interaction of S278/279A HDAC5 mutant with HDAC3, led us to speculate that these phosphorylation sites may play a role in the localization of HDAC5. To test this hypothesis, we used transient transfections and immunofluorescence analysis in both U2OS and HEK293 cells to evaluate the effect of individual phosphorylation sites. Wild type HDAC5-EGFP was mainly localized to the nucleus, with a smaller subset localized to the cytoplasm (Fig. 5). The observed punctate appearance was previously reported following transient transfections of class IIa deacetylases, including HDAC4 and HDAC7 (15, 32), and may derive from the overexpression of the protein. Immunofluorescence analysis showed that although the S278A mutant did not alter HDAC5 localization, the S278/279A mutant displayed a striking shift in localization (Fig. 5A). Approximately 60% of cells transfected with S278/279A HDAC5-EGFP displayed cytoplasmic localization compared with 15% for wild type HDAC5 as observed in both U2OS (Fig. 5B) and HEK293 cells (Supplemental Fig. S2C). Although the S1108A and S755A mutants did not display a significant change in localization (Fig. 5A and Supplemental Fig. S3A), this does not exclude the possibility that these mutants affect protein associations within the nucleus.
Fig. 5.
Phosphorylation within the NLS affects HDAC5 localization. A, Cellular localization of transiently transfected wild type HDAC5, and S278/279A, S278A, and S1108A mutants was assessed by direct immunofluorescence microscopy in U2OS cells (bars, 10 μm); green (EGFP), blue (DAPI). B, wild type and mutant HDAC5-EGFP localization was classified as cytoplasmic, nuclear, or both (pan-cellular) (% ± S.D., n ≥ 220 cells from triplicate experiments). C, WT and S278/279A mutants were transiently transfected in U2OS cells, treated with leptomycin B, and visualized by live cell imaging for up to 4 h after treatment. Images are representative of three independent experiments.
Phosphorylation Within the NLS is Critical for Efficient Nuclear Import of HDAC5
To investigate if the enhanced cytoplasmic localization of the S278/279A mutant resulted from an increased nuclear export or reduced nuclear import, we inhibited nuclear export of transiently transfected cells by leptomycin B (LepB) treatment (Fig. 5C). For wild type HDAC5-EGFP, we focused on the ∼15% of cells with cytoplasmic HDAC5 localization (Fig. 5B). Live cell imaging showed that wild type HDAC5-EGFP was already accumulating in the nucleus following 40 min of LepB treatment, with nearly complete import at 3 h, whereas the S278/279A mutant showed minimal accumulation even following 4 h of treatment. Therefore, phosphorylation within the NLS is critical for the efficient nuclear import of HDAC5. Further supporting the importance of the phosphorylations at Ser278/279, the corresponding unmodified peptide was not detected in our analyses, suggesting these sites are extensively modified. Taking into account these relatively stable phosphorylations, it is tempting to hypothesize that these sites have a possible structural role as opposed to one in transient signaling events. Altogether, these data identify a new point of regulating HDAC5 steady-state cellular distribution that requires phosphorylation within the NLS.
Phosphorylation-dependent Associations of HDAC5
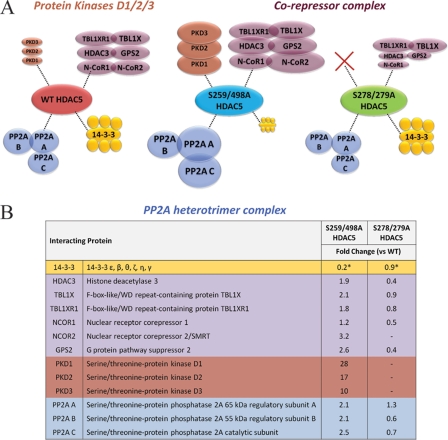
Having established that mutations of specific phosphorylation sites affect HDAC5 localization, we next assessed whether cellular redistribution results in a change of HDAC5 interactions by performing immunoisolations of HDAC5 from HEK293 cell lines stably expressing S259/498A (predominant nuclear localization) and S278/279A (increased cytoplasmic localization) mutant HDAC5-EGFP. HDAC5 protein interactions were compared by performing parallel isolations from wild type and mutant cell lines (representative one-dimensional SDS-PAGE gels shown in Supplemental Fig. S5). Changes in relative levels of interacting proteins were assessed using spectral counting from two biological replicates. As a correction for the amount of isolated HDAC5 complexes, spectral counts for associated proteins identified in mutant HDAC5 were normalized by wild type HDAC5-EGFP spectral counts (Supplemental Tables S4 and S5).
Assessment of previously characterized HDAC5 interactions, including the corepressor complex and 14–3-3 proteins, revealed changes in protein interactions that were consistent with the cellular redistribution of HDAC5-EGFP mutants (Fig. 6). In the S259/498A mutant, the association with the corepressor complex members (HDAC3, GPS2, N-CoR1, N-CoR2, TBL1XR1, and TBL1X) was increased, whereas the 14–3-3 isoforms were decreased, in agreement with the predominant nuclear localization of this mutant and its inability to bind 14–3-3 proteins. For the S278/279A mutant, the 14–3-3 proteins were present at similar levels as in wild type isolations, whereas the corepressor complex displayed, on average, a reduction in association, opposite to the trend observed for S259/498A (Fig. 6). These results are consistent with the indirect transcriptional regulation by HDAC5 via localization-dependent mechanisms.
Fig. 6.
Mutation of 14–3-3-binding sites and NLS phosphorylations leads to changes in HDAC5 interactions. Wild type, S259/498A and S278/279A HDAC5-EGFP were immunoaffinity purified from HEK293 stable cell lines with associating proteins, resolved by 1-D SDS-PAGE (Fig. S5) and analyzed by mass spectrometry. Changes in protein interactions were assessed using spectral counting (n = 2 biological replicates). A, The relative size of the circles associated with the mutant and WT (wild type) HDAC5 reflect increased or decreased relative abundance as assessed by the fold change in spectral counts for mutant versus WT. Color-coding illustrates functional clustering according to protein families or complexes: yellow, 14-3-3 protein isoforms, blue, PP2A subunits, purple, corepressor complex, orange, PKD enzymes. B, The average fold changes in spectral counts for each interaction illustrated in (A) are shown for the isolated mutant versus the wild type HDAC5.* indicates fold change for 14-3-3 isoforms representing the average of six isoforms (S.D. < ± 0.1).
HDAC5 association with PKD1, PKD2, and PKD3 showed interesting changes; while present at low levels in wild type isolations, these proteins were substantially increased in the isolation of the S259/498A mutant. In contrast, PKD proteins were not detected with the S278/279A mutant. Supporting these results, previous studies showed that PKD1-GFP is localized to the nucleus and that its overexpression leads to a cytoplasmic localization of HDAC5 via the phosphorylation of the two 14–3-3-binding sites (30). Furthermore, the identification of PKD proteins in the S259/298A mutant demonstrated that these mutations do not disrupt their interaction with HDAC5. It remains to be elucidated if the increase in PKD interactions in the S259/498A mutant is a direct consequence of reduced 14–3-3 protein interaction or represents a 14–3-3-independent functional recruitment. An increase in the association with PP2A subunits was observed for the S259/498A mutant. A previous study observed that 14–3-3 and PP2A can compete for binding to the class IIa HDAC7 and proposed that, in the absence of extracellular stimuli, these competitive signals may function to dynamically regulate its cellular localization (34). Our observations are consistent with these reports suggesting that a similar phosphorylation-dependent regulation of HDAC5 may occur. Interestingly, the changes observed for the S278/279A mutant were recapitulated in an isolation of a S279A HDAC5-EGFP mutant from a HEK293 stably expressing cell line (Supplemental Table S5). For the S279A mutant, the maintenance of 14–3-3 and absence of PKD proteins was accompanied by an even more pronounced reduction in binding of several members of the corepressor complex (compared with the S278/279A mutant) and a slight reduction in association of PP2A subunits. Overall, these results provide for the first time a link between phosphorylation outside the known 14–3-3 sites and downstream changes in protein interactions.
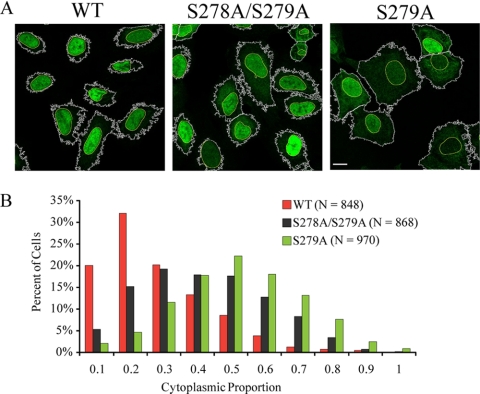
Stable Expression of Ser279Ala HDAC5-EGFP Impairs Its Cellular Localization
Phosphorylation within the NLS was identified by mass spectrometry as either a single Ser278 or double Ser278/279 phosphorylation (Table I, Fig. 2G, and Supplemental Fig. S2), without localization of Ser279 phosphorylation. However, the profound impact of the double S278/279A mutant on HDAC5 localization, the lack of this effect for the single S278A mutant, and the decrease in association with the corepressor complex for both the S278/279A and S279A mutants motivated us to investigate whether phosphorylation of Ser279 alone was involved in HDAC5 localization. Stable U2OS cell lines expressing wild type, S278/279A, or S279A HDAC5-EGFP were generated and analyzed by immunofluorescence to assess the cellular localization of HDAC5. These stable cell lines express HDAC5-EGFP at a lower level than our transient transfections, significantly reducing the punctate staining pattern previously observed (Fig. 5A). In contrast to transient transfections, cellular localization of HDAC5-EGP in the stable cell lines could not be classified into distinct groups (i.e. nuclear, cytoplasmic, or both), likely as a result of the reduced expression of HDAC5-EGFP. Therefore, the proportion of cytoplasmic HDAC5 within each cell was quantified by immunofluorescence image analysis using CellProfiler (23) (Fig. 7). Consistent with the transient transfections studies (Fig. 5), an increase in the cytoplasmic localization of HDAC5 was observed for the S278/279A mutant compared with wild type-expressing cells (median cytoplasmic proportion = 0.36 versus 0.20, n = 868 and 848 cells, respectively) (Fig. 7B). Interestingly, increased cytoplasmic-localized HDAC5 was also observed for the single S279A mutant (median cytoplasmic proportion = 0.46, n = 970 cells). This result strongly implicates Ser279 as a critical phosphorylation site within the NLS that influences HDAC5 cellular localization.
Fig. 7.
Preclusion of serine 279 phosphorylation alters HDAC5 localization. A, Cellular localization of stably transfected wild type (WT), S278A/S279A, and S279A HDAC5-EGFP was assessed by direct immunofluorescence microscopy in U2OS cells (bar, 20 μm). Increased cytoplasmic accumulation of HDAC5 (EGFP) was observed because of Ser-to-Ala mutations. Nuclear and HDAC5 (EGFP) boundaries used for the quantification of HDAC5 cellular localization are shown. B, Distribution plot of the percent of HDAC5-EGFP-expressing U2OS cells as a function of the fraction of total HDAC5-EGFP within the cytoplasm (cytoplasmic proportion). Cytoplasmic proportion of HDAC5 was calculated by dividing the integrated intensity of the EGFP signal within the cytoplasm by the total integrated intensity of EGFP within each cell (see Experimental Procedures). WT HDAC5 (red bars; n = 848), S278A/S279A (black bars; n = 868), and S279A (green bars; n = 970) cells showed a median cytoplasmic proportion of 0.20, 0.36, and 0.45, respectively.
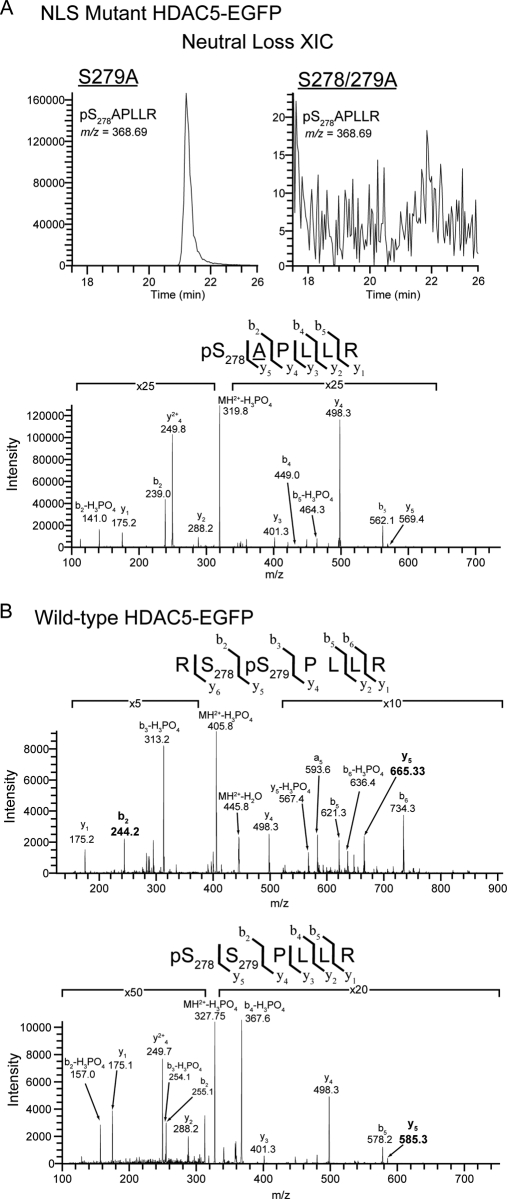
Phosphorylation at Ser278 Exists Independently of Ser279 Phosphorylation
A change in HDAC5 localization was observed for both the double S278/279A and single S279A mutants. Nevertheless, if the mutation of Ser279 affects the phosphorylation at Ser278, the single S279A mutant would mimic the double S278/279A mutant phenotype. To test this hypothesis, we performed an isolation of the S279A HDAC5 mutant from stably expressing HEK293 cell line and used targeted MS/MS analyses to determine if Ser278 is still phosphorylated in this mutant. Our results clearly demonstrate the presence of the phosphorylated Ser278 in the S279A mutant as shown by the precursor-neutral loss extracted ion chromatogram (XIC) and CID fragmentation of pS278APLLR NLS tryptic peptide (m/z = 368.69) (Fig. 8A). Importantly, this ion was not detected from the isolated S278/279A HDAC5 mutant, further supporting the identification of Ser278 phosphorylation in the S279A mutant. These data indicate that the phosphorylation at Ser278 exists independently of Ser279 phosphorylation, suggesting that the changes in HDAC5 localization for the S279A and S278/279A mutants are likely to be in both cases triggered by the mutation of Ser279. Therefore, we performed targeted MS/MS analyses on the isolated wild type HDAC5-EGFP to determine if Ser279 exists as a single phosphorylation, but at a level below the threshold of our initial analysis. As shown in Fig. 8B, data-independent CID MS/MS confirmed the presence of both peptides singly phosphorylated at Ser279 or Ser278. Altogether, our results suggest that Ser279 can solely affect the nuclear import of HDAC5, being required for its nuclear localization.
Fig. 8.
Targeted CID-MS2 analysis of NLS region demonstrated the independence of Ser278 phosphorylation from Ser279 phosphorylation. A, Neutral loss extracted ion chromatogram (NL-XIC) (top) and CID MS/MS spectrum (bottom) of the tryptic phosphopeptide pS278APLLR from the NLS region demonstrated the presence of Ser278 phosphorylation in S279A HDAC5-EGFP. This peptide ion (m/z = 368.69) was absent in the equivalent NL-XIC from S278/279A HDAC5-EGFP. B, Phosphopeptide CID spectra corresponding to doubly charged tryptic peptides from the NLS region of wild-type HDAC5 localized a single phosphorylation to Ser279 (top) and confirmed the previous identification of Ser278 phosphorylation by ETD (bottom). y and b ion fragments are indicated above each spectra, with site determining ions (b2 and y5) highlighted in bold.
CONCLUSIONS
In summary, to our knowledge, we report the first comprehensive phosphorylation study focusing on a class IIa enzyme—HDAC5. Although phosphorylation is known to play a key role in regulating these enzymes, this understanding mainly derives from studies on the two 14–3-3 interacting sites. Here we have extended this knowledge with the identification of 13 novel in vivo phosphorylation sites, including sites within the NLS, NES, and deacetylation domain. Noteworthy, many of these sites are conserved among the majority of the class IIa human HDACs (Supplemental Fig. S6). The previously reported Ser259, Ser498, and Ser661, and the novel Ser368 sites are conserved among all members, the next most conserved sites being those localized within the NLS. In fact, Ser278/279 and the surrounding residues are conserved in HDAC4 and HDAC9, but not in HDAC7, where the two Ser residues are replaced by Lys-Asn (Supplemental Fig. S6). Six novel phosphorylation sites are unique to HDAC5, including Ser755 and Ser1108 within the deacetylation domain and the NES, respectively. It remains to be determined if these unique sites represent potential functions or binding sites specific to HDAC5 that are conserved among vertebrate species.
Our characterization of NLS phosphorylation sites identified an in vivo phosphorylation at Ser279 that was present both with and without the neighboring phosphorylation at Ser278. Serine to alanine mutation of the NLS sites (S278/279A and S279A) resulted in a greater proportion of cytoplasmically localized HDAC5 that was accompanied by a decrease in corepressor complex association and loss of PKD interactions. Taken together, phosphorylation-dependent cellular localization and protein interactions, and targeted MS/MS analyses identified Ser279 phosphorylation as a critical point of regulation that could function independent of traditional extracellular stimuli to control HDAC5 nucleo-cytoplasmic shuttling, thereby indirectly regulating transcriptional activity. During the course of our study, Ha and coworkers (16) published an elegant study describing a novel PKA-dependent mechanism for inhibition of HDAC5 nuclear export in Cos7 and cardiomyocytes that involved phosphorylation of Ser280 (analogous to Ser279 in our study). Although our results demonstrated a role for Ser279 in nuclear import, this difference could result from: (1) the current study examined the regulation of HDAC5 in the absence of external stimuli, whereas the former focused on phenylephrine/PKA-mediated regulation; and (2) steady-state nucleo-cytoplasmic shuttling of HDAC5 could be differentially regulated in HEK293 and U2OS cells compared with Cos7 and neonatal rate ventricular myocytes. Overall, Ser279 may have multiple roles that impart distinct phosphorylation-dependent regulatory control mechanisms depending upon the cellular environment. The dynamic addition and removal of phosphate groups is known to provide protein conformational changes that indirectly affect enzymatic activity or protein interactions, as well as provide direct interacting sites for phosphosite-binding proteins (42). This is of particular interest with regard to the regulation of class IIa HDACs that undergo nucleo-cytoplasmic shuttling. Our study suggests that HDAC5 phosphorylation may not only encompass stimulus-dependent regulation of protein interactions and repressor activity, but also stimulus-independent control of steady-state localization. Future studies will be required to delineate the biological roles of these phosphorylations, particularly as they relate to the regulation of other class IIa histone deacetylases.
Acknowledgments
We are grateful to Rosa Viner (Thermofisher Scientific) for assistance with sample analysis on the LTQ Orbitrap Velos mass spectrometer, and to Joe Glavy (Stevens Institute) for the Mab414 antibody. We thank Tuo Li, Yang Luo and Mark Amirault for MS assistance, Yana Miteva for the GFP control isolation, and J. Goodhouse and C. DeCoste (Microscopy and Flow Cytometry Core Facilities, Princeton University).
Footnotes
* This work was supported by NIDA grant DP1DA026192 and HFSPO award RGY0079/2009-C to IMC.
 This article contains supplemental Figs. 1–6 and Tables 1–5.
This article contains supplemental Figs. 1–6 and Tables 1–5.
1 The abbreviations used are:
- ETD
- electron transfer dissociation
- HCD
- higher-energy C-trap dissociation
- HDAC
- histone deacetylase
- HEK
- human embryonic kidney cells
- NES
- nuclear export signal
- NLS
- nuclear localization signal
- PP2A
- protein phosphatase 2A.
REFERENCES
- 1. Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature. 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 2. Yang X. J., Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Renthal W., Maze I., Krishnan V., Covington H. E., 3rd, Xiao G., Kumar A., Russo S. J., Graham A., Tsankova N., Kippin T. E., Kerstetter K. A., Neve R. L., Haggarty S. J., McKinsey T. A., Bassel-Duby R., Olson E. N., Nestler E. J. (2007) Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 56, 517–529 [DOI] [PubMed] [Google Scholar]
- 4. Bossuyt J., Helmstadter K., Wu X., Clements-Jewery H., Haworth R. S., Avkiran M., Martin J. L., Pogwizd S. M., Bers D. M. (2008) Ca2+/calmodulin-dependent protein kinase IIdelta and protein kinase D overexpression reinforce the histone deacetylase 5 redistribution in heart failure. Circ. Res. 102, 695–702 [DOI] [PubMed] [Google Scholar]
- 5. Takami Y., Nakayama T. (2000) N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J. Biol. Chem. 275, 16191–16201 [DOI] [PubMed] [Google Scholar]
- 6. Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. (2002) Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 9, 45–57 [DOI] [PubMed] [Google Scholar]
- 7. Zhou X., Richon V. M., Rifkind R. A., Marks P. A. (2000) Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. U.S.A. 97, 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McKinsey T. A., Zhang C. L., Olson E. N. (2001) Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21, 6312–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kao H. Y., Verdel A., Tsai C. C., Simon C., Juguilon H., Khochbin S. (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276, 47496–47507 [DOI] [PubMed] [Google Scholar]
- 10. McKinsey T. A., Zhang C. L., Olson E. N. (2000) Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14–3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. U.S.A. 97, 14400–14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paroni G., Cernotta N., Dello, Russo C., Gallinari P., Pallaoro M., Foti C., Talamo F., Orsatti L., Steinkühler C., Brancolini C. (2008) PP2A regulates HDAC4 nuclear import. Mol. Biol. Cell. 19, 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 408, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang A. H., Kruhlak M. J., Wu J., Bertos N. R., Vezmar M., Posner B. I., Bazett-Jones D. P., Yang X. J. (2000) Regulation of histone deacetylase 4 by binding of 14–3-3 proteins. Mol. Cell. Biol. 20, 6904–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 15. Harrison B. C., Huynh K., Lundgaard G. L., Helmke S. M., Perryman M. B., McKinsey T. A. (2010) Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 584, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 16. Ha C. H., Kim J. Y., Zhao J., Wang W., Jhun B. S., Wong C., Jin Z. G. (2010) PKA phosphorylates histone deacetylase 5 and prevents its nuclear export, leading to the inhibition of gene transcription and cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 107, 15467–15472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chawla S., Vanhoutte P., Arnold F. J., Huang C. L., Bading H. (2003) Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 85, 151–159 [DOI] [PubMed] [Google Scholar]
- 18. Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. (2002) Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 110, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang S., Bezprozvannaya S., Li S., Olson E. N. (2005) An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 102, 8120–8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 21. Roy S., Shor A. C., Bagui T. K., Seto E., Pledger W. J. (2008) Histone deacetylase 5 represses the transcription of cyclin D3. J. Cell. Biochem. 104, 2143–2154 [DOI] [PubMed] [Google Scholar]
- 22. Moorman N. J., Sharon-Friling R., Shenk T., Cristea I. M. (2010) A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 9, 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., Sabatini D. M. (2006) CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo Y., Li T., Yu F., Kramer T., Cristea I. M. (2010) Resolving the composition of protein complexes using a MALDI LTQ orbitrap. J. Am. Soc. Mass Spectrom. 21, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bailey C. M., Sweet S. M., Cunningham D. L., Zeller M., Heath J. K., Cooper H. J. (2009) SLoMo: Automated site localization of modifications from ETD/ECD mass spectra. J Proteome Res. 8, 1965–1971 [DOI] [PubMed] [Google Scholar]
- 26. Searle B. C. (2010) Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 27. Yoon H. G., Chan D. W., Huang Z. Q., Li J., Fondell J. D., Qin J., Wong J. (2003) Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 22, 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J., Kalkum M., Chait B. T., Roeder R. G. (2002) The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell. 9, 611–623 [DOI] [PubMed] [Google Scholar]
- 29. Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. (2004) Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ha C. H., Wang W., Jhun B. S., Wong C., Hausser A., Pfizenmaier K., McKinsey T. A., Olson E. N., Jin Z. G. (2008) Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J. Biol. Chem. 283, 14590–14599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huynh Q. K., McKinsey T. A. (2006) Protein kinase D directly phosphorylates histone deacetylase 5 via a random sequential kinetic mechanism. Arch. Biochem. Biophys. 450, 141–148 [DOI] [PubMed] [Google Scholar]
- 32. Grozinger C. M., Schreiber S. L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14–3-3-dependent cellular localization. Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Janssens V., Longin S., Goris J. (2008) PP2A holoenzyme assembly: In cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33, 113–121 [DOI] [PubMed] [Google Scholar]
- 34. Martin M., Potente M., Janssens V., Vertommen D., Twizere J. C., Rider M. H., Goris J., Dimmeler S., Kettmann R., Dequiedt F. (2008) Protein phosphatase 2A controls the activity of histone deacetylase 7 during T cell apoptosis and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 4727–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Juan L. J., Shia W. J., Chen M. H., Yang W. M., Seto E., Lin Y. S., Wu C. W. (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275, 20436–20443 [DOI] [PubMed] [Google Scholar]
- 36. Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeyakumar M., Liu X. F., Erdjument-Bromage H., Tempst P., Bagchi M. K. (2007) Phosphorylation of thyroid hormone receptor-associated nuclear receptor corepressor holocomplex by the DNA-dependent protein kinase enhances its histone deacetylase activity. J. Biol. Chem. 282, 9312–9322 [DOI] [PubMed] [Google Scholar]
- 38. Götz J. (2001) Tau and transgenic animal models. Brain Res. Brain Res. Rev. 35, 266–286 [DOI] [PubMed] [Google Scholar]
- 39. Taouatas N., Drugan M. M., Heck A. J., Mohammed S. (2008) Straightforward ladder sequencing of peptides using a lys-N metalloendopeptidase. Nat Methods. 5, 405–407 [DOI] [PubMed] [Google Scholar]
- 40. Carabetta V. J., Li T., Shakya A., Greco T. M., Cristea I. M. (2010) Integrating lys-N proteolysis and N-terminal guanidination for improved fragmentation and relative quantification of singly-charged ions. J. Am. Soc. Mass Spectrom. 21, 1050–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bottomley M. J., Lo, Surdo P., Di, Giovine P., Cirillo A., Scarpelli R., Ferrigno F., Jones P., Neddermann P., De Francesco R., Steinkühler C., Gallinari P., Carfi A. (2008) Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 283, 26694–26704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Page R. D. (1996) TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]