Abstract
The transmembrane subunits of viral envelope proteins are thought to perform all of the functions required for membrane fusion during entry of enveloped viruses. However, changes in a conserved SPHQ motif near the N terminus of the receptor binding subunit of a murine leukemia virus (MLV) envelope protein block infection and induction of cell-cell fusion but not receptor binding. Here we report evidence that a histidine-to-arginine change at position 8 (H8R) in the SPHQ motif of Moloney MLV blocks infection by arresting virus-cell fusion at the hemifusion state. In cell-cell fusion assays, H8R envelope protein induced mixing of membrane outer leaflet lipids but did not lead to content mixing, a finding indicative of fusion pore formation. Kinetic studies of virus-cell fusion showed that lipid mixing of H8R virus membranes begins much later than for wild-type virus. The length of the delay in lipid mixing decreased upon addition of two second-site changes that increase H8R virus infection to 100-fold less than the wild-type virus. Finally, chlorpromazine, dibucaine, and trifluoperazine, agents that induce pores in an arrested hemifusion state, rescued infection by H8R virus to within 2.5-fold of the level of wild-type virus infection and cell-cell fusion to half that mediated by wild-type envelope protein. We interpret these results to indicate that fusion progressed to the hemifusion intermediate but fusion pore formation was inhibited. These results establish that membrane fusion of Moloney MLV occurs via a hemifusion intermediate. We also interpret these findings as evidence that histidine 8 is a key switch-point residue between the receptor-induced conformation changes that expose fusion peptide and those that lead to six-helix bundle formation.
Viral envelope proteins act as a catalyst for fusion of the viral and host cell membranes during infection by enveloped viruses (21). Most envelope proteins are synthesized as a single precursor polypeptide chain that is cleaved into a soluble N-terminal protein containing the receptor-binding domain and a C-terminal transmembrane protein. The transmembrane protein appears to be the catalytic subunit. Mutations that prevent exposure of the hydrophobic fusion peptide on the N terminus of influenza A virus hemagglutinin (HA) or lower the hydrophobicity of its internal residues abolish membrane outer leaflet lipid mixing (15, 20). A point mutation that changes glycine 1 to serine (G1S) at the N terminus of the HA fusion peptide or replacement of its transmembrane anchor with a glycosylphosphatidylinositol (GPI-HA) anchor allow outer leaflet mixing but abolish fusion pore formation, arresting fusion at an intermediate called hemifusion (11, 21, 33).
In retroviral envelope proteins (Env), the association of the binding subunit (SU) with the transmembrane protein (TM) is thought to constrain TM to a metastable conformation that is incompetent for fusion. During infection, the interaction of SU with its cellular receptor induces conformation changes that release the constraint so that the fusion peptide is exposed and new intramolecular interactions of TM domains form the six-helix bundles that are thought to catalyze viral and cellular membrane fusion. In contrast to influenza virus HA, replacement of the transmembrane anchor of human immunodeficiency virus type 1 (HIV-1) Env with a GPI anchor did not give lipid mixing (39). Thus, there is no retroviral Env mutant that causes a hemifusion intermediate.
The SUs of gammaretroviruses contain a highly conserved sequence, SPHQ, near their N termini that influences viral fusion. Replacement or deletion of the histidine at position 8 in the SPHQ motif of ecotropic Moloney murine leukemia virus (MLV) SU almost completely abolished infection (4, 41). The altered Env showed no change in receptor binding but lost the ability to induce cell-cell fusion. H8R does not inhibit R-peptide cleavage in virions (41), indicating that the defect was not due to retention of this fusion modulating sequence. The addition of two second-site changes, glutamine 227 to arginine (Q227R) and aspartate 243 to tyrosine (D243Y), increased infection by H8R virus to 100-fold less than wild-type virus, indicating that these changes partially suppress the fusion defect resulting from an H8R change (41).
Here we report evidence that Moloney MLV fusion proceeds via a hemifusion intermediate and that an H8R change blocks infection by arresting virus-cell fusion at the hemifusion state. In cell-cell fusion assays, wild-type Env mediated mixing of membrane outer leaflet lipids and of cell contents. In contrast, an H8R mutant Env mediated outer leaflet lipid mixing but not content mixing, suggesting that fusion pore formation was blocked. Kinetic studies of virus-cell fusion showed that H8R virus lipid mixing is delayed, taking twice as long to begin as wild-type virus. We interpret the longer delay until lipid mixing as evidence that the H8R change arrests fusion at the hemifusion state. In agreement with this interpretation, the length of the delay was shortened upon addition of the two second-site changes that increase H8R virus infection. Finally, chlorpromazine (CPZ), dibucaine (DB), and trifluoperazine (TFP), agents that induce pores in an arrested hemifusion state, restored infection by H8R virus to within 2.5-fold of the wild-type virus level, indicating that the majority of bound virus had progressed to the hemifusion intermediate but no further. CPZ also increased H8R-mediated cell-cell fusion to half that mediated by wild-type envelope protein. These results establish that membrane fusion of Moloney MLV proceeds through a hemifusion intermediate. We also interpret them as evidence that histidine 8 is a key switch-point residue in the receptor-induced conformation changes that give six-helix bundle formation after fusion peptide exposure.
MATERIALS AND METHODS
Cell-cell fusion assays.
In three-color experiments, 100-mm dishes of 80% confluent human 293 cells were transiently transfected with 40 μg of an R-less form (34, 35) of wild-type or mutant Moloney MLV Env expression plasmids by standard calcium phosphate coprecipitation. The next day, each culture was split into two dishes; one dish was used for quantification of cell surface expression by using flow cytometry (see below for methods), and the other was used as a source of effector cells in the fusion assays. The cytoplasmic contents of the effector cells were labeled with the blue fluorescent dye CMAC CellTracker Blue (Molecular Probes) as follows: adherent effector cells were washed with serum-free OptiMEM (Life Technologies) and then incubated for 40 min at 37°C in 20 μM CMAC diluted in serum-free OptiMEM, washed again with regular growth medium (Dulbecco modified Eagle medium and 8% donor calf serum), and incubated at 37°C for an additional 2 to 3 h. In control experiments, effector cells were 293 cells transfected with wild-type or mutant influenza A virus HA expression plasmids (a gift of Judy White). These cells were labeled with CMAC CellTracker Blue as described above; the cells were then detached, HA activation was initiated, and interactions with sialic acid receptors on effector cell surfaces were eliminated by incubating about 1 million detached cells in 1 ml of 10 mg of trypsin (TPCK [tolylsulfonyl phenylalanyl chloromethyl ketone] treated; Sigma) and 250 mg of neuraminidase (Sigma)/ml at room temperature for 20 min, followed by quenching with regular medium and three washes in phosphate-buffered saline (PBS).
In most experiments, target cells were a previously described transfectant of human 293 cells stably expressing the cDNA of the ecotropic MLV receptor (24) and displaying endogenous sialic acid Lewis receptors for the HA controls. In some experiments, rat XC sarcoma cells that express endogenous ecotropic MLV receptor were used as the target cells. The cytoplasmic contents and membrane outer leaflet lipids of target cells were labeled as follows: adherent target cells were incubated in regular medium (high-glucose Dulbecco modified Eagle medium and 10% fetal bovine serum) containing 0.5 μM calcein AM (Molecular Probes) for 20 min at 37°C, washed three times with regular growth medium, and washed three times with PBS; after which the cells were detached from the plates by gentle pipetting. Detached cells were washed once with serum-free OptiMEM, incubated in sterile siliconized microfuge tubes with serum-free OptiMEM containing 4 μM DiI (a red fluorescent, lipid-soluble dye [Molecular Probes]) in 0.6% dimethyl sulfoxide for 5 min at 37°C, and then shifted to 4°C for an additional 20 min. Excess dye was removed by three 10-min washes in regular growth meduim on ice.
Equal numbers of labeled effector and target cells were mixed, applied to poly-l-lysine (Sigma)-coated plates to promote adherence and incubated on ice for 30 min to allow cell attachment and binding, after which cells were shifted to 37°C for membrane fusion. In control experiments with HA-expressing effectors, fusion was activated by adding 2 ml of isotonic solution (pH 4.9; 100 mM NaCl, 2.5 mM CaCl2, 1 mg of glucose/ml, 20 mM succinate) to cell mixtures that were in 1 ml of regular medium. After 2 min, the acidic solution was replaced by a pH 7.4 solution (110 mM NaCl, 1.5 mM KCl, 2 mM MgCl2, 1 mg of glucose/ml, 20 mM HEPES), and then the cells were shifted to 37°C. After a 30-min incubation at 37°C, representative fluorescent and phase micrographs of live cells were captured by using Axiovision imaging software (Zeiss) and a Zeiss Axiocam digital camera on a Zeiss Axiphot epifluorescent microscope. All images were collected within 60 min of mixing effector and target cells to obtain data representing specific, Env-mediated dye transfer. Leakage of calcein AM began to be observed at 2 h after cells were mixed in negative control effector cells expressing vector alone or 595* Env, a binding-competent, fusion-defective Env shown to be blocked before fusion peptide exposure (27). No nonspecific transfer of DiI or CMAC dyes was observed even after 2 h.
In some experiments, effector cells expressed wild-type and mutant Env containing R-peptide, and target cells were receptor positive rat XC cells (ATCC CCL-165). In these experiments, the membrane outer leaflets of effector cells expressing Env containing R-peptide were labeled with the lipid-soluble, fluorescent dye R-18 (octadecylrhodamine-B; Molecular Probes), and the cytoplasmic contents of target cells were labeled with green fluorescent CellTracker Green (Molecular Probes) as follows: cells were detached by using 0.02% EDTA, incubated with 0.1 mg of R-18/ml for 15 min at room temperature, washed to remove unincorporated dye, and immediately applied to adherent target cells whose cytoplasm was labeled with 7 μM CellTracker Green CMFDA (Molecular Probes) according to the manufacturer's instructions, and the cell mixture was incubated at 37°C. In other experiments, effector cell contents were labeled with calcein AM, and target cell contents were labeled with CellTracker Orange (Molecular Probes). Representative images were captured at between 30 and 60 min after effector cells were applied to the target cells. Nonspecific, spontaneous transfer of R-18 began to be observed after a 90-min incubation of the samples containing control pcDNA3 vector expressing effector cells mixed with target cells.
Fluorescence-activated cell sorting analysis of Env expression on effector cells.
Negative control (untransfected) human 293 and transfected effector cells were detached from culture dishes with 0.02% EDTA and incubated with polyclonal goat anti-Rauscher SU (serum ID 80S000018; Quality Biotech, Inc.) that cross-reacts with the surface or SU subunit of Moloney MLV Env, in the presence of 0.02% sodium azide to prevent internalization of immune complexes, for 1 h at 4°C. Cells were washed thrice, incubated with mouse anti-goat conjugated to fluorescein isothiocyanate for an additional hour on ice, and then washed thrice to remove unbound antibodies, and the mean fluorescence intensity of 5,000 cells was measured by flow cytometry.
Kinetics of virus-cell fusion.
Infectious virions were precipitated from virus-containing supernatants in 50% ammonium sulfate, solubilized in 10 mM Tris (pH 8.0)-1 mM EDTA-100 mM NaCl, and purified by size-exclusion chromatography on Sepharose 4B (Sigma) columns essentially as described by C. Cepko (3). Fractions containing virus were identified as those exhibiting an absorbance ratio (260 nm/280 nm) of ca. 1.0 to 1.1 (1) and concentrated further on CentriPlus-100 devices (Millipore). The reverse transcriptase activity in 10 μl of each concentrated virus preparation was used to adjust the volume of the stock to achieve comparable particle concentrations prior to labeling with R-18. Typically, the wild-type adjusted stock contained 107 infection-forming units (IFU)/ml. Viruses were labeled with R-18 as described for HIV-1 (37) with the following modifications: 50 μl of adjusted virus stocks were mixed with 50 μl of 20 μg of R-18/ml, and then unincorporated R-18 was removed by size-exclusion chromatography. Viruses exhibiting a 5- to 10-fold increase in mean fluorescence intensity after incubation in 0.2% Triton X-100 were used immediately for virus-cell fusion experiments. A total of 106 receptor-positive XC or mouse NIH 3T3 cells were incubated in the absence or presence of R-18-labeled virus and 5 μg of Polybrene/ml for 1 h at 0°C and then washed twice to remove unbound virus. Nonspecific virus adsorption and spontaneous R-18 transfer from virus to cell membranes was quantified by incubating each virus with receptor-negative 293 cells. Virus-cell complexes and negative control cells were placed in separate wells of a white 96-well fluorimetry plate (Costar) on ice, and then plates were quickly warmed to 37°C and the relative fluorescence intensity (RFI) was measured at equal intervals at 37°C in a plate-reading fluorimeter (550-nm excitation and 585-nm emission; HTS 7000 Bioassay Reader with incubator [Perkin-Elmer]). At the end of kinetics measurements, complete dequenching was achieved by addition of Triton X-100, and the RFI measured was taken to be F100%. As a measure of virus-cell fusion, the percent increase in dequenching at time point t was calculated for each sample as described by Blumenthal et al. (9), except that the RFI of negative control 293 cells incubated with virus was subtracted at each time point.
Effect of membrane-curving agents on infection and cell-cell fusion.
Env pseudotypes of Moloney MLVs were produced by transient transfection of a prepackaging cell line, H1 BAG, as described previously (41). Fresh stock solutions of 200 mM CPZ, 1 M DB, and 50 mM oleic acid in ethanol and of 50 mM TFP in PBS were prepared for each experiment. Cell culture medium lacking sodium bicarbonate was prepared from powdered medium with 50 mM BES (Sigma) adjusted to pH 6.0, 6.5, 7.0, or 7.5. Quadruplicate wells of XC cells were exposed to 10-fold serial dilutions of wild-type or H8R virus with 20 μg of Polybrene (Sigma/ml) for 1 h at 37°C and then washed and incubated with each agent in BES-buffered medium at the indicated pH for 1 min (CPZ, DB, or TFP) or 5 min (oleic acid). Cells were quickly washed, returned to drug-free medium at the same pH, and scored for β-galactosidase activity transduced by virus infection as previously described (41). In experiments with murine NIH 3T3 cells as host, 0.3 mM CPZ was applied to cells for 5 min.
To determine the effect of CPZ on cell-cell fusion, mixtures of CMAC CellTracker Blue labeled effector cells expressing wild-type or mutant Moloney MLV Env or influenza A virus HA were prepared and mixed with DiI and calcein AM-labeled 293 cells expressing the ecotropic MLV receptor cDNA as described above. Fluorescence and phase micrographs were captured by using a ×10 immersible objective lens after an initial 30-min incubation at 37°C, and then the medium was replaced with 0.4 mM CPZ. After 1 min, the CPZ was removed, the cells were incubated in fresh BES-buffered medium at 37°C for an additional 30 min, and a second set of fluorescence micrographs were captured.
RESULTS
H8R Env mediated outer leaflet lipid mixing, but no content mixing was observed in cell-cell fusion.
Initially, we sought to determine whether H8R Env mediates outer leaflet lipid mixing and fusion pore formation. Our hypothesis was that H8R prevented the conformation change that exposes fusion peptide and, since the loss of fusion peptide exposure results in a loss of detectable mixing in other enveloped viruses (20, 21), H8R Env was not expected to produce lipid mixing. As in the study of Melikyan et al. (27) on Moloney Env cell fusion, wild-type and mutant Env were expressed as C-terminal truncations that lack R-peptide (R-less). The cell surface expression of Env was quantified on parallel cultures of effector cells by using flow cytometry and an anti-SU antiserum (Table 1). The mean fluorescence intensities of H8R- and 595* Env-expressing cells were comparable to that of wild-type Env effector cells (Table 1), indicating that effector cells displayed comparable levels of Env.
TABLE 1.
Cell surface expression of Env protein
Values shown for percent cells expressing Env are the percentage of total cells analyzed that showed fluorescence above that of negative control parental 293 cells incubated with primary and secondary antibodies.
Relative surface expression was calculated as 100 times the ratio of the mean fluorescence intensity of H8R- or 595*-expressing cells to the mean fluorescence intensity of wild-type Env-expressing cells.
The cytoplasmic contents of Env-expressing 293 effectors were labeled with CMAC Blue, and ecotropic MLV receptor-expressing 293 target cells contents were labeled with calcein AM. Both dyes become fluorescent upon hydrolysis by esterases in the cell cytoplasm and rapidly convert into dye-protein adducts that do not readily leak out of the cell. In addition, the membrane of target cells was labeled with DiI, a red fluorescing lipid-soluble dye. Labeled effector and target cells were mixed and then applied to culture dishes coated with poly-l-lysine to promote adherence so that cells lay in a single focal plane for image capture. This incubation was performed on ice to restrict membrane fluidity. After attachment, cell mixtures were then rapidly warmed to 37°C. In studies of HIV cell-cell fusion, short incubation on ice prior to incubation at 37°C did not affect the kinetics of membrane fusion (39). Since fusion by Moloney MLV Env is not induced by low pH, we relied on the normal events to initiate cell-cell fusion.
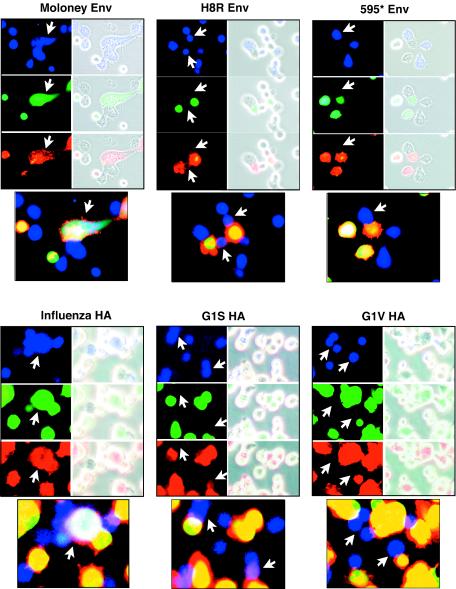
The micrographs shown in Fig. 1 present close-up views that illustrate representative events from each sample. Effector cells are identified by blue fluorescence in their cytoplasm, whereas target cells have green fluorescent cytoplasmic content. Fusion events between the wild-type Moloney Env (WT) effector cells and target cells typically became evident by 15 min after a shift to 37°C and rapidly progressed to syncytia by 20 min after the shift. Transfer of red fluorescence from the target cell membrane to the effector cell and reciprocal transfer of green and blue fluorescent cytoplasm indicated that membrane lipids mixed and that a conduit for cell content mixing was established (Fig. 1). These results are similar to those previously reported by Melikyan et al. (27) for wild-type R-less Moloney MLV Env-induced cell-cell fusion.
FIG. 1.
The H8R envelope protein mediates lipid mixing but not fusion pore formation. The cytoplasm of receptor-negative human 293 cells transiently transfected with Moloney Env R-less, histidine 8-changed-to-arginine (H8R Env) R-less, negative control 595* Env, influenza virus HA, glycine 1-changed-to-serine HA (G1S HA), or glycine 1-changed-to-valine HA (G1V HA) expression plasmids was labeled with CMAC CellTracker Blue, and the cytoplasm of target human 293 cells stably expressing ecotropic MLV receptor was labeled with calcein AM. In addition, the membrane lipids of target cells were labeled with DiI. Equal numbers of fluorescently labeled effector and target cells were mixed, applied to poly-l-lysine-coated plates to promote adherence, and incubated on ice for 30 min to allow cell attachment and binding, after which cells were shifted to 37°C for membrane fusion. Control wild-type and mutant HA-expressing effector cells were activated prior to mixing, and fusion was initiated by a brief exposure to low pH as described in Materials and Methods. Representative micrographs of live cells were captured after 30 min. Fluorescent images of Env-expressing effectors (blue) and receptor-positive target cells (red and green) are shown on the left side of each panel and are merged with a phase-contrast image on the right side. Below each set is a panel of the merged fluorescence images. Arrows point to example effector cells paired with target cells.
H8R Env-positive effector cells also showed membrane lipid mixing with target cells, but it typically began after 25 to 30 min. There was no transfer of cytoplasmic content dye, and no syncytia formed, indicating that the lipid mixing was not accompanied by fusion pore formation (Fig. 1). These observations suggested that an H8R change caused a hemifusion phenotype. Samples of target cells mixed with negative control effectors expressing Env mutant 595* were also observed. Env mutant 595* is a binding-competent, fusion-defective Env shown to be blocked before lipid mixing, presumably because fusion peptide does not become exposed (27). Lipid mixing was rare, and no content mixing was observed in these samples even though there were numerous cell pairs with extensive cell-cell contacts (Fig. 1). After 2 h, nonspecific leakage of green fluorescing cytoplasm began to be observed. No nonspecific transfer of DiI or CMAC CellTracker Blue was observed.
Comparison of H8R with the previously characterized G1S mutant of influenza virus HA mutant.
Qiao et al. (33) concluded that the G1S HA mutant arrests cell-cell fusion at the hemifusion state based on three findings from cell-cell fusion assays: (i) lipid dye transferred but content dye does not; (ii) confirmation of that finding by using electrophysical measurements; and (iii) the membrane-curving agent CPZ induced redistribution of content dye, indicating complete fusion. We compared the characteristics of H8R Env and G1S HA by using the first and last of these assays. The G1V HA mutant (glycine 1 to valine), which shows no appreciable hemifusion or fusion activity (33), and HA that had not been activated by trypsin cleavage were used as negative controls.
Figure 1 shows close-up views of representative cell pairs for the wild-type and mutant HA control samples. Table 2 shows values of the percentage of hemifused and fused cell pairs calculated from quantification of the number of cell pairs showing hemifusion (scored as lipid dye transfer in the absence of content dye transfer) and fusion (scored as lipid and content dye transfer). At least 250 cell pairs were scored from low-magnification micrographs of each sample. Moloney MLV Env or activated influenza virus HA did not show hemifusion, but full fusion was abundant (Table 2). In contrast, H8R Moloney Env gave 8% hemifusion, and G1S HA gave 20% hemifusion (Table 2).
TABLE 2.
Quantification of cell-cell fusion
| Env protein | % Fusiona |
|||
|---|---|---|---|---|
| No CPZ |
0.4 mM CPZ |
|||
| Hemifusion | Fusion | Hemifusion | Fusion | |
| Moloney MLV | 0 | 53 | 0 | 63 |
| Influenza HA | 0 | 31 | 0 | 34 |
| H8R Moloney | 8 | 3 | 0 | 29 |
| G1S HA | 20 | 4 | 21 | 13 |
| 595* Moloney | 4 | 5 | 9 | 11 |
| G1V HA | 4 | 4 | 4 | 3 |
| HA, no trypsin | 3 | 2 | 4 | 4 |
Values shown are the percent hemifusion or fusion calculated as the number of cell pairs showing hemifusion or fusion/total number of cell pairs × 100, where hemifusion was defined as lipid mixing in the absence of content mixing and fusion was defined as lipid and content mixing.
Lipid mixing is delayed for H8R virus.
To quantify lipid mixing during virus-cell membrane fusion, we used a fluorescence-dequenching assay based on the incorporation of a fluorescent probe, R-18, into the membrane outer leaflet of intact virions in amounts great enough to self-quench (9). R-18-labeled virus was incubated with target cells (NIH 3T3 and XC cells) at 4°C to allow virus binding but not membrane fusion. If outer leaflet mixing between viral and cell membranes occurs after shifting to 37°C, the R-18 is diluted, producing in an increase in the RFI called dequenching. Since the RFI is measured in arbitrary units, the extent of lipid mixing was determined by calculating the percent of total fluorescence released as described by Blumenthal et al. (9), with the following modification: R-18 transfer resulting from nonspecific virus binding, as well as from nonspecific transfer, i.e., nonfusion (31, 38), was quantified by using receptor-negative 293 cells incubated with each labeled virus. The RFI value of this control at each time point was subtracted from the RFI of the virus plus receptor-positive cell samples for the same time point prior to the calculation of the percent dequenching.
No increase in fluorescence dequenching occurred during the first few minutes after shifting wild-type Moloney MLV-cell complexes to 37°C, indicating that little if any fusion events were completed immediately (Fig. 2). This initial lag was followed by a steady increase, reaching a maximum on both target cell types typically after 3 h. Others have reported that HIV, influenza virus, paramyxovirus SV5, Rous sarcoma virus, vesicular stomatitis virus, and bovine leukemia virus showed maximal R-18 dequenching in comparable times after a shift to 37°C (5, 6, 16, 21, 32, 37, 40).
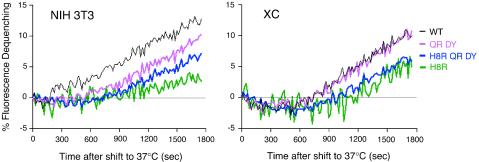
FIG. 2.
Lipid mixing is delayed with H8R virus. The membrane lipid of purified virions containing either wild-type Env or mutant Env were labeled to self-quenching levels with R-18, incubated at 4°C with receptor-positive cells (NIH 3T3 or XC) or cells lacking receptor (293) for 1 h, and then shifted to 37°C, and the RFI was monitored continuously. The data are representative of two independent experiments. Colors: blue, wild-type virus; magenta, H8R virus; purple, triple-mutant H8R QR DY virus; cyan, double-mutant QR DY virus.
H8R virus also exhibited an increase in fluorescence, in agreement with the outer leaflet mixing observed in the cell-cell fusion assays. However, the initial increase occurred after a lag period of twice that observed with wild-type virus (Fig. 2). The triple mutant containing an H8R substitution and the two second-site suppressor mutations exhibited an intermediate lag time.
Normally, outer leaflet lipid mixing is restricted to the fusion site until after fusion pores open and stabilize (10, 29, 42). Thus, the lipid mixing observed in wild-type virus fusion may represent continuity of the outer leaflets after fusion was completed. The low level of H8R virus lipid mixing immediately after the delay might represent completion of a few successful fusion events since it correlates with the low level of infection consistently observed for H8R virus in XC cells (41). The increase in the percentage of bound virus that mixed lipids during the initial period when the second-site mutations are added to Env is consistent with this interpretation. We do not know what the increase in H8R virus dequenching represents at later times. We cannot rule out that the high levels of dequenching at late times represents spontaneous R-18 transfer.
H8R cell pairs fuse when exposed to CPZ, an agent that induces fusion pore formation in hemifusion intermediates.
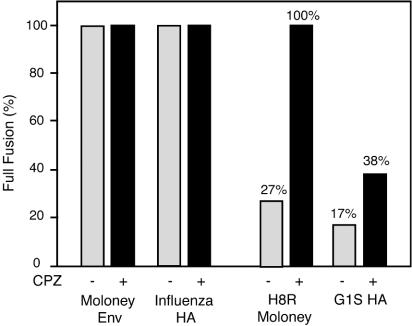
We next compared the effect of the membrane-curving agent CPZ on H8R and G1S HA cell-cell fusion by using the three-color cell fusion assay. CPZ is a lipid analog that partitions preferentially into the inner leaflet (36) and induces curvature that favors pore formation in a membrane bilayer (18). Brief exposure to CPZ induces holes in the hemifusion intermediate captured by GPI-anchored HA (26) but did not induce cell-cell fusion with the Moloney MLV mutant 595* (27). H8R cell fusion increased 10-fold after exposure to CPZ, and G1S HA fusion increased 3-fold (Table 2). Similar fold increases were observed in a replicate experiment. For comparison to published quantification of G1S HA and GPI-HA mutants, Fig. 3 shows the percent full fusion, a parameter defined by Qiao et al. (33) and calculated as the percentage of lipid dye transfer events that also showed content dye transfer.
FIG. 3.
Stability of the hemifusion intermediates caused by H8R Moloney Env and G1S HA. Cell-cell fusion assays were performed as described in the legend to Fig. 1 except that, after the initial set of micrographs were captured, effector-target cell mixtures were briefly exposed to CPZ and incubated an additional 30 min, and then a second set of micrographs were captured. The total number of cell pairs, the number of cell pairs showing lipid dye transfer alone, and the number of pairs showing lipid and content dye transfer were counted from the micrographs, with a total of at least 250 cell pairs scored for each sample. Values shown are the percent full fusion as defined in Qiao et al. (33) and calculated as follows: (the number of fully fused cell pairs/total number of cell pairs showing lipid transfer) × 100. The identity of the envelope protein expressed on the effector cells is indicated below the horizontal axis. Bars: ░⃞, no CPZ; ▪, 0.4 mM CPZ.
Rescue of H8R virus infection by CPZ and similar agents.
DB and TFP are lipid analogs of CPZ that also prefer the inner leaflet (36), induce curvature favorable to pore formation (18) and fusion by GPI-anchored HA (26). Oleic acid induces curvature that promotes the formation of a hemifusion intermediate but is unable to relieve the block to GPI-HA fusion (10, 19, 26). We sought to determine whether any of these agents could rescue H8R virus infection. Since prolonged exposure is toxic to cells, the agents were briefly added to virus-cell complexes formed after a 1-h incubation.
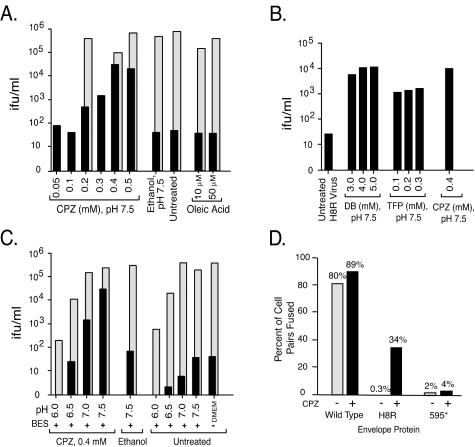
Oleic acid did not increase the infectivity of H8R virus (Fig. 4A). In contrast, brief exposure to CPZ enhanced H8R virus infection in a concentration-dependent manner. CPZ at 0.2 mM increased infection from <100 to >500 IFU/ml on average, whereas 0.4 mM CPZ increased it to an average of 20-fold less than wild-type virus infection. In two of four independent titrations, the higher concentrations of CPZ increased H8R infection to within 2.5-fold less than wild-type (wild-type and H8R virus titers were 4 × 105 and 2 × 104, 105 and 4 × 104, 105 and 4 × 104, and 4 × 105 and 2 × 104, respectively). No infection was detected in cells exposed to virions lacking Env, indicating that the increase was dependent on interaction with the receptor. The lowest concentration of DB was as effective at relieving the block to H8R virus entry as was CPZ, whereas TFP was slightly less effective (Fig. 4B). At an acidic pH CPZ becomes protonated and is unable to relieve a block to fusion pore formation (26). When virus-cell complexes were treated with CPZ at pH 7.5, the average titer of H8R virus was 10-fold less than the average of wild-type virus, but at pH 7.0 it was 100-fold less, and at pH 6.5 and 6.0 it was 200-fold less (Fig. 4C). The CPZ-induced increase in H8R virus infection correlated with an increase in cell-cell fusion when XC were the target cells (Fig. 4D).
FIG. 4.
CPZ, DB, and TFP rescued infection by H8R virus. XC cells were exposed to 10-fold serial dilutions of H8R or wild-type virus for 1 h at 37°C and then washed and incubated briefly with the membrane-curving agents or with the solvent ethanol alone in BES-buffered medium at the indicated pH. Cells were scored 48 h later for virus infection, and the titers were calculated from the endpoint dilutions. The values shown are averages from two independent experiments of each type. No infection was detected in cells exposed to virions lacking Env (data not shown). (A) Dose response to CPZ and oleic acid. For comparison, wild-type virus was titrated for 0.2, 0.4, and 0.5 mM CPZ and for 10 and 50 μM oleic acid at pH 7.5. (B) DB and TFP also rescue H8R virus infection. (C) pH dependence of rescue by CPZ. For comparison, wild-type virus was titrated under all conditions. Bars (A and C): ▪, H8R virus; ░⃞, wild-type Moloney MLV. (D) Effect of CPZ on cell-cell fusion with XC target cells. Cell-cell fusion assays were performed as described in the legend to Fig. 3, except that XC cells were used as target cells. The values shown were calculated as follows: (number of cell pairs showing lipid and content dye transfer/the total number of cell pairs) × 100. Bars: , no CPZ; ▪, 0.4 mM CPZ.
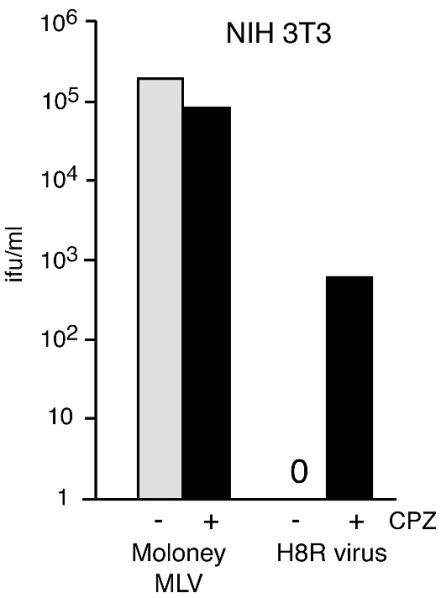
CPZ also rescued H8R infection of NIH 3T3 cells, causing an increase from none to 600 IFU/ml (Fig. 5). In the NIH 3T3 experiments, 0.3 mM CPZ was applied to cells for 5 min (Fig. 5) because a 1-min exposure to 0.4 mM CPZ caused the fibroblasts to detach from the culture plate, whereas a 1-min exposure to 0.3 mM CPZ was not toxic but resulted in very little increase in H8R infection. In XC cells in which fusion appears to occur at the cell surface, a brief pulse of CPZ would be expected to give the agent immediate access to virus-receptor complexes. However, in NIH 3T3, virus-receptor complexes should be internalized and thus out of immediate access. We reasoned that a longer exposure might give the opportunity for recycling membrane vesicles to bring the CPZ into the intracellular compartments. The observed increase is consistent with this notion, but alternative explanations are also possible.
FIG. 5.
CPZ rescued H8R virus infection of NIH 3T3 cells. Quadruplicate wells of NIH 3T3 cells were exposed to serial 10-fold dilutions of wild-type Moloney MLV or H8R Env psuedotype virus stock for 1 h at 37°C, after which fresh medium containing 0.3 mM CPZ was applied for 5 min at 37°C, followed by rapid washes with regular culture medium. Cells were scored 48 h later for virus transduction of β-galactosidase activity. Titers were calculated from the endpoint dilutions.
DISCUSSION
We conclude that the failure to resolve the hemifusion state into stable fusion pores is the primary cause of the loss of H8R virus infection. In cell-cell fusion assays, H8R Env-target cell pairs mixed their lipids but not cell contents, a finding consistent with the formation of a hemifusion intermediate that does not proceed to stable fusion pore formation. In addition, the kinetics of H8R-mediated lipid mixing differed from those of the wild type; the onset of virus-cell lipid mixing occurred much later than in wild-type virus-cell complexes. We propose that the lipid of H8R virus was delayed in merging with that of the cell because fusion was arrested at the hemifusion state.
In agreement with this interpretation, the delay in lipid mixing was reduced upon addition of the two second-site changes that increase H8R virus infection by 10,000-fold. The rescue of H8R virus infection by the membrane-curving agents CPZ, DB, and TFP also strongly supports this interpretation. These agents elevated H8R infection in a range of 5 to 40% of wild type, a finding consistent with many particles having reached the hemifusion intermediate. The increase in infection correlated with an increase in H8R Env mediated cell-cell fusion when CPZ was added. Although the effect of CPZ on infection by influenza virus hemifusion mutants has not been reported, CPZ was shown to induce 20 to 25% of HA hemifusion mutant-expressing cells to fuse (2, 10), with levels as high as 60% reported (26) at the concentration of CPZ at which we observed the greatest H8R MLV infection and the increase in G1S HA cell fusion. Our observations are also consistent with the possibility that H8R Env opens pores that are very small or unstable. Others have observed transient pore formation not accompanied by sustained pore opening (28) or pore enlargement (25). Detailed conductance measurements will be important for determining whether this is the case.
To our knowledge this is the first evidence that gammaretroviral Env-mediated fusion involves a hemifusion intermediate and the first mutation in a retroviral Env shown to cause hemifusion. Previous attempts to capture retroviral hemifusion with an Env mutant were inconclusive. Unlike the hemifusion observed with GPI-anchored influenza virus HA, GPI-anchored HIV Env did not give outer leaflet merging with susceptible CD4+ lymphocytes (39).
Qiao et al. (33) compared the stability of the hemifusion diaphragm in G1S HA and GPI-HA by calculating the percent full fusion after exposure to 0.4 mM CPZ. These authors defined the percent full fusion as the percentage of total lipid transfer events that also showed content dye transfer; thus, the more stable the intermediate, the less CPZ should destabilize it to give full fusion. Since both HA mutants showed 40% full fusion, Qiao et al. concluded that the stability of their hemifusion diaphragms was comparable (33). In our studies, full fusion occurred in 38% of G1S HA lipid transfer events, whereas 100% of H8R lipid transfer events represented full fusion (Fig. 3), suggesting that H8R hemifusion intermediates are less stable than G1S HA intermediates.
Histidine 8 appears to be involved in a common step of the membrane fusion mechanism of gammaretroviruses. A histidine in the sequence Ser/Asn-Pro-His-Gln is conserved near the N terminus of Env (8). The histidines at position 5 of the amphotropic MLV Env and position 8 of gibbon ape leukemia virus Env are critical for virus infection (13, 23; Z. Qian and L. M. Albritton, unpublished data). In addition, exposure to CPZ, or placing substitutions corresponding to the second-site mutations in Moloney MLV in the amphotropic Env, rescues infection by amphotropic MLV carrying a histidine 5-to-arginine change (Qian and Albritton, unpublished), suggesting that this change also arrests fusion at the hemifusion state. Moreover, the first 234 residues of the Moloney MLV binding subunit can be replaced with the corresponding region of amphotropic Env without loss of infection, although the receptor used for virus entry does change (8). Lastly, addition of the purified binding fragment from amphotropic MLV resulted in infection of Moloney MLV virus carrying a deletion of histidine 8 (H8del) but only if the cells displayed both the Moloney MLV and amphotropic virus receptors (7, 23).
How does the H8R substitution arrest fusion in the hemifusion state? Reducing the numbers of exposed fusion peptides induces a hemifusion phenotype (10). Thus, one possibility is that H8R does not prevent fusion peptide exposure but decreases or delays it. Alternatively, substitution of H8 may prevent or delay six-helix bundle formation, the step thought to catalyze fusion pore formation, by preventing dissociation of SU from TM. For example, dissociation of gp120 from HIV-1 particles was induced by incubation with a soluble form of CD4, the high-affinity receptor for HIV Env, suggesting that receptor interaction leads to SU dissociation (17, 30). However, HIV fusion requires a second interaction of gp120 with a coreceptor molecule (14), and cell-cell fusion is not inhibited by soluble CD4 (12). Thus, membrane fusion may involve dissociation of SU after fusion peptide exposure to remove a physical constraint to six-helix bundle formation (Fig. 6). We favor this possibility as the principal defect in H8R infection, while decreased or delayed fusion peptide exposure may be a less-significant contributing factor. We further propose that residue 8 is one of the key switch-points for the conformation changes that dissociate the SU-receptor complexes from TM whose fusion peptides have intercalated in the host cell membrane.
FIG. 6.
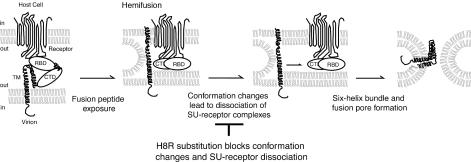
Schematic representation of a model for the block to membrane fusion in an H8R mutant. Receptor binding induces conformation changes that expose the fusion peptide on the N terminus of TM. Membrane fusion begins with local joining of the outer leaflets of juxtaposed membranes (22). As the outer leaflets merge, the inner leaflets remain separate, forming an intermediate called hemifusion that does not allow mixing of the contents of the fusing compartments. Additional conformation changes involve the conserved N-terminal histidine. These changes lead to dissociation of SU from TM, removing the physical constraint on six-helix bundle formation. Hairpin formation merges the inner membrane leaflets to complete fusion and open pores between the virion interior and the host cell cytoplasm. RBD, receptor-binding domain of SU; CTD, carboxy-terminal domain of SU.
Acknowledgments
We thank Alan Rein for providing a plasmid encoding the R-less form of Moloney MLV Env, Eric Hunter for pointing out the possibility that histidine 8 changes may block dissociation of SU, and Judy White for the wild-type and mutant HA plasmids, as well as for helpful discussion on their use.
This study was supported by grant AI33410 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health to L.M.A.
REFERENCES
- 1.Adams, R. M., M. Wang, D. Steffen, and F. D. Ledley. 1995. Infection by retroviral vectors outside of their host range in the presence of replication-defective adenovirus. J. Virol. 69:1887-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, R. T., A. S. Kushnir, and J. M. White. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151:425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 4.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagai, S., and R. A. Lamb. 1997. A glycine to alanine substitution in the paramyxovirus SV5 fusion peptide increases the initial rate of fusion. Virology 238:283-290. [DOI] [PubMed] [Google Scholar]
- 6.Bagai, S., and R. A. Lamb. 1996. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J. Cell Biol. 135:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 262:13614-13619. (Erratum, 263: 588, 1988.) [PubMed] [Google Scholar]
- 10.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrov, D. S., K. Hillman, J. Manischewitz, R. Blumenthal, and H. Golding. 1992. Correlation between kinetics of soluble CD4 interactions with HIV-1-Env-expressing cells and inhibition of syncytia formation: implications for mechanisms of cell fusion and therapy for AIDS. AIDS 6:249-256. [DOI] [PubMed] [Google Scholar]
- 13.Farrell, K. B., Y. T. Ting, and M. V. Eiden. 2002. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J. Virol. 76:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 15.Gething, M. J., R. W. Doms, D. York, and J. White. 1986. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J. Cell Biol. 102:11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert, J. M., D. Mason, and J. M. White. 1990. Fusion of Rous sarcoma virus with host cells does not require exposure to low pH. J. Virol. 64:5106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart, T. K., R. Kirsh, H. Ellens, R. W. Sweet, D. M. Lambert, S. R. J. Petteway, J. Leary, and P. J. Bugelski. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 88:2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornby, A. P., and P. R. Cullis. 1981. Influence of local and neutral anaesthetics on the polymorphic phase preferences of egg yolk phosphatidylethanolamine. Biochim. Biophys. Acta 647:285-292. [DOI] [PubMed] [Google Scholar]
- 19.Kamp, F., D. Zakim, F. Zhang, N. Noy, and J. A. Hamilton. 1995. Fatty acid flip-flop in phospholipid bilayers is extremely fast. Biochemistry 34:11928-11937. [DOI] [PubMed] [Google Scholar]
- 20.Kemble, G. W., D. L. Bodian, J. Rose, I. A. Wilson, and J. M. White. 1992. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 66:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemble, G. W., T. Danieli, and J. M. White. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76:383-391. [DOI] [PubMed] [Google Scholar]
- 22.Koslov, M. M., and V. S. Markin. 1983. Possible mechanism of membrane fusion. Biofizika 28:255-261. [PubMed] [Google Scholar]
- 23.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malhotra, S., A. G. Scott, T. Zavorotinskaya, and L. M. Albritton. 1996. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J. Virol. 70:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markosyan, R. M., F. S. Cohen, and G. B. Melikyan. 2000. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11:1143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melikyan, G. B., W. D. Niles, V. A. Ratinov, M. Karhanek, J. Zimmerberg, and F. S. Cohen. 1995. Comparison of transient and successful fusion pores connecting influenza hemagglutinin expressing cells to planar membranes. J. Gen. Physiol. 106:803-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 31.Ohki, S., T. D. Flanagan, and D. Hoekstra. 1998. Probe transfer with or without membrane fusion in a fluorescence fusion assay. Biochemistry 37:7496-7503. [DOI] [PubMed] [Google Scholar]
- 32.Puri, A., J. Winick, R. J. Lowy, D. Covell, O. Eidelman, A. Walter, and R. Blumenthal. 1988. Activation of vesicular stomatitis virus fusion with cells by pretreatment at low pH. J. Biol. Chem. 263:4749-4753. [PubMed] [Google Scholar]
- 33.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheetz, M. P., and S. J. Singer. 1974. Biological membranes as bilayer couples: a molecular mechanism of drug-erythrocyte interactions. Proc. Natl. Acad. Sci. USA 71:4457-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinangil, F., A. Loyter, and D. J. Volsky. 1988. Quantitative measurement of fusion between human immunodeficiency virus and cultured cells using membrane fluorescence dequenching. FEBS Lett. 239:88-92. [DOI] [PubMed] [Google Scholar]
- 38.Stegmann, T., P. Schoen, R. Bron, J. Wey, I. Bartoldus, A. Ortiz, J. Nieva, and J. Wilschut. 1993. Evaluation of viral membrane fusion assays. Comparison of the octadecylrhodamine dequenching assay with the pyrene excimer assay. Biochemistry 32:11330-11337. [DOI] [PubMed] [Google Scholar]
- 39.Weiss, C. D., S. W. Barnett, N. Cacalano, N. Killeen, D. R. Littman, and J. M. White. 1996. Studies of HIV-1 envelope glycoprotein-mediated fusion using a simple fluorescence assay. AIDS 10:241-246. [DOI] [PubMed] [Google Scholar]
- 40.Zarkik, S., F. Defrise-Quertain, D. Portetelle, A. Burny, and J. M. Ruysschaert. 1997. Fusion of bovine leukemia virus with target cells monitored by R18 fluorescence and PCR assays. J. Virol. 71:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zavorotinskaya, T., and L. M. Albritton. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J. Virol. 73:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerberg, J., R. Blumenthal, D. P. Sarkar, M. Curran, and S. J. Morris. 1994. Restricted movement of lipid and aqueous dyes through pores formed by influenza hemagglutinin during cell fusion. J. Cell Biol. 127:1885-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]