Abstract
Wild-type herpes simplex virus type 1 (HSV-1) induces apoptosis in human epithelial HEp-2 cells, but infected cell proteins produced later in infection block the process from killing the cells. Thus, HSV-1 infection in the presence of the translational inhibitor cycloheximide (CHX) results in apoptosis. Our specific goal was to gain insight as to the viral feature(s) responsible for triggering apoptosis during HSV-1 infection. We now report the following. (i) No viral protein synthesis or death factor processing was detected after infection with HSV-1(HFEMtsB7) at 39.5°C; this mutant virus does not inject its virion DNA into the nucleus at this nonpermissive temperature. (ii) No death factor processing or apoptotic morphological changes were detected following infection with UV-irradiated, replication-defective viruses possessing transcriptionally active incoming VP16. (iii) Addition of the transcriptional inhibitor actinomycin D prevented death factor processing upon infection with the apoptotic, ICP27-deletion virus HSV-1(vBSΔ27). (iv) Apoptotic morphologies and death factor processing were not observed following infection with HSV-1(d109), a green fluorescent protein-expressing recombinant virus possessing deletions of all five immediate-early (IE) (or α) genes. (v) Finally, complete death factor processing was observed upon infection with the VP16 transactivation domain-mutant HSV-1(V422) in the presence of CHX. Based on these findings, we conclude that (vi) the expression of HSV-1 α/IE genes is required for the viral induction of apoptosis and (vii) the transactivation activity of VP16 is not necessary for this induction.
Herpes simplex virus type 1 (HSV-1) is an important pathogen that causes a variety of clinical manifestations in humans. It has the ability to remain latent in host neurons for life and reactivate to cause lesions at or near the site of initial infection. Reactivation from the latent state results in productive infection that ultimately leads to the lytic destruction of distal epithelial cells. During the lytic cycle of the virus in cultured cells, regulation of HSV-1 replication occurs mainly at the transcriptional level and involves the coordination of three phases of gene expression (reviewed in reference 57). Viral genes are expressed in a tightly regulated, ordered cascade (27, 28), which begins with the production of the immediate-early (IE), or α, genes. The resulting α/IE proteins, which include infected cell proteins ICP0, ICP4, ICP22, and ICP27, are responsible for regulating viral gene expression during subsequent phases of the replication cycle (57). Transcription of the α genes occurs in the absence of de novo viral protein synthesis (8) and is highly stimulated by the α trans-inducing activity of virion VP16 (13, 50, 52). The early (β) gene products, for example, the viral thymidine kinase (TK), are synthesized next and are principally involved in viral DNA synthesis (reviewed in reference 12). The last set of genes expressed are the late (γ) genes, which encode proteins involved in virion structure and assembly, such as VP16, the virion host shutoff (vhs) protein, VP22, and glycoprotein C (gC) (reviewed in reference 17).
Productive HSV-1 replication induces major biochemical changes in infected cells that are collectively referred to as cytopathic effect (CPE). This cytopathology is characterized by (i) rounding up of cells caused by cytoskeletal destabilizations and loss of matrix binding proteins, (ii) nucleolar alterations and chromatin margination or damage, and (iii) an overall decrease in cellular macromolecular synthesis (5, 25, 26, 55, 57, 58). Although cytolysis due to viral replication is generally believed to occur through a necrotic route, recent studies have indicated that one aspect of HSV-1-induced CPE is programmed cell death, or apoptosis (1, 3, 19, 32, 35). Apoptosis is a highly regulated process of cell suicide that is induced by death signals received from either a cell surface death receptor or the mitochondria (22, 23, 47, 67). The process of apoptosis involves a family of aspartate-specific cysteinyl proteases, or caspases, which are activated by proteolytic cleavage. Regardless of the source from which a death signal originates, downstream “executioner” caspases, such as caspase-3, are common to both pathways (22, 59, 63, 68). This subclass of caspases is responsible for the processing of various cytoplasmic and nuclear substrates, such as the DNA repair enzyme poly(ADP-ribose) polymerase (PARP), a 116-kDa protein which generates an 85-kDa product upon processing (59). The cascade of events culminating in caspase activation ultimately leads to the morphological and biochemical features characteristic of apoptosis, including cell shrinkage, membrane blebbing, chromatin condensation, chromosomal DNA fragmentation, and apoptotic body formation (reviewed in reference 30).
Koyama et al. originally showed that wild-type HSV-1 infection could induce apoptosis in the absence of de novo protein synthesis (32). Studies from our laboratory (1, 4) and others (35) demonstrated that infection by replication-defective HSV-1 viruses that do not make late proteins results in apoptosis in infected human epithelial cells. Infected cell proteins synthesized between 3 and 6 h postinfection (hpi) (i.e., during the so-called apoptosis prevention window) are responsible for blocking apoptosis (3, 4). While the process of apoptosis during HSV-1 infection has now been clearly demonstrated (reviewed in reference 2), the specific mechanism(s) of its induction remain unknown. The induction of apoptosis by HSV-1 occurs early, and de novo protein synthesis is not required to induce the process (1, 3, 32).
The goal of this study was to define more precisely the features of viral replication which are involved in apoptosis induction during HSV-1 infection of human epithelial cells. We show that no viral protein synthesis or death factor processing was detected after infection with HSV-1(HFEMtsB7) at the nonpermissive temperature of 39.5°C. Following infection with UV-treated HSV-1 viruses shown to contain transcriptionally functional incoming VP16, no death factor processing was detected. No death factor processing was detected following infection with an apoptotic, ICP27 deletion virus in the presence of the transcription inhibitor actinomycin D. In addition, upon infection with HSV-1(d109), death factor processing and apoptotic morphologies were not observed. Finally, complete death factor processing was observed upon infection with the VP16 transactivation domain-mutant HSV-1(V422) in the presence of cycloheximide (CHX). After consideration of these findings, we conclude that the expression of HSV-1 α/IE genes is essential for the viral induction of apoptosis and the transactivation activity of VP16 is not required for this induction.
(F. N. W. Chirimuuta performed these studies in partial fulfillment of her “Sandwich Year” requirement at the University of the West of England, Bristol, United Kingdom.)
MATERIALS AND METHODS
Cells and viruses.
All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS). Human epithelial (HEp-2) and monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Manassas, Va.). Vero 2.2 is a derivative Vero cell line expressing ICP27 under its own promoter (62). FO6 is a derivative Vero cell line expressing ICP4, ICP27, and ICP0 under their own promoters (61). KOS1.1 is the strain of wild-type HSV-1 used in this study. vBSΔ27 (66) is the ICP27-null mutant virus used in this analysis; it contains a replacement of the α27 gene with the Escherichia coli lacZ gene and therefore must be propagated and titrated on an ICP27-complementing cell line, such as Vero 2.2 (62). Vero 2.2 cells, HSV-1(KOS1.1), and HSV-1(vBSΔ27) were generously provided by Saul Silverstein (Columbia University). HSV-1(HFEMtsB7), obtained from Bernard Roizman (University of Chicago), contains a temperature-sensitive mutation in the unique long 36 (UL36) gene (7, 31, 57). Viral DNA is not released from capsids in HSV-1(HFEMtsB7)-infected cells maintained at the nonpermissive temperature of 39.5°C (7). HSV-1(HFEM), generously provided by Patricia Spear (Northwestern University), is the wild-type strain used for comparison in the temperature sensitivity studies. HSV-1(d109) and complementing FO6 cells were generously provided by Neal DeLuca (University of Pittsburgh). HSV-1(d109), a mutant virus inactivated for all five α/IE genes, contains the green fluorescent protein (GFP) gene under the control of the human cytomegalovirus IE promoter in place of the deletion in ICP27 (60). HSV-1(V422) was generously provided by James R. Smiley (University of Alberta) and contains a truncation of the C-terminal transcriptional activation domain of VP16 at amino acid 422 (34). V422 was propagated and titrated on Vero cells in the presence of 3 mM hexamethylene bisacetamide (HMBA) (Sigma) as described previously (65). All viral titers were determined at 48 hpi by standard dilution techniques on Vero, Vero 2.2, or FO6 cells, and all values are the means of results of duplicate infections. Unless otherwise specified, cell monolayers were infected with multiplicities of infection (MOI) between 5 and 20 PFU per cell and the infections were maintained at 37°C in DMEM containing 5% newborn calf serum for the times indicated in the text. All tissue culture reagents were purchased from Life Technologies.
UV treatment of virus and CAT assays.
Previous preliminary studies from our laboratory indicated that 10 min of UV light exposure was sufficient to reproducibly decrease viral titers by greater than 5 logs compared to those of untreated controls (3). Further analyses have determined that this original treatment protocol is quite excessive. An optimized UV treatment method was established using HSV-1(KOS1.1), and this is now our recommended procedure. UV-treated HSV-1(KOS1.1) and HSV-1(vBSΔ27) stocks were made as follows. Virus stocks (109 PFU) in 2 ml of 199V medium (Life Technologies) containing 1% FBS were placed in a 10-mm-diameter sterile petri dish (Falcon) on ice at a standard (3) distance of 10 cm from a germicidal lamp (UVP Multiple-Ray 8-W UV lamp [60 Hz]; Fisher). The lamp was allowed to warm up for 5 min, and the amount of energy radiating from the lamp was then measured by a Fisherbrand traceable UV light radiometer (dosimeter) and recorded. The dosimeter readings were consistently between 3,300 and 3,600 ergs/s · cm2. Virus particles were exposed to UV light for various time intervals with mixing every 2 min. After removal of 100 μl for titer purposes, the UV-treated mixture was then divided into aliquots and stored at −80°C until use. Virus aliquot volumes were calculated based the amount of virus necessary for an MOI of 20 PFU/cell prior to UV treatment. Viral titers following UV treatment were determined on Vero cells for HSV-1(KOS1.1) or Vero 2.2 cells for HSV-1(vBSΔ27). A UV treatment time of 4 min was found to be sufficient to reproducibly decrease viral titers to <10 PFU/ml yet contain maximally functional VP16 as indicated by chloramphenicol acetyltransferase (CAT) assays.
pIGA53(CAT) is a promoterless control plasmid containing a CAT cassette, and pIGA65(α0 CAT) is a plasmid containing the HSV-1 α0/IE-1 complete promoter fused to a CAT cassette (20, 21). Both plasmids were generously provided by Irwin Gelman (Mount Sinai School of Medicine) and Saul Silverstein. HEp-2 cells were seeded at 8 × 105 cells/10-mm-diameter dish in DMEM containing 5% FBS and transfected 20 to 24 h later (50 to 60% confluence) with the DOTAP lipid transfection agent (Boehringer Mannheim) containing 2.5 μg of plasmid DNA in 20 mM HEPES buffer, pH 7.4. The DOTAP-containing medium was replaced with fresh DMEM plus 5% FBS 24 h posttransfection. At 48 h posttransfection, cells were infected with untreated or UV-treated HSV-1 at an MOI of 20 PFU/cell. At 6 hpi, cells were scraped into the medium, collected by low-speed centrifugation, and lysed in 0.1 ml of 0.25 M Tris-HCl, pH 7.5, by freezing and thawing them three times and then centrifuging them. Serially diluted supernatant samples (50 μl) were added to a mixture (final volume, 150 μl) containing 0.25 M Tris-HCl (pH 7.50), acetyl coenzyme A (70 mg/ml), and 0.05 μCi of [14C]-labeled chloramphenicol (NEN). Acetylated-14C-labeled chloramphenicol (Ac-Cm) forms were separated from unacetylated chloramphenicol on a silica gel thin-layer chromatography plate (Macherey Nagel) in an equilibrated mixture of chloroform and methanol (1:19, vol/vol) and exposed to Kodak autoradiographic film for 72 h. Signals on the thin-layer chromatography plates were digitized using a Molecular Dynamics Storm 860 PhosphorImager; intensities were calculated using ImageQuant (version 1.2) software, with analysis areas being held constant; and percent CAT activity was determined. Percent CAT activity was defined as the percent conversion of chloramphenicol to Ac-Cm forms: [Ac-Cm/(Ac-Cm + chloramphenicol)] × 100. To normalize for any (background) signals on the screen, densities obtained in the mock-transfected lane were subtracted from the densities at corresponding locations in all of the other lanes. All values were standardized to the amount of mock-infected α0 CAT activity and were expressed in terms of relative CAT activity. Each experiment was performed a minimum of two separate times with essentially identical results.
Maintenance of infected cells in the presence of drugs and at elevated temperatures. (i) Protein synthesis inhibition by CHX.
To inhibit de novo protein synthesis and, therefore, allow apoptosis in infected cells (1, 32), CHX (Sigma) was added to the medium of HEp-2 monolayers at a final concentration of 10 μg/ml. This concentration of CHX was previously shown to be sufficient to completely block viral protein synthesis in HSV-1-infected HEp-2 cells (1). Cells were pretreated with CHX for 1 h at 37°C prior to infection, and CHX was maintained in the medium until 24 hpi, at which time whole-cell extracts were prepared as described below.
(ii) Infection at 39.5°C.
Confluent HEp-2 cells (4 × 106) in 25-cm2 flasks were placed on ice in a 4°C cold room for 30 min and then exposed to 5 PFU of HSV-1(KOS1.1), HSV-1(HFEMtsB7), or HSV-1(vBSΔ27) per cell in 199V medium containing 1% FBS. After 1 h of adsorption at 4°C, the inoculum was replaced with cold (4°C) DMEM containing 5% newborn calf serum, and the flasks were immediately placed into either 34°C (permissive) or 39.5°C (nonper- missive) incubators with 5% CO2. Cells were harvested at 24 hpi, and whole-cell extracts were prepared.
(iii) Transcription inhibition by actinomycin D.
Actinomycin D (Sigma) was dissolved in dimethyl sulfoxide (DMSO) (Sigma) to make a 1-mg/ml stock solution for use in transcription inhibition studies. Actinomycin D was then added to the medium of HEp-2 monolayers at final concentrations of either 125 or 250 ng/ml. Cells were pretreated with either actinomycin D or equal volumes of DMSO (either 125 or 250 nl/ml) for 1 h at 37°C prior to infection with HSV-1(vBSΔ27), and both actinomycin D and DMSO were maintained in the medium until 12 or 15 hpi, at which time whole-cell extracts were prepared as described below.
Preparation of infected-cell extracts, denaturing gel electrophoresis, and transblotting.
Whole extracts of infected cells were obtained as follows. Cells were scraped into the medium and collected following low-speed centrifugation. After a single wash (3 ml) with phosphate-buffered saline containing protease inhibitors [0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM l-1-chlor-3-(4-tosylamido)-4-phenyl-2-butanone (TPCK), 0.01 mM l-1-chlor-3-(4-tosylamido)-7-amino-2-heptanon-hydrochloride (TLCK)], the infected cells were lysed in a solution containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 0.4% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM TPCK, and 0.01 mM TLCK and sonicated with a Branson Sonifier. The protein concentrations of all cell extracts were determined by a modified Bradford assay (Bio-Rad) as recommended by the vendor. Equal amounts of infected cell proteins (50 μg) were separated in denaturing N,N′-diallyltartardiamide-15% acrylamide gels (11) and electrically transferred to nitrocellulose membranes in a tank apparatus (Bio-Rad) prior to immunoblotting with various primary antibodies. Unless otherwise noted in the text, all biochemical reagents were obtained from Sigma. Nitrocellulose was obtained directly from Schleicher & Schuell. Prestained protein molecular weight markers were purchased from Life Technologies.
Quantitation of apoptotic cells with condensed chromatin.
The numbers of apoptotic cells were determined as previously described (4). Cells were stained with Hoechst dye, and the exact amount with condensed chromatin DNA and fragmented nuclei (1), as well as the total number of cells in representative fields, was determined using an Olympus model IX70 fluorescence microscope. The percentage of apoptotic cells was calculated as follows: (number of apoptotic cells/total number of cells) × 100.
Immunological reagents.
The following primary antibodies were used to detect viral proteins: (i) 1113, mouse anti-ICP27 monoclonal antibody (Goodwin Institute for Cancer Research, Plantation, Fla.); (ii) 1114, mouse anti-ICP4 monoclonal antibody (Goodwin); (iii) rabbit anti-TK polyclonal antibody (obtained from B. Roizman); (iv) 1104, mouse anti-gC monoclonal antibody (Goodwin); and (v) RGST49, rabbit polyclonal antibody directed against a glutathione S-transferase-VP22 fusion protein. Affinity-purified RGST49 antibody was generated as described previously (51). Immunoblotting experiments were performed to detect cellular apoptotic proteins by using mouse anti-caspase-3 monoclonal antibody (Transduction Laboratories Inc.) and mouse anti-PARP monoclonal antibody (Pharmingen). Secondary goat anti-rabbit and goat anti-mouse antibodies conjugated with alkaline phosphatase were purchased from Southern Biotechnology (Birmingham, Ala.).
Microscopy and computer graphics.
The phenotypes of the infected cells were documented by fluorescence and phase-contrast light microscopy. For analyses of chromatin condensation, cells were incubated at 23 hpi with the DNA dye Hoechst 33258 (Sigma) at a final concentration of 0.05 μg/ml for 15 min. Live cells were observed with an Olympus IX70/IX-FLA inverted fluorescent microscope, and images were acquired using a Sony DKC-5000 digital photo camera at a resolution of 600 to 800 dots per in. linked to a PowerMac G3 and processed through Adobe Photoshop (version 4.0). Immunoblots and autoradiographs were digitized (600 to 800 dots per in.) with an AGFA Arcus II scanner. Raw digital images were saved as TIFF image files with Adobe Photoshop (version 5.0) and organized into figures with Adobe Illustrator (version 7.0.1).
RESULTS
HSV-1(HFEMtsB7) does not induce apoptosis at its nonpermissive temperature.
The goal of this study was to test the hypothesis that incoming virion structural proteins might facilitate the apoptosis induction process. To this end, we performed three series of experiments using the previously characterized (7, 8, 31) temperature-sensitive mutant virus HSV-1(HFEMtsB7), termed tsB7. In tsB7-infected cells maintained at the nonpermissive temperature (39°C), capsids accumulate at nuclear pores and are unable to release the viral DNA core into nuclei (7). Our first set of experiments were carried out to confirm that tsB7 behaves as a wild-type virus at the standard temperature of 37°C and was compared to HSV-1(KOS1.1), termed KOS1.1, for its ability to induce apoptosis in the absence of viral protein synthesis. HEp-2 cells were mock infected or infected with 5 PFU of KOS1.1 or tsB7 per cell in the presence or absence of the protein synthesis inhibitor CHX. The rationale of this experiment is that, in the presence of CHX, viral and cellular survival factors are not produced and the cells die as a result of the triggering of apoptosis by the virus (2). In mock-infected, CHX-treated cells, low (background) levels of apoptosis are also observed (4, 19, 32). However, this is not due to active triggering of apoptosis by CHX. Even under ideal conditions, cells in culture will readily die within various time frames, depending on the nature of the line, if essential growth factors are withdrawn. Thus, cultured cells will undergo cell death simply in response to routine incubation if survival factors become depleted (e.g., due to hyperthermia) or are not continuously provided and/or produced. If CHX is then added to these cells, no new factors are generated to replace preexisting survival factors that become depleted during the experimental time period. This physiological background level of cell death, which usually occurs in less than 10% of the total cells (4, 19, 32, 35), should not be mistaken for that triggered by virus.
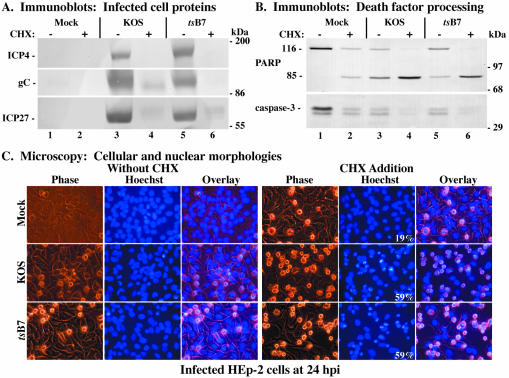
Whole-cell extracts were prepared at 24 hpi, and immunoblotting assays were performed using anti-ICP4, anti-gC, anti-ICP27, anti-PARP, and anti-caspase-3 antibodies. As stated earlier, the processing of PARP, a 116,000-molecular-weight protein, generates an 85,000-molecular-weight product which is detected by the anti-PARP antibody used in this study. Apoptosis-induced processing of caspase-3 (molecular weight of 32,000) results in loss of reactivity with our anti-caspase-3 antibody. Prior to the preparation of whole-cell extracts, cell morphologies were documented by phase-contrast and fluorescence microscopy as described in Materials and Methods. The results (Fig. 1) from this control experiment were as follows.
FIG. 1.
Immune reactivities of infected cell proteins (A) and death factors (B) and cellular and nuclear morphologies (C) in infected HEp-2 cells. Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(KOS1.1) or HSV-1(HFEMtsB7) in the absence (−) or presence (+) of CHX, separated in a denaturing gel, transferred to nitrocellulose, and probed with anti-ICP4, -gC, and -ICP27 (A) or -PARP and -caspase-3 (B) antibodies as described in Materials and Methods. Full-length (116,000-molecular-weight) (116) PARP and processed (85,000-molecular-weight) (85) PARP are shown. The locations of prestained molecular weight markers are indicated in the right margin. Phase-contrast, corresponding fluorescence (Hoechst), and merged (Overlay) images were obtained for mock-, HSV-1(KOS1.1)-, and HSV-1(HFEMtsB7)-infected HEp-2 cells in the absence (Without CHX) and presence (CHX Addition) of CHX; stained with Hoechst H33258 DNA dye; and visualized at 24 hpi as described in Materials and Methods. These cells were used to prepare the whole-cell extracts shown in panels A and B. White numbers in panels refer to the percentages of cells showing apoptotic condensed chromatin. Magnification, ×40.
In the absence of CHX, similar levels of the viral proteins ICP4, gC, and ICP27 were detected in cells infected with tsB7 or KOS1.1 (Fig. 1A, compare lane 3 with lane 5). In the KOS1.1-and tsB7-infected cells treated with CHX, little to no viral proteins were detected (Fig. 1A, lanes 4 and 6), as expected since CHX blocks their synthesis. Following KOS1.1, or tsB7 infection without CHX, minimal levels of caspase-3 and PARP processing were detected (Fig. 1B, lanes 3 and 5), as expected for wild-type HSV-1 strains (3). Interestingly, the level of PARP processing with tsB7 was equal to that of mock-infected cells, while KOS1.1 was above this level, as previously observed (3). It should be noted that the doublet observed with the anti-caspase-3 antibody is most likely a degradation product of caspase-3, as the lower band disappears concomitantly with caspase-3 activation. Treatment of mock-, KOS1.1-, or tsB7-infected cells with CHX resulted in (i) lower levels of caspase-3 and (ii) larger amounts of the processed 85,000-molecular-weight cleavage product of PARP than observed with untreated cells (Fig. 1B, compare lanes 2, 4, and 6 with lane 1), consistent with previously published results (3). This finding confirms previous observations that apoptosis occurs when infections with wild-type HSV-1 proceed in the absence of de novo protein synthesis (1, 3, 32). Thus, both KOS1.1- and tsB7-infected cells treated with CHX showed almost complete PARP and caspase-3 processing (Fig. 1B, lanes 4 and 6), suggesting that tsB7 is able to induce apoptosis when protein synthesis is inhibited similar to the way wild-type KOS1.1 is inhibited under standard conditions.
Phase-contrast and fluorescence microscopy were used to follow cell morphology changes during the above-described infections (Fig. 1C). Infected cell nuclei were observed at 24 hpi following staining with the Hoechst 33258 dye as described in Materials and Methods. Cells undergoing apoptosis show characteristic morphological changes, such as cell shrinkage, membrane blebbing, chromatin condensation, and nuclear fragmentation (30). In the absence of CHX, both KOS1.1 and tsB7 infection led to typical CPE phenotypes, as expected (1, 56), including a slightly rounded morphology, enlarged nuclei, and chromatin margination (Fig. 1C, without CHX). When protein synthesis was inhibited by the addition of CHX, 59% of KOS1.1- and tsB7-infected cells presented the previously described apoptotic morphologies (1, 3) of small size, irregular shape, condensed chromatin, and nuclear fragmentation. Even though the presence of CHX induced some apoptotic features in a few (19%) mock-infected cells, consistent with earlier findings (1), infection with KOS1.1 or tsB7 under the same conditions greatly increased the number of cells with apoptotic characteristics by a significant amount (Fig. 1C, CHX addition). These results confirm those shown in Fig. 1B and indicate that tsB7 at the standard temperature of 37°C exhibits wild-type HSV-1 behavior since tsB7 is able to induce apoptosis when viral protein synthesis is inhibited.
The biochemical and microscopic findings shown in Fig. 1 demonstrate that the tsB7 virus acted in a manner similar to that of the wild-type KOS1.1 virus during infection under standard conditions (37°C). Therefore, we initially used KOS1.1 as the control wild-type virus in the next set of experiments. These studies were designed to focus on the first steps of viral infections, i.e., virion binding, envelope fusion, and tegument and nucleocapsid entry into the cell, to assess their role in triggering apoptosis. Replicate cultures of HEp-2 cells were either mock infected or infected with 5 PFU of tsB7, KOS1.1, or vBSΔ27 virus per cell on ice as described in Materials and Methods. After the low-temperature adsorption step, one set of cultures was incubated at a permissive temperature (34°C), while the other set was incubated at the nonpermissive temperature (39.5°C). Following the preparation of whole-cell extracts at 24 hpi, immunoblotting analyses were performed using antibodies specific for the viral proteins ICP27, VP22 and gC, as well as for the cellular death factors PARP and caspase-3. The results from this experiment are presented in Fig. 2A.
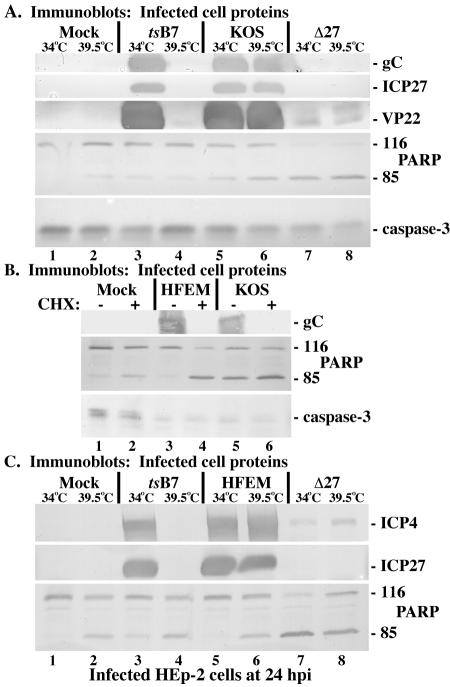
FIG. 2.
Immune reactivities of infected cell proteins and death factors in infected HEp-2 cells. (A) Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(HFEMtsB7), HSV-1(KOS1.1), or HSV-1(vBSΔ27) (Δ27) at the permissive (34°C) and nonpermissive (39.5°C) temperatures as described in Materials and Methods. Immunoblot analyses utilized anti-gC, -ICP27, -VP22, -PARP, and -caspase-3 antibodies. (B) Whole-cell extracts were prepared 24 hpi from mock-infected cells or cells infected with HSV-1(HFEM) or HSV-1(KOS1.1) in the absence (−) or presence (+) of CHX. Immunoblot analyses utilized anti-gC, -PARP, and -caspase-3 antibodies. (C) Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(HFEMtsB7), HSV-1(HFEM), or HSV-1(vBSΔ27) at the permissive (34°C) and nonpermissive (39.5°C) temperatures. Immunoblot analyses utilized anti-ICP4, -ICP27, or -PARP antibodies. 116 and 85, full-length and processed PARP, respectively.
Examination of infected-cell proteins showed high levels of gC, ICP27, and VP22 detected in the KOS1.1-infected cells, regardless of the incubation temperature (Fig. 2A, lanes 5 and 6). In vBSΔ27-infected cells at either temperature, no ICP27 was produced and no late protein synthesis occurred, as indicated by the absence of gC, which is as expected (4). Slight but detectable amounts of VP22 were observed in the vBSΔ27-infected cells, which likely reflects incoming tegument protein and serves as a convenient positive marker of infection. Cells infected with tsB7 synthesized gC, ICP27, and VP22 at 34°C at levels comparable to those in KOS1.1-infected cells (Fig. 2A, compare lane 3 with lanes 5 and 6), while no newly synthesized viral proteins were detected in tsB7-infected cells incubated at 39.5°C (lane 4). However, low but detectable levels of (presumably) incoming virion VP22 indicated that the tsB7 infection proceeded as expected (Fig. 2A, compare lane 4 with lanes 5 and 6).
In vBSΔ27-infected cells (Fig. 2A, lanes 7 and 8), we detected almost complete PARP cleavage and caspase-3 processing at both 34 and 39.5°C. No processed forms of these death factors were observed in mock-infected cells incubated at the permissive temperature (Fig. 2A, lane 1). However, in mock-infected cells at 39.5°C (Fig. 2A, lane 2), we detected slight levels of PARP and caspase-3 processing, most likely a consequence of incubation at the increased temperature (35). Low background levels of PARP and caspase-3 processing were seen in cells infected with both tsB7 and KOS at 34°C (Fig. 2A, lanes 3 and 5), consistent with our earlier observations concerning wild-type viruses (Fig. 1 and 3). In KOS1.1-infected cells incubated at 39.5°C, levels of PARP and caspase-3 cleavage were increased compared to levels in cells infected with KOS1.1 at the permissive temperature (Fig. 2A, compare lanes 5 and 6). This finding agrees with the observation that incubation at 39.5°C induces minor levels of apoptosis, independent of virus infection. In cells infected with tsB7 at the nonpermissive temperature, the amounts of PARP and caspase-3 processing did not surpass that of mock-infected cells under the same conditions (Fig. 2A, compare lanes 4 and 2). Taken together, the results from this second set of experiments confirm that viral proteins are not synthesized in tsB7-infected HEp-2 cells at 39.5°C, as the viral DNA is not released from nucleocapsids at this nonpermissive temperature (7) and, more importantly, indicate that the only apoptosis observed in tsB7-infected cells at 39.5°C is due to maintenance of the cells at this elevated temperature.
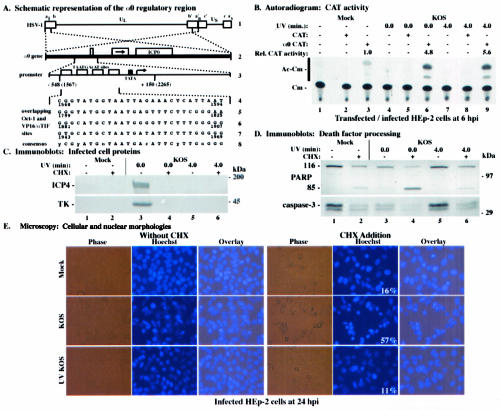
FIG. 3.
(A) Schematic diagram of the HSV-1 α0 promoter region contained in the α0 CAT plasmid. (Line 1) HSV-1 DNA genome, indicating the unique sequences (thin lines) flanked by the inverted repeats (boxes). The letters above line 1 designate the a sequence (al) and b sequence (b) of the long component, the unique sequence of the long component (UL), the repetitions of the b and of a variable (n) number of a sequences (an), the inverted c sequence, the unique sequence of the short component (US), and finally the terminal a sequence (as) of the short component (41, 42, 57). (Line 2) α0 gene, present in two copies (dotted lines) in the UL inverted repeats of the HSV-1 DNA (thick line). The three exons encoding the ICP0 protein are indicated by boxes, separated by introns (thin lines). The arrow in line 2 indicates the direction of protein translation. (Line 3) α0 promoter region (dotted lines). Open boxes indicate the four TAATGArAT sequences, which are the cis sites for the induction of α genes by αTIF (18, 38, 39). The cellular TATA-binding protein (TBP) binding site (TATA sequence) is designated by a filled box. The arrow indicates the transcription start site. Vertical lines below line 3 show the section of the α0 promoter region contained in the α0 CAT plasmid. Nucleotide positions are shown with reference to both the transcriptional start site and coordinates in the HSV-1 genome (in parentheses) (41, 42). (Lines 4 to 7) Nucleotide sequences of the overlapping Oct-1 and VP16/αTIF binding sites. The numbers below each line give the genome positions of each TAATGArAT sequence. (Line 8) Consensus TAATGArAT sequence. y is pyrimidine, r is purine, and n is any nucleotide. (B) Autoradiographic image depicting CAT activity in HEp-2 cells infected with HSV-1(KOS1.1). HEp-2 cells were transfected with 2.5 μg of either CAT or α0 CAT plasmid and infected 48 h posttransfection with untreated or UV-treated HSV-1(KOS1.1). Cytoplasmic extracts were prepared at 6 hpi and assayed for CAT activity as described in Materials and Methods. Relative CAT activity refers to the level of increase in CAT activity compared to that in mock-infected α0 CAT-transfected cells. Unmodified (Cm) and acetylated (Ac-Cm) forms of chloramphenicol are indicated on the left. (C and D) Immune reactivities of infected cell proteins (C) and death factors (D) in infected HEp-2 cells. Whole-cell extracts prepared at 24 hpi from mock-infected cells or cells infected with untreated or UV-treated HSV-1(KOS1.1) in the absence (−) or presence (+) of the protein synthesis inhibitor CHX (10 mg/ml) were separated in a denaturing gel, transferred to nitrocellulose, and probed with the anti-ICP4 or -TK (C) or -PARP or -caspase-3 (D) antibodies, as described in Materials and Methods. UV exposure times refer to untreated and UV-treated viruses, respectively. 116 and 85, full-length and processed PARP, respectively. The locations of prestained molecular weight markers are indicated in the right margins. (E) Phase-contrast, corresponding fluorescence (Hoechst), and merged (Overlay) visualizations of infected HEp-2 cells. HEp-2 cells at 24 h after mock infection or infection with untreated or UV-treated HSV-1(KOS1.1) in the absence (Without CHX) or presence (CHX Addition) of CHX were visualized as described in Materials and Methods. Numbers in panels (white) refer to the percentages of cells showing apoptotic condensed chromatin. Magnification, ×40. These cells were used to prepare the whole-cell extracts shown in panels C and D.
In the next part of this study (Fig. 2B and C), we expanded on the above results and used the parental strain HSV-1(HFEM), from which tsB7 is derived, for comparison. As above, to assess whether HFEM was able to induce apoptosis similar to that of KOS1.1 when protein synthesis is inhibited, HEp-2 cell monolayers were mock infected or infected with 5 PFU of either HFEM or KOS1.1 per cell in both the absence and presence of CHX. Immunoblot analyses using anti-gC, -PARP, and -caspase-3 antibodies were performed following whole-cell extract preparation at 24 hpi. Similar levels of the viral protein gC and comparable amounts of processed PARP and caspase-3 were detected in cells infected with HFEM or KOS1.1 in the absence of CHX (Fig. 2B, compare lanes 3 and 5). The background level of PARP cleavage observed with KOS is consistent with our earlier observations showing differences between KOS and HSV-1(F) (3). Interestingly, it now appears that both tsB7 (Fig. 2A) and HFEM do not have this level of background apoptosis and are more like HSV-1(F). A possible explanation for this is that KOS is a passaged laboratory strain which may have acquired mutations in viral preventers of apoptosis. In contrast, treatment of HFEM- or KOS1.1-infected cells with CHX resulted in (i) little or no viral proteins produced and (ii) almost complete PARP and caspase-3 processing (Fig. 2B, lanes 4 and 6), indicating that the tsB7 parental strain, HSV-1(HFEM), behaves like wild-type KOS1.1 in its ability to induce apoptosis in the absence of de novo protein synthesis.
Finally, replicate cultures of HEp-2 cells were either mock-infected or infected with 5 PFU of tsB7, HFEM, or vBSΔ27 virus per cell and incubated at either the permissive (34°C) or nonpermissive (39.5°C) temperature. Whole-cell extracts were prepared at 24 hpi and immunoblotting analyses were performed using antibodies specific for ICP4, ICP27, and PARP. High levels of ICP4 and ICP27 were detected in HFEM-infected cells, regardless of the incubation temperature (Fig. 2C, lanes 5 and 6). While cells infected with tsB7 synthesized these viral proteins at 34°C at levels comparable to HFEM (Fig. 2C, compare lane 3 with lanes 5 and 6), no newly synthesized ICP4 or ICP27 was detected in tsB7-infected cells incubated at 39.5°C (Fig. 2C, lane 4), consistent with previous data (Fig. 2A) (7). In vBSΔ27 virus-infected cells at either temperature, little ICP4 and no ICP27 were produced and almost complete PARP cleavage was detected (Fig. 2C, lanes 7 and 8). In contrast, cells infected with tsB7 at 39.5°C had levels of PARP processing comparable to mock-infected cells under the same conditions (Fig. 2C, compare lanes 4 and 2). This confirms the results in Fig. 2A. The implication is that the tsB7 virus is unable to induce apoptosis when its virion DNA does not enter host cell nuclei.
Based on these results, we conclude that tsB7 infection of HEp-2 cells at the nonpermissive temperature does not induce apoptosis. This finding is consistent with previously reported, but not shown, data (19). Under its nonpermissive conditions, tsB7 is unable to synthesize viral mRNA or proteins since viral DNA is not released from its capsids (7). Taken together, these data suggest that virion binding, envelope fusion, and nucleocapsid/tegument protein entry into the cell do not play significant roles in inducing apoptosis during HSV-1 infection.
UV-treated, replication-defective HSV-1 contains functional VP16 yet is unable to induce apoptosis.
The results from Fig. 1 and 2 suggest that incoming viral proteins are not responsible for the induction of apoptosis. However, since the integrities of each of these proteins were not directly measured, we could not eliminate the possibility that incubation of tsB7-infected cells at the nonpermissive temperature affects the function of other viral structural proteins in addition to the UL36 gene product. In a previous preliminary study from our laboratory (3), we showed that no apoptosis occurs upon infection of HEp-2 cells with UV-inactivated HSV-1, suggesting that virus binding and entry are not required for the induction process. In that study, the virion protein VP16 but not the viral IE protein ICP22 was detected in the nuclei of infected cells at 2 hpi. The absence of ICP22 confirmed that following UV treatment of HSV-1, no viral IE gene translation occurred.
However, at least two potential confounders exist in this preliminary experiment. First, during our later use of this technique, we realized that the UV treatment time we originally used (10 min) is extremely excessive and may generate unwanted damage. Second, while VP16 derived from UV-irradiated HSV-1 retained its ability to translocate to the nucleus, it was unknown whether the αTIF function of VP16 remained functional for the transactivation of viral IE genes. Thus, it was our desire to rigorously reevaluate our system. The description of the methodology we used to optimize our UV irradiation protocol is detailed in Materials and Methods. Our goal is to test that the representative incoming structural protein VP16 is active following the UV treatment of HSV-1 virions.
It was previously shown that reporter constructs containing IE (α) promoters linked to the CAT gene were induced following HSV-1 infection, suggesting that VP16 (Vmw65) acted to stimulate their expression (21, 49). We therefore selected one such construct, α0 CAT, for use in the following studies as an assay for VP16 transactivation activity. The exact promoter sequence contained in the α0 CAT construct is depicted in Fig. 3A. Subconfluent HEp-2 cells were transfected with the α0 CAT plasmid or the promoterless CAT plasmid and infected with, per cell, 20 PFU of untreated or UV-treated HSV-1(KOS1.1) at 48 h posttransfection. Cytoplasmic extracts of infected cells were then prepared at 6 hpi for use in a CAT assay as described in Materials and Methods. The results (Fig. 3B) were as follows.
In UV-treated KOS1.1-infected cells transfected with the α0 CAT plasmid (Fig. 3B, lane 9), we detected a 5.6-fold increase in CAT activity compared to that in mock-infected, α0 CAT-transfected cells (Fig. 3B, lane 3). This amount of CAT activity was comparable to the 4.8-fold increase observed in control KOS1.1-infected cells transfected with the α0 CAT construct (Fig. 3B, lane 6). As expected, no acetylated chloramphenicol forms were detected in cells transfected with the control promoterless CAT plasmid (Fig. 3B, lanes 2, 5, and 8). We observed a small amount of background CAT activity in mock-infected cells transfected with the α0 CAT construct (Fig. 3B, lane 3). This finding was not unexpected, as a basal level of CAT activity was detected in other cell types (21). These data confirm earlier findings with Vero cells (21, 49) and show that virion VP16 from wild-type HSV-1(KOS1.1) transcriptionally transactivates the α0/IE1 promoter contained in α0 CAT plasmid in HEp-2 cells. Accordingly, we conclude that incoming VP16 remains functional for the stimulation of viral α/IE genes following UV irradiation of the virus. These results suggest that the UV inactivation system we developed and optimized consistently yields virus particles containing functional VP16. By extension, we predict that other incoming tegument proteins retain their respective functions and are not rendered inactive by UV treatment.
The results presented in Fig. 3B demonstrated that UV-irradiated KOS1.1 contains VP16 that is able to transactivate the α0 promoter. This indicates that our UV-treated viruses are able to enter cells since their VP16 translocates to the nucleus, as previously reported (3). However, it was not known whether the KOS1.1 genome was rendered defective for replication and transcription by exposure to UV light. To determine whether UV-irradiated KOS1.1 containing functional VP16 is replication defective and whether this inactivated virus is able to induce apoptosis, four series of experiments were performed. In the first portion of the study, replication by UV-treated KOS1.1 virus was examined by preparing viruses which were exposed to UV light by our optimized system, as described in Materials and Methods. After these treatment conditions, we were able to reproducibly decrease UV-treated KOS1.1 titers to <10 PFU/ml (data not shown), indicating that our UV-irradiated virus is unable to replicate. Since UV treatment damages the viral DNA genome in the virion, it was also expected that viral transcription would be prevented and no IE gene products would be synthesized.
We next (Fig. 3C) examined the levels of viral proteins produced in infected cells. HEp-2 cells were mock infected and infected with, per cell, 10 PFU of either untreated or UV-treated KOS1.1 in the presence or absence of CHX. Whole-cell extracts were prepared at 24 hpi, and immunoblotting analyses were performed using anti-ICP4 and anti-TK antibodies as described in Materials and Methods. While high levels of ICP4 and TK were detected in untreated KOS1.1-infected cells in the absence of CHX (Fig. 3C, lane 3), these viral proteins were not detected in either KOS1.1-infected cells treated with CHX (lane 4), or in cells infected with UV-inactivated KOS1.1 either in the absence (Fig. 3C, lane 5) or presence (Fig. 3C, lane 6) of CHX. These findings indicate that UV treatment prevents the production of viral gene products. This is likely the consequence of the loss of transcription of viral DNA due to UV-induced damage to the DNA.
In the third part of this study (Fig. 3D), the processing of cellular death factors in the above-described (Fig. 3C) infected cells was analyzed. To assess the ability of UV-irradiated KOS1.1 viruses to induce apoptosis, the whole-cell extracts from the previous experiment were used in an immunoblotting assay with anti-PARP and anti-caspase-3 antibodies. When control infections were performed with untreated KOS1.1 (Fig. 3D, lane 3), a low level of caspase-3 and PARP processing was detected, as expected (3). As shown and discussed previously (Fig. 1B and 2B) (3), treatment of mock-infected cells with CHX resulted in low background levels of PARP and caspase-3 processing (Fig. 3D, lane 2). While KOS1.1 infection in the presence of CHX generated almost complete PARP and caspase-3 processing (Fig. 3D, lane 4), the amount of processed death factors from UV-treated KOS1.1-infected cells resembled that of mock-infected cells under the same conditions (Fig. 3D, compare lanes 5 and 6 with lanes 1 and 2). This result suggests that replication-defective UV-inactivated KOS1.1 is unable to induce apoptosis in human HEp-2 cells.
In our final set of experiments (Fig. 3E), we complemented the results described above (Fig. 3D) by examining cell morphology changes using phase-contrast and fluorescence microscopy. Prior to the preparation of the whole-cell extracts utilized above (Fig. 3C and D), cells were stained with Hoechst DNA dye and visualized at 24 hpi. Comparisons of the cell morphologies of untreated KOS1.1- or UV-treated KOS1.1-infected cells with and without CHX were made and quantitated as described in Materials and Methods. Without CHX treatment, fluorescence images of KOS1.1-infected cells showed typical CPE phenotypes, while the UV-inactivated KOS1.1-infected cells showed a morphology very similar to that of the confluent monolayer of flat cells observed in mock-infected cells (Fig. 3E, without CHX). In the presence of CHX (Fig. 3E, CHX addition), as many as 57% of KOS1.1-infected cells exhibited characteristic apoptotic features of chromatin condensation and nuclear fragmentation, compared to 16% of mock-infected cells (background due to the presence of the drug). In contrast, only 11% of cells infected with UV-treated KOS1.1 showed apoptotic morphologies (Fig. 3E, CHX addition). These results confirm those shown in Fig. 3D and indicate that UV-inactivated KOS1.1 virus has lost its ability to induce apoptosis in HEp-2 cells.
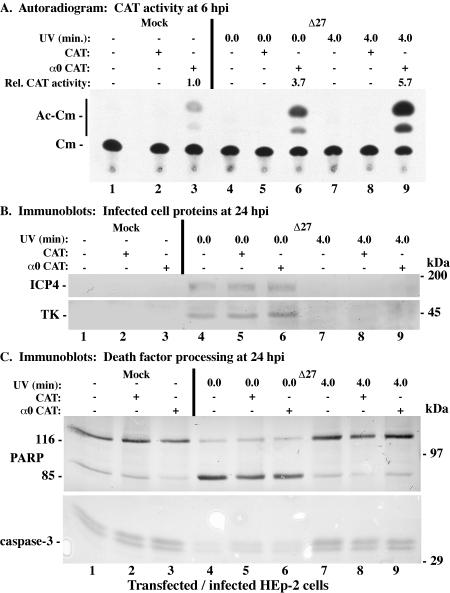
We previously reported that the ICP27-null recombinant HSV-1 strain vBSΔ27 (66), or the vBSΔ27 virus, induces apoptosis in infected HEp-2 cells similarly to wild-type KOS1.1, when total protein synthesis is inhibited during the KOS1.1 infection (1). The goal of this experiment was to examine whether the vBSΔ27 virus is still able to induce apoptosis following UV inactivation. Replicate HEp-2 cell monolayers were transfected with the promoterless CAT or α0 CAT plasmids and infected at 48 h posttransfection with, per cell, 20 PFU of untreated vBSΔ27 or UV-treated vBSΔ27 as described in Materials and Methods. One set of cultures was harvested at 6 hpi to make cytoplasmic extracts for CAT assays, while the second set was kept for 24 h before whole-cell extracts were prepared. Four techniques were used to characterize the UV-irradiated vBSΔ27 virus and assess its ability to induce apoptosis in infected HEp-2 cells and the results are shown in Fig. 4. First, a CAT assay was used to examine the transactivation activity of virion VP16. In UV-treated, vBSΔ27-infected cells transfected with α0 CAT, a 5.7-fold increase in CAT activity above that seen in mock-infected, α0 CAT-transfected cells was observed (Fig. 4A, compare lane 9 with lane 3). When cells transfected with the α0 CAT plasmid were infected with untreated vBSΔ27, a similar increase (3.7-fold) in CAT activity was seen (Fig. 4A, compare lane 6 with 3). In cells transfected with the promoterless CAT construct (lane 2, 5, and 8), no acetylated chloramphenicol forms were detected, as shown above (Fig. 3B). These results suggest that virion VP16 from the vBSΔ27 virus is functional for the transactivation of α promoters following UV irradiation.
FIG. 4.
Autoradiographic images (A) and immune reactivities (B and C) of HEp-2 cells infected with untreated or UV-treated HSV-1(vBSΔ27) (Δ27). HEp-2 cells were transfected with 2.5 μg of either CAT or α0 CAT plasmid and infected 48 h posttransfection with untreated or UV-treated HSV-1(vBSΔ27). (A) CAT activity from infected HEp-2 cell cytoplasmic extracts prepared at 6 hpi. Relative CAT activity refers to the increase (n-fold) in CAT activity compared to that of mock-infected α0 CAT-transfected cells. Unmodified and acetylated forms of chloramphenicol are indicated in the left margin. Whole-cell extracts prepared from infected HEp-2 cells at 24 hpi were separated in a denaturing gel, transferred to nitrocellulose, and probed with the anti-ICP4 and -TK (B) or -PARP and -caspase-3 (C) antibodies. UV exposure times refer to untreated and UV-treated viruses, respectively. The locations of prestained molecular mass markers are indicated in the right margins. 116 and 85, full-length and processed PARP, respectively.
We next assessed the ability of UV-treated vBSΔ27 to replicate in complementing cells in the second part of this study. UV-treated vBSΔ27 virus stocks were prepared as described in Materials and Methods, and their titers were reproducibly decreased to <10 PFU/ml compared to the >107-PFU/ml titers of the untreated samples (data not shown) when they were tested on Vero 2.2 cells. This finding implies that UV light rendered these viruses unable to replicate their viral DNA. To determine whether UV-irradiated vBSΔ27 is defective for transcription, whole-infected-HEp-2-cell extracts obtained 24 hpi were subjected to immunoblotting analyses using anti-ICP4 and anti-TK antibodies. Regardless of transfection status, cells infected with UV-treated vBSΔ27 (Fig. 4B, lanes 7 to 9) produced no measurable amounts of viral proteins, while ICP4 and TK were detected in untreated vBSΔ27-infected cells (Fig. 4B, lanes 4 to 6). These findings suggest that UV-treated vBSΔ27 is transcriptionally defective due to DNA damage induced by the UV light.
The final portion of this experiment involved analyzing the induction of apoptosis by UV-irradiated vBSΔ27, using anti-PARP and anti-caspase-3 antibodies to examine the levels of death factor processing. High levels of the PARP and caspase-3 processing were observed in cells infected with untreated vBSΔ27 (Fig. 4C, lanes 4 to 6), as expected (3). In contrast, the amounts of PARP and caspase-3 processing detected in cells infected with UV-inactivated vBSΔ27 were similar to the levels observed in mock-infected cells (Fig. 4C, compare lanes 7 to 9 with lanes 1 to 3). While minimal processing was detected in mock-infected cells (Fig. 4C, lanes 1 to 3), most likely the result of exposure to the transfection agent (see Materials and Methods), infection with UV-treated vBSΔ27 did not increase the extent of apoptosis by any measurable amount above that of mock-infected cells. These results demonstrate that UV-inactivated vBSΔ27 is incapable of inducing apoptosis in infected HEp-2 cells.
Based on the results presented in Fig. 3 and 4, we conclude the following. (i) Our UV-treated HSV-1 strains, inactivated by our optimized system, contain VP16 that is functional for transcriptional transactivation. These viruses are defective for replication, as indicated by significantly reduced viral titers and the absence of viral protein synthesis. (ii) The UV-inactivated HSV-1 viruses have lost their ability to induce apoptosis, as evidenced by the lack of cellular death factor processing and the failure of the cells to exhibit characteristic apoptotic morphologies. These results suggest that (iii) the processes of HSV-1 binding, membrane fusion, capsid translocation, and viral DNA injection into the nucleus and the virion VP16 protein, and presumably other viral structural proteins, are not required for the apoptotic induction process in infected human epithelial HEp-2 cells.
HSV-1(vBSΔ27) is unable to induce apoptosis in the presence of actinomycin D.
The results presented in Fig. 1 through 4 suggest two things concerning the induction of apoptosis during HSV-1 infection. First, the induction event occurs prior to the translation of viral proteins since apoptosis is detected in cells infected with wild-type HSV-1 when protein synthesis is inhibited (Fig. 1 to 3) (1, 3, 32). Second, the induction event occurs at some point following virus binding, membrane fusion, tegument dissociation, and nucleocapsid translocation to the nuclear pore, inasmuch as tsB7 infection at the nonpermissive temperature or UV-treated virus infection is unable to induce apoptosis. At this point, it was our proposal that viral α/IE gene expression is required for the induction of apoptosis during HSV-1 infection. To test this hypothesis, we made use of the transcription inhibitor actinomycin D. Because this compound is known to be toxic to cells (10), it was our desire to first find a concentration of actinomycin D which induced the least possible levels of death factor processing yet inhibited viral protein synthesis in infected cells.
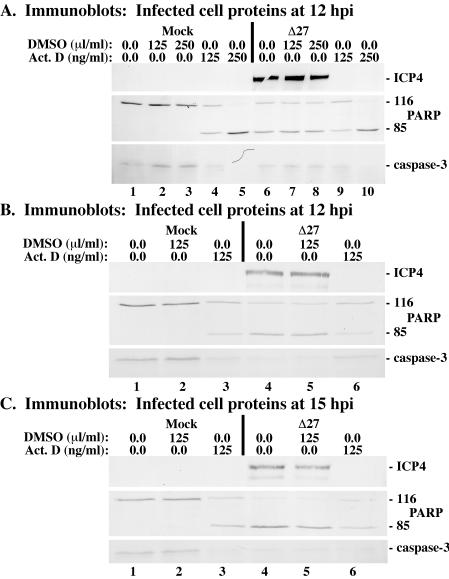
Since previous studies from our laboratory indicated that significant PARP and caspase-3 processing could be observed in HEp-2 cells as early as 12 h following infection with HSV-1(vBSΔ27) (3), we initially selected this time point for analysis. In the first portion of this study, HEp-2 cells were either mock-infected or infected with 5 PFU of vBSΔ27 per cell in the presence or absence of actinomycin D at either 125 or 250 ng/ml as described in Materials and Methods. Whole-cell extracts were prepared at 12 hpi, and immunoblot analyses using anti-ICP4, -PARP, and -caspase-3 antibodies were performed. The results of two independent experiments (Fig. 5A and B) were as follows.
FIG. 5.
Immune reactivities of infected cell proteins and death factors at 12 hpi (A and B) or 15 hpi (C) from vBSΔ27-infected HEp-2 cells plus actinomycin D. Whole-cell extracts were prepared from mock-infected cells or cells infected with HSV-1(vBSΔ27) (Δ27) in the absence or presence of the transcription inhibitor actinomycin D or DMSO solvent as described in Materials and Methods. Immunoblot analyses utilized anti-ICP4, -PARP, and -caspase-3 antibodies.
High levels of ICP4 were observed in both untreated and DMSO-treated cells following infection with vBSΔ27 (Fig. 5A, lanes 6 to 8; Fig. 5B, lanes 4 and 5). However, no ICP4 was detected in vBSΔ27-infected cells in the presence of either concentration of actinomycin D (Fig. 5A, lanes 9 and 10, and B, lane 6). While high levels of PARP and caspase-3 processing were observed in cells infected with vBSΔ27 in the absence of actinomycin D (Fig. 5A, lanes 6 to 8, and B, lanes 4 and 5), the addition of DMSO to either mock- or vBSΔ27-infected cells did not increase the amounts of death factor processing by any measurable amount above that of untreated cells (Fig. 5A, compare lanes 2 and 3 with lane 1 and compare lanes 7 and 8 with lane 6; Fig. 5B, compare lane 2 with lane 1 and lane 5 with lane 4). We also observed that, regardless of infection status, treatment of cells with actinomycin D (250 ng/ml) resulted in complete death factor processing (Fig. 5A, lanes 5 and 10). In contrast, the levels of PARP and caspase-3 processing observed in vBSΔ27-infected cells in the presence of actinomycin D (125 ng/ml) (i) did not surpass those seen in mock-infected cells under the same conditions (Fig. 5A, compare lane 9 with lane 4; Fig. 5B, compare lane 6 with lane 3) and (ii) were decreased in comparison to levels in vBSΔ27-infected cells in the absence of actinomycin D treatment (Fig. 5A, compare lane 9 with lane 6, 7, or 8; Fig. 5B, compare lane 6 with lane 4 or 5). These findings indicate that vBSΔ27-infected cells treated with actinomycin D (125 ng/ml) do not synthesize detectable levels of viral proteins and induce lower levels of apoptosis than untreated, vBSΔ27-infected cells.
In the final part of this study, we expanded on the above results (Fig. 5A and B) by examining viral protein synthesis and death factor processing at a later time point. HEp-2 cells were pretreated with either actinomycin D (125 ng/ml) or an equal concentration of DMSO (125 nl/ml) and then either mock infected or infected with 5 PFU of vBSΔ27 per cell. Immunoblot analyses were performed using antibodies against ICP4, PARP, and caspase-3 following the preparation of whole-cell extracts at 15 hpi. The results (Fig. 5C) confirmed those seen in Fig. 5A and B. Cells infected with vBSΔ27 synthesized ICP4 and showed complete death factor processing in the absence of actinomycin D treatment (Fig. 5C, lanes 4 and 5). However, in vBSΔ27-infected cells in the presence of actinomycin D, ICP4 was not detected and the levels of PARP and caspase-3 processing observed were at background levels, noticeably less than that seen in untreated, vBSΔ27-infected cells (Fig. 5C, compare lane 6 with lanes 3 to 5).
Together, these results (Fig. 5) show that the addition of actinomycin D at 125 ng/ml sufficiently inhibits the production of viral proteins and blocks the ability of vBSΔ27 to induce apoptosis. Although the presence of actinomycin D induced minor levels of apoptosis in uninfected cells, infection with vBSΔ27 did not increase the extent of death factor processing by any measurable amount. Even though actinomycin D is difficult to work with and it was necessary for us to perform titration experiments, it is clear that apoptosis is not so extensive in vBSΔ27-infected cells when actinomycin D is present. Thus, we conclude that HSV-1 α/IE viral transcription is required for the induction of apoptosis in HEp-2 cells.
HSV-1(d109) is unable to induce apoptosis.
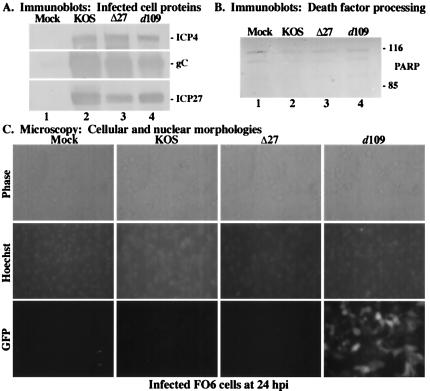
The results above (Fig. 1 to 5) suggest that α/IE transcription is required to trigger the induction process. However, since we did not directly measure the RNA levels in the presence of actinomycin D, we could not eliminate the possibility that some, perhaps abortive, α/IE transcription occurs following actinomycin D treatment. The goal of this study was to test the hypothesis that α/IE gene expression was essential for the induction of apoptosis in the context of viral infection. We made use of the recombinant virus HSV-1(d109), a GFP-expressing virus which contains deletions in all five α/IE genes and has been previously shown to be nontoxic to Vero and HEL cells (60). In our first set of experiments, we examined the phenotypes of infected FO6 cells, the d109-complementing cell line which expresses ICP4, ICP0, and ICP27. FO6 cell monolayers were infected with either KOS1.1, vBSΔ27, or d109 for 24 h, and both immunoblot and microscopy analyses were performed as described in Materials and Methods. The results (Fig. 6) are as follows.
FIG. 6.
Immune reactivities of infected cell proteins (A) and death factors (B) and cell morphologies (C) of infected FO6 cells. Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(KOS1.1), HSV-1(vBSΔ27) (Δ27), or HSV-1(d109) as described in Materials and Methods. Immunoblot analyses utilized anti-ICP4, -gC, and -ICP27 antibodies (A) or -PARP antibodies (B). 116 and 85, full-length and processed PARP, respectively. (C) Phase-contrast and fluorescence (Hoechst and GFP) images of corresponding infected FO6 cells. Mock-, HSV-1(KOS1.1)-, HSV-1(vBSΔ27)-, and HSV-1(d109)-infected FO6 cells were visualized at 24 hpi as described in Materials and Methods. Magnification, ×40. These cells were used to prepare the whole-cell extracts shown in panels A and B.
KOS1.1-infected cells synthesized high levels of ICP4, ICP27, and gC (Fig. 6A, lane 2). In addition, cells infected with either vBSΔ27 or d109 produced amounts of viral proteins similar to those produced by wild-type KOS1.1, as expected since their mutations are effectually complemented in FO6 cells (Fig. 6A, lanes 3 and 4). Upon examination of death factor processing, no PARP cleavage product was detected following infection with any virus (Fig. 6B, lanes 2 to 4), indicating that FO6 cells do not undergo apoptosis following infection with KOS1.1, vBSΔ27, or d109. Before cells in the previous experiment (Fig. 6A and B) were harvested for immunoblotting assays, we used phase-contrast and fluorescence microscopy to compare cellular morphology changes as described in Materials in Methods. As d109 contains a GFP reporter cassette, we were able to observe the delivery of the GFP gene to infected cells and this provided a convenient positive control for infection. Cells infected with KOS1.1, vBSΔ27, or d109 all exhibited characteristic features of HSV-1-induced CPEs, including a rounded morphology and marginated chromatin, while only d109-infected cells showed expression of the GFP transgene (Fig. 6C). These data confirm the results shown in Fig. 6A and B, demonstrating that when infection is carried out in complementing cells, d109 acts in a manner similar to that of wild-type KOS1.1, as indicated by high levels of viral proteins synthesis, the absence of death factor processing, and the manifestations of typical HSV-1 CPEs.
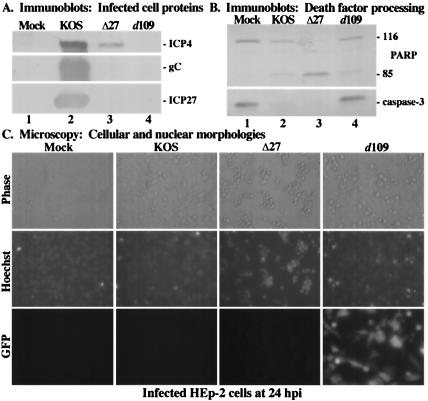
We were then interested in determining whether d109 was able to induce apoptosis in HEp-2 cells. Following 24 h of infection of HEp-2 cells with KOS1.1, vBSΔ27, or d109, immunoblot and microscopy analyses were performed (Fig. 7). High levels of viral proteins were detected in cells infected with KOS1.1 (Fig. 7A, lane 2). While vBSΔ27-infected cells synthesized minimal amounts of ICP4, as expected at this time point (1), we did not observe production of the late protein gC (Fig. 7A, lane 3). In contrast, we detected no viral proteins following d109 infection (Fig. 7A, lane 4), consistent with previously published results (60). When the levels of PARP and caspase-3 processing were examined, we observed that, as shown previously (Fig. 1 to 3) (3), cells infected with KOS1.1 showed larger amounts of PARP and caspase-3 cleavage than that of mock-infected cells (Fig. 7B, compare lanes 1 and 2). Additionally, complete death factor processing was detected in vBSΔ27-infected cells, as expected. However, in cells infected with d109, the amounts of PARP and caspase-3 processing were comparable to that seen in mock-infected cells (Fig. 7B, compare lanes 4 and 1), indicating that d109 is unable to induce apoptosis in HEp-2 cells. We also performed an important control to eliminate the unlikely possibility that the effects of CHX and virion components might have some sort of an additive effect on the extent of apoptosis. We repeated the above-described experiment in the presence of CHX and observed that the level of PARP cleavage in d109-infected HEp-2 cells was the same as that of mock-infected cells plus CHX (data not shown). These results indicate that the induction of apoptosis by HSV-1 is independent of CHX treatment and that virion factors do not augment any effects of CHX.
FIG. 7.
Immune reactivities of infected cell proteins (A) and death factors (B) and cell morphologies (C) of infected HEp-2 cells. Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(KOS1.1), HSV-1(vBSΔ27) (Δ27), or HSV-1(d109) as described in Materials and Methods. Immunoblot analyses utilized anti-ICP4, -gC, and -ICP27 antibodies (A) or -PARP and -caspase-3 antibodies (B). 116 and 85, full-length and processed PARP, respectively. (C) Phase-contrast and fluorescence (Hoechst and GFP) images of corresponding infected HEp-2 cells. Mock-, HSV-1(KOS1.1)-, HSV-1(vBSΔ27)-, and HSV-1(d109)-infected HEp-2 cells were visualized at 24 hpi as described in Materials and Methods. Magnification, ×40. These cells were used to prepare the whole-cell extracts shown in panels A and B.
We also observed the infected cell morphologies by phase-contrast and fluorescence microscopy in order to complement the above immunoblotting results (Fig. 7C). While control KOS1.1-infected cells showed expected CPE characteristics, cells infected with vBSΔ27 displayed the characteristic apoptotic features, consistent with earlier findings (1). d109-infected cells exhibited little to no condensed chromatin as visualized by Hoechst staining. Again, the GFP reporter cassette contained in d109 provided a convenient positive control for infection. Additionally, typical apoptotic hallmarks, such as cell shrinkage and membrane blebbing, were not observed upon examination of d109-infected cells by phase-contrast microscopy. Interestingly, we did detect distinct cellular morphology alterations following infection with d109, particularly cell enlargement and rounding, which are not characteristic of apoptosis. It appears unlikely that this unique morphology in the d109-infected cells is due to the presence of GFP since GFP is detected in both flat and rounded cells. Nonetheless, these microscopic images confirm the results from the above-described immunoblot analyses (Fig. 7B), indicating that d109 does not induce apoptosis in HEp-2 cells due to the absence of α/IE gene expression.
HSV-1(V422) is able to induce apoptosis when protein synthesis is inhibited.
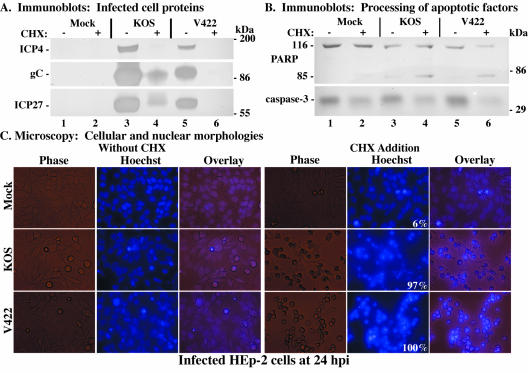
The data shown in Fig. 5 and 7 illustrate that viral α/IE gene expression is required for the induction of apoptosis in human epithelial cells. Although we previously demonstrated that virion VP16 did not play a direct role in the induction process using the UV-treated viruses (Fig. 3 and 4), it was unknown whether sufficient levels of α/IE gene expression for induction would be achieved in the absence of VP16 transcriptional transactivation. Thus, the goal of this study was to determine whether the transactivating activity of VP16 is absolutely required for the induction process to occur in infected HEp-2 cells. It was previously shown (34) that cells infected with HSV-1(V422), an HSV-1(KOS) derivative in which the acidic activation domain of VP16 is truncated following codon 422, exhibit an overall reduction in viral protein synthesis. While this virus is able to replicate on noncomplementing cells, albeit with an approximately 10- to 100-fold reduction in viral titers (34), the defect of this VP16 mutant can be largely overcome by the addition of HMBA to the culture medium (40, 65). Thus, high-titer virus stocks of HSV-1(V422), termed V422, were prepared in the presence of HMBA as described in Materials and Methods. As it was our desire to examine the effect of this VP16 mutation on the induction of apoptosis, HMBA was not included in this subsequent study since HMBA efficiently compensates for the VP16 transactivation domain mutation. HEp-2 cells were mock infected or infected with 10 PFU of KOS1.1 or V422 per cell in both the presence and the absence of CHX. At 24 hpi, immunoblotting experiments were performed using anti-ICP4, anti-gC, anti-ICP27, anti-PARP, and anti-caspase-3 antibodies, and infected cell morphologies were documented using fluorescence and phase-contrast microscopy.
In the first portion of the study (Fig. 8A), the levels of viral protein accumulation in infected cells were analyzed. As shown previously (Fig. 1 to 3), KOS1.1-infected cells in the absence of CHX (Fig. 8A, lane 3) synthesized large amounts of ICP4 and overwhelming amounts of gC and ICP27. In contrast, reduced expression levels of these proteins were seen in cells infected with V422 under the same conditions (Fig. 8A, lane 5), consistent with earlier findings (34). In the V422-infected cells treated with CHX (Fig. 8A, lane 6), ICP4, gC, and ICP27 were not detected, as expected since CHX blocks their synthesis. Upon CHX treatment of KOS1.1-infected cells (lane 4), no ICP4 and minimal amounts of ICP27 and gC were observed. Interestingly, KOS1.1-infected cells treated with CHX (Fig. 8A, lane 4), showed at least three different slow-migrating proteins that reacted with the anti-ICP27 antibody. It should be noted that these ICP27 moieties were also observed in Fig. 1A, lanes 4 and 6, as well as during our earlier studies (M. Aubert and J. A. Blaho, unpublished observations). The origin of these factors is unknown and is currently under investigation. Together, these results confirm previous findings that cells infected with V422 express reduced levels of viral proteins.
FIG. 8.
Immune reactivities of infected cell proteins (A) and death factors (B) and cell morphologies (C) of infected HEp-2 cells. Whole-cell extracts were prepared at 24 hpi from mock-infected cells or cells infected with HSV-1(KOS1.1) or HSV-1(V422) in the absence (−) or presence (+) of CHX as described in Materials and Methods. Immunoblot analyses utilized anti-ICP4, -gC, and -ICP27 antibodies (A) or -PARP and -caspase-3 antibodies (B). The locations of prestained molecular mass markers are indicated in the right margins. 116 and 85, full-length and processed PARP, respectively. (C) Phase-contrast, fluorescence (Hoechst), and merged (Overlay) images of corresponding infected HEp-2 cells. Mock-, HSV-1(KOS1.1)-, and HSV-1(V422)-infected HEp-2 cells in the absence (Without CHX) and presence (CHX Addition) of CHX were visualized at 24 hpi as described in Materials and Methods. Numbers in panels (white) refer to the percentage of cells showing apoptotic condensed chromatin. Magnification, ×40. These cells were used to prepare the whole-cell extracts shown in panels A and B.
In the second part of this experiment (Fig. 8B), we used an immunoblotting assay to monitor the processing of cellular death factors in V422-infected cells. A low level of PARP and caspase-3 processing was observed in control KOS1.1-infected cells in the absence of CHX (Fig. 8B, lane 3), as expected. Further processing of these proteins was detected in cells infected with KOS1.1 upon CHX treatment (Fig. 8B, lane 4). Although PARP cleavage and caspase-3 processing were not complete, the levels of these processed proteins were distinctly higher than those observed in both mock-infected cells with CHX treatment and KOS1.1-infected cells in the absence of CHX (Fig. 8B, compare lane 4 with lanes 2 and 3). In cells infected with V422 (Fig. 8B, lane 5), we observed levels of PARP and caspase-3 processing which were slightly lower than those seen in KOS1.1-infected cells (Fig. 8B, compare lanes 5 and 3). This slight variation likely reflects the replication efficiency differences between the two viruses, as mentioned above. However, when cells were infected with the V422 virus in the presence of CHX, more PARP and caspase-3 processing was observed than with either mock-infected cells plus CHX or cells infected with V422 in the absence of the inhibitor (Fig. 8B, compare lane 6 with lanes 2 and 5). These results suggest that the apoptotic induction process does not require the transactivation function of VP16.
In the last part of this study, fluorescence and phase-contrast microscopy were used to observe cell morphologies following infection with the V422 virus. HEp-2 cells were stained with the Hoechst 33258 dye and visualized at 24 hpi as described in Materials and Methods, prior to the preparation of cell extracts (Fig. 8A and B). The results (Fig. 8C) were as follows. In the absence of CHX, nearly all of the KOS1.1- and V422-infected cells were slightly rounded, possessed enlarged nuclei and marginated chromatin, and had few apoptotic features (Fig. 8C, without CHX). It should be noted that the CPE phenotype we observed in cells infected with V422 was not as pronounced as that seen in wild-type KOS1.1-infected cells. When protein synthesis was inhibited by the addition of CHX, KOS1.1-infected cells (97%) presented characteristic apoptotic morphologies, including smaller sizes and condensed chromatin (Fig. 8C, CHX addition). In the presence of CHX, the number of V422-infected cells exhibiting apoptotic features reached 100%. These results confirm those obtained for Fig. 8B and support the theory that the transactivation domain of VP16 is not necessary for the induction of apoptosis.
Based on the results shown in Fig. 8, we conclude that HEp-2 cells infected with the VP16 transactivation domain-mutant virus HSV-1(V422) synthesize reduced levels of viral proteins in infected cells, consistent with previous observations using other cell types (34). We also conclude that V422 is able to induce apoptosis when viral protein synthesis is inhibited. Taken together, these results suggest that while HSV-1 IE gene expression is a necessary step in the induction of apoptosis, the transactivation function of VP16 is not required for its induction.
DISCUSSION
Human HEp-2 cells infected with either (i) viruses containing a deletion in the key viral regulator ICP4 or ICP27 or (ii) wild-type virus with the addition of CHX at the time of infection die by apoptosis (1, 32, 35). While most recent efforts have focused on identifying viral gene products involved in apoptosis prevention during infection (1, 3, 4, 9, 24, 29, 35, 36, 45, 46, 69-72), little is known of the role that incoming viral components play in the induction process. In this study, we used (i) UV-inactivated, (ii) temperature-sensitive mutant, and (iii) recombinant mutant viruses as tools to investigate the process of the induction of apoptosis during HSV-1 infection. The significant findings of our study can be summarized as follows.
(i) Virion binding, membrane fusion, tegument dissociation, nucleocapsid translocation, and genome uncoating are not sufficient to induce apoptosis in HSV-1-infected human HEp-2 cells.
This conclusion is based on our findings that no death factor processing or apoptotic cell morphologies were observed following HEp-2 cell infection with either UV-inactivated viruses or the tsB7 DNA injection-mutant virus at the nonpermissive temperature. In their original report, Koyama and Adachi proposed that a structural protein of the HSV-1 may be responsible for the induction of apoptosis in cells treated with CHX (32). Since virion VP16 is functional for transactivation following UV irradiation (discussed further below), other tegument proteins presumably remained structurally intact and functional following the UV treatment. The possibility that incoming virion components contribute to apoptosis induction has been a major focus of our study. Our data using UV-irradiated wild-type virus in the presence of CHX showing no apoptosis induction argue strongly that the effects of virion functions and CHX do not have an additive effect on the extent of apoptosis. The similar CAT activities observed in cells infected with both untreated and UV-treated HSV-1 are an indication that UV exposure did not perturb the ability of the virus to bind and enter cells. In both the UV irradiation and tsB7 studies, viral gene expression did not ensue and no apoptosis was observed. Thus, the activities of viral glycoproteins and proteins packaged in virions and introduced into the cell during infection can be excluded as initiating factors in the induction event.
Additionally, the UV inactivation of viruses was evidenced by an absence of viral protein synthesis and an dramatic reduction in viral titers. Since the actual consequences of UV irradiation on the virion particle were unknown at the onset of our study, various UV exposure intervals were assessed and the respective amounts of radiating energy were carefully recorded in an attempt to develop an optimized system. A UV treatment time of 4 min yielded maximal inactivation with our equipment and experimental conditions. Using this optimized UV inactivation system, it should be possible to investigate further the damaging effects that UV irradiation might have on virion structure and the integrities of various structural proteins. Incoming VP16 from UV-irradiated HSV-1 stimulated the α0 promoter-driven transcription of a CAT reporter gene at levels comparable to that of untreated virus. An inverse correlation existed between the relative amount of CAT activity and the time period of UV exposure (data not shown), suggesting that excessive uncontrolled irradiation might indeed damage virion particles. UV-irradiated virus-infected cells yielded slightly, but reproducibly, larger amounts of CAT activity than that observed with the unexposed virus. Two possible explanations exist for this observation. The first is that at 6 hpi, virion-derived VP16 from untreated virus is prevented from binding TAATGARAT sequences due to some form of negative regulation mediated by viral proteins from later kinetic classes, while VP16 from UV-irradiated HSV-1 continues to stimulate transcription from α promoters. Previous results from our laboratory showed that newly synthesized VP16 can be detected in infected HEp-2 cells as early as 6 hpi (1, 3). However, this newly synthesized VP16 and vhs form a complex during infection that prevents VP16 from binding the TAATGARAT sequence and transactivating α promoters (64). The second, and most likely, explanation is that UV-inactivated viruses have lost negative regulation of α promoters mediated by members of the α/IE protein class (14, 20, 49). Consistent with this view is our observation that UV-inactivated viruses do not produce de novo-synthesized viral proteins.
(ii) The transactivating activity of VP16 is not required for the induction of apoptosis in HSV-1-infected human cells.
UV-inactivated HSV-1 with functional VP16 does not induce apoptosis, indicating that the virion-derived VP16 protein does not trigger apoptosis. However, the present data are insufficient to ascertain whether residual transcripts or truncated or partial viral transcripts are produced by the UV-treated viruses. That the V422 virus in the presence of CHX induced amounts of death factor processing similar to those produced by wild-type KOS1.1 under the same conditions suggests that in the context of an intact and replication-competent viral genome, the transcriptional transactivating ability of VP16 is not required for the induction of apoptosis to occur. One of our more puzzling findings was that at least three distinct factors that react with an anti-ICP27 antibody were observed when certain infections were carried out in the presence of CHX. Since these species were consistently observed (data not shown), additional studies focusing on the compositions and origins of these entities should help clarify this point.
(iii) HSV-1 α/IE gene expression is required for the induction of apoptosis in HEp-2 cells.
Our findings indicate that events and protein functions that occur prior to α/IE gene transcription are not sufficient to stimulate the apoptotic process during HSV-1 infection. That apoptosis occurs when HSV-1 infects cells in the presence of CHX (1, 3, 32) indicates that de novo viral protein synthesis is not required to trigger the cell death process. The fact that CHX has been shown to increase HSV IE gene expression supports our conclusion (53). The rapid kinetics of apoptosis induction during wild-type HSV-1 infection in the presence of CHX strongly argues that the trigger under these conditions is of viral origin. One apparent misconception that may exist is that it is only the CHX treatment that is the trigger in such studies. CHX treatment alone is not a potent inducer of apoptosis. Rather, the low-level, background amounts of apoptosis detected in mock-infected cells arises due to the depletion of preexisting cellular survival factors which are not replenished when protein synthesis is blocked. One could simply imagine CHX acting in the same manner as a double-stranded RNA induced protein kinase R-dependent termination of translation. In both cases, the kinetics of cell death are extremely long compared to the rapid replication of HSV-1 and result from the secondary effects of the cells' inability to replenish needed factors, not active induction. Thus, CHX addition experiments remain a valid methodology for analyzing the rapid induction of apoptosis by HSV-1. Still, our need to use CHX in this study is unfortunate. Although it is very useful in that it allows for analyses in the absence of protein synthesis, we cannot ignore other effects of CHX. For example, CHX treatment has been reported to activate signaling cascades, including the Jun N-terminal protein kinase/mitogen-activated protein kinase pathways (6, 33, 37). The present data are insufficient to assess whether such signaling pathways participate in the rapid apoptotic induction by HSV-1.
Two additional key pieces of evidence support our conclusion. First, the finding that apoptosis above mock levels was not observed in vBSΔ27-infected cells treated with actinomycin D implies that the act of transcription of viral α/IE genes is necessary to trigger cell death. This is an important finding since it suggests that the incoming viral genomic DNA molecule is not the signal for induction, as has been proposed for adeno-associated virus (54). However, since actinomycin D is an intercalating agent (10), we cannot at this time exclude the possibility that a potential triggering structure may exist in viral DNA which is then “melted out” due to binding of the drug. Second, the strongest evidence that the expression of α/IE genes is required for the induction of apoptosis in HSV-1-infected human HEp-2 cells is the finding that cells infected with the d109 strain did not express viral IE genes or HSV proteins and did not present any apoptotic features. Note that these data were generated under conditions which did not include CHX addition, UV irradiation, or actinomycin D addition. Both the tsB7 infections at the nonpermissive temperature and the d109 infections do not allow viral gene expression and protein synthesis. Thus, in these cases the trigger of apoptosis is absent. This finding is in contrast to those from infections with wild-type HSV-1 in the presence of CHX where the trigger is present but apoptotic preventers are not made and apoptosis occurs. These data are consistent with and strongly support previous findings showing that the abolition of all IE gene expression was necessary to eliminate the toxicity associated with HSV infection (60).
One interesting finding was the manifestation of an altered cellular morphology in d109-infected HEp-2 cells. One possible explanation for this observation may be due to the establishment of a host antiviral state upon HSV-1 infection. Previous studies have shown that HSV-1 induces a host antiviral response and that inhibition of this response is dependent on the production of ICP0 (15, 43, 44, 48). Because ICP0 is not synthesized in d109-infected cells, the activation of interferon-stimulated genes may bring about morphology changes in HEp-2 cells. However, the reason why this effect is not seen in HEp-2 cells infected with UV-irradiated HSV-1, which has been shown to be capable of triggering this antiviral response, eludes us at this time. Further experimentation on the cellular biochemical and morphological alterations caused by HSV-1-induced antiviral activity may offer some clarification on this issue.
It remains unclear whether α/IE gene mRNA itself is sufficient for the induction event. It is conceivable that each individual α/IE gene may not have the same potential for activating apoptosis. Since α/IE genes are expressed with differing kinetics and accumulate to different levels (16, 57), investigations into the role of each gene in the triggering process is now required. Other potential viral stimuli beyond the actual mRNA molecules themselves might involve any one of the steps associated with the expression of the α/IE genes. These possible induction mechanisms include splicing of the gene transcripts, export of the mRNAs from the nucleus, initial interactions of these RNAs with the cellular translational machinery, and the general stability of transcripts in infected cells. Development of appropriate biochemical and molecular genetic systems are necessary to elucidate the exact molecular mechanism(s) responsible for the induction of apoptosis in HSV-1-infected HEp-2 cells.
Acknowledgments
We thank Martine Aubert for encouragement, advice, and discussions and Renee Baranin and Margot Goodkin for technical advice regarding hyperthermia infection and CAT assay protocols, respectively. We also thank (i) J. Smiley (University of Alberta) for the HSV-1(V422) virus, (ii) S. Silverstein (Columbia University) for the pIGA53 and pIGA65 plasmids, (iii) P. Spear (Northwestern University) for the HSV-1(HFEM) virus, (iv) B. Roizman (University of Chicago) for the HSV-1(HFEMtsB7) virus, and (v) N. DeLuca (University of Pittsburgh) for the HSV-1(d109) virus and complementing FO6 cells.
These studies were supported in part by grants from the USPHS (AI 38873 and AI 48582). C.M.S. is a predoctoral trainee and was supported in part by a USPHS Institutional Research Training Award (AI 07647).
REFERENCES
- 1.Aubert, M., and J. A. Blaho. 1999. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J. Virol. 73:2803-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, M., and J. A. Blaho. 2001. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 3:859-866. [DOI] [PubMed] [Google Scholar]
- 3.Aubert, M., J. O'Toole, and J. A. Blaho. 1999. Induction and prevention of apoptosis in human HEp-2 cells by herpes simplex virus type 1. J. Virol. 73:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubert, M., S. A. Rice, and J. A. Blaho. 2001. Accumulation of herpes simplex virus type 1 early and leaky-late proteins correlates with apoptosis prevention in infected human HEp-2 cells. J. Virol. 75:1013-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr, R. K., and M. A. Bogoyevitch. 2001. The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs). Int. J. Biochem. Cell Biol. 33:1047-1063. [DOI] [PubMed] [Google Scholar]
- 7.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benetti, L., J. Munger, and B. Roizman. 2003. The herpes simplex virus 1 US3 protein kinase blocks caspase-dependent double cleavage and activation of the proapoptotic protein BAD. J. Virol. 77:6567-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaho, J. A., C. Mitchell, and B. Roizman. 1993. Guanylylation and adenylylation of the α regulatory proteins of herpes simplex virus require a viral β or γ function. J. Virol. 67:3891-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaho, J. A., and B. Roizman. 1998. Analyses of HSV proteins for posttranslational modifications and enzyme functions. Methods Mol. Med. 10:237-256 [DOI] [PubMed]
- 12.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 14.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshiekh, N. A., E. Harris-Hamilton, and S. L. Bachenheimer. 1991. Differential dependence of herpes simplex virus immediate-early gene expression on de novo-infected cell protein synthesis. J. Virol. 65:6430-6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1998. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 18.Gaffney, D. F., J. McLauchlan, J. L. Whitton, and J. B. Clements. 1985. A modular system for the assay of transcription regulatory signals: the sequence TAATGARAT is required for herpes simplex virus immediate early gene activation. Nucleic Acids Res. 13:7847-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gelman, I. H., and S. Silverstein. 1986. Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J. Mol. Biol. 191:395-409. [DOI] [PubMed] [Google Scholar]
- 21.Gelman, I. H., and S. Silverstein. 1987. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J. Virol. 61:2286-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green, D. R. 1998. Apoptotic pathways: the roads to ruin. Cell 94:695-698. [DOI] [PubMed] [Google Scholar]
- 23.Green, D. R., and G. I. Evan. 2002. A matter of life and death. Cancer Cell 1:19-30. [DOI] [PubMed] [Google Scholar]
- 24.Hagglund, R., J. Munger, A. P. W. Poon, and B. Roizman. 2002. US3 protein kinase of herpes simplex virus 1 blocks caspase 3 activation induced by the products of US1.5 and UL13 genes and modulates expression of transduced US1.5 open reading frame in a cell type-specific manner. J. Virol. 76:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampar, B., and S. A. Elison. 1961. Chromosomal aberrations induced by an animal virus. Nature 192:145-147. [DOI] [PubMed] [Google Scholar]
- 26.Heeg, U., H. P. Dienes, S. Muller, and D. Falke. 1986. Involvement of actin-containing microfilaments in HSV-induced cytopathology and the influence of inhibitors of glycosylation. Arch. Virol. 91:257-270. [DOI] [PubMed] [Google Scholar]
- 27.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, US5 and US3. J. Virol. 73:8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerr, J. F. R., and B. V. Harmon. 1991. Definition and incidence of apoptosis: an historical perspective, p. 5-29. In L. D. Tomei and F. O. Cope (ed.), Apoptosis: the molecular basis of cell death. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koyama, A. H., and A. Adachi. 1997. Induction of apoptosis by herpes simplex virus type 1. J. Gen. Virol. 78:2909-2912. [DOI] [PubMed] [Google Scholar]
- 33.Kyriakis, J. M., and J. Avruch. 1990. pp54 microtubule-associated protein 2 kinase. A novel serine/threonine protein kinase regulated by phosphorylation and stimulated by poly-L-lysine. J. Biol. Chem. 265:17355-17363. [PubMed] [Google Scholar]
- 34.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 35.Leopardi, R., and B. Roizman. 1996. The herpes simplex virus major regulatory protein ICP4 blocks apoptosis induced by the virus or by hyperthermia. Proc. Natl. Acad. Sci. USA 93:9583-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, W. W., and Y. W. Hsu. 2000. Cycloheximide-induced cPLA(2) activation is via the MKP-1 down-regulation and ERK activation. Cell. Signal. 12:457-461. [DOI] [PubMed] [Google Scholar]
- 38.Mackem, S., and B. Roizman. 1982. Differentiation between alpha promoter and regulator regions of herpes simplex virus 1: the functional domains and sequence of a movable alpha regulator. Proc. Natl. Acad. Sci. USA 79:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackem, S., and B. Roizman. 1982. Regulation of α genes of herpes simplex virus: the α 27 gene promoter-thymidine kinase chimera is positively regulated in converted L cells. J. Virol. 43:1015-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFarlane, M., J. I. Daksis, and C. M. Preston. 1992. Hexamethylene bisacetamide stimulates herpes simplex virus immediate early gene expression in the absence of trans-induction by Vmw65. J. Gen. Virol. 73:285-292. [DOI] [PubMed] [Google Scholar]
- 41.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 42.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 43.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munger, J., and B. Roizman. 2001. The US3 protein kinase of herpes simplex virus 1 mediates the posttranslational modification of BAD and prevents BAD-induced programmed cell death in the absence of other viral proteins. Proc. Natl. Acad. Sci. USA 98:10410-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagata, S. 1997. Apoptosis by death factor. Cell 88:355-365. [DOI] [PubMed] [Google Scholar]
- 48.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 49.O'Hare, P., and G. S. Hayward. 1987. Comparison of upstream sequence requirements for positive and negative regulation of a herpes simplex virus immediate-early gene by three virus-encoded trans-acting factors. J. Virol. 61:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 82:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 53.Preston, C. M., A. Rinaldi, and M. J. Nicholl. 1998. Herpes simplex virus type 1 immediate early gene expression is stimulated by inhibition of protein synthesis. J. Gen. Virol. 79:117-124. [DOI] [PubMed] [Google Scholar]
- 54.Raj, K., P. Ogston, and P. Beard. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412:914-917. [DOI] [PubMed] [Google Scholar]
- 55.Roizman, B. 1962. Polykaryocytosis induced by viruses. Proc. Natl. Acad. Sci. USA 48:228-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology. Plenum, New York, N.Y.
- 57.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In P. M. Howley and D. M. Knipe (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 58.Roizman, B., and P. R. Roanne. 1964. Multiplication of herpes simplex virus. II. The relationship between protein synthesis and the duplication of viral DNA in infected HEp-2 cells. Virology 22:262-269. [DOI] [PubMed] [Google Scholar]
- 59.Salvesen, G. S., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 60.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekulovich, R. E., K. Leary, and R. M. Sandri-Goldin. 1988. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 62:4510-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi, Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell 9:459-470. [DOI] [PubMed] [Google Scholar]
- 64.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 71:6191-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun, X. M., M. MacFarlane, J. Zhuang, B. B. Wolf, D. R. Green, and G. M. Cohen. 1999. Distinct caspase cascades are initiated in receptor-mediated and chemical-induced apoptosis. J. Biol. Chem. 274:5053-5060. [DOI] [PubMed] [Google Scholar]
- 68.Villa, P., S. H. Kaufmann, and W. C. Earnshaw. 1997. Caspases and caspase inhibitors. Trends Biochem. Sci. 22:388-393. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, G., E. Avitabile, G. Campadelli-Fiume, and B. Roizman. 2003. The domains of glycoprotein D required to block apoptosis induced by herpes simplex virus 1 are largely distinct from those involved in cell-cell fusion and binding to nectin1. J. Virol. 77:3759-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, G., and B. Roizman. 2001. The domains of glycoprotein D required to block apoptosis depend on whether glycoprotein D is present in the virions carrying herpes simplex virus 1 genome lacking the gene encoding the glycoprotein. J. Virol. 75:6166-6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou, G., and B. Roizman. 2002. Truncated forms of glycoprotein D of herpes simplex virus 1 capable of blocking apoptosis and of low-efficiency entry into cells form a heterodimer dependent on the presence of a cysteine located in the shared transmembrane domains. J. Virol. 76:11469-11475. [DOI] [PMC free article] [PubMed] [Google Scholar]