Abstract
Evidence for human immunodeficiency virus type 1 (HIV-1) superinfection was sought among 37 HIV-1-positive street-recruited active injection drug users (IDUs) from the San Francisco Bay area. HIV-1 sequences from pairs of samples collected 1 to 12 years apart, spanning a total of 215 years of exposure, were generated at p17 gag, the V3-V5 region of env, and/or the first exon of tat and phylogenetically analyzed. No evidence of HIV-1 superinfection was detected in which a highly divergent HIV-1 variant emerged at a frequency >20% of the serum viral quasispecies. Based on the reported risk behavior of the IDUs and the HIV-1 incidence in uninfected subjects in the same cohort, a total of 3.4 new infections would have been expected if existing infection conferred no protection from superinfection. Adjusted for risk behaviors, the estimated relative risk of superinfection compared with initial infection was therefore 0.0 (95% confidence interval, 0.00, 0.79; P = 0.02), indicating that existing infection conferred a statistically significant level of protection against superinfection with an HIV-1 strain of the same subtype, which was between 21 and 100%.
Recent case reports of human immunodeficiency virus type 1 (HIV-1) superinfection demonstrate that an established infection does not confer complete protection against reinfection with a divergent strain (3, 28, 31, 61).
Lentivirus superinfection experiments with nonhuman primate systems using intravenous challenges with high-viral-titer inoculums have shown variable results. One of two chimpanzees could be superinfected with another strain of the same HIV-1 subtype (20). Superinfection with a heterologous HIV-2 strain was restricted to the first 2 months postinfection in pig-tailed macaques (54). Six of seven baboons infected with HIV-2 resisted superinfection with heterologous HIV-2 strains (42), while four coculture-negative HIV-2-infected rhesus macaques could be superinfected with pathogenic simian immunodeficiency virus (SIV) strain SIVmac251 and became persistently viremic (57). Attenuated strains of SIV could protect rhesus macaque against superinfection (10), but such protection decreased with more-attenuated vaccine strains (27) and more-divergent and -pathogenic SIV challenge strains (57, 77, 84). One of two (57) and three of three (60) cynomologous macaques infected with HIV-2 could be superinfected with pathogenic SIVsm but showed reduced rates of disease progression.
The ability of an established lentivirus infection and associated immune responses to protect against superinfection may depend in part on the degree of sequence divergence between the two viruses and the state of immunocompetence of the host. The greater the degree of divergence between the resident and the challenge strains the less the immune responses to the first virus may recognize a later challenge strain. Within the HIV-1 family a wide range of sequence divergence exists. Strains within a single subtype of group M HIV-1 can vary by up to 20% in envelope amino acid sequence, while different group M subtypes vary by up to 35% (21). Even greater divergence between the different HIV-1 groups (M, N, and O) and between HIV-1 and HIV-2 is seen. In humans, prior infection with HIV-2 does not appear to protect against subsequent infection with HIV-1 (82). An individual infected with both group M and group O HIV-1 viruses was identified (71), although it is not known whether the infections were nearly simultaneous or sequential. Indirect evidence for infections with more then one subtype of HIV-1 group M viruses is seen in the large number of distinct intersubtype recombinants, each with a presumably independent origin (8, 12, 24, 25, 36, 46, 56, 66, 73, 74, 76), and in numerous cases of dual-subtype infections (4, 6, 24-26, 62). It is not known with what time intervals such dual infections are acquired.
The ability of HIV-1 to infect seropositive hosts previously infected with different subtypes of group M HIV-1 has been recently reported for two injection drug users (IDUs) and a homosexual man (28, 61). The two IDUs were untreated and became superinfected with a different HIV-1 subtype 3 to 11 months after their initial infections (61). The homosexual man was successfully treated for a period of over 2 years with a regimen of highly active antiretroviral therapy (HAART) initiated during primary infection. He became infected with a different subtype of HIV-1 3 to 4 months after discontinuing HAART (28). These three cases indicate that any protection against HIV superinfection that may have been provided by a preexisting infection was only partial when the challenge strains belonged to a different HIV-1 group M subtype.
Two cases of superinfection with a virus belonging to the same HIV-1 subtype have been reported (3, 31). A patient who had initiated effective HAART treatment at the time of acute infection was superinfected while undergoing structured treatment interruption, followed by rapidly rising viremia (3). In the second case an untreated subject infected with a drug-resistant strain was superinfected with a wild-type strain within 4 months of infection (31). No other cases of same-subtype superinfection have been reported among the numerous longitudinal sequence analysis studies of HIV-1 (15, 16, 35, 45, 47, 50, 51, 67-69, 83a), although it is conceivable that prior cases were dismissed as PCR contamination (38). Cases of same-subtype coinfection involving strains from different sources have been reported, but such coinfections occurred either simultaneously (18), nearly simultaneously (41, 89), or at unknown intervals (64, 65).
As only two clear cases of same-subtype HIV-1 superinfection have been reported to date, it remains possible that some level of protection against superinfection is provided by an established infection. To measure the frequency of same-subtype superinfection in a highly exposed and largely untreated population, we phylogenetically analyzed HIV-1 sequences in longitudinally collected serum from 37 street-recruited active IDUs.
MATERIALS AND METHODS
Subject recruitment.
The viral sequences analyzed in this study were amplified from paired serum specimens drawn from IDUs participating in the Urban Health Study from 1987 through 2000. The Urban Health Study has conducted semiannual seroepidemiological surveys of IDUs recruited from street settings in inner-city communities in the San Francisco Bay area since 1985 (32, 33, 81). IDUs were recruited every 6 months in each of six bay area neighborhoods by using targeted sampling methods (79, 80). Study neighborhoods were selected for their high concentrations of injection drug use based on review of drug treatment admission data, police arrest data, direct observation, and ethnographic research. IDUs were recruited from street settings by experienced outreach workers. Eligibility for participation was based on injection drug use in the past 30 days or previous participation in the Urban Health Study. New respondents were screened for visible signs of recent subcutaneous or intravenous drug use. Trained interviewers collected demographic and risk behavior information using a standardized questionnaire. Blood was collected at the field site for HIV antibody testing, and aliquots of serum were frozen and stored at −70°C for future use. Respondents were given pre- and post-HIV test counseling, referred to medical and social services as needed, and paid $15 to $20 at each visit for their contribution to the study. All subjects provided written informed consent each time they participated. All study procedures were approved by the Committee on Human Research at the University of California, San Francisco. Thirty-seven participants were selected for the present study based on the availability of frozen serum specimens, long sampling interval between earliest and latest time point available, and self-reports of continued high-risk behavior.
RNA extraction and nested RT-PCR.
All RNA extractions, reverse transcription, and setting up of the first-round PCR tubes were performed in a preamplification room free of amplified HIV products. Viral RNA was extracted from 280 μl of serum with the Qiagen (Valencia, Calif.) viral RNA kit. Ten units of RNase inhibitor was added to 30 μl of eluted RNA. Ten microliters of RNA was combined with 50 μg of random 6-nucleotide oligomers (Life Technologies, Carlsbad, Calif.) and 1 μl of 10 mM deoxynucleoside triphosphate (dNTP; New England Biolabs, Beverly, Mass.). The mixture was heated at 65°C for 5 min and rapidly cooled on ice. A reverse transcriptase (RT) mixture comprising 4 μl of 5× first-strand buffer (Life Technologies), 2 μl of 0.1 M dithiothreitol (Life Technologies), 1 μl of 200-U/μl Moloney murine leukemia virus RT (Life Technologies), and diethyl pyrocarbonate-treated water was added to a final volume of 25 μl. The mixture was incubated for an hour at 37°C and heated for 15 min at 70°C. Nested PCR involved a first-round mixture with externally annealing primers followed by a second-round mixture containing internally annealing primers. The primers for the V3-to-V5 region of env were previously described (17). Sixty-eight env sequences were determined. HXB2 positions 7042 to 7644 were used in the env alignment. The first-round primers for p17 gag were JA152 and JA155, and the second round primers were JA153 and JA154, as previously described (39). One hundred fifty-six p17 sequences were determined. HXB2 positions 707 to 1176 were used in the p17 alignment. First-round primers for tat were TatED1 (5′GCAGGAGTGGAAGCCATAATAAG3′; HXB2 positions 5721 to 5743) and TatED2 (5′TTCTATGAATACTATGGTCCACACAACTAT3′; 6119 to 6148). Second-round primers were TatED3 (5′GAATTCTGCAACAACTGCTGTTTAT3′; 5743 to 5767) and TatED4 (5′ATTGCTGCTACTACTAATGCTACTATTGC3′; 6083 to 6111). A total of 50 tat PCR products were sequenced. HXB2 positions 5768 to 6072 were used in the tat alignment. Five microliters of cDNA was added to a PCR mixture composed of 10× PCR buffer (Promega, Madison, Wis.), 2.5 mM MgCl, 1 mM dNTP, 0.2 μM sense and antisense primers, 1 U of Taq (Promega), and water to a final volume of 50 μl. Amplification was carried out as follows: 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 45 s at 57°C, and 2 min at 72°C and a final extension for 5 min at 72°C. PCR fragments were purified with the Qiagen kit. The env V3-to-V5 region and p17 gag PCR products were subcloned into the plasmid vector pCR2.1 of the TA cloning kit (Invitrogen, Carlsbad, Calif.).
HMA of p17 sequence variants.
Subcloned p17 inserts were classified into different clonotypes by heteroduplex mobility analysis (HMA) prior to sequencing (78). An average of 17.5 subcloned plasmids (range, 6 to 25) were analyzed per subject. One p17 subclone variant per time point and per subject was randomly selected, and 2.5 μl of its second-round PCR product was reannealed with 2.5 μl of the PCR product derived from subclones from the other time point for the same subject. The resulting DNA heteroduplexes were electrophoretically resolved in 8% polyacrylamide gels (acrylamide/bisacrylamide ratio, 29:1) at 235 V for 4.5 h in 1× Tris-borate-EDTA buffer. Gels were stained with ethidium bromide, and the UV fluorescence was recorded with a charge-coupled device camera.
Sequencing.
Subcloned env and gag regions were sequenced with standard vector primers, an automated capillary sequencer (ABI 3700), and the ABI sequence viewer program EditView. tat PCR amplicons were directly sequenced with the second-PCR-round antisense primer.
Phylogenetic analysis.
All sequences were aligned in BioEdit with the CLUSTAL W sequence alignment tool. Sequences were further manually edited to preserve in-frame insertions and deletions in the final alignments. All phylogenetic analyses were performed with PAUP*, version 4.0 beta 10.
Initial trees were generated via the neighbor-joining algorithm. For each alignment, from this initial tree, a maximum-likelihood (ML) tree, under the general time-reversible model, with previously estimated six-term rate matrix, base frequencies, gamma distribution alpha term, and proportion of invariant sites, was generated with the tree bisection resection algorithm (72). Initial parameters were determined with ModelTest, version 3.06 (58). Each tree was allowed to rearrange 10,000 times before the final tree, and the lowest observed likelihood score (most likely tree) was chosen. To provide statistical support for observed branching patterns, a bootstrap analysis was performed. Starting from a neighbor-joining tree and with the previously estimated tree parameters for the ML trees, 100 bootstrap replicates were grown, and each was allowed to rearrange 1,000 times under the tree bisection resection algorithm. Bootstrap values were applied to the corresponding ML tree from the central to terminal nodes. Bootstrap values under 50% are not reported. If a tree branch structure in the final bootstrap tree differed from that observed in the ML tree, as happened at a few terminal nodes, bootstrap values were not applied.
Statistical analysis.
To determine whether the observed rate of superinfection reflected protection against superinfection, we conducted an analysis to test the null hypothesis that superinfection occurs as easily in HIV-infected persons as initial infection occurs in HIV-uninfected persons with the same exposures. We first constructed an exponential-regression model for the risk of initial infection using the SAS Lifereg procedure (SAS/STAT user's guide, version 8, SAS Institute Publishing, Cary, N.C., 2000) and data from Urban Health Study participants who were initially uninfected and who returned at least once for follow-up testing. Time-dependent variables for demographic characteristics, calendar time, and injection and sexual risk behaviors were included in the model. Participants who seroconverted were assumed to have been infected at the midpoint between their last negative and first positive tests. This model was constructed with data from 3,549 study participants interviewed on 15,462 occasions over 10,738 person-years of observation. We then used this model of the risk of initial infection to calculate a predicted risk of superinfection for each of the HIV-infected subjects in the present study, accounting for their demographic characteristics, risk behaviors, and calendar time. The expected number of superinfections was calculated as the sum of the predicted probabilities of superinfection in the 37 studied subjects. The P value for an observation is the probability of arriving at that observation if the null hypothesis is true. Since no superinfections were observed, the P value for rejecting the null hypothesis that superinfection occurs as easily as initial infection, when risk behaviors and other risk factors are held constant, was the probability of observing no superinfections in the 37 subjects, assuming that the model correctly predicted their superinfection risk. The probability of observing no superinfections among all participants was calculated as the product of the probabilities of no superinfection for the 37 subjects. The probability of no superinfection for each subject was 1 − the probability of superinfection predicted by the model. Since no superinfections were detected, the estimated relative risk (r) of superinfection compared with that of initial infection was zero. To determine the upper 95% confidence bound for this estimate, we determined r such that the probability of observing no superinfections was 0.05 if the risk of superinfection for each subject was r times the risk predicted by the model. Thus the upper confidence bound was the value of r for which the product of the 37 values of 1 − (r × risk) was 0.05.
Nucleotide sequence accession numbers. The GenBank accession numbers of nucleotide sequences analyzed in this study are AY450962 through AY451235.
RESULTS
IDU population.
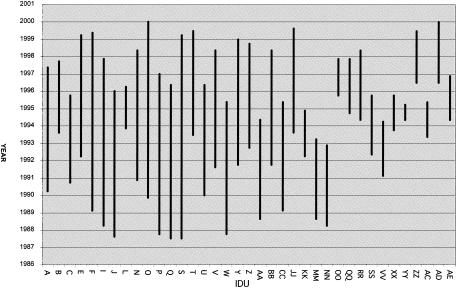
The 37 IDUs were men and women of diverse ethnicity, predominately in their fourth and fifth decades of life (Table 1). The samples analyzed were collected between 1987 and 2000 (Fig. 1). The total duration of time between the baseline and follow-up samples for all 37 IDUs combined was 215 person-years. Exposure to HAART was infrequent; 170 (79%) of the 215 person-years of observation occurred before the beginning of the HAART era in mid-1996, and study participants reported currently taking HAART regimens at only 8 (23%) of the 35 interviews conducted from 1997 through 2000. We therefore estimate that participants were taking HAART regimens during less than 5% of the total follow-up time.
TABLE 1.
Characteristics of 37 HIV-infected, street-recruited IDUs, San Francisco Bay area, 1987 to 2000
| Characteristic | Value |
|---|---|
| Age at baseline (yr) (median [IQRa]) | 37 (30, 42) |
| Sex (no. [%]) | |
| Men | 25 (68) |
| Women | 12 (32) |
| Race or ethnicity (no. [%]) | |
| African-American | 16 (43) |
| White | 15 (41) |
| Other | 6 (16) |
| Time between baseline and follow-up samples (yr) (median [IQR]) | 6.0 (3.3, 7.6) |
| Time between baseline and follow-up samples (yr) (total) | 214.8 |
| No. of visits, including baseline and follow-up (median [IQR]) | 5 (3, 8) |
| No. (%) that injected: | |
| Heroin | 33 (89) |
| Cocaine | 25 (68) |
| “Speedball” (mixture of heroin and cocaine) | 29 (78) |
| Amphetamines | 21 (57) |
| Risk behaviors | |
| Injections/day (median [IQR]) | 3 (2,4) |
| Reported use of a syringe previously used by another IDU (no. [%]) | 29 (78) |
| MSMb (no. [%]) | 11 (30) |
| No. of male sex partners in the last 6 mo (women and MSM; n = 23) (median [IQR]) | 6 (1, 15) |
| Received money or drugs in exchange for sex (no. [%]) | 15 (41) |
IQR, interquartile range.
MSM, men who had sex with men.
FIG. 1.
Time span analyzed for HIV-1 superinfection in 37 IDUs.
Generation of HIV-1 sequence data.
Viral quasispecies were analyzed at two time points to determine whether highly divergent variants emerged, indicating possible superinfection. Viral sequences were analyzed in the env V3-to-V5 region for 6 subjects, in the first tat exon for 25 subjects, and in the gag p17 region for 16 subjects. Sequences from two subjects were analyzed in env and tat, those from two were analyzed in env and p17, and those from six were analyzed in p17 and tat. Differences in the locus analyzed were the consequence of changes in the PCR methodology during the course of this study. Analyses at two loci were performed to test conclusions reached on the basis of the first studied locus. All samples were PCR positive with the particular primer sets used. Following reverse transcription initiated with random primers, the different regions were amplified by nested PCR (see Materials and Methods).
For the env region analysis 10 to 13 randomly selected env plasmid subclones were sequenced per subject.
The tat region PCR amplicons were directly sequenced. To measure the ability of tat population sequencing to detect a highly divergent, coamplified, variant, we performed the following analyses. Reconstituted mixtures of tat PCR amplicons from different subjects were directly sequenced. A tat variant present at a frequency ≥20% was readily detectable through mixed nucleotide base peaks at greater than 4% of the positions in the sequencing electrophoregrams (data not shown). A pairwise substitution analysis showed that tat variants from different San Francisco Bay area IDUs differed at ≥4% of nucleotide positions in the region amplified in >93% of pairwise comparisons. Because the mixed nucleotide positions seen in direct tat amplicon sequencing of the IDU samples studied here were present in less than 4% of the nucleotide positions, we concluded that these mixed bases reflected the quasispecies nature of HIV rather than the presence of highly divergent coamplified tat variants. In one subject a PCR product resulted in almost completely mixed sequencing base peaks. The PCR product was subcloned, and six plasmids were sequenced. Alignments showed that four of six variants carried a 3-bp deletion in the region of tat where population sequencing produced mixed sequencing bases. Subcloned sequences all clustered phylogenetically (data not shown). The mixed base positions were therefore the result of sequencing coamplified variants of different lengths rather than superinfection (data not shown).
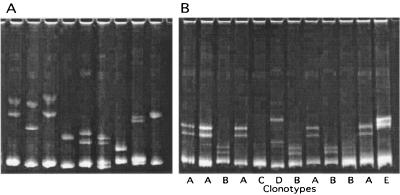
To increase p17 sequence variant sampling while minimizing plasmid sequencing, multiple p17 subclones from each subject were first classified into clonotypes by using DNA HMA. The electrophoretic mobility of DNA heteroduplexes is affected by both the number and the positions of mismatched and unmatched nucleotide base pairs (14, 16, 29, 75). The PCR product from one subcloned p17 variant from each of both time points was reannealed to PCR amplicons from plasmid subclones from the other time point. The resulting DNA heteroduplexes were then electrophoretically separated through polyacrylamide (Fig. 2). Subclones that produced identical HMA gel patterns were considered to belong to the same clonotype group, and only a single representative plasmid from each group was sequenced. An average of 17.5 p17 plasmids (range, 6 to 25) per subject were subjected to HMA clonotype analysis and plasmid sequencing.
FIG. 2.
Example of HMA clonotype analysis of p17 sequence variants. PCR products from plasmid subclones were reannealed, and DNA heteroduplexes were resolved through a polyacrylamide gel. (A) Inter-IDU HMA. A clonal PCR product from a subclone from one IDU was reannealed to clonal PCR amplicons from nine other IDUs, showing that different sequence variants produced distinct electrophoretic mobilities. (B) Intra-IDU HMA. A clonal PCR product from one subclone from an IDU was reannealed to PCR products derived from subclones from the other time point. Subclones producing the same mobility heteroduplexes were classified as members of the same clonotype groups (A through E).
Phylogenetic analysis of the V3-to-V5 envelope.
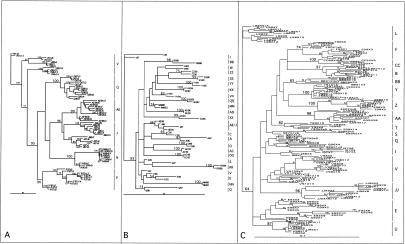
The env phylogenetic analysis showed that every sequence variant from each of the six IDUs clustered by subject (Fig. 3A). Five of the six intrasubject clusters were supported with high bootstrap values (≥90). Every sequence variant from subject J also clustered together but did so with bootstrap values <50. Two J subclusters were observed; they consisted mostly of variants from each of the time points analyzed collected 9 years apart. AE and J variants clustered together with a bootstrap value of 96%, while AE variants alone clustered with a 99% bootstrap value. When the phylogenetic analysis was repeated in the absence of AE variants, all J variants clustered together with a bootstrap value of 100 (data not shown). The high degree of similarity between AE and J sequences therefore reduced the bootstrap support for the J variants alone. We therefore attribute the low bootstrap values for the J sequence variant cluster to its close phylogenetic relationship with subject AE variants. Both AE and J were sampled in San Francisco and, according to their closely related HIV-1 sequences, may be directly or indirectly epidemiologically linked.
FIG. 3.
ML phylogenetic analysis. (A) V3-to-V5 region of env. (B) First exon of tat. (C) p17. The IDU subjects are represented by letters followed by years of sampling and, following the first “x,” the clone number. Bootstrap values for the nodes holding all sequence variants from any one IDU are shown in large font, while other bootstrap values are shown in smaller fonts. For p17 the total number of sequence variants belonging to the same HMA clonotype group is noted following the second “x”; e.g., Y91x3x4 indicates subject Y sampled in 1991, subclone number 3, belonging to an HMA clonotype group consisting of four different subclones.
Phylogenetic analysis of tat.
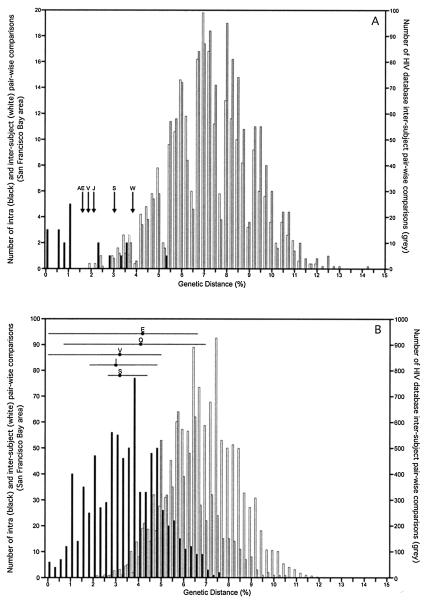
All sequence pairs from the 25 IDUs analyzed in the tat region also clustered by individuals (Fig. 3B). Bootstrap analysis statistically supported intrasubject pair clusters for 20 IDUs. Intrasubject clusters with bootstrap value below 50 were seen for W, S, V, AE, and J. As in the env region analysis, tat variants from AE and J clustered together. The phylogenetic analysis was repeated with either AE or J sequences omitted. The bootstrap values for the AE and J clusters were then 77 and 76, respectively. The AE/J tat sequence cluster therefore also reflects a direct or indirect epidemiological linkage between these two IDUs. To determine if the intrasubject sequence pairs from subjects W, S, and V (whose analyzed samples were collected 7 to 12 years apart) showed a degree of sequence divergence characteristic of unrelated HIV strains rather than long-term intrapatient HIV-1 evolution, we performed the following analysis. The intra- and interpatient sequence pair genetic distances for the 20 IDUs whose variants clustered together with bootstrap values >50 were separately plotted along with the genetic distances between unrelated HIV-1 sequences from the HIV-1 database (Fig. 4A). The intrasubject genetic distances for subjects W, S, V, AE, and J all fell within the range of intrapatient distances whose clustering was supported by bootstrap values >50 (Fig. 4A). This result indicated that the low bootstrap values (<50) seen for these five IDUs were the result of either strong sequence similarities (AE/J) or intrapatient evolution during the long intervals between sampling of 7 to 12 years (W, S, and E) rather than the result of superinfection with a highly divergent strain.
FIG. 4.
Intra- and interpatient distribution of pairwise genetic distances for the tat (A) and p17 (B) regions. The intrasubject (black lines) and intersubject (white lines) pairwise HIV-1 genetic distances for San Francisco Bay area IDUs whose variants all clustered with bootstrap values >50, as well as the intersubject distances from unrelated sequences from the HIV database (grey lines), are plotted. Arrows, intrasubject tat genetic distances for the two time points sequenced which clustered phylogenetically together with bootstrap values below 50; horizontal lines, means and ranges of the p17 intrapatient genetic distances for variants that clustered phylogenetically together with bootstrap values below 50. The phylogenetically linked subject pair AE and J was omitted from the intersubject tat distance pairs.
Phylogenetic analysis of p17.
All intrasubject sequences from the 16 IDUs analyzed in the p17 region also clustered together phylogenetically (Fig. 3C). Bootstrap analysis statistically supported the intrasubject sequence clusters for 11 IDUs. For the five subjects whose p17 sequences were not all held together with bootstrap values >50 (S, V, Q, E, and I) the tat region was also analyzed. All five sequence pairs grouped together phylogenetically in tat (Fig. 3B). Of these five, Q, E, and I sequences showed significant bootstrap values in the tat region (73, 54, and 83, respectively) while sequences from the other two subjects (S and V) had bootstrap values <50. Q and V sequences were also analyzed at the env loci, where their intrasubject variants clustered with bootstrap values of 100 and 95, respectively. To determine if the intrasubject p17 sequences from subjects S, V, Q, E, and I showed a degree of sequence divergence characteristic of unrelated HIV strains, we also compared their genetic distances to the intra- and intersubject genetic distances for the 11 subjects whose variants clustered phylogenetically with bootstrap values >50. The genetic distance distribution between epidemiologically unrelated strains from the HIV database was also included (Fig. 4B). The means as well as the ranges of intrasubject genetic distances for the five IDUs all fell within the range of intrapatient distances supported by bootstrap analysis (Fig. 3C). The range for some subjects (i.e., Q and E) reached 6.5%, reflecting the extensive sequence divergence between time points and the diversification of their quasispecies. This result indicated that the low bootstrap values (<50) for clusters holding intrasubject sequence variants from S, V, Q, E, and I IDUs were the result of within-subject evolution during the 7- to 12-year intervals between samplings rather than superinfection with a highly divergent strain.
HIV database search.
The two env, tat, and p17 sequence variants from each IDU that showed the greatest genetic distance were used for BLAST searches against the HIV database. The five database sequences most similar to each query sequence were then used in neighbor-joining phylogenetic analyses that included every sequence variant from the query sequence IDU. In no case was an intrapatient cluster interspersed with a database sequence (data not shown).
Therefore, while we observed extensive sequence evolution occurring during the long sampling intervals, in none of the 37 tested IDUs did we find convincing evidence of superinfection.
HIV-1 exposure.
The 37 HIV-1-infected subjects included in this study reported high levels of sexual and injection risk behaviors (Table 1). The predicted likelihood of superinfection, based on the incidence of seroconversion observed in seronegative IDUs reporting the same risk behaviors, ranged from 0.01 to 0.61 for the individual subjects. The expected number of superinfections among the 37 subjects, assuming superinfection in HIV-infected IDUs was as likely as an initial infection in uninfected IDUs with the same risk behaviors (i.e., no superinfection protection), was 3.38. The summary likelihood that none of the 37 IDUs became superinfected as a result of chance alone (the null hypothesis) was 0.02. The null hypothesis was therefore rejected. The estimated relative risk of superinfection compared with risk of initial infection was 0.0, with a 95% confidence interval of (0.00, 0.79), indicating that the level of protection against superinfection conferred by existing HIV infection measured in this study was between 21 and 100%.
DISCUSSION
Conclusion.
In this study, we examined paired HIV-1 serum samples from 37 IDUs spanning 215 person-years of high-risk behavior and found no evidence for superinfection with a highly divergent strain of the same subtype. HAART was taken during approximately 5% of the 215 years of exposure by the 37 IDUs analyzed, such that HAART's potential protective effect against superinfection was minimal. The IDU population studied here is therefore distinct from the predominantly treated patient population studied by Gonzales et al., in which protease and RT drug resistance genotyping was performed and in whom no superinfection was detected over a total of 1,072 years of follow-up (22).
While superinfecting viruses may be considered by the already-infected host as a form of immune escape variant (1, 2, 5, 7, 23, 30, 48, 52, 53) and may therefore be difficult to recognize immunologically, several factors may argue against this scenario and explain why infected persons may be at least partially protected against superinfection. The resident viral quasispecies presumably adapted to an initially highly focused but gradually widening and eventually very high level of immune responses aimed at multiple epitopes through the stepwise selection of immune escape mutants (1, 2, 5, 7, 23, 30, 48, 52, 53). A superinfecting strain, facing a high level of preexisting immunity aimed at multiple epitopes in a chronic infection, may find it difficult to escape through the simultaneous selection of multiple escape mutations. A superinfecting strain may therefore not compete successfully against the multitude of already-host-adapted resident viruses. The strong HIV-1 genetic bottleneck often experienced during many, although not all, HIV transmissions (13, 15, 37, 43, 49, 55, 59, 67, 83, 83a, 86-88) and its anticipated detrimental effect on viral fitness (85) may also put superinfecting strains at a further disadvantage. Nonetheless, a recent case of same-subtype superinfection occurred despite the presence of up to nine targeted cytotoxic T-lymphocyte epitopes in the superinfecting strain (3). In this case susceptibility to superinfection may have been affected by the concomitant structured treatment interruption, possibly by decreasing resident strain viremia or as a result of increased target cell availability. In the second case of same-subtype superinfection the 4 months following primary infection in this untreated patient (31) may not have provided sufficient time for a highly cross-reactive immune response to develop and/or the wild-type superinfecting strain genotype may have had a replication fitness advantage over the resident drug-resistant genotype (11). An increasing frequency of primary infection with drug-resistant strains (40, 63, 70) may therefore similarly increase the likelihood of future superinfection with fitter wild-type viruses. Conversely, a highly drug-resistant superinfecting strain may have a selective advantage over resident wild-type variants in patients on antiviral drug therapy. Last, a large and growing level of viral genetic diversity, both within HIV-1 subtype B (19, 34, 44) and worldwide (21), and increasing circulation of different subtypes and their recombinants may all lead to increasing frequencies of superinfection.
Although superinfection was not detected in this study, several caveats need to be attached to this conclusion. It remains possible that HIV-1 superinfection went undetected because the superinfecting strain remained a minority variant below our detection limit, was compartmentalized, only replicated transiently, or most of its genome was deleted through recombination. Among the three published cases of superinfection with a different subtype, the superinfecting strains reached different frequencies in the plasma quasispecies. In one case the superinfecting strain completely displaced the prior resident strain within 2 months of superinfection (28). In the other two cases, using subtype-specific primers, both subtypes were seen cocirculating in the plasma. When the PCR products were subcloned and plasmid was sequenced, the superinfecting strain was dominant in one case (73% of subclones) and was a minority variant (33% of subclones) in the other case (61). In the same-subtype cases of superinfection the second strains appeared to completely replace the initial strains (3, 31). Because of the extensive population sampling at the p17 and env loci (i.e., the multiple subclones analyzed per sample) and the ability of direct tat amplicon sequencing to detect minority divergent strains, we conclude that in none of the subjects analyzed did a highly divergent strain reach a frequency greater than 20% of the later serum quasispecies. Because a significant proportion of HIV-1 transmission in this cohort is related to sexual activity (32), restriction of a new HIV-1 strain to an unsampled site such as mucosal tissues cannot also be excluded. As precedent, attenuated-SIV-infected macaques protected from intrarectal challenge with a more virulent strain did not show the presence of the challenge strain in the rectum or lymph nodes (9). The possibility that a transient episode of viremia with a superinfecting strain occurred cannot be discounted in this study, as only two time points were analyzed. It is also conceivable that recombination between resident and superinfecting strains deleted most of a superinfecting variant's genome. In the three previously reported cases of superinfection with a different subtype, the superinfecting viruses were analyzed at multiple loci with no sign of recombination (28, 61). Similarly, no recombination was detected in the two cases of same-subtype superinfection (3, 31). Nonetheless because of the longer time interval between samplings in this study (1 to 12 years) the opportunities for recombination to obscure superinfection events were greater here than in these prior cases.
In summary, using samples collected from 37 IDUs whose behavior put them at risk of superinfection during 215 person-years, we did not detect highly divergent HIV-1 strains in their later quasispecies. The reported cases of same-subtype superinfection (3, 31) have shown that existing infection does not confer absolute protection against superinfection. Based on the expected level of exposures of our subjects, however, the absence of superinfection indicates that some degree of protection (21 to 100%) against superinfection with a virus of the same subtype was provided by prior HIV-1 infection (P = 0.02).
Acknowledgments
We thank the CDC (U64/CCU917889-01) for financial support to R.M.G., J.D.B., R.T., B.L.H., and E.L.D and NIH (AI-447320) for financial support to R.T., B.L.H., and E.L.D.
We thank Michael Busch for helpful discussions.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nyström, and E. M. Fenyö. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 4.Artenstein, A. W., T. C. VanCott, J. R. Mascola, J. K. Carr, P. A. Hegerich, J. Gaywee, E. Sanders-Buell, M. L. Robb, D. E. Dayhoff, S. Thitivichianlert, et al. 1995. Dual infection with human immunodeficiency virus type 1 of distinct envelope subtypes in humans. J. Infect. Dis. 171:805-810. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 6.Becker-Pergola, G., J. L. Mellquist, L. Guay, F. Mmiro, C. Ndugwa, P. Kataaha, J. B. Jackson, and S. H. Eshleman. 2000. Identification of diverse HIV type 1 subtypes and dual HIV type 1 infection in pregnant Ugandan women. AIDS Res. Hum. Retrovir. 16:1099-1104. [DOI] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. A. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-217. [DOI] [PubMed] [Google Scholar]
- 8.Carr, J. K., M. Avila, M. Gomez Carrillo, H. Salomon, J. Hierholzer, V. Watanaveeradej, M. A. Pando, M. Negrete, K. L. Russell, J. Sanchez, D. L. Birx, R. Andrade, J. Vinoles, and F. E. McCutchan. 2001. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS 15:F41-F47. [DOI] [PubMed] [Google Scholar]
- 9.Cranage, M. P., A. M. Whatmore, S. A. Sharpe, N. Cook, N. Polyanskaya, S. Leech, J. D. Smith, E. W. Rud, M. J. Dennis, and G. A. Hall. 1997. Macaques infected with live attenuated SIVmac are protected against superinfection via the rectal mucosa. Virology 229:143-154. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 12.Delgado, E., M. M. Thomson, M. L. Villahermosa, M. Sierra, A. Ocampo, C. Miralles, R. Rodriguez-Perez, J. Diz-Aren, R. Ojea-de Castro, E. Losada, M. T. Cuevas, E. Vazquez-de Parga, R. Carmona, L. Perez-Alvarez, L. Medrano, L. Cuevas, J. A. Taboada, and R. Najera. 2002. Identification of a newly characterized HIV-1 BG intersubtype circulating recombinant form in Galicia, Spain, which exhibits a pseudotype-like virion structure. J. Acquir. Immune Defic. Syndr. 29:536-543. [DOI] [PubMed] [Google Scholar]
- 13.Delwart, E., M. Magierowska, M. Royz, B. Foley, L. Peddada, R. Smith, C. Heldebrant, A. Conrad, and M. Busch. 2002. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS 16:189-195. [DOI] [PubMed] [Google Scholar]
- 14.Delwart, E. L., and C. J. Gordon. 1997. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 12:348-354. [DOI] [PubMed] [Google Scholar]
- 15.Delwart, E. L., H. Pan, H. W. Sheppard, D. Wolpert, A. U. Neumann, B. Korber, and J. I. Mullins. 1997. Slower evolution of HIV-1 quasispecies during progression to AIDS. J. Virol. 71:7498-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart, E. L., H. W. Sheppard, B. D. Walker, J. Goudsmit, and J. I. Mullins. 1994. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 68:6672-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delwart, E. L., E. G. Shpaer, F. E. McCutchan, J. Louwagie, M. Grez, H. Rübsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 18.Diaz, R. S., E. C. Sabino, A. Mayer, J. W. Mosley, and M. P. Busch. 1995. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J. Virol. 69:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foley, B., H. Pan, S. Buchbinder, and E. L. Delwart. 2000. Apparent founder effect during the early years of the San Francisco HIV type 1 epidemic (1978-1979). AIDS Res. Hum.Retrovir. 16:1463-1469. [DOI] [PubMed] [Google Scholar]
- 20.Fultz, P. N., A. Srinivasan, C. R. Greene, D. Butler, R. B. Swenson, and H. M. McClure. 1987. Superinfection of a chimpanzee with a second strain of human immunodeficiency virus. J. Virol. 61:4026-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 22.Gonzales, M. J., E. Delwart, S. Y. Rhee, R. Tsui, A. R. Zolopa, J. Taylor, and R. W. Shafer. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J Infect. Dis. 188:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goulder, P. J. R., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 24.Hoelscher, M., W. E. Dowling, E. Sanders-Buell, J. K. Carr, M. E. Harris, A. Thomschke, M. L. Robb, D. L. Birx, and F. E. McCutchan. 2002. Detection of HIV-1 subtypes, recombinants, and dual infections in east Africa by a multi-region hybridization assay. AIDS 16:2055-2064. [DOI] [PubMed] [Google Scholar]
- 25.Iversen, A. K., G. H. Learn, L. Fugger, J. Gerstoft, J. I. Mullins, and P. Skinhoj. 1999. Presence of multiple HIV subtypes and a high frequency of subtype chimeric viruses in heterosexually infected women. J. Acquir. Immune Defic. Syndr. 22:325-332. [DOI] [PubMed] [Google Scholar]
- 26.Janini, L. M., D. Pieniazek, J. M. Peralta, M. Schechter, A. Tanuri, A. C. Vicente, N. de la Torre, N. J. Pieniazek, C. C. Luo, M. L. Kalish, G. Schochetman, and M. A. Rayfield. 1996. Identification of single and dual infections with distinct subtypes of human immunodeficiency virus type 1 by using restriction fragment length polymorphism analysis. Virus Genes 13:69-81. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, R. P., J. D. Lifson, S. C. Czajak, K. S. Cole, K. H. Manson, R. Glickman, J. Yang, D. C. Montefiori, R. Montelaro, M. S. Wyand, and R. C. Desrosiers. 1999. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J. Virol. 73:4952-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jost, S., M. C. Bernard, L. Kaiser, S. Yerly, B. Hirschel, A. Samri, B. Autran, L. E. Goh, and L. Perrin. 2002. A patient with HIV-1 superinfection. N. Engl. J. Med. 347:731-736. [DOI] [PubMed] [Google Scholar]
- 29.Kitrinos, K. M., N. G. Hoffman, J. A. Nelson, and R. Swanstrom. 2003. Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection. J. Virol. 77:6811-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klenerman, P., Y. Wu, and R. Phillips. 2002. HIV: current opinion in escapology. Curr. Opin. Microbiol. 5:408-413. [DOI] [PubMed] [Google Scholar]
- 31.Koelsch, K. K., D. M. Smith, S. J. Little, C. C. Ignacio, T. R. Macaranas, A. J. Brown, C. J. Petropoulos, D. D. Richman, and J. K. Wong. 2003. Clade B HIV-1 superinfection with wild-type virus after primary infection with drug-resistant clade B virus. AIDS 17:F11-F16. [DOI] [PubMed] [Google Scholar]
- 32.Kral, A. H., R. N. Bluthenthal, J. Lorvick, L. Gee, P. Bacchetti, and B. R. Edlin. 2001. Sexual transmission of HIV-1 among injection drug users in San Francisco, USA: risk-factor analysis. Lancet 357:1397-1401. [DOI] [PubMed] [Google Scholar]
- 33.Kral, A. H., J. Lorvick, L. Gee, P. Bacchetti, B. Rawal, M. Busch, and B. R. Edlin. 2003. Trends in human immunodeficiency virus seroincidence among street-recruited injection drug users in San Francisco, 1987-1998. Am. J. Epidemiol. 157:915-922. [DOI] [PubMed] [Google Scholar]
- 34.Kuiken, C., G. Zwart, E. Baan, R. Coutinho, J. van den Hoek, and J. Goudsmit. 1993. Increasing antigenic and genetic diversity of the HIV-1 V3 domain in the course of the AIDS epidemic. Proc. Natl. Acad. Sci. USA 90:9061-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiken, C. L., J.-J. DeJong, E. Baan, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Evolution of the V3 envelope domain in proviral sequences and isolates of human immunodeficiency virus type 1 during transition of the viral biological phenotype. J. Virol. 66:4622-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laukkanen, T., J. K. Carr, W. Janssens, K. Liitsola, D. Gotte, F. E. McCutchan, E. Op de Coul, M. Cornelissen, L. Heyndrickx, G. van der Groen, and M. O. Salminen. 2000. Virtually full-length subtype F and F/D recombinant HIV-1 from Africa and South America. Virology 269:95-104. [DOI] [PubMed] [Google Scholar]
- 37.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Learn, G. H., Jr., B. T. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leitner, T., D. Escanilla, C. Franzen, M. Uhlen, and J. Albert. 1996. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl. Acad. Sci. USA 93:10864-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 41.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Locher, C. P., D. J. Blackbourn, S. W. Barnett, K. K. Murthy, E. K. Cobb, S. Rouse, G. Greco, G. Reyes-Teran, K. M. Brasky, K. D. Carey, and J. A. Levy. 1997. Superinfection with human immunodeficiency virus type 2 can reactivate virus production in baboons but is contained by a CD8 T cell antiviral response. J. Infect. Dis. 176:948-959. [DOI] [PubMed] [Google Scholar]
- 43.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 44.Lukashov, V. V., and J. Goudsmit. 2002. Recent evolutionary history of human immunodeficiency virus type 1 subtype B: reconstruction of epidemic onset based on sequence distances to the common ancestor. J. Mol. Evol. 54:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martins, L. P., N. Chenciner, B. Åsjö, A. Meyerhans, and S. Wain-Hobson. 1991. Independent fluctuation of human immunodeficiency virus type 1 rev and gp41 quasispecies in vivo. J. Virol. 65:4502-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCutchan, F. E., J. K. Carr, M. Bajani, E. Sanders-Buell, T. O. Harry, T. C. Stoeckli, K. E. Robbins, W. Gashau, A. Nasidi, W. Janssens, and M. L. Kalish. 1999. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 254:226-234. [DOI] [PubMed] [Google Scholar]
- 47.McDonald, R. A., D. L. Mayers, R. C. Chung, K. F. Wagner, S. Ratto-Kim, D. L. Birx, and N. L. Michael. 1997. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J. Virol. 71:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMichael, A. J., and R. E. Phillips. 1997. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 15:271-296. [DOI] [PubMed] [Google Scholar]
- 49.McNearney, T., Z. Hornickova, R. Markham, A. Birdwell, M. Arens, A. Saah, and L. Ratner. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA 89:10247-10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyerhans, A., R. Cheynier, J. Albert, M. Seth, S. Kwok, J. Sninsky, L. Morfeldt-Månson, B. Asjö, and S. Wain-Hobson. 1989. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell 58:901-910. [DOI] [PubMed] [Google Scholar]
- 51.Michael, N. L., G. Chang, L. A. d'Arcy, C. J. Tseng, D. L. Birx, and H. W. Sheppard. 1995. Functional characterization of human immunodeficiency virus type 1 nef genes in patients with divergent rates of disease progression. J. Virol. 69:6758-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 53.O'Connor, D., T. Friedrich, A. Hughes, T. M. Allen, and D. Watkins. 2001. Understanding cytotoxic T-lymphocyte escape during simian immunodeficiency virus infection. Immunol. Rev. 183:115-126. [DOI] [PubMed] [Google Scholar]
- 54.Otten, R. A., D. L. Ellenberger, D. R. Adams, C. A. Fridlund, E. Jackson, D. Pieniazek, and M. A. Rayfield. 1999. Identification of a window period for susceptibility to dual infection with two distinct human immunodeficiency virus type 2 isolates in a Macaca nemestrina (pig-tailed macaque) model. J. Infect. Dis. 180:673-684. [DOI] [PubMed] [Google Scholar]
- 55.Pang, S., Y. Shlesinger, E. S. Daar, T. Moudgil, D. D. Ho, and I. S. Y. Chen. 1992. Rapid generation of sequence variation during primary HIV-1 infection. AIDS 6:453-460. [DOI] [PubMed] [Google Scholar]
- 56.Paraskevis, D., M. Magiorkinis, V. Paparizos, G. N. Pavlakis, and A. Hatzakis. 2000. Molecular characterization of a recombinant HIV type 1 isolate (A/G/E/?): unidentified regions may be derived from parental subtype E sequences. AIDS Res. Hum. Retrovir. 16:845-855. [DOI] [PubMed] [Google Scholar]
- 57.Petry, H., U. Dittmer, C. Stahl-Hennig, C. Coulibaly, B. Makoschey, D. Fuchs, H. Wachter, T. Tolle, C. Morys-Wortmann, F. J. Kaup, E. Jurkiewicz, W. Lüke, and G. Hunsmann. 1995. Reactivation of human immunodeficiency virus type 2 in macaques after simian immunodeficiency virus SIVmac superinfection. J. Virol. 69:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 59.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. de Vange Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Putkonen, P., L. Walther, Y. J. Zhang, S. L. Li, C. Nilsson, J. Albert, P. Biberfeld, R. Thorstensson, and G. Biberfeld. 1995. Long-term protection against SIV-induced disease in macaques vaccinated with a live attenuated HIV-2 vaccine. Nat. Med. 1:914-918. [DOI] [PubMed] [Google Scholar]
- 61.Ramos, A., D. J. Hu, L. Nguyen, K. O. Phan, S. Vanichseni, N. Promadej, K. Choopanya, M. Callahan, N. L. Young, J. McNicholl, T. D. Mastro, T. M. Folks, and S. Subbarao. 2002. Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J. Virol. 76:7444-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramos, A., A. Tanuri, M. Schechter, M. A. Rayfield, D. J. Hu, M. C. Cabral, C. I. Bandea, J. Baggs, and D. Pieniazek. 1999. Dual and recombinant infections: an integral part of the HIV-1 epidemic in Brazil. Emerg. Infect. Dis. 5:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roland, M. E., J. N. Martin, R. M. Grant, N. S. Hellmann, J. D. Bamberger, M. H. Katz, M. Chesney, K. Franses, T. J. Coates, and J. O. Kahn. 2001. Postexposure prophylaxis for human immunodeficiency virus infection after sexual or injection drug use exposure: identification and characterization of the source of exposure. J. Infect. Dis. 184:1608-1612. [DOI] [PubMed] [Google Scholar]
- 64.Sala, M., E. Pelletier, and S. Wain-Hobson. 1995. HIV-1 gp120 sequences from a doubly infected drug user. AIDS Res. Hum. Retrovir. 11:653-655. [DOI] [PubMed] [Google Scholar]
- 65.Sala, M., G. Zambruno, J. P. Vartanian, A. Marconi, U. Bertazzoni, and S. Wain-Hobson. 1994. Spatial discontinuities in human immunodeficiency virus type 1 quasispecies derived from epidermal Langerhans cells of a patient with AIDS and evidence for double infection. J. Virol. 68:5280-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salminen, M. O., J. K. Carr, D. L. Robertson, P. Hegerich, D. Gotte, C. Koch, E. Sanders-Buell, F. Gao, P. M. Sharp, B. H. Hahn, D. S. Burke, and F. E. McCutchan. 1997. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J. Virol. 71:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 68.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmonds, P., L. Q. Zhang, F. McOmish, P. Balfe, C. A. Ludlam, and A. J. Leigh-Brown. 1991. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J. Virol. 65:6266-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon, V., J. Vanderhoeven, A. Hurley, B. Ramratnam, M. Louie, K. Dawson, N. Parkin, D. Boden, and M. Markowitz. 2002. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS 16:1511-1519. [DOI] [PubMed] [Google Scholar]
- 71.Takehisa, J., L. Zekeng, T. Miura, E. Ido, M. Yamashita, I. Mboudjeka, L. G. Gurtler, M. Hayami, and L. Kaptue. 1997. Triple HIV-1 infection with group O and group M of different clades in a single Cameroonian AIDS patient. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 14:81-82. [DOI] [PubMed] [Google Scholar]
- 72.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomson, M. M., E. Delgado, I. Herrero, M. L. Villahermosa, E. Vazquez-de Parga, M. T. Cuevas, R. Carmona, L. Medrano, L. Perez-Alvarez, L. Cuevas, and R. Najera. 2002. Diversity of mosaic structures and common ancestry of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Argentina revealed by analysis of near full-length genome sequences. J. Gen. Virol. 83:107-119. [DOI] [PubMed] [Google Scholar]
- 74.Thomson, M. M., E. Delgado, N. Manjon, A. Ocampo, M. L. Villahermosa, A. Marino, I. Herrero, M. T. Cuevas, E. Vazquez-de Parga, L. Perez-Alvarez, L. Medrano, J. A. Taboada, and R. Najera. 2001. HIV-1 genetic diversity in Galicia, Spain: BG intersubtype recombinant viruses circulating among injecting drug users. AIDS 15:509-516. [DOI] [PubMed] [Google Scholar]
- 75.Upchurch, D. A., R. Shankarappa, and J. I. Mullins. 2000. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 28:E69. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vidal, N., M. Peeters, C. Mulanga-Kabeya, N. Nzilambi, D. Robertson, W. Ilunga, H. Sema, K. Tshimanga, B. Bongo, and E. Delaporte. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in central Africa. J. Virol. 74:10498-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakrim, L., R. Le Grand, B. Vaslin, A. Cheret, F. Matheux, F. Theodoro, P. Roques, I. Nicol-Jourdain, and D. Dormont. 1996. Superinfection of HIV-2-preinfected macaques after rectal exposure to a primary isolate of SIVmac251. Virology 221:260-270. [DOI] [PubMed] [Google Scholar]
- 78.Wang, Y. M., S. C. Ray, O. Laeyendecker, J. R. Ticehurst, and D. L. Thomas. 1998. Assessment of hepatitis C virus sequence complexity by electrophoretic mobilities of both single- and double-stranded DNAs. J. Clin. Microbiol. 36:2982-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watters, J. K. 1993. The significance of sampling and understanding hidden populations. Drugs Soc. 7:13-21. [Google Scholar]
- 80.Watters, J. K., and P. Biernacki. 1989. Targeted sampling: options for the study of hidden populations. Soc. Probl. 36:416-430. [Google Scholar]
- 81.Watters, J. K., M. J. Estilo, G. L. Clark, and J. Lorvick. 1994. Syringe and needle exchange as HIV/AIDS prevention for injection drug users. JAMA 271:115-120. [PubMed] [Google Scholar]
- 82.Wiktor, S. Z., J. N. Nkengasong, E. R. Ekpini, G. T. Adjorlolo-Johnson, P. D. Ghys, K. Brattegaard, O. Tossou, T. J. Dondero, K. M. De Cock, and A. E. Greenberg. 1999. Lack of protection against HIV-1 infection among women with HIV-2 infection. AIDS 13:695-699. [DOI] [PubMed] [Google Scholar]
- 83.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103-110. [DOI] [PubMed] [Google Scholar]
- 83a.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 84.Wyand, M. S., K. Manson, D. C. Montefiori, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 73:8356-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuste, E., S. Sanchez-Palomino, C. Casado, E. Domingo, and C. Lopez-Galindez. 1999. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J. Virol. 73:2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang, L., L. Rowe, T. He, C. Chung, J. Yu, W. Yu, A. Talal, M. Markowitz, and D. D. Ho. 2002. Compartmentalization of surface envelope glycoprotein of human immunodeficiency virus type 1 during acute and chronic infection. J. Virol. 76:9465-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Leigh-Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 89.Zhu, T., N. Wang, A. Carr, S. Wolinsky, and D. D. Ho. 1995. Evidence for coinfection by multiple strains of human immunodeficiency virus type 1 subtype B in an acute seroconverter. J. Virol. 69:1324-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]